Abstract
Abnormal expression of sialylated Thomsen–Friedenreich antigen (Neu5Acα2-3Galβ1-3GalNAcα-O-Ser/Thr, sialyl-T) has a strong relationship with various types of human cancers and many other diseases. However, the size and structural complexity, and relatively lower abundance of sialyl-T have posed a significant challenge to its detection. Therefore, details about the role of sialyl-T in a variety of physiological and pathological processes are still poorly understood. Here, a one-step chemoenzymatic labeling strategy to probe sialyl-T is described. This approach enables the sensitive, selective, and rapid detection of sialyl-T, and global profiling and identification of unknown sialyl-T-attached glycoproteins, which are potential therapeutic targets or biomarkers. The use of one-step labeling strategy not only has a higher sensitivity than a typical two-step reporter strategy but also avoids undergoing an additional chemical reaction step to introduce a reporter group after the labeling reaction, making it particularly useful for detecting low-abundance glycan epitopes on living cells.
Short abstract
A one-step chemoenzymatic labeling strategy is described for the selective and rapid detection of sialyl-T, and global profiling and identification of unknown sialyl-T-attached glycoproteins.
Introduction
It is well established that truncated mucin-type O-glycans located on the cell surface, such as Tn antigen, sialyl-Tn antigen, T antigen, and sialyl-T antigen (Figure 1), are a hallmark feature of many human cancers.1,2 In humans, addition of a sialic acid to Galβ1-3GalNAcα-O-Ser/Thr by ST3Gal sialyltransferases produces sialyl-T antigen and inhibits any further elongation of the glycan except for possible addition of another α(2,6)-linked sialic acid to the GalNAc by α-N-acetylgalactosaminide sialyltransferase (ST6GalNAc) to form disialyl-T (Figure 1).3 Sialyl-T is expressed at low levels in some normal cells such as epithelium and leukocytes,4,5 but is more highly expressed on the cell surface of many human cancers such as breast, cervix, and bladder cancer, and other diseases such as GNE myopathy.6,7 Recent studies indicate sialyl-T might be produced during tumorigenesis, and its prevalence might be reduced when tumors become metastatic.8 It was also reported that sialyl-T could enhance tumor growth9,10 and modulate the tumor immunological microenvironment through engagement of the lectin Siglec-9.11 These discoveries distinguish sialyl-T as an important disease-associated glycan epitope.
Figure 1.
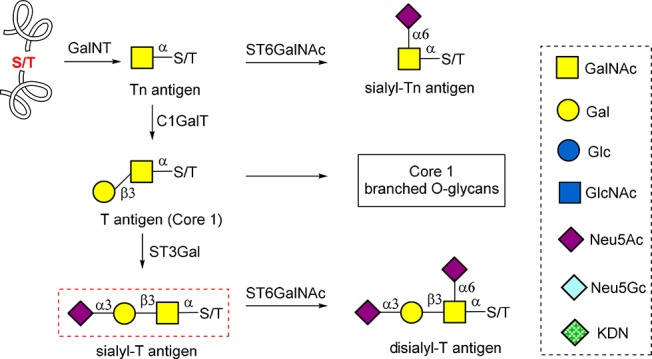
Biosynthesis and structures of truncated mucin-type O-glycans.
Although there is growing evidence that sialyl-T is a good biomarker for cancer and many other diseases,6,7 and its utility for immunotherapy and clinical diagnosis has also been suggested,11,12 details about its role in a variety of physiological and pathological processes are still largely unknown due to the lack of an effective analytical technique. Unlike other truncated O-glycans, there are no specific monoclonal antibodies or lectins for the direct detection of sialyl-T. An indirect method that combines neuraminidase treatment and the use of a T-antigen-binding lectin is the primary method for sialyl-T analysis.13,14 Additionally, gene expression level or enzymatic assay of ST3Gal1,15 and mass spectrometry based methods16 are also used to evaluate the expression of sialyl-T. Nevertheless, all these methods are time-consuming with relatively low accuracy.
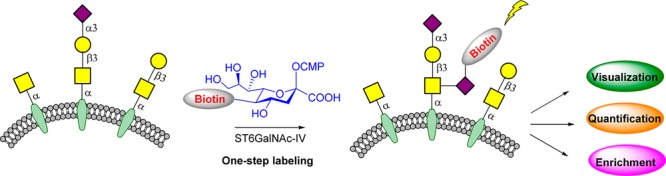
Here, we report a simple one-step chemoenzymatic labeling strategy to probe cell surface sialyl-T (Scheme 1). Inspired by the biosynthetic pathway of sialyl-T, this approach capitalizes on the relaxed donor specificity and strict acceptor specificity of human sialyltransferase ST6GalNAc-IV, which selectively and rapidly labels the sialyl-T with sialic acid derivative functionalized with triazole-linked biotin in vitro for further visualization, quantification, and enrichment analysis of sialyl-T (Scheme 1). This approach expands the technologies available for understanding the role of sialyl-T in a variety of biological and pathological processes.
Scheme 1. Biosynthesis-Inspired Design for Probing Sialyl-T.
(A) Two-step labeling strategy. (B) One-step labeling strategy.
Results and Discussion
Design of Chemoenzymatic Labeling Strategy
In recent years, the development of bioorthogonal chemistry has provided powerful tools for the analysis of glycans, proteins, lipids, nucleic acids, and other metabolites in living systems.17−23 Typically, bioorthogonal functional groups are metabolically incorporated into target molecules, allowing covalent conjugation by corresponding biorthogonal chemical reactions with either fluorescent or affinity tags for subsequent visualization or enrichment. As a complementary strategy to remodel glycans with unnatural functionalities, an chemoenzymatic labeling strategy has offered exciting possibilities to interrogate structure-defined glycan epitopes.24−30 This strategy takes advantage of glycosyltransferases to tag target epitopes with biorthogonal reactive groups. However, we and others31 have found that many glycosyltransferases that are active in assembling oligosaccharides in the test tube or even recognize standard glycoproteins failed to label glycan epitopes on the cell surfaces. This requires careful investigations on a variety of glycosyltransferases for their capability to work on complex samples to develop a practical chemoenzymatic labeling tool.
In the sialyl-T biosynthetic pathway, the ST6GalNAc family is the only enzyme that can directly modify sialyl-T.32 Among the reported six ST6GalNAc isoenzymes, ST6GalNAc-IV exhibits a strict acceptor specificity toward sialyl-T, while other ST6GalNAc isoenzymes show broader specificity with other structures such as Tn antigen, T-antigen, and glycolipids.33−35 Inspired by this biosynthetic process, we choose ST6GalNAc-IV as a candidate for further studies. Recombinant human ST6GalNAc-IV was expressed in baculovirus insect cells (Figure S1), and enzymatic activity was tested using Neu5Acα2-3Galβ1-3GalNAcα-O-Bn and CMP-Neu5Ac. A metal ion effect study indicated that the activity of ST6GalNAc-IV does not rely on metal ions such as Mg2+ or Mn2+ (Figure S2), which is different from the previous assays,34,35 in which high concentration of Mn2+ was added. This is crucial for the chemoenzymatic labeling reaction as a high concentration of Mn2+ is toxic toward living cells and will lead to a high background labeling signal (data not shown).
Evaluation of Donor and Acceptor Specificity of ST6GalNAc-IV
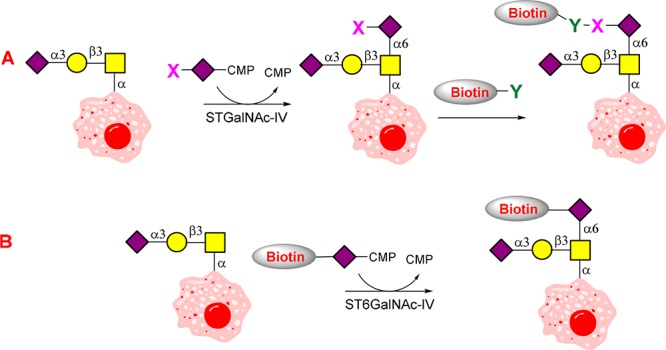
A donor and acceptor specificity study is an important step to develop a practical chemoenzymatic labeling tool. Relaxed donor specificity is the prerequisite for glycosyltransferases to carry biorthogonal functional groups, while acceptor specificity determines what kinds of epitopes can be labeled. It has been reported that many sialyltransferases tolerate modifications, even large moieties like biotin at C-5 or C-9 position.36,37 To find a good bio-orthogonal functional group carrier for ST6GalNAc-IV, we designed and synthesized 10 CMP-Neu5Ac analogues (2–11 in Figure 2). Preparative scale synthesis of 1–9 was from ManNAc or Neu5Ac analogues using sialic acid aldolase from Escherichia coli K12 and CMP-sialic acid synthetase from Neisseria meningitis (Schemes S1 and S2).38 The products were purified using the method reported previously.39,4010 and 11 was synthesized from 2 and 3 by CuAAC ligation, respectively (Scheme S3). The products were confirmed by NMR and MS analysis (see Supporting Information). These analogues contain azide, alkyne, alkene, ketone, and triazole biotin at the C-5 or C-9 position, and can be effectively probed by well-established methods such as copper-catalyzed azide–alkyne cycloaddition (CuAAC),41 strain promoted alkyne–azide cycloaddition (SPACC),41 Staudinger–Bertozzi ligation,17 Pd-catalyzed bioorthogonal elimination reaction,42 Diels–Alder reaction,43 dual labeling strategy,44 or one-step selective exoenzymatic labeling (SEEL) strategy.45 It is worth mentioning that one-step SEEL is a newly developed strategy by the Boons group for probing glycoproteins, which has a much better sensitivity than traditional methods such as the use of NHS-activated biotin and DBCO-biotin.45 Donor specificity study of ST6GalNAc-IV with Neu5Acα2-3Galβ1-3GalNAcα-O-Bn showed that all these CMP-Neu5Ac derivatives could be well accepted by ST6GalNAc-IV (Figure 2). Interestingly, a slightly higher activity was even observed toward substrate with a large modified group at the C-5 position (10).
Figure 2.
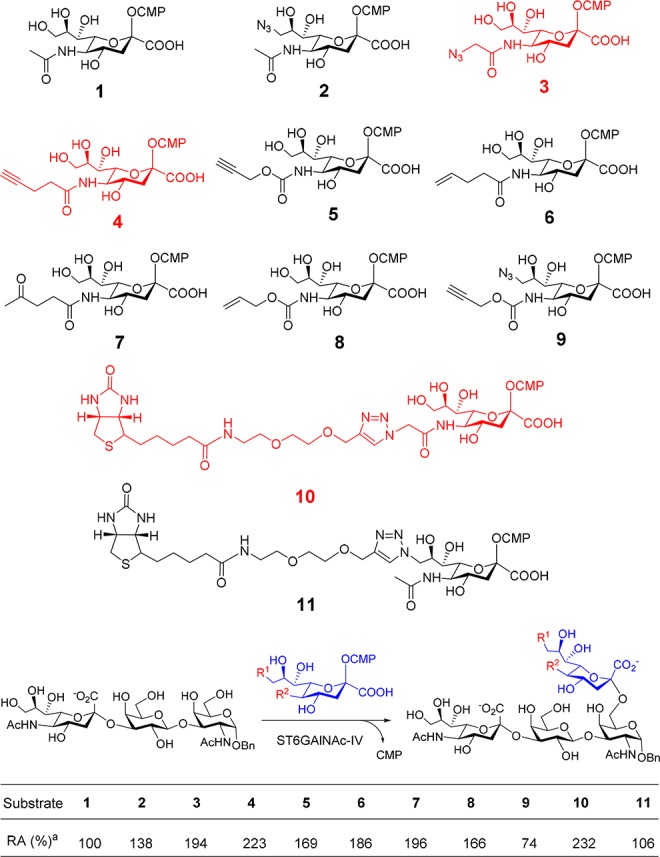
Structures of the CMP-Neu5Ac analogues and donor specificity of ST6GalNAc-IV. aRA: Relative activity. The activity with natural substrate CMP-Neu5Ac (1) was set as 100. See Supporting Information for experimental details.
To study the acceptor specificity of ST6GalNAc-IV, a sugar library including 12 representative structures listed in Table 1 was prepared as described in Supporting Information. Acceptor specificity study using CMP-Neu5Ac analogues 10 showed that ST6GalNAc-IV is highly specific toward sialyl-T. Although relative high activities toward Neu5Gcα2-3Galβ1-3GalNAcα-O-Bn and Kdnα2-3Galβ1-3GalNAcα-O-Bn were also observed, Kdn and Neu5Gc are rarely present in humans.46 These studies indicate the potential of ST6GalNAc-IV for use in the versatile labeling of cell surface sialyl-T antigen.
Table 1. Acceptor Specificity of ST6GalNAc-IV with Compound 10a.
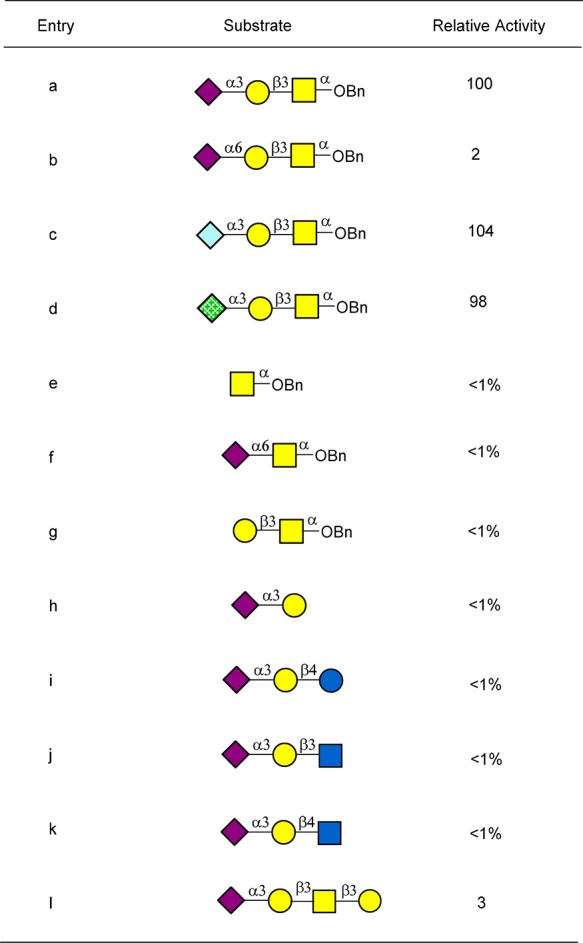
The activity with entry a was set as 100. The reaction was carried at 37 °C for 30 min. The activity was quantified by HPLC analysis.
Chemoenzymatic Labeling Sialyl-T on Living Cells
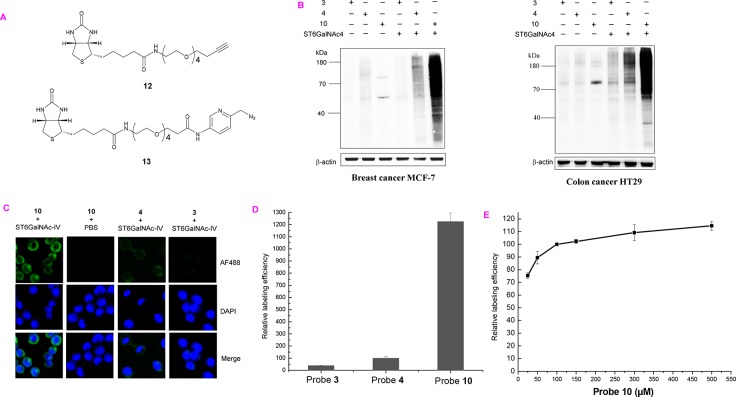
To test whether ST6GalNAc-IV can be used for cell surface labeling, we chose azido, alkynyl, and biotinylated probes (3, 4, and 10) for further experiments as they have a higher relative activity and can be easily detected by commercially available reagents. Azido and alkynyl probes (3 and 4 in this work) are the most popular two probes that are widely used in typical two-step bio-orthogonal reporter strategy.21 Since compound 10 contains a biotin group, it can be used to directly biotinylate the target epitopes by a simpler one-step reaction.45 Breast cancer MCF7 (∼5 × 107) was incubated in suspension with 200 μM of probe 3, 4, or 10 in the presence of 80 μg of ST6GalNAc-IV at 37 °C for 1 h, while the same labeling reactions without enzyme were performed in parallel as controls. After the labeling reaction, CuAAC (1 h, RT) was performed to introduce biotin group: 3 was biotinylated with 12, and 4 was biotinylated with 13 (Figure 3A). It is worth mentioning that 13 is a newly developed picolyl azide reporter probe for the sensitive detection of the alkyne group.47,48 The labeling efficiency of the various labeling probes (3, 4, and 10) was determined by SDS–PAGE of cell lysates followed by Western blotting using streptavidin conjugated with HRP. As shown in Figure 3B, the successful cell labeling requires both enzyme ST6GalNAc-IV and labeling probes. We found that the popular two-step clickable approach as the use of reporter pairs of 3 & 12 and 4 & 13 does not work well for detecting sialyl-T (Figure 3B). This is probably due to the relatively lower abundance of sialyl-T on cell surface compared to other structures such as total sialylated glycans and Neu5Acα-(2,3)-Gal glycans.29,45 The conventional azido probe 3 requires a longer exposure time to get a better signal but also with a higher background noise (Figure S3). The 4 and 13 reporter pair does give a better signal than 3 and 12, but it is still much lower (around 4 times, Figure S4) than probe 10 as the use of probe 10 does not need to undergo an additional click reaction step. In fact, experiment showed that 100 μM of 10 was sufficient for robust labeling (Figure 3E). Furthermore, the similar results were also observed when above-mentioned labeling probes were used to label another sialyl-T positive cell line colon cancer HT29 (Figure 3B). Indeed, even for other truncated mucin-type O-glycans such as T antigen and Tn antigen, it has been difficult to detect them by using the Western blot method as their relatively lower abundance and weak affinity of the currently available antibodies or lectins. Therefore, the described strategy provides a sensitive way to detect sialyl-T.
Figure 3.
(A) Structures of the reporter probes used for biotinylation 3 and 4. (B) Western blot analysis of chemoenzymatic labeling of breast cancer MCF7 and colon cancer HT29 cells (result represents at least three repeats). (C) Imaging of cell surface sialyl-T on MCF7 cells (green) by fluorescence microscopy. 200 ms acquisition time for 10 + ST6GalNAc-IV group and 500 ms acquisition time for the rest groups. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). (D) Comparison study of the relative labeling efficiency across probes 3, 4, and 10. (E) MCF7 cells (∼5 × 107) were labeled with ST6GalNAc-IV using variable concentrations of probe 10 (25–500 μM). See Supporting Information for experimental details.
We next investigated whether the chemoenzymatic strategy could be used to image sialyl-T glycans in living cells. Adherent MCF7 was labeled with probe 3, 4, or 10 as described above. Then, CuAAC (10 min, RT) was performed to biotinylate 3 and 4 with probe 12 and 13, respectively. The biotinylated samples were then stained with Alexa Fluor 488 streptavidin (30 min, 4 °C) to install a fluorescent reporter onto the sialyl-T. Strong fluorescence labeling was observed in cells labeled with ST6GalNAc-IV and 10, while no labeling signal was observed in the absence of ST6GalNAc-IV, confirming the specificity of the chemoenzymatic reaction in situ (Figure 3C). Meanwhile, only a very weak fluorescence signal was observed when using probe 3 or 4 even with a longer acquisition time (2.5 times longer than probe 10). As quantified by flow cytometry, the use of probe 10 afforded around 12-fold higher labeling efficiency than probe 4, or 30-fold higher than probe 3 (Figure 3D). Therefore, the use of probe 10 provides a sensitive way to image cell surface sialyl-T antigen. More importantly, the chemoenzymatic labeling strategy is independent of metabolic machinery pathway, making it particularly useful in staining tissue biopsies.
Next, this new labeling method was used to detect and compare the expression levels of sialyl-T across different cell lines. MCF7, T47D (breast cancer cell line), MDA-mb-231 (highly invasive breast cancer cell line), HT29, Raw 264.7 (macrophage cell line), HEK293 (human embryonic kidney cell line) cells were chemoenzymatically labeled in suspension by ST6GalNAc-IV and 10, and stained with the streptavidin conjugated with Alexa Fluor 488. The cells were untreated or labeled in the absence of ST6GalNAc-IV were performed as controls. As shown by flow cytometry analysis, all the well-known sialyl-T positive cell lines including MCF7, T47D, and HT29 shows robust staining compared to the control cells (Figure 4), highlighting the feasibility of the described strategy for use in living cell labeling. At the same time, much lower expression of sialyl-T was observed in Raw 264.7 and HEK293. Interestingly, MDA-mb-231 displayed around a 3-fold decrease in sialyl-T expression on the surface compared to MCF7 and T47D, indicating lower expression sialyl-T may be an invasive biomarker of breast cancers. These results also demonstrate that the described chemoenzymatic labeling approach can readily discriminate different cell lines.
Figure 4.
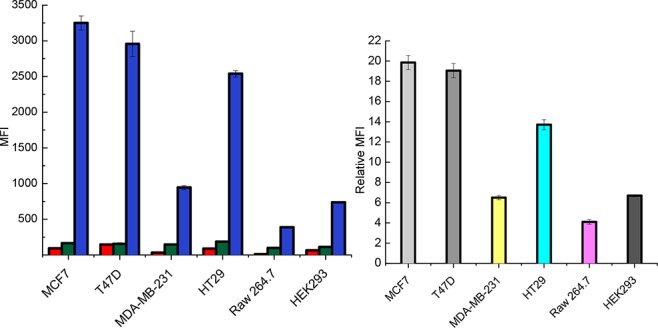
Flow cytometry analysis of the relative expression levels of sialyl-T across various cancer cell lines. Cells were untreated (red) or chemoenzymatically labeled in the absence (green) or presence (blue) of ST6GalNAc-IV. Quantification of the mean fluorescence intensity (MFI) relative to cells labeled in the absence of ST6GalNAc-IV is shown on the right. Error bars represent data from triplicate experiments.
Global Profiling Sialyl-T-Attached Glycoproteins on Cell Surface
We next used the described labeling strategy to globally profile sialyl-T-attached glycoproteins from cancer cell lines. It is well established that sialyl-T-attached glycoproteins that located on cell surface play important roles in living cells,11 where these glycoproteins are the potential therapeutic targets or biomarkers. However, there has been relatively little research to identify or profile sialyl-T-attached glycoproteins due to the lack of effective enrichment method. Taking advantage of the described one-step labeling strategy, sialyl-T-attached proteins that located on the cell surface (MCF7 and HT29) were biotinylated by ST6GalNAc-IV and 10 (Figure 5). The biotinylated proteins were captured using avidin agarose, which has an extremely strong affinity toward biotin group (Kd = 10–14 to 10–15 M). After digestion with trypsin, the peptide fragments were analyzed by tandem mass spectrometry. We successfully identified 78 cell surface proteins in breast cancer MCF7 and 43 proteins in colon cancer HT29 (Table S2 and Table S3). Among the total number of proteins, 14 are identified in both cell lines (Table S3). The classification of the identified proteins by PANTHER system showed that proteins with binding activity and catalytic activity are the two main classes in both cell lines (Figure 5). This is different from the previous classification, in which proteins with catalytic activity and transporter activity are two main classes of total Neu5Acα-(2,3)-Gal glycans.29 Many well-known sialylated cell adhesion molecules, such as integrin and CD44,49 are also identified in this study. This finding may imply that sialyl-T plays certain roles in protein binding and cell adhesion and migration. This result also accords with the previous observations, in which truncated mucin-type O-glycans have a strong relationship with tumor cell adhesion and migration.50
Figure 5.
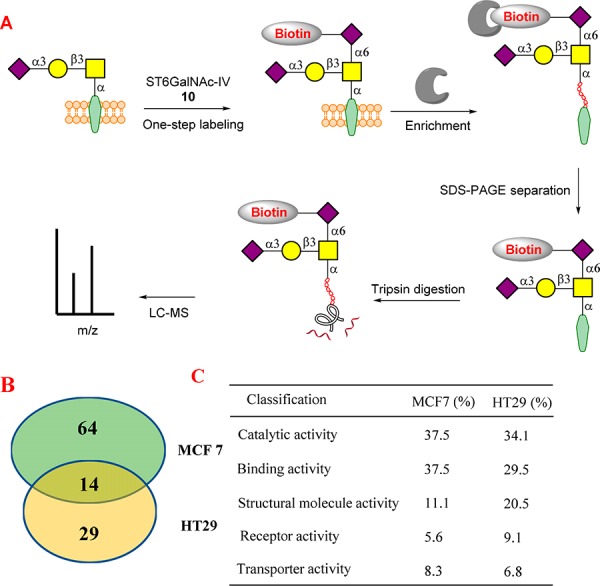
(A) Scheme of global profiling of sialyl-T-attached glycoproteins. (B) Number of the identified proteins from MCF7 and HT29. (C) Classification of the identified proteins by the PANTHER system.
Conclusions
In summary, on the basis of relaxed donor specificity and strict acceptor substrate specificity of human ST6GalNAc-IV, we have developed a practical labeling method for visualization, quantification, and enrichment analysis of sialyl-T antigen with excellent selectivity and sensitivity. This method is far superior to the traditional lectin-based detection method, which is limited by a time-consuming process and relatively low accuracy. We found the popular two-step labeling approach employing the click chemistry reaction does not work well to detect sialyl-T as the relatively lower abundance of sialyl-T on cell surface, while the one-step labeling strategy using CMP-Neu5Ac derivative 10 containing a triazole-linked biotin group provides a supersensitive way to detect sialyl-T. We anticipate this work will accelerate the study of investigating the function of sialyl-T epitope in a variety of biological and pathological processes. Moreover, we found tolerance of a large modification group such as biotin is not a proprietary nature of some randomly selected enzymes. We have tested all the sialyltransferases in our hand including various bacterial sialyltransferases (α-2,3-, α-2,6-, and α-2,8-sialyltransferase)51−53 and many human sialyltransferases (α-2,3- and α-2,6-sialyltransferase).54,55 All the tested sialyltransferases can well accept substrate with a large modification group such as compounds 10 and 11 (results will be reported in due course), indicating acceptance of large modification group is the common nature of the sialyltransferase family. In addition, protein engineering of key amino acids will also allow some other glycosyltransferases to accept substrates with a large modification group.56 This will make a one-step labeling strategy very popular in the future for use in analysis of kinds of glycan epitopes by either in vitro chemoenzymatic labeling or in vivo metabolic labeling method.
Acknowledgments
We thank the National Institutes of Health (U01GM116263) for financial support of this work. We thank Dr. Donald L. Jarvis from University of Wyoming for providing the baculovirus vector used to produce ST6GalNAC-IV for this study. This vector was produced as part of the glycorepository project, which was supported by NIH P41GM103390. We thank Dr. Yuan Liu and Dr. Ritu Aneja from Georgia State University for kindly providing breast cancer cell lines.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00573.
Experimental details, supporting figures, supporting tables, 1H NMR and 13C NMR spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Chia J.; Goh G.; Bard F. Short O-GalNAc glycans: regulation and role in tumor development and clinical perspectives. Biochim. Biophys. Acta, Gen. Subj. 2016, 1860, 1623–1639. 10.1016/j.bbagen.2016.03.008. [DOI] [PubMed] [Google Scholar]
- Dziadek S.; Brocke C.; Kunz H. Biomimetic synthesis of the tumor-associated (2,3)-sialyl-T antigen and its incorporation into glycopeptide antigens from the mucins MUC1 and MUC4. Chem. - Eur. J. 2004, 10, 4150–4162. 10.1002/chem.200400228. [DOI] [PubMed] [Google Scholar]
- Burchell J. M.; Mungul A.; Taylor-Papadimitriou J. O-linked glycosylation in the mammary gland: changes that occur during malignancy. J. Mammary Gland Biol. Neoplasia 2001, 6, 355–364. 10.1023/A:1011331809881. [DOI] [PubMed] [Google Scholar]
- Videira P. A.; Amado I. F.; Crespo H. J.; Alguero M. C.; Dall’Olio F.; Cabral M. G.; Trindade H. Surface alpha 2–3- and alpha 2–6-sialylation of human monocytes and derived dendritic cells and its influence on endocytosis. Glycoconjugate J. 2008, 25, 259–268. 10.1007/s10719-007-9092-6. [DOI] [PubMed] [Google Scholar]
- Priatel J. J.; Chui D.; Hiraoka N.; Simmons C. J.; Richardson K. B.; Page D. M.; Fukuda M.; Varki N. M.; Marth J. D. The ST3Gal-I sialyltransferase controls CD8+ T lymphocyte homeostasis by modulating O-glycan biosynthesis. Immunity 2000, 12, 273–283. 10.1016/S1074-7613(00)80180-6. [DOI] [PubMed] [Google Scholar]
- Videira P. A.; Correia M.; Malagolini N.; Crespo H. J.; Ligeiro D.; Calais F. M.; Trindade H.; Dall’Olio F. ST3Gal.I sialyltransferase relevance in bladder cancer tissues and cell lines. BMC Cancer 2009, 9, 357. 10.1186/1471-2407-9-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leoyklang P.; Malicdan M. C.; Yardeni T.; Celeste F.; Ciccone C.; Li X.; Jiang R.; Gahl W. A.; Carrillo-Carrasco N.; He M.; Huizing M. Sialylation of Thomsen-Friedenreich antigen is a noninvasive blood-based biomarker for GNE myopathy. Biomarkers Med. 2014, 8, 641–652. 10.2217/bmm.14.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S. A.; Bshara W.; Morrison C.; Chandrasekaran E. V.; Matta K. L.; Neelamegham S. Overexpression of alpha2,3sialyl T-antigen in breast cancer determined by miniaturized glycosyltransferase assays and confirmed using tissue microarray immunohistochemical analysis. Glycoconjugate J. 2014, 31, 509–521. 10.1007/s10719-014-9548-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungul A.; Cooper L.; Brockhausen I.; Ryder K.; Mandel U.; Clausen H.; Rughetti A.; Miles D. W.; Taylor-Papadimitriou J.; Burchell J. M. Sialylated core 1 based O-linked glycans enhance the growth rate of mammary carcinoma cells in MUC1 transgenic mice. Int. J. Oncol. 2004, 25, 937–943. [PubMed] [Google Scholar]
- Picco G.; Julien S.; Brockhausen I.; Beatson R.; Antonopoulos A.; Haslam S.; Mandel U.; Dell A.; Pinder S.; Taylor-Papadimitriou J.; Burchell J. Over-expression of ST3Gal-I promotes mammary tumorigenesis. Glycobiology 2010, 20, 1241–1250. 10.1093/glycob/cwq085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatson R.; Tajadura-Ortega V.; Achkova D.; Picco G.; Tsourouktsoglou T. D.; Klausing S.; Hillier M.; Maher J.; Noll T.; Crocker P. R.; Taylor-Papadimitriou J.; Burchell J. M. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 2016, 17, 1273–1281. 10.1038/ni.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D. M.; Byrne S. N.; Payne R. J. Synthetic self-adjuvanting glycopeptide cancer vaccines. Front. Chem. 2015, 3, 60. 10.3389/fchem.2015.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrilho C.; Cantel M.; Gouveia P.; David L. Simple mucin-type carbohydrate antigens (Tn, sialosyl-Tn, T and sialosyl-T) and gp 230 mucin-like glycoprotein are candidate markers for neoplastic transformation of the human cervix. Virchows Arch. 2000, 437, 173–179. 10.1007/s004280000218. [DOI] [PubMed] [Google Scholar]
- Ju T.; Lanneau G. S.; Gautam T.; Wang Y.; Xia B.; Stowell S. R.; Willard M. T.; Wang W.; Xia J. Y.; Zuna R. E.; Laszik Z.; Benbrook D. M.; Hanigan M. H.; Cummings R. D. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008, 68, 1636–1646. 10.1158/0008-5472.CAN-07-2345. [DOI] [PubMed] [Google Scholar]
- Dall’Olio F.; Chiricolo M. Sialyltransferases in cancer. Glycoconjugate J. 2001, 18, 841–850. 10.1023/A:1022288022969. [DOI] [PubMed] [Google Scholar]
- Muller S.; Hanisch F. G. Recombinant MUC1 probe authentically reflects cell-specific O-glycosylation profiles of endogenous breast cancer mucin. High density and prevalent core 2-based glycosylation. J. Biol. Chem. 2002, 277, 26103–26112. 10.1074/jbc.M202921200. [DOI] [PubMed] [Google Scholar]
- Saxon E.; Bertozzi C. R. Cell surface engineering by a modified Staudinger reaction. Science 2000, 287, 2007–2010. 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- Laughlin S. T.; Baskin J. M.; Amacher S. L.; Bertozzi C. R. In vivo imaging of membrane-associated glycans in developing zebrafish. Science 2008, 320, 664–667. 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescher J. A.; Bertozzi C. R. Chemistry in living systems. Nat. Chem. Biol. 2005, 1, 13–21. 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- Sletten E. M.; Bertozzi C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem., Int. Ed. 2009, 48, 6974–6998. 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaniappan K. K.; Bertozzi C. R. Chemical Glycoproteomics. Chem. Rev. 2016, 116, 14277–14306. 10.1021/acs.chemrev.6b00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. T.; Robinson P. V.; Spencer C. A.; Bertozzi C. R. Ultrasensitive Antibody Detection by Agglutination-PCR (ADAP). ACS Cent. Sci. 2016, 2, 139–147. 10.1021/acscentsci.5b00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G.; Li J.; Xie R.; Fan X.; Liu Y.; Zheng S.; Ge Y.; Chen P. R. Bioorthogonal Chemical Activation of Kinases in Living Systems. ACS Cent. Sci. 2016, 2, 325–331. 10.1021/acscentsci.6b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidekel N.; Arndt S.; Lamarre-Vincent N.; Lippert A.; Poulin-Kerstien K. G.; Ramakrishnan B.; Qasba P. K.; Hsieh-Wilson L. C. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J. Am. Chem. Soc. 2003, 125, 16162–16163. 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- Chaubard J. L.; Krishnamurthy C.; Yi W.; Smith D. F.; Hsieh-Wilson L. C. Chemoenzymatic probes for detecting and imaging fucose-alpha(1–2)-galactose glycan biomarkers. J. Am. Chem. Soc. 2012, 134, 4489–4492. 10.1021/ja211312u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng T. Q.; Jiang H.; Gros M.; Soriano del Amo D.; Sundaram S.; Lauvau G.; Marlow F.; Liu Y.; Stanley P.; Wu P. Tracking N-Acetyllactosamine on Cell-Surface Glycans In Vivo. Angew. Chem., Int. Ed. 2011, 50, 4113–4118. 10.1002/anie.201100265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q.; Li Z.; Duan X.; Yi W. A tandem enzymatic approach for detecting and imaging tumor-associated Thomsen-Friedenreich antigen disaccharide. J. Am. Chem. Soc. 2014, 136, 12536–12539. 10.1021/ja5054225. [DOI] [PubMed] [Google Scholar]
- Mbua N. E.; Li X.; Flanagan-Steet H. R.; Meng L.; Aoki K.; Moremen K. W.; Wolfert M. A.; Steet R.; Boons G. J. Selective exo-enzymatic labeling of N-glycans on the surface of living cells by recombinant ST6Gal I. Angew. Chem., Int. Ed. 2013, 52, 13012–13015. 10.1002/anie.201307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L.; Zheng Y.; Jiang K.; Zhang M.; Kondengaden S. M.; Li S.; Huang K.; Li J.; Song J.; Wang P. G. Two-Step Chemoenzymatic Detection of N-Acetylneuraminic Acid-alpha(2–3)-Galactose Glycans. J. Am. Chem. Soc. 2016, 138, 11473–11476. 10.1021/jacs.6b07132. [DOI] [PubMed] [Google Scholar]
- Capicciotti C. J.; Zong C.; Sheikh M. O.; Sun T.; Wells L.; Boons G. J. Cell-Surface Glyco-Engineering by Exogenous Enzymatic Transfer Using a Bifunctional CMP-Neu5Ac Derivative. J. Am. Chem. Soc. 2017, 139, 13342–13348. 10.1021/jacs.7b05358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Aguilar A.; Briard J. G.; Yang L.; Ovryn B.; Macauley M. S.; Wu P. Tools for Studying Glycans: Recent Advances in Chemoenzymatic Glycan Labeling. ACS Chem. Biol. 2017, 12, 611–621. 10.1021/acschembio.6b01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harduin-Lepers A.; Mollicone R.; Delannoy P.; Oriol R. The animal sialyltransferases and sialyltransferase-related genes: a phylogenetic approach. Glycobiology 2005, 15, 805–817. 10.1093/glycob/cwi063. [DOI] [PubMed] [Google Scholar]
- Dimitroff C. J. Galectin-Binding O-Glycosylations as Regulators of Malignancy. Cancer Res. 2015, 75, 3195–3202. 10.1158/0008-5472.CAN-15-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harduin-Lepers A.; Stokes D. C.; Steelant W. F.; Samyn-Petit B.; Krzewinski-Recchi M. A.; Vallejo-Ruiz V.; Zanetta J. P.; Auge C.; Delannoy P. Cloning, expression and gene organization of a human Neu5Ac alpha 2–3Gal beta 1–3GalNAc alpha 2,6-sialyltransferase: hST6GalNAcIV. Biochem. J. 2000, 352 (Pt 1), 37–48. 10.1042/bj3520037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z. L.; Huang X.; Burton A. J.; Swift K. A. Probing sialoglycans on fetal bovine fetuin with azido-sugars using glycosyltransferases. Glycobiology 2016, 26, 329–334. 10.1093/glycob/cwv109. [DOI] [PubMed] [Google Scholar]
- Skretas G.; Carroll S.; DeFrees S.; Schwartz M. F.; Johnson K. F.; Georgiou G. Expression of active human sialyltransferase ST6GalNAcI in Escherichia coli. Microb. Cell Fact. 2009, 8, 50. 10.1186/1475-2859-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.; Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem. Biol. 2010, 5, 163–176. 10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Yu H.; Karpel R.; Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg. Med. Chem. 2004, 12, 6427–6435. 10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Wen L.; Huang K.; Liu Y.; Wang P. G. Facile Enzymatic Synthesis of Phosphorylated Ketopentoses. ACS Catal. 2016, 6, 1649–1654. 10.1021/acscatal.5b02234. [DOI] [Google Scholar]
- Wen L.; Huang K.; Wei M.; Meisner J.; Liu Y.; Garner K.; Zang L.; Wang X.; Li X.; Fang J.; Zhang H.; Wang P. G. Facile Enzymatic Synthesis of Ketoses. Angew. Chem., Int. Ed. 2015, 54, 12654–12658. 10.1002/anie.201505714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwe K.; Brechbiel M. W. Growing applications of ″click chemistry″ for bioconjugation in contemporary biomedical research. Cancer Biother.Radiopharm. 2009, 24, 289–302. 10.1089/cbr.2008.0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Cheng B.; Li J.; Zhang Z. Y.; Hong W. Y.; Chen X.; Chen P. R. Chemical Remodeling of Cell-Surface Sialic Acids through a Palladium-Triggered Bioorthogonal Elimination Reaction. Angew. Chem., Int. Ed. 2015, 54, 5364–5368. 10.1002/anie.201409145. [DOI] [PubMed] [Google Scholar]
- Niederwieser A.; Spate A. K.; Nguyen L. D.; Jungst C.; Reutter W.; Wittmann V. Two-color glycan labeling of live cells by a combination of Diels-Alder and click chemistry. Angew. Chem., Int. Ed. 2013, 52, 4265–4268. 10.1002/anie.201208991. [DOI] [PubMed] [Google Scholar]
- Feng L.; Hong S.; Rong J.; You Q.; Dai P.; Huang R.; Tan Y.; Hong W.; Xie C.; Zhao J.; Chen X. Bifunctional unnatural sialic acids for dual metabolic labeling of cell-surface sialylated glycans. J. Am. Chem. Soc. 2013, 135, 9244–9247. 10.1021/ja402326z. [DOI] [PubMed] [Google Scholar]
- Sun T.; Yu S. H.; Zhao P.; Meng L.; Moremen K. W.; Wells L.; Steet R.; Boons G. J. One-Step Selective Exoenzymatic Labeling (SEEL) Strategy for the Biotinylation and Identification of Glycoproteins of Living Cells. J. Am. Chem. Soc. 2016, 138, 11575–11582. 10.1021/jacs.6b04049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangvoranuntakul P.; Gagneux P.; Diaz S.; Bardor M.; Varki N.; Varki A.; Muchmore E. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 12045–12050. 10.1073/pnas.2131556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uttamapinant C.; Tangpeerachaikul A.; Grecian S.; Clarke S.; Singh U.; Slade P.; Gee K. R.; Ting A. Y. Fast, Cell-Compatible Click Chemistry with Copper-Chelating Azides for Biomolecular Labeling. Angew. Chem., Int. Ed. 2012, 51, 5852–5856. 10.1002/anie.201108181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Zheng T. Q.; Lopez-Aguilar A.; Feng L.; Kopp F.; Marlow F. L.; Wu P. Monitoring Dynamic Glycosylation in Vivo Using Supersensitive Click Chemistry. Bioconjugate Chem. 2014, 25, 698–706. 10.1021/bc400502d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S. H.; Zhao P.; Sun T.; Gao Z.; Moremen K. W.; Boons G. J.; Wells L.; Steet R. Selective Exo-Enzymatic Labeling Detects Increased Cell Surface Sialoglycoprotein Expression upon Megakaryocytic Differentiation. J. Biol. Chem. 2016, 291, 3982–3989. 10.1074/jbc.M115.700369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazet A.; Julien S.; Bobowski M.; Burchell J.; Delannoy P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010, 12, 204. 10.1186/bcr2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J.; Yu H.; Lau K.; Huang S.; Chokhawala H. A.; Li Y.; Tiwari V. K.; Chen X. Multifunctionality of Campylobacter jejuni sialyltransferase CstII: characterization of GD3/GT3 oligosaccharide synthase, GD3 oligosaccharide sialidase, and trans-sialidase activities. Glycobiology 2008, 18, 686–697. 10.1093/glycob/cwn047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.; Chokhawala H.; Karpel R.; Yu H.; Wu B.; Zhang J.; Zhang Y.; Jia Q.; Chen X. A multifunctional Pasteurella multocida sialyltransferase: a powerful tool for the synthesis of sialoside libraries. J. Am. Chem. Soc. 2005, 127, 17618–17619. 10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Yu H.; Huang S.; Chokhawala H.; Sun M.; Zheng H.; Chen X. Highly efficient chemoenzymatic synthesis of naturally occurring and non-natural alpha-2,6-linked sialosides: a P. damsela alpha-2,6-sialyltransferase with extremely flexible donor-substrate specificity. Angew. Chem., Int. Ed. 2006, 45, 3938–3944. 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitroff C. J. Galectin-Binding O-Glycosylations as Regulators of Malignancy. Cancer Res. 2015, 75, 3195–3202. 10.1158/0008-5472.CAN-15-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.; Matta K. L.; Neelamegham S. A systematic analysis of acceptor specificity and reaction kinetics of five human alpha(2,3)sialyltransferases: Product inhibition studies illustrate reaction mechanism for ST3Gal-I. Biochem. Biophys. Res. Commun. 2016, 469, 606–612. 10.1016/j.bbrc.2015.11.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan B.; Qasba P. K. Structure-based Design of β1, 4-Galactosyltransferase I (β4Gal-T1) with Equally EfficientN-Acetylgalactosaminyltransferase Activity POINT MUTATION BROADENS β4Gal-T1 DONOR SPECIFICITY. J. Biol. Chem. 2002, 277, 20833–20839. 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.