Abstract
The aim of the present study was to investigate the expression of transactivation response RNA-binding protein (TARBP)1 and its clinical significance in human non-small-cell lung cancer (NSCLC). TARBP1 expression at the mRNA level was detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) in 10 NSCLC tissues and paired adjacent normal tissues. TARBP1 protein expression was analyzed in 90 paraffin-embedded NSCLC tissue samples and paired adjacent normal tissues by immunohistochemistry. Statistical analyses were performed to assess the clinicopathological significance of TARBP1 expression. The expression of TARBP1 mRNA was higher in the 10 NSCLC samples than in the paired adjacent non-tumor tissues (P=0.0017). In the paraffin-embedded tissue samples, the expression level of TARBP1 was higher in the cancer tissues than in the adjacent non-cancerous tissues. TARBP1 expression was detected in 76.67% (69/90) of the NSCLC samples and in 22.22% (20/90) of the adjacent normal lung tissues (P<0.001). The expression of TARBP1 was significantly associated with histological grade (P<0.001), clinical stage (P=0.024) and pathological type (P<0.001), along with a decreased overall survival (OS) rate (P<0.001). On multivariate analysis, the expression of TARBP1 was an independent prognostic factor for hazard ratio (OS, 2.729; 95% confidence interval, 1.471–5.061; P=0.003). TARBP1 is overexpressed in NSCLC, and the expression of TARBP1 is associated with pathological grade, clinical stage and pathological type. Thus, TARBP1 may be an independent prognostic marker in patients with NSCLC.
Keywords: non-small-cell lung cancer, TARBP1, expression, prognosis
Introduction
Lung cancer is the leading cause of cancer-related mortality in male patients and the second leading cause in female patients worldwide (1). Non-small-cell lung cancer (NSCLC) accounts for ~85% of those mortalities (2). It was predicted that 222,500 individuals would be diagnosed with lung cancer, while 155,870 would die of the disease in America in 2017 (3). Despite rapid progress in clinical and experimental oncology practices in recent years, the prognosis of the majority of lung cancer patients is still not optimistic, and the 5-year overall survival (OS) rate of such patients is ~15% (4,5). Moreover, the survival rate of patients with metastatic NSCLC is ~12 months, while the median progression-free survival (PFS) is between 3 and 6 months (6). Therefore, there is an urgent need to discover novel molecular biomarkers related to NSCLC as therapeutic targets for the prevention, diagnosis and treatment of NSCLC.
Human transactivation response RNA-binding proteins (TARBPs) were initially identified as proteins that bind with HIV type-1 (HIV-1) transactivation response RNA to activate long terminal repeat (LTR) expression in the presence or absence of the viral transactivator Tat (7). TARBP1 is a double-stranded RNA (dsRNA) binding protein that associates with certain other RNA interference factors to form a minimal RNA-induced silencing complex (RISC) (8,9), which serves as a Dicer co-factor in the processing of ~70-nucleotide pre-microRNAs (miRNAs) to 21–25-nucleotide mature miRNAs (10). Furthermore, TARBP1, being primarily located in cell nuclei (11), may play a crucial role in regulating the process of transcription. In this study, we detected the expression of TARBP1 in NSCLC and assessed its clinicopathological and prognostic significance.
Materials and methods
Data mining
Analysis of TARBP1 mRNA expression in NSCLC tissue samples and normal controls (meta-analysis of TARBP1 genes) was performed through the online cancer microarray database Oncomine (www.oncomine.org; Compendia Biosciences, Ann Arbor, MI, USA), which collects published cancer microarray data and processes these data with the same criteria (12). The data regarding mRNA expression status were organized into different types of human cancer tissue samples defined by the original publications. Sixteen sets of published mRNA expression data were chosen in this study. The mining strategy of these datasets was performed according to a method formulated by Oncomine (12). Significantly upregulated TARBP1 in cancer tissue samples compared with normal controls (>2-fold) were first selected. Next, concept filters of Oncomine were employed to identify the different expression of TARBP1 in lung carcinoma. Median rank was defined as the estimation of unreliability values based on the failure order number and the cumulative binomial distribution and was calculated using the formula BetaInv (0.5, k,N-k+1), where N represents sample size and k represents order (13).
Patients and specimens
Analyses were conducted on a total of 90 paraffin-embedded NSCLC samples from the archives of the First Affiliated Hospital of Guangzhou Medical University. All the patients were histopathologically diagnosed with primary lung cancer and underwent curative surgery between July 2004 and June 2009. None of the patients had received any type of neoadjuvant therapy. The median age of the patients was 61.5 years (range, 30–84 years), and the median tumor size was 4.6 cm (range, 1.5–9.0 cm). Among these patients, 81 of them were diagnosed adenocarcinoma, two were diagnosed large cell carcinoma, and seven squamous cell carcinoma. The demographic features and clinicopathological information of the patients are summarized in Table I. The follow-up time for the NSCLC cohort ranged from 1 to 121 months, and the median follow-up time was 39 months. Both tumor tissue and adjacent non-cancerous tissue (defined as tissue at least 1-cm distance from the tumor edge) were obtained during surgery. The expression of TARBP1 protein was detected by immunohistochemistry (IHC) in all 90 matched tissues. Another 20 paired NSCLC samples and adjacent non-cancerous tissues were collected immediately after operation for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis (11 male and 9 female). All the patients were histopathologically diagnosed with primary lung cancer between January 2015 and December 2016. The histological type of all the 20 patients were NSCLC. The median age of the patients was 63 years (range, 35–79 years), and the median tumor size was 5.2 cm (range, 4.1–8.5 cm).
Table I.
Correlation of TARBP1 expression with clinicopathological features.
| TARBP1 (%) | ||||
|---|---|---|---|---|
| Characteristics | Total (%) | Positive | Negative | P-value |
| Sex | 0.828 | |||
| Male | 49 (54.4) | 38 (77.6) | 11 (22.4) | |
| Female | 41 (45.6) | 31 (75.6) | 10 (24.4) | |
| Age, years | 0.189 | |||
| ≥60 | 55 (61.1) | 40 (72.7) | 15 (27.3) | |
| <60 | 35 (38.9) | 29 (82.9) | 6 (17.1) | |
| Lymphatic metastasis | 0.110 | |||
| Positive | 48 (53.3) | 40 (83.3) | 8 (16.7) | |
| Negative | 42 (46.7) | 29 (69.0) | 13 (31.0) | |
| Tumor size (cm) | 0.596 | |||
| ≥5 | 21 (23.3) | 17 (81.0) | 4 (19.0) | |
| <5 | 69 (76.7) | 52 (75.4) | 17 (24.6) | |
| Histological grade | <0.001 | |||
| 1 | 12 (13.3) | 9 (75.0) | 3 (25.0) | |
| 2 | 57 (63.3) | 42 (73.7) | 15 (26.3) | |
| 3 | 21 (23.3) | 18 (85.7) | 3 (14.3) | |
| Clinical stage | 0.024 | |||
| I–II | 50 (55.6) | 35 (70.0) | 15 (30.0) | |
| III | 30 (33.3) | 26 (86.7) | 4 (13.3) | |
| IV | 10 (11.1) | 8 (80.0) | 2 (20.0) | |
| Pathological type | <0.001 | |||
| Adenocarcinoma | 81 (90.0) | 61 (75.3) | 20 (24.7) | |
| Other | 9 (10.0) | 1 (11.1) | 8 (88.9) | |
TARBP, transactivation response RNA-binding protein.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University, and all patients provided written informed consent for the use of their clinical samples in the present study.
RT-qPCR analysis
Total RNA was extracted from the tissue samples with TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol, and pretreated with RNase-free DNase. cDNA synthesis was performed with 1 µg of total RNA from each sample and the use of a First-Strand Synthesis System (Fermentas; Thermo Fisher Scientific, Inc.). Real-time PCR was performed with a CFX96 Real-Time System and 2× SYBR-Green Master Mix (both from Bio-Rad, Hercules, CA, USA) in a total volume of 10 µl. The primer sequences were as follows: TARBP1, sense, 5′-TGCAACATTTCACCCACTCAA-3′ and antisense, 5′-CCCGCAGCTAAAGGAACATC-3′; and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, internal control), sense, 5′-TGTTGCCATCAATGACCCC-3′ and antisense, 5′-CTCCACGACGTACTCAGC-3′. All the reactions were performed in triplicate in three independent experiments and were quantified using the 2−ΔΔCq method (14).
IHC
IHC staining was performed according to the manufacturer's protocol (Zymed®; Life Technologies; Thermo Fisher Scientific, Inc.). Briefly, the paraffin-embedded samples were heated at 60°C for 1 h and deparaffinized with xylene. The slides were then rehydrated through a graded ethanol series, submerged in sodium citrate buffer and heated in a microwave for antigen retrieval. The sections were treated with 0.3% H2O2 for 10 min to inhibit endogenous peroxidase activity, and then incubated with normal goat serum at room temperature for 30 min to block nonspecific staining. Following blocking, the sections were incubated with rabbit polyclonal anti-TARBP1 antibody (dilution, 1:100; LSBIO) at 4°C overnight. Normal goat serum was used as a negative control. After three washes with phosphate-buffered saline (PBS), the slides were incubated with a biotinylated anti-rabbit secondary antibody, and then with streptavidin-horseradish peroxidase (both from Zymed; Thermo Fisher Scientific, Inc.) at 37°C for 30 min. After three washes with PBS, the tissue sections were incubated in diaminobenzidine (DAB) for color development.
The percentage and degree of positive staining were assessed and recorded separately by two independent pathologists, who were entirely blinded to the clinical characteristics of the samples. Scores assigned by the two independent pathologists were then averaged for the evaluation of TARBP1 expression. The staining intensity of TARBP1 was classified according to the following standards: no staining was marked as 0; weak staining (light yellow) was marked as 1; moderate staining (yellow brown) was marked as 2; and strong staining (brown) was marked as 3. The proportion of positively stained tumor cells was classified according to the following standards: no positive tumor cells was marked as 0; 1–25% positive tumor cells was marked as 1; 26–50% positive tumor cells was marked as 2; 51–75% positive tumor cells was marked as 3; and >75% positive tumor cells was marked as 4.
The final score was calculated as the product of the percentage and intensity scores. Cut-off values for TARBP1 were chosen on account of the heterogeneity using log-rank test concerning overall survival (OS). The optimal cut-off value was calculated as follows: A staining index score over 8 was used to define tumors with high TARBP1 expression and under 8 indicated low TARBP1 expression.
Statistical analysis
All the statistical analyses were conducted using the Statistical Software Package for the Social Sciences 20.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA). TARBP1 mRNA levels in the NSCLC and paired normal tissues were compared using a paired-samples t-test. The difference in the rate of TARBP1 positive expression between the NSCLC and adjacent non-tumor tissues was analyzed using a Chi-square test. Survival curves were plotted by the Kaplan-Meier method and compared with a log-rank test. The time span from the date of each patient's randomization to either the date of the patient's death for any reason or the date of the last follow-up was defined as OS. The relationships between TARBP1 expression and other clinicopathological features were evaluated using Chi-square tests and Fisher's exact tests. The prognostic value of clinicopathological features was analyzed by univariate and multivariate Cox regression analyses; the enter method of the Cox model was used for univariate analysis, while the forward method was used for multivariate analysis. All statistical tests were two-sided and P<0.05 was considered to indicate statistical significance.
Results
TARBP1 is overexpressed in NSCLC patients
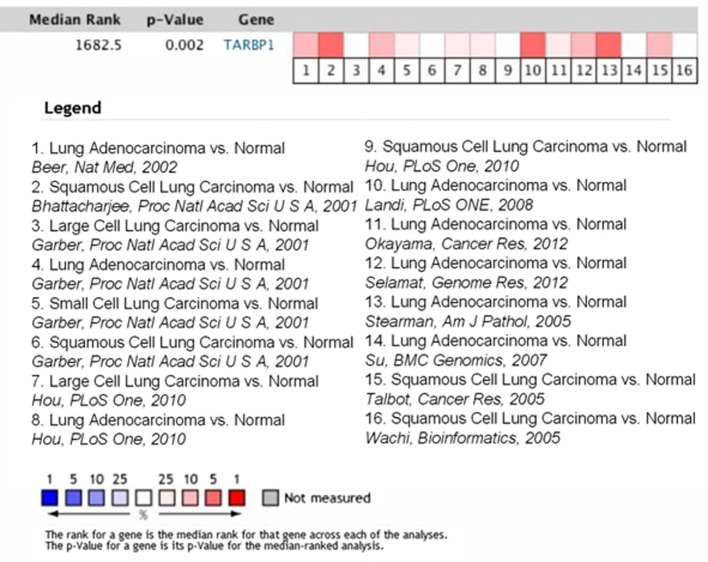
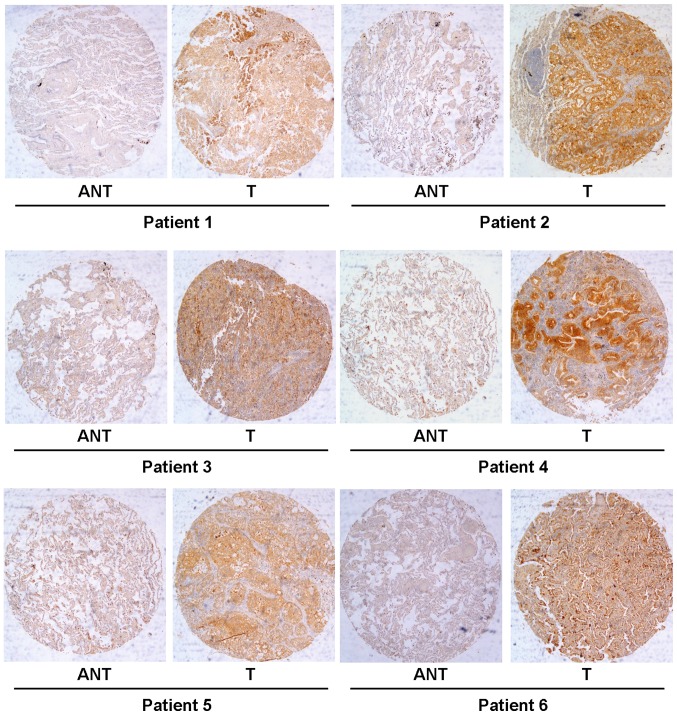
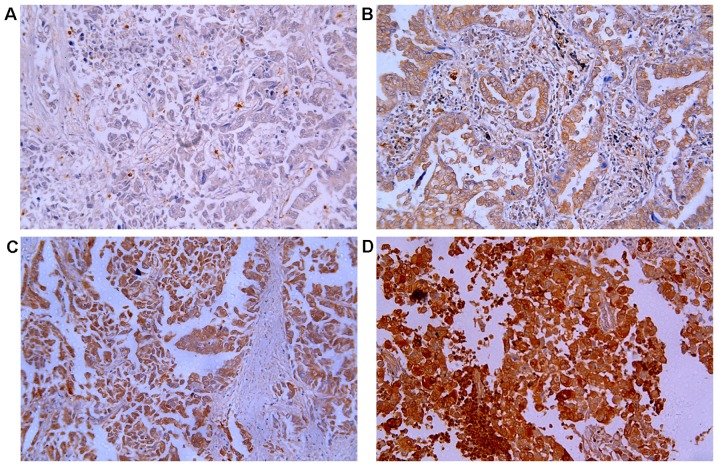
To determine whether TARBP1 expression was elevated in human NSCLC samples, we first conducted a meta-analysis of the Oncomine database, and found that TARBP1 expression was significantly higher in lung cancer than in matched adjacent non-tumor tissues, with a median rank of 1862.5 and a P-value of 0.002 (Fig. 1) (15–25). To verify this result, we performed RT-qPCR analysis on 10 NSCLC tissues and paired adjacent normal lung tissues. As illustrated in Fig. 2, the level of TARBP1 mRNA in the 10 NSCLC samples was significantly higher than that in the adjacent normal lung tissues (P=0.0017). Meanwhile, on IHC, the positive expression of TARBP1 was detected in 69 out of 90 (76.67%) of the NSCLC samples, while staining of TARBP1 protein was weak or absent in the adjacent non-cancerous lung tissues, with a positive expression detection rate of only 22.2% (20/90) (Figs. 3 and 4). This difference in the rate of TARBP1 positive expression between the NSCLC samples and non-cancerous lung tissues was statistically significant (χ2=53.362, P<0.001).
Figure 1.
TARBP1 expression is upregulated in human lung cancer samples in the Oncomine database. Oncomine heat map of TARBP1 gene expression in clinical lung cancer samples compared with the normal lung tissues (www.oncomine.org). Sixteen sets of published mRNA expression data were chosen in this study. Significantly upregulated TARBP1 in cancer tissue samples compared with normal controls (>2-fold) were first selected. In the meta-analysis, TARBP1 expression was identified to be significantly higher in lung cancer than in the corresponding normal tissues with a median rank of 1862.5 and a P-value of 0.002. The degree of colour correlates to the gene rank percentile of the highest ranking analyses. The red colour represents overexpression of TARBP1 genes, and the blue colour represents underexpression of TARBP1 genes.
Figure 2.
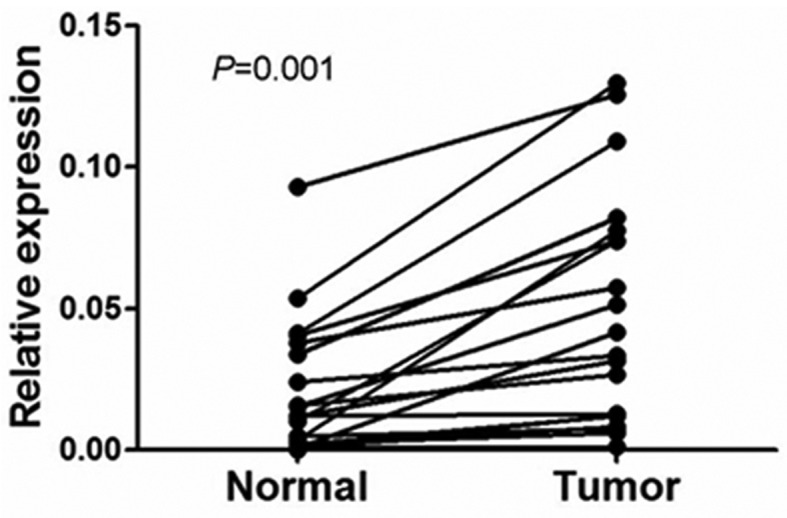
Expression levels of TARBP1 mRNA in lung cancer and adjacent non-cancerous tissues. Expression levels of TARBP1 mRNA in 20 paired lung cancer tissues measured by real-time PCR. Normal, para carcinoma (normal) lung tissues. Tumor, lung cancer tissues.
Figure 3.
Expression status of TARBP1 in paired lung cancer and adjacent normal tissues. Immunohistochemical assay of TARBP1 protein expression in paired lung cancer and normal lung tissues. T, lung cancer tissues; ANT, matched adjacent non-tumor lung tissues (×40).
Figure 4.
Expression of TARBP1 protein measured by immunohistochemistry. TARBP1 expression was mainly localized in the cytoplasm of tumor cells. Negative expression of TARBP1 (A), low (B), medium (C) and high (D) expression of TARBP1 in lung cancer tissues (×200).
TARBP1 expression correlates with clinicopathological features in NSCLC
To better understand the potential functions of TARBP1 in the development and progression of NSCLC, IHC staining was used to investigate the expression status of TARBP1 in 90 paraffin-embedded archived NSCLC tissue samples (31 stage I tumors, 19 stage II tumors, 30 stage III and 10 stage IV tumors) (Table I). Among the 90 samples, the positive expression of TARBP1 protein was observed in 69 cases (76.67%), while no obvious signals were detected in the remaining 21 tumor tissue samples (23.33%) (Table I). By contrast, no staining or only weak staining were observed in the adjacent normal tissue samples, and the positive expression of TARBP1 was significantly higher in the NSCLC tissues (Figs. 3 and 4). The predominant subcellular location of TARBP1 was the cytoplasm.
We further investigated the relationship between TARBP1 expression and clinicopathological indices of NSCLC patients. As summarized in Table I, there was no significant correlation between TARBP1 protein expression and patient age, patient sex, lymphatic metastasis, pathological type or tumor size. However, TARBP1 expression was significantly associated with histological grade (P<0.001), clinical stage (P=0.024), and pathological type (P<0.001).
Association between TARBP1 expression and patient OS
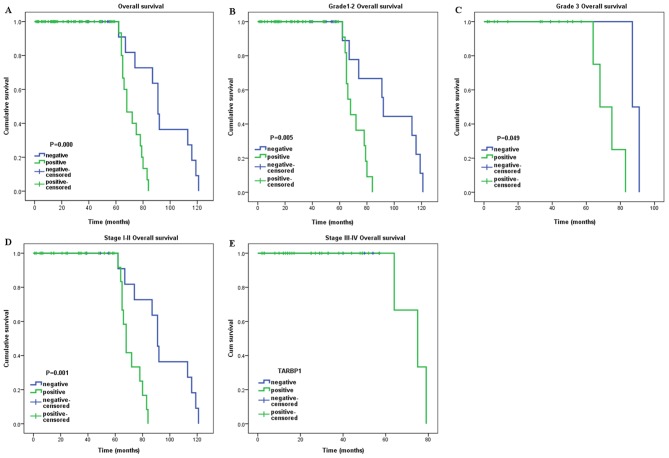
The survival analysis identified a significant negative correlation between the expression level of TARBP1 protein and the OS of the NSCLC patients (P<0.001) (Fig. 5A). Additionally, Cox regression revealed that the expression of TARBP1 was an independent prognostic factor for the OS of the NSCLC patients (Table II).
Figure 5.
Kaplan-Meier curves with univariate analysis (log-rank). (A) OS rates for cases with TARBP1 positive expression vs. those with TARBP1 negative expression in all patients. (B) OS rate for grade 1–2 cases with TARBP1 positive expression vs. those with TARBP1 negative expression. (C) OS rate for grade 3 cases with TARBP1 positive expression vs. those with TARBP1 negative expression. (D) OS rate for early clinical stage cases (stage I/II) with TARBP1 positive expression vs. those with TARBP1 negative expression. (E) OS rate for late stage cases (stage III/IV) with TARBP1 positive expression vs. those with TARBP1 negative expression.
Table II.
Cox-regression analysis of various prognostic parameters in patients for all patients.
| Univariate | Multivariate | |||
|---|---|---|---|---|
| Factor | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Sex | ||||
| Male | Reference | |||
| Female | 0.592 (0.261–1.344) | 0.210 | – | – |
| Age, years | ||||
| ≥60 | Reference | |||
| <60 | 0.835 (0.549–1.270) | 0.399 | – | – |
| Lymphatic metastasis | ||||
| Positive | Reference | |||
| Negative | 0.565 (0.197–1.619) | 0.288 | – | – |
| Tumor size, cm | ||||
| ≥5 | Reference | |||
| <5 | 4.514 (1.515–13.453) | 0.007 | – | – |
| Histological grade | ||||
| 1 | Reference | |||
| 2 | 0.879 (0.355–2.175) | 0.781 | – | – |
| 3 | 1.167 (0.404–3.370) | 0.775 | – | – |
| Clinical stage | ||||
| I–II | Reference | |||
| III | 2.698 (0.925–7.866) | 0.069 | – | – |
| IV | 2.832 (0.729–11.001) | 0.133 | – | – |
| Pathological type | ||||
| Adenocarcinoma | Reference | |||
| Others | 1.034 (0.350–3.048) | 0.952 | – | – |
| TARBP1 | ||||
| Positive | Reference | Reference | ||
| Negative | 0.133 (0.036–0.484) | 0.002 | 2.729 (1.471–5.061) | 0.003 |
TARBP, transactivation response RNA-binding protein; HR, hazard ratio; CI, confidence interval.
We further investigated the prognostic value of TARBP1 in selective patient subgroups classified by histological grade and clinical stage, respectively. TARBP1 expression was significantly associated with the OS of patients in the grade 1–2 subgroup (log-rank test, P=0.005) and the grade 3 subgroup (log-rank test, P=0.049) (Fig. 5B and C). However, when evaluated on the basis of clinical stage, the influence of TARBP1 expression on patient outcome continued to be significant only in the stage I–II subgroup (log-rank test, P=0.001) (Fig. 5D), but not in the stage III–IV subgroup (P>0.05) (Fig. 5E).
Discussion
Lung cancer is among the most prevalent malignant tumors, with a high economic impact and rate of premature death worldwide. In China, lung cancer is the most common type of cancer and the leading cause of cancer-associated mortality in men (26,27). In America, lung cancer was predicted to be the second most common type of cancer in males and females, as well as the leading cause of cancer-related mortality in patients of either sex (3). Smoking tobacco is associated with a large proportion of all lung cancer cases (28,29). Over the last several decades, developments in early diagnostic and treatment strategies have considerably increased the average survival rate of cancer patients, and some types of cancer are now considered to be curable. However, lung cancer is typically diagnosed at a late age (47% of patients were over 70-years-old at the time of diagnosis) and late stage (50% of patients were diagnosed at advanced stage) (30,31). Thus, the prognosis of lung cancer is not optimistic. Due to the lack of apparent symptoms in early-stage lung cancer, the majority of patients are diagnosed at an advanced stage when the treatment options are limited (32–34). The 5-year estimated survival rates are ~6–14% in male patients and 7–18% in female patients (35). NSCLC is the most common type of lung cancer (36,37). The prognosis of NSCLC patients remains poor, and NSCLC accounts for ~85% of all lung cancer-associated mortalities (2). Therefore, there is a great need to identify novel biomarkers and genetic risk factors of lung cancer, in order to improve prognostic predictions and guide therapy.
The cellular dsRNA binding protein TARBP1 has recently been shown to promote the replication of HIV-1 and −2 (38). Additionally, it has been identified as a component of RISC, which serves as a Dicer cofactor in the formation of 21–25 nucleotide mature miRNAs from ~70-nucleotide pre-miRNAs (10). In accordance with the notion that viral infection of mammalian cells may be restricted by miRNA regulation, the sequestration of TARBP1 by TAR RNA results in increased replication of HIV-1 in human cells (39). Thus, TARBP1, which contains two dsRNA-binding domains (RBD), is established as a multifarious protein with two prominent roles (8): First, it serves as a pre-requisite for the formation of Dicer-containing complexes; and second, it gives assistance to Dicer in generating miRNA (9,40).
Recently, truncating mutations of TARBP1 were found in several human malignant cancers with microsatellite instability (41). In Kaposi's sarcoma-associated herpesvirus [KSHV, also known as human herpesvirus-8 (HHV-8)], the expression of Dicer and TARBP1 is upregulated following KSHV infection, which may promote an increase in the amount of the miRNA-RISC loading complex available for both host and viral miRNAs (42). In cardiomyopathy patients, TARBP2 could regulate heart function through miRNA-mediated Sox6 repression (43). Meanwhile, Chen et al reported that SUMOylation of TARBP2 may suppress the progression of tumors (44). Furthermore, knockdown of TARBP1 could reduce the accumulation of hepatitis C virus RNA (45), and TARBP1 has recently been proposed as a target for antiviral therapies (38,46,47). The function of TARBP1 may be regulated by phosphorylation via the JNK-MAPK axis (48). TARBP1 has a biological function in spermatogenesis and growth control during development. It can also bind the interferon-induced dsRNA-activated protein kinase (PKR) and with the PKR activator (PACT), as well as with tumor suppressors (47). More recently, several diseases have been associated with TARBP1, such as cardiac disease (43) and hepatocellular carcinoma (49), among others. In this study, we investigated for the first time the expression of TARBP1 and its associated clinical significance in NSCLC.
Our results clearly showed that TARBP1 was upregulated at the mRNA level in 10 NSCLC tissue samples. In addition, IHC staining identified a TARBP1 positive staining rate of 76.67% (69/90) in tumor tissues, which was markedly higher than that for adjacent normal tissues. Analogous observations have been reported for other diseases, including HIV (47) and epithelial skin cancer (50). The differential expression of TARBP1 between tumor tissues and adjacent normal tissues indicates that TARBP1 may be an important molecular biomarker in tumorigenesis, which could aid to improve the precision of diagnoses. However, at present, the exact roles of TARBP1 in malignant cancer remain unclear, and thus further study is required to understand the associated signaling pathways of TARBP1 in NSCLC. Our research did not include the investigation of TARBP1 expression in tissue sample of inflammatory lesions, because the patients with pneumonia usually receive conservative treatment and we seldom do surgery or biopsy for these kind of patients.
We further explored the relationship between TARBP1 expression and certain clinical indices of patients with NSCLC. TARBP1 expression was found to be significantly correlated with histological grade (P<0.001), clinical stage (P=0.024), and pathological type (P<0.001). However, no associations were identified between TARBP1 expression and patient age, patient sex, lymphatic metastasis, pathological type or tumor size. Meanwhile, Cox regression demonstrated that the expression of TARBP1 was an independent prognostic factor in NSCLC patients after curative resection. This finding indicates the potential of TARBP1 expression level as a predictor for patient prognosis and potential therapeutic target. Interestingly, sub-group analysis implied that TARBP1 positive expression was significantly associated with a poor prognosis among well-differentiated and poorly-differentiated cases, and among patients with early-stage disease. Nonetheless, the molecular mechanism underlying the association of TARBP1 with patient prognosis and survival remains obscure and yet to be explored.
To conclude, to the best of our knowledge, this is the first report on TARBP1 expression and its clinicopathological and prognostic significance in NSCLC. Our findings indicate that TARBP1 is upregulated and closely associated with histological grade, clinical stage, and pathological type in NSCLC. Based on these findings, we infer that TARBP1 is associated with aggressive clinical features in NSCLC. Notably, multivariate analysis revealed that TARBP1 might be an independent molecular biomarker for the prediction of patient prognosis and survival. Combined with the above findings, high expression of TARBP1 may cause worse prognosis in NSCLC. Comprehensive evaluation of the expression of TARBP1 will help the clinicians to identify patients with a worse prognosis, paving a way of discovering effective strategies targeting TARBP1 in the treatment of NSCLC patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Chinese National Natural Science Foundation (no. 81672661), Guangdong Province Natural Science Foundation (no. 2015A030310126) and Guangzhou Medical and Health Technology Program (no. 20161A010077).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LX designed the experiments. JY and NZ carried out the experiments. JW analyzed experimental results. YL and LT collected the clinical samples, performed the statistical analysis and wrote the manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangzhou Medical University, and all patients provided written informed consent for the use of their clinical samples in the present study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Crowley J, Chansky K, Giroux DJ, Groome PA, Rami-Porta R, Postmus PE, Rusch V, Sobin L. International Association for the Study of Lung Cancer International Staging Committee; Participating Institutions: The IASLC lung cancer staging project: Proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM Classification of malignant tumours. J Thorac Oncol. 2007;2:706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.DeSantis CE, Lin CC, Mariotto AB, Siegel RL, Stein KD, Kramer JL, Alteri R, Robbins AS, Jemal A. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64:252–271. doi: 10.3322/caac.21235. [DOI] [PubMed] [Google Scholar]
- 5.Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32:669–692. doi: 10.1016/j.ccm.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Khan AJ, Dicker AP. On the merits and limitations of whole-brain radiation therapy. J Clin Oncol. 2013;31:11–13. doi: 10.1200/JCO.2012.46.0410. [DOI] [PubMed] [Google Scholar]
- 7.Kozak CA, Gatignol A, Graham K, Jeang KT, McBride OW. Genetic mapping in human and mouse of the locus encoding TRBP, a protein that binds the TAR region of the human immunodeficiency virus (HIV-1) Genomics. 1995;25:66–72. doi: 10.1016/0888-7543(95)80110-8. [DOI] [PubMed] [Google Scholar]
- 8.Daniels SM, Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress and cancer. Microbiol Mol Biol Rev. 2012;76:652–666. doi: 10.1128/MMBR.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi T, Zenno S, Ishibashi O, Takizawa T, Saigo K, Ui-Tei K. Interactions between the non-seed region of siRNA and RNA-binding RLC/RISC proteins, Ago and TRBP, in mammalian cells. Nucleic Acids Res. 2014;42:5256–5269. doi: 10.1093/nar/gku153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi YH, Semmes OJ, Jeang KT. A proteomic study of TAR-RNA binding protein (TRBP)-associated factors. Cell Bios. 2011;1:9. doi: 10.1186/2045-3701-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci USA. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landi MT, Dracheva T, Rotunno M, Figueroa JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 21.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su LJ, Chang CW, Wu YC, Chen KC, Lin CJ, Liang SC, Lin CH, Whang-Peng J, Hsu SL, Chen CH, Huang CY. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, O-charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein R, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 25.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Zheng R, Zeng H, Zhang S. The updated incidences and mortalities of major cancers in China, 2011. Chin J Cancer. 2015;34:502–507. doi: 10.1186/s40880-015-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen W, Zheng R, Zeng H, Zhang S. The incidence and mortality of major cancers in China, 2012. Chin J Cancer. 2016;35:73. doi: 10.1186/s40880-016-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.No authors listed: Lung cancer. Breathe (Sheff) 2016;12:392–399. doi: 10.1183/20734735.ELF124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 30.Owonikoko TK, Ragin CC, Belani CP, Oton AB, Gooding WE, Taioli E, Ramalingam SS. Lung cancer in elderly patients: An analysis of the surveillance, epidemiology, and end results database. J Clin Oncol. 2007;25:5570–5577. doi: 10.1200/JCO.2007.12.5435. [DOI] [PubMed] [Google Scholar]
- 31.Ramalingam SS, Owonikoko TK, Khuri FR. Lung cancer: New biological insights and recent therapeutic advances. CA Cancer J Clin. 2011;61:91–112. doi: 10.3322/caac.20102. [DOI] [PubMed] [Google Scholar]
- 32.Pirozynski M. 100 years of lung cancer. Respir Med. 2006;100:2073–2084. doi: 10.1016/j.rmed.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz AG, Prysak GM, Bock CH, Cote ML. The molecular epidemiology of lung cancer. Carcinogenesis. 2007;28:507–518. doi: 10.1093/carcin/bgl253. [DOI] [PubMed] [Google Scholar]
- 34.Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med. 2005;172:523–529. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- 35.Marshall AL, Christiani DC. Genetic susceptibility to lung cancer-light at the end of the tunnel? Carcinogenesis. 2013;34:487–502. doi: 10.1093/carcin/bgt016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldstraw P, Ball D, Jett JR, Le Chevalier T, Lim E, Nicholson AG, Shepherd FA. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. doi: 10.1016/S0140-6736(10)62101-0. [DOI] [PubMed] [Google Scholar]
- 37.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 38.Christensen HS, Daher A, Soye KJ, Frankel LB, Alexander MR, Lainé S, Bannwarth S, Ong CL, Chung SW, Campbell SM, et al. Small interfering RNAs against the TAR RNA binding protein, TRBP, a Dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J Virol. 2007;81:5121–5131. doi: 10.1128/JVI.01511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem. 2006;281:27674–27678. doi: 10.1074/jbc.C600072200. [DOI] [PubMed] [Google Scholar]
- 40.Redfern AD, Colley SM, Beveridge DJ, Ikeda N, Epis MR, Li X, Foulds CE, Stuart LM, Barker A, et al. RNA-induced silencing complex (RISC) Proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci USA. 2013;110:6536–6541. doi: 10.1073/pnas.1301620110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Happel C, Ramalingam D, Ziegelbauer JM. Virus-mediated alterations in miRNA factors and degradation of viral miRNAs by MCPIP1. PLoS Biol. 2016;14:e2000998. doi: 10.1371/journal.pbio.2000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding J, Chen J, Wang Y, Kataoka M, Ma L, Zhou P, Hu X, Lin Z, Nie M, Deng ZL, et al. Trbp regulates heart function through microRNA-mediated Sox6 repression. Nat Genet. 2015;47:776–783. doi: 10.1038/ng.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C, Zhu C, Huang J, Zhao X, Deng R, Zhang H, Dou J, Chen Q, Xu M, Yuan H, et al. SUMOylation of TARBP2 regulates miRNA/siRNA efficiency. Nat Commun. 2015;6:8899. doi: 10.1038/ncomms9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C, Huys A, Thibault PA, Wilson JA. Requirements for human Dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virology. 2012;433:479–488. doi: 10.1016/j.virol.2012.08.039. [DOI] [PubMed] [Google Scholar]
- 46.Eekels JJ, Geerts D, Jeeninga RE, Berkhout B. Long-term inhibition of HIV-1 replication with RNA interference against cellular co-factors. Antiviral Res. 2011;89:43–53. doi: 10.1016/j.antiviral.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 47.Sanghvi VR, Steel LF. The cellular TAR RNA binding protein, TRBP, promotes HIV-1 replication primarily by inhibiting the activation of double-stranded RNA-dependent kinase PKR. J Virol. 2011;85:12614–12621. doi: 10.1128/JVI.05240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura T, Kunz RC, Zhang C, Kimura T, Yuan CL, Baccaro B, Namiki Y, Gygi SP, Hotamisligil GS. A critical role for PKR complexes with TRBP in Immunometabolic regulation and eIF2α phosphorylation in obesity. Cell Rep. 2015;11:295–307. doi: 10.1016/j.celrep.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye J, Wang J, Tan L, Yang S, Xu L, Wu X, Deng H, Tan H. Expression of protein TARBP1 in human hepatocellular carcinoma and its prognostic significance. Int J Clin Exp Pathol. 2015;8:9089–9096. [PMC free article] [PubMed] [Google Scholar]
- 50.Sand M, Skrygan M, Georgas D, Arenz C, Gambichler T, Sand D, Altmeyer P, Bechara FG. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51:916–922. doi: 10.1002/mc.20861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.