Abstract
The actin family members, consisting of actin and actin-related proteins (ARPs), are essential components of chromatin remodeling complexes. ARP6, one of the nuclear ARPs, is part of the Snf-2-related CREB-binding protein activator protein (SRCAP) chromatin remodeling complex, which promotes the deposition of the histone variant H2A.Z into the chromatin. In this study, we showed that ARP6 influences the structure and the function of the nucleolus. ARP6 is localized in the central region of the nucleolus, and its knockdown induced a morphological change in the nucleolus. We also found that in the presence of high concentrations of glucose ARP6 contributed to the maintenance of active ribosomal DNA (rDNA) transcription by placing H2A.Z into the chromatin. In contrast, under starvation, ARP6 was required for cell survival through the repression of rDNA transcription independently of H2A.Z. These findings reveal novel pleiotropic roles for the actin family in nuclear organization and metabolic homeostasis.
Keywords: Actin-related protein, ARP6, Histone H2A.Z, Nucleolus, Wndchrm
1. Introduction
Gene functions are regulated by the modulation of chromatin structure as well as by the spatial association of genes with nuclear regions, including the nuclear lamina and the nucleolus. Chromatin remodeling complexes play central roles in the change of chromatin structure through their enzymatic activity and their regulatory subunits.
The actin family consists of conventional actin and of actin-related proteins (ARPs). The ARPs share the evolutionarily conserved basal structure with actin, which is called the actin fold. In addition, each ARP has individual features. ARPs are classified into ten subfamilies according to the degree of similarity to actin. Among these subfamilies, ARPs 4, 5, 6, 7, 8, and 9 are predominantly localized in the nucleus and were found in chromatin remodeling and histone modification complexes [1]. For example, vertebrate ARP6 is an essential subunit of the Snf-2-related CREB-binding protein activator protein (SRCAP) chromatin remodeling complexes together with β-actin and ARP4 [2,3]. The yeast homolog of the SRCAP complex, the SWR1 chromatin remodeling complex, also includes the same actin family molecules. We have previously shown that nuclear ARPs are indispensable for the function of chromatin remodeling complexes [3]. Particularly, the vertebrate ARP6 is essential for the activity of the SRCAP complex [3]. The SRCAP complex incorporates the histone variant H2A.Z into nucleosomes at promoter regions. H2A.Z plays important roles in epigenetic regulation and chromosome architecture and is associated with a variety of cellular mechanisms, including stem cell maintenance, cellular proliferation, and stress response, through transcriptional regulation and chromosome architecture [4–6].
Nuclear actin family proteins have additional roles in nuclear domain organization independently of the chromatin remodeling complexes. Actin directly interacts with lamins [7], and the dynamics of nuclear actin contribute to nuclear morphology [8,9]. In budding yeast, a fraction of ARP6 is free from the SWR1 complex and regulates genes encoding ribosomal proteins by relocating the gene loci to the nuclear periphery [10]. In addition, vertebrate ARP6 is involved in the spatial organization of chromosome territories in a SRCAP complex-dependent and -independent manner [11].
The nucleolus is composed of three compartments. The fibrillar center (FC) and the dense fibrillar component (DFC) occupy the central region, where ribosomal DNA (rDNA) and pre-ribosomal RNA (pre-rRNA) are transcribed. The granular component (GC) surrounds the FC-DFC component and consists of ribosomal proteins assembled into pre-ribosomes. Ribosomal biogenesis consists of multiple steps, starting from transcription and processing of pre-rRNAs and ending with ribosome assembly. These processes are required for protein synthesis, which consume substantial amounts of cellular energy [12]. Therefore, rDNA transcription and ribosome assembly are linked to cell proliferation [13–16]. Under nutrient shortage, cells preserve cellular energy by downregulating ribosome biosynthesis, thus protecting cells from energy deprivation–induced apoptosis [14,17]. Therefore, the nucleolus supplies sufficient amount of ribosomes for cell growth in the presence of glucose, whereas this activity is switched off upon nutrient deprivation. Although several studies have been performed [14,18], a molecule linking nucleolar organization and function has not been fully investigated.
Here, we analyzed the involvement of vertebrate ARP6 in the organization of the nucleolus and in the transcriptional regulation of rDNA. Loss of function analyses suggested that ARP6 contributes to the morphological organization and functions of the nucleolus. In the presence of high concentrations of glucose, ARP6 maintains rDNA transcription possibly through the incorporation of histone H2A.Z into the chromatin. Instead, upon glucose deprivation, ARP6 contributes to the repression of rDNA transcription to avoid cell death, independently of H2A.Z. From these results, we propose a dual role for ARP6 in rDNA transcription homeostasis, one dependent and the other independent of H2A.Z.
2. Materials and methods
2.1. Cell culture and treatment
DT40 cells were cultured at 38.5 °C under 5% CO2 in DMEM (Wako) supplemented with 10% fetal bovine serum, 1% chicken serum, penicillin-streptomycin (Gibco) and 10 µM 2-mercaptoethanol (Wako). HeLa cells were maintained as described previously [19]. For glucose deprivation, DT40 cell pellet was washed twice with 10 mL of phosphate-buffered saline PBS (−) and cultured in DMEM medium without glucose (Gibco), supplemented as described above. For the selective inhibition of the Pol I activity, HeLa cells were treated with actinomycin D (Wako) at 5 nM for 3 h. Dead cells were counted by Trypan blue staining. For RNAi-mediated ARP6 knockdown, HeLa cells were transfected with siRNAs using RNAiMAX (Invitrogen). The cells were analyzed 48 h after transfection. The siRNA duplexes used are: HSS148894 for siARP6 (i) and HSS148895 for siARP6 (ii) (Invitrogen). The target sequence for the control (siControl) is: CUUACGCUGAGUACUUCGA (GL3, Nippon EGT). To repress the expression of the tetracycline-responsive transgenes in DT40 cells, ARP6 and H2A.Z conditional knockout cell lines (ARP6-KO and H2A.Z-KO) were treated with tetracycline (Wako) at a final concentration of 2 µg/mL [11]. The cells were analyzed 4–6 days (ARP6-KO) and 3–4 days (H2A.Z-KO) after the tetracycline treatment, when the proteins were almost absent [11].
2.2. Immunofluorescence microscopy
To investigate the subcellular localization of ARP6, immunofluorescence analyses were performed as follows. HeLa cells grown on coverslips were transfected with a Flag-hARP6 cDNA construct [20] using Lipofectamine 2000 (Invitrogen). The cells were fixed with freshly prepared 4% paraformaldehyde/PBS for 15 min at room temperature and then permeabilized with 0.5% Triton X-100 for 10 min. The monoclonal anti-Flag M2 antibody (F1804, Sigma–Aldrich) and polyclonal anti-RPA194 antibody (Santa Cruz) were used for the detection of Flag-hARP6 and RPA194, respectively. Nuclei were visualized with DAPI and the coverslips were mounted with vector shield (Vector Laboratories Inc.). Images were acquired with a confocal laser scanning microscope FV1000 (Olympus). To investigate nucleolar morphology, HeLa cells were analyzed by immunofluorescence as described above using anti-Arp6 (Sigma–Aldrich, A1857), anti-upstream binding factor (UBF) (Santa Cruz, sc-13125), anti-fibrillarin (Santa Cruz, ab4566), and anti-B23 (Santa Cruz, sc-6013-R) antibodies. The images were obtained with an IX-71 microscope (Olympus).
2.3. Quantification of nucleolar morphology
To measure differences, we performed supervised machine learning of nucleolar images using pattern recognition software, wndchrm (weighted neighbor distance using a computed hierarchy of algorithms representing morphology) ver. 1.31 [21–23]. We trained a machine with 70 nucleolar images for each cell type/class. Nuclear regions were pre-excised from immunofluorescent images to be placed in the center of the area in 150 × 150 pixels using the Image J1 program [24]. Cross-validation tests were automatically repeated for 20 times with 56 training/14 test image data set. The options used for the image analysis were a large feature set of 2873 (−l) and multi-processors (−m). To measure pairwise class dissimilarity, morphological distances were calculated as the euclidean distances (d = √Σ(A−B)2) from the values in class probability matrix obtained from the cross-validations [25].
2.4. RNA preparation and reverse transcription–quantitative PCR (RT-qPCR)
RNA from HeLa cells or DT40 cells was extracted with the RNeasy Mini kit (QIAGEN) according to the manufacturer's protocol. The RNA was converted into cDNA by reverse transcriptase using random primers (Applied Biosystems). RT-qPCR was performed with ABI Prism 7300 instrument and SYBR Green PCR Master MIX (Applied Biosystems). Values were normalized against β-actin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene expression before calculating relative fold changes. The sequences of the primers used are: human pre-ribosomal RNA (pre-rRNA): GAACGGTGGTGTGTCGTTC and GCGTCTCGTCTCGTCTCACT; human ARP6 (hARP6): ATGACGACCTTAGTGCTGGA and GTGCTGTTTTT-GACCGGAAC; human β-actin: ATCGTCCACCGCAAATGCTTCTA and AGCCATGCCAATCTCATCTTGTT; gallus pre-rRNA: TCGTCTGTAG-GAGCGAGTGAG and CCTTAGCCCAGGACAGAGC; gallus GAPDH: GGTGGTGCTAAGCGTGTTA and CCCTCCACAATGCCAA; human RPL5: CACTGGCAATAAAGTTTTTGGTG and AACCAGGGAATCGTTTGGT; human RPL11: CCGCAAACTCTGTCTCAACA and TGCCAAAGGATCT-GACAGTG; human RPL21: GAGCCGAGATAGCTTCCTGA and CTCCTTCCCATTGGTTCTCA. The primers for gallus pre-rRNA were designed according to the database sequence (Genebank: AADN02004142.1 and GenBank: DQ018752.1).
3. Results and discussion
3.1. Localization of ARP6 in the nucleolus
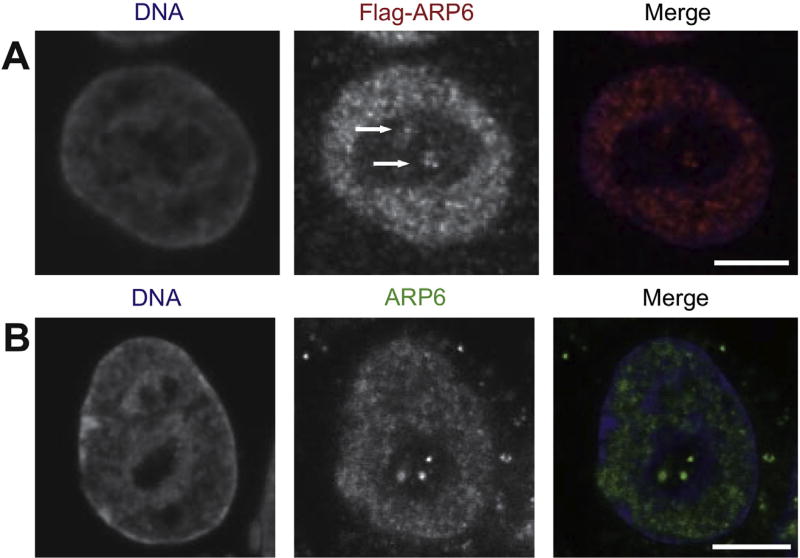
To analyze the involvement of the nuclear actin family in nuclear organization, we first examined the subnuclear localization of ARP6 in HeLa cells by expressing Flag-tagged human ARP6, which was observed in the nucleoplasm and in the nucleolus where DAPI density was low, (Fig. 1A, white arrows). The nucleolar localization of ARP6 was also confirmed by the detection of endogeneous ARP6 with a specific antibody (Fig. 1B). Therefore, we concluded that a part of ARP6 is localized in the nucleolus.
Fig. 1.
Subcellular localization of human ARP6 in HeLa cells. (A) Flag-ARP6 was transiently expressed in HeLa cells and its localization was detected by immunofluorescence with anti-Flag antibodies. ARP6 in the nucleolus is indicated by arrows. (B) Endogenous ARP6 molecules were detected by immunofluorescence using an anti-ARP6 antibody. As in (A), dot-like signals were observed in the nucleolus. Scale bar, 5 µm.
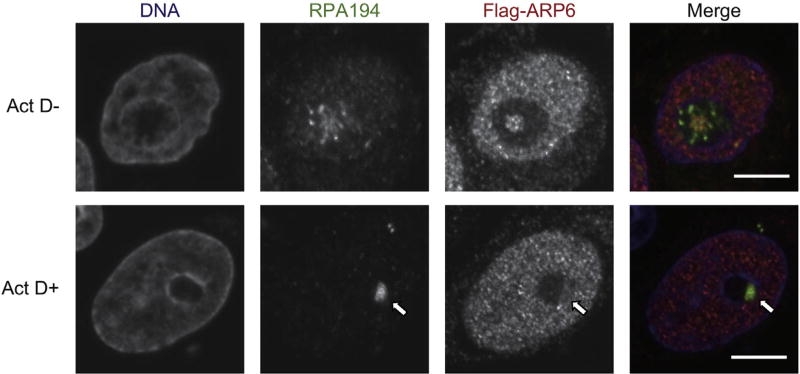
Since ARP6 localizes in the central region of the nucleolus where rDNA is transcribed, we further investigated whether ARP6 was involved in rDNA transcription, a nucleolar function. Immunofluorescence analysis showed that the nucleolar ARP6 (Flag-ARP6) partially colocalized with RPA194, the catalytic subunit of RNA polymerase I (Pol I) (Fig. 2, upper panels). When Pol I transcription was inhibited with a low dose of actinomycin D, ARP6 relocated from the nucleolus to the nucleolar periphery and formed a unique structure, called nucleolar cap, where RPA194 was accumulated (Fig. 2, lower panels). Since many nucleolar factors that relocalize to the nucleolar cap after Pol I inhibition are involved in rDNA transcription [26], ARP6 may also play a role in regulation of rDNA transcription.
Fig. 2.
Relocation of ARP6 and RPA194, a catalytic subunit of RNA polymerase I, to the nucleolar cap after the treatment with a low dose of actinomycin D (Act D+). Flag-ARP6 was detected as in Fig. 1A, and RPA194 was detected with a specific antibody. The nuclear cap by the treatment of actinomycin D was shown by a white arrow. Scale bar, 5 µm.
3.2. ARP6 is involved in morphological organization of the nucleolus
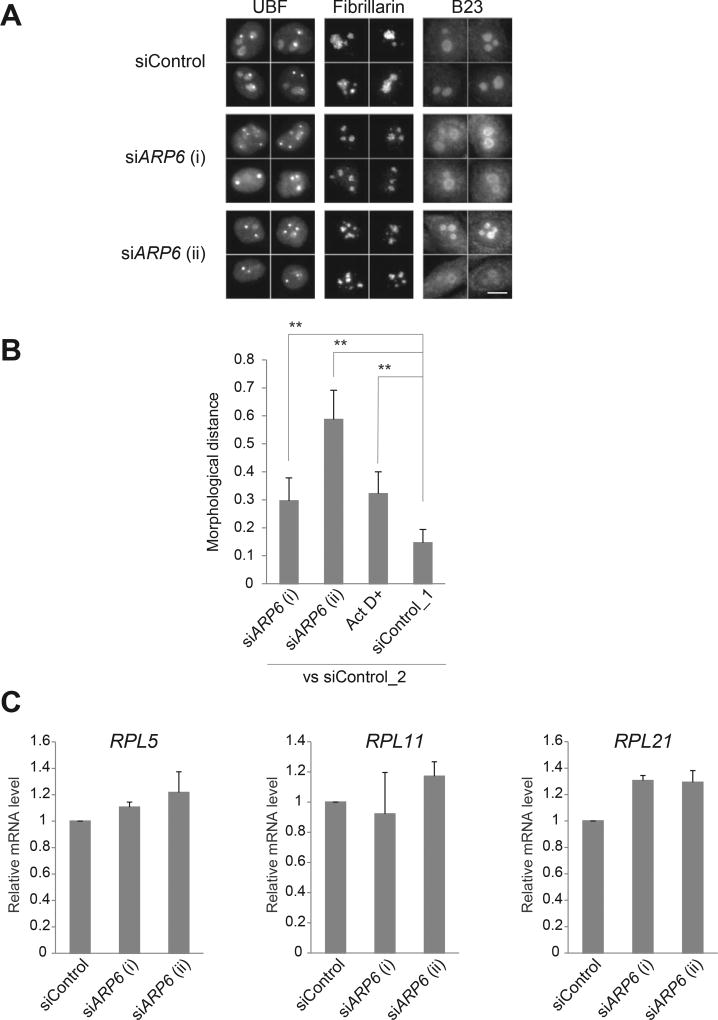
To further test the contribution of ARP6 to the nucleolar structure, we depleted ARP6 in HeLa cells with specific siRNAs and visualized the nucleolus by immunofluorescence using antibodies against UBF, fibrillarin, and B23, all markers of the three sub-nucleolar compartments, FC, DFC and GC, respectively. After 48 h from the transfection of the siRNA, ARP6 mRNA levels were efficiently reduced (Supplementary Fig. 1). ARP6 depletion changed the structures of both the FC and the DFC, where it localizes. The FC (visualized by UBF) in the ARP6 knockdown cells became more condensed, and DFC (visualized by fibrillarin) became smaller and round (Fig. 3A, Supplementary Figs. 2 and 3). ARP6 depletion also had an effect on GC (visualized by B23), which appeared a ring-like shape (Fig. 3A).
Fig. 3.
Morphological changes of the nucleolus upon ARP6 depletion. (A) HeLa cells were transfected with siRNA targeting ARP6 and immunostained with antibodies against UBF, fibrillarin, and B23, specific markers for the nucleolar subcompartments FC, DFC, and GC, respectively. Four representative images for each staining are shown. Scale bar, 10 µm. (B) Quantification of morphological difference of the UBF-stained nucleoli was performed with wndchrm, to compute degree of morphological distance from siControl_2. A large value of the morphological distance indicates that the morphology of the cells is different from the morphology of siControl_2 cells. siControl_1 was used as a negative control. Image sets used in this analysis are shown in Supplementary Fig. 2. Values are means ± standard error of 20 cross validation tests; **, p < 0.01. (C) Ribosomal protein gene expressions in ARP6 knockdown cells. The mRNA was measured with RT-qPCR. GAPDH mRNA expression was used as the internal control. Expression levels of control cells are set to 1. Values are represented as means ± standard error of triplicates.
To quantitatively assess nucleolar morphology changes, we used a supervised machine learning algorithm, wndchrm, which is useful for image classification and detection of morphological differences [21,22,29]. To train the machine, we created five image libraries: siARP6 (i), siARP6 (ii), Act D+, siControl_1 and siControl_2, each containing 70 nucleolar immunofluorescence images of UBF (Supplementary Fig. 2). Two control image classes, siControl_1 and siControl_2, were intentionally created from the cells that were treated with the same siRNA control; one serves as a reference for image comparison and the other as a negative control, since induces no morphological differences. A positive control class, Act D+ contains cells treated with low-dose actinomycin D, a Pol I inhibitor that induces changes in nucleolar morphology [18].
The image classification test among the five classes provided marginal class probabilities, from which we calculated pairwise distances that reflect degree of morphological differences (see Materials and method) [25]. As expected, a comparison between control cells (siControl_2 vs siControl_1) showed a low level of morphological differences, while ARP6 knockdown cells (siControl_2 vs siARP6 [i] and siARP6 [ii]) showed significant difference from control cells (siControl_2), similarly to or more than actinomycin D-treated cells (Act D+) (Fig. 3B). Similar results were obtained with fibrillarin-stained nucleoli, which identify DFC (Supplementary Fig. 3).
Unlike Arp6 deletion in yeast [10], ARP6 knockdown in mammalian cells had a small or no effect on the expression of ribosomal protein genes (Fig. 3C), suggesting that the nucleoli changed their morphologies without modifying the level of ribosomal proteins. Since the proteins are assembled in the nucleolus and expected to contribute to the nucleolar structure, the effect of ARP6 on the nucleolus can be direct. In summary, ARP6 may influence the formation and/or maintenance of the proper nucleolar structure and function.
3.3. ARP6 is required for the maintenance of active rDNA transcription through H2A.Z
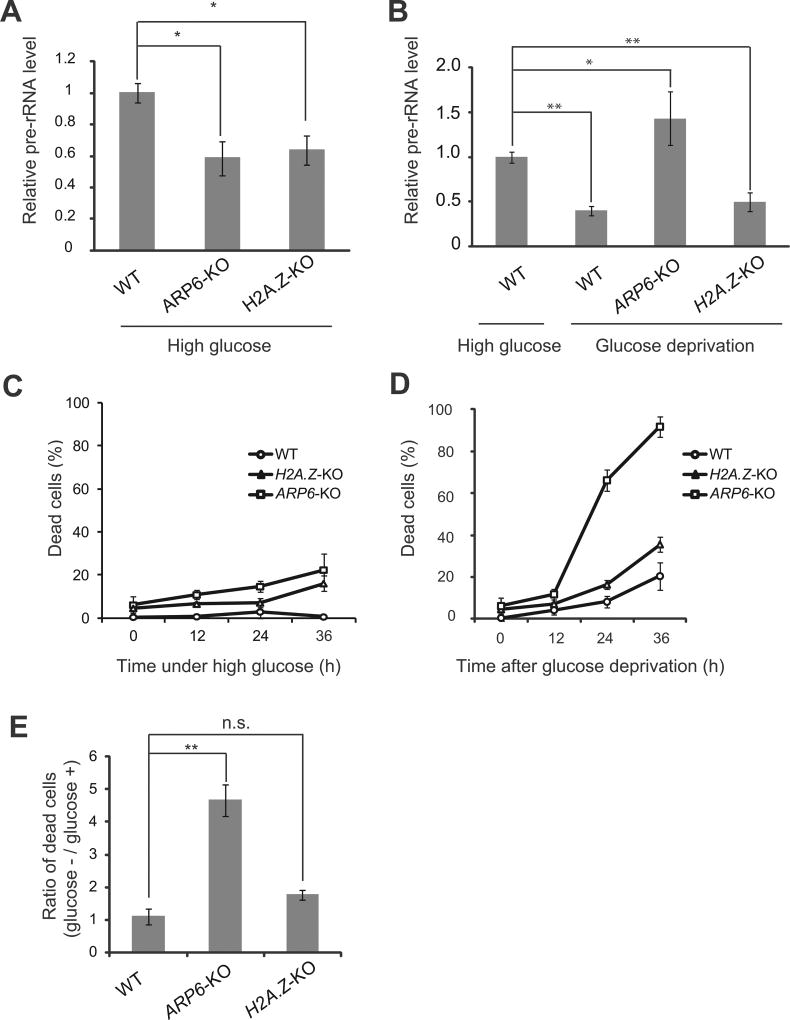
To investigate the effect of ARP6 dysfunction in rDNA transcription, we used chicken DT40 cell lines (ARP6-KO and H2A.Z-KO) where tetracycline was used to suppress ARP6 and H2A.Z gene expression [11]. To test whether ARP6 was involved in rDNA transcription in the presence of high glucose, we quantified the relative level of pre-rRNA in ARP6-KO cells by RT-qPCR. The rDNA transcription decreased in the absence of ARP6 (Fig. 4A). A portion of vertebrate ARP6 is an essential component of the SRCAP chromatin remodeling complex [3], which deposits H2A.Z into the chromatin. H2A.Z was previously shown to be incorporated in the rDNA promoter [30]. To test the ARP6 function on rDNA chromatin as a component of the SRCAP, we analyzed rDNA transcription in H2A.Z-KO cells [11]. Our RT-qPCR experiment showed that pre-rRNA decreased in H2A.Z-KO cells to a similar level as in ARP6-KO cells, suggesting that H2A.Z plays a significant role in rDNA chromatin (Fig. 4A). These observations suggested that in a high glucose environment ARP6 is involved in the maintenance of active rDNA transcription through the SRCAP complex.
Fig. 4.
ARP6 is involved in the activation and repression of rDNA transcription. (A) The comparison of rDNA transcription among wild-type (WT), ARP6-KO, and H2A.Z-KO cells in the presence of high glucose. GAPDH was used as the internal control. (B) Analysis of rDNA transcription in WT cells under high glucose (the left bar) and in WT, ARP6-KO, and H2A.Z-KO cells after glucose deprivation for 24 h (three right bars). GAPDH was used as the internal control. (C and D) Cells were cultured with a continuous supply of high-dose glucose (C), or in the absence of glucose (D). Dead cells (%) were counted every 12 h by trypan blue staining. (E) Ratio between dead cells at 24 h after glucose deprivation and cells under high glucose. For (A), (B) and (E), values represent means ± standard error of triplicates; *, p < 0.05; **, p < 0.01; n.s. not significant.
3.4. ARP6 contributes to the repression of rDNA transcription under glucose starvation with a SRCAP complex-independent mechanism
The rDNA transcription is regulated by environmental stresses including limited nutrient availability [18]. Under glucose deprivation, cells reduce the rDNA transcription to save energy derived by glucose metabolism [14,17,31], otherwise the cells go through apoptosis due to lack of energy. To test the contribution of ARP6 and H2A.Z to the repression of rDNA transcription, we measured pre-rRNA synthesis in wild-type, ARP6-KO, and H2A.Z-KO cells under glucose starvation (Fig. 4B). Whereas glucose starvation for 24 h in wild-type cells reduced rDNA transcription (Fig. 4B, two left bars), in ARP6-KO cells rDNA transcription was not repressed (Fig. 4B, ARP6-KO). On the other hand, under glucose starvation in H2A.Z-KO cells rDNA transcription was repressed to a similar level as in wild-type cells (Fig. 4B, H2A.Z-KO and the second bar).
Because pre-rRNA transcription was dysregulated in the absence of ARP6 (Fig. 4A and B), we measured cell viability. ARP6-KO cells showed a significantly impaired survival under glucose deprivation (Fig. 4C and D). In contrast, cell death was only slightly increased in H2A.Z-KO cells. When the relative increase of dead cells was calculated based on their growth in a high glucose environment, ARP6-KO cells, but not H2A.Z-KO cells, showed a significantly impaired survival under glucose deprivation (Fig. 4E). Therefore, ARP6 may contribute to the repression of rDNA transcription in a nutrient-limited environment independently of the deposition of H2A.Z by the SRCAP complex.
3.5. ARP6 is involved in nucleolar function and organization
In this study, we showed that ARP6 is localized in the nucleolus and is required for its structure and function. Our intensive morphological measurement clearly demonstrated that the FC region of the nucleolus, where rDNA transcription takes place, changed its shape upon ARP6 knockdown. ARP6 has a dual role in the regulation of rDNA transcription. In the presence of high glucose, when cells grow steadily, ARP6 maintains active rDNA transcription through H2A.Z deposition (Fig. 4). Under glucose starvation, ARP6 is required for the repression of rDNA transcription, and this function may be independent of H2A.Z-deposition. ARP6 plays a structural role for the nucleolus, either directly or through the regulation of rDNA transcription.
Whereas ARP6 deletion in yeasts leads to derepression of genes encoding ribosomal proteins [10], upregulation of ribosomal protein genes was not observed in ARP6-knock down HeLa cells in this study (Fig. 3). The discrepancy might be due to the different spatial localization of these genes in the nucleus; in the yeast, ribosomal protein genes are associated with the nuclear pore complex, but in the vertebrate they are not [10], thus suggesting different roles of yeast and vertebrate ARP6 in the regulation of ribosomal protein genes.
Besides the ribosomal roles, the nucleolus functions also as a stress sensor [13,18,32]. Many of the cell stress signal pathways linked to the nucleolus efficiently block rRNA synthesis, stabilize and activate tumor suppressors, and lead to cell cycle arrest [18,33,34]. Perturbation of ribosomal biogenesis or of nucleolar architecture results in the so-called nucleolar stress, which has been implicated in several diseases [35–38]. Since our study suggests that ARP6 could link nucleolar architecture and function, further analyses on ARP6 might contribute to the elucidation of the relationship between the nucleolus and diseases.
Supplementary Material
Acknowledgments
We thank all the members of the laboratory for the discussions. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (25116009) (M.H. and N.S.) and the Human Frontier Science Program (RGP0017) (M.H.). H. K. and H. M. thank the Japan Society for the Promotion of Science (JSPS) for Young Scientist Fellowships.
P. H. was supported by Human Frontier in Science programe (RGP0017/2013), A. K. by Ministry of Education, Youth and Sports of the Czech Republic (CZ.1.07/2.3.00/30.0050). This publication is supported by the project “BIOCEV – Biotechnology and Biomedicine Centre of the Academy of Sciences and Charles University” (CZ.1.05/1.1.00/02.0109), from the European Regional Development Fund and with institutional support (RVO68378050).
IGG is supported by the Intramural Research Program of the U.S. National Institutes of Health, National Institute on Aging.
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.bbrc.2015.07.005.
Transparency document
Transparency document related to this article can be found online at http://dx.doi.org/10.1016/j.bbrc.2015.07.005.
References
- 1.Oma Y, Harata M. Actin-related proteins localized in the nucleus: from discovery to novel roles in nuclear organization. Nucleus. 2011;2:38–46. doi: 10.4161/nucl.2.1.14510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda R, Hori T, Kitamura H, Takeuchi K, Fukagawa T, Harata M. Identification and characterization of the two isoforms of the vertebrate H2A.Z histone variant. Nucleic Acids Res. 2010;38:4263–4273. doi: 10.1093/nar/gkq171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallant-Behm CL, Ramsey MR, Bensard CL, Nojek I, Tran J, Liu M, Ellisen LW, Espinosa JM. DeltaNp63alpha represses anti-proliferative genes via H2A.Z deposition. Genes Dev. 2012;26:2325–2336. doi: 10.1101/gad.198069.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon DN, Zastrow MS, Wilson KL. Direct actin binding to A- and B-type lamin tails and actin filament bundling by the lamin A tail. Nucleus. 2010;1:264–272. doi: 10.4161/nucl.1.3.11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cibulka J, Fraiberk M, Forstova J. Nuclear actin and lamins in viral infections. Viruses. 2012;4:325–347. doi: 10.3390/v4030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verboon JM, Rincon-Arano H, Werwie TR, Delrow JJ, Scalzo D, Nandakumar V, Groudine M, Parkhurst SM. Wash interacts with lamin and affects global nuclear organization. Curr. Biol. 2015;25:804–810. doi: 10.1016/j.cub.2015.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida T, Shimada K, Oma Y, Kalck V, Akimura K, Taddei A, Iwahashi H, Kugou K, Ohta K, Gasser SM, Harata M. Actin-related protein Arp6 influences H2A.Z-dependent and -independent gene expression and links ribosomal protein genes to nuclear pores. PLoS Genet. 2010;6:e1000910. doi: 10.1371/journal.pgen.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maruyama EO, Hori T, Tanabe H, Kitamura H, Matsuda R, Tone S, Hozak P, Habermann FA, von Hase J, Cremer C, Fukagawa T, Harata M. The actin family member Arp6 and the histone variant H2A.Z are required for spatial positioning of chromatin in chicken cell nuclei. J. Cell Sci. 2012;125:3739–2743. doi: 10.1242/jcs.103903. [DOI] [PubMed] [Google Scholar]
- 12.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 14.Grummt I. The nucleolus-guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- 15.Grummt I, Voit R. Linking rDNA transcription to the cellular energy supply. Cell Cycle. 2010;9:225–226. doi: 10.4161/cc.9.2.10614. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka Y, Okamoto K, Teye K, Umata T, Yamagiwa N, Suto Y, Zhang Y, Tsuneoka M. JmjC enzyme KDM2A is a regulator of rRNA transcription in response to starvation. EMBO J. 2010;29:1510–1522. doi: 10.1038/emboj.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoppe S, Bierhoff H, Cado I, Weber A, Tiebe M, Grummt I, Voit R. AMP-activated protein kinase adapts rRNA synthesis to cellular energy supply. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17781–17786. doi: 10.1073/pnas.0909873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol. Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitayama K, Kamo M, Oma Y, Matsuda R, Uchida T, Ikura T, Tashiro S, Ohyama T, Winsor B, Harata M. The human actin-related protein hArp5: nucleo-cytoplasmic shuttling and involvement in DNA repair. Exp. Cell Res. 2009;315:206–217. doi: 10.1016/j.yexcr.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 20.Ohfuchi E, Kato M, Sasaki M, Sugimoto K, Oma Y, Harata M. Vertebrate Arp6, a novel nuclear actin-related protein, interacts with heterochromatin protein 1. Eur. J. Cell. Biol. 2006;85:411–421. doi: 10.1016/j.ejcb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Orlov N, Shamir L, Macura T, Johnston J, Eckley DM, Goldberg IG. WNDCHARM: multi-purpose image classification using compound image transforms. Pattern Recognit. Lett. 2008;29:1684–1693. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shamir L, Delaney JD, Orlov N, Eckley DM, Goldberg IG. Pattern recognition software and techniques for biological image analysis. PLoS Comput. Biol. 2010;6:e1000974. doi: 10.1371/journal.pcbi.1000974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eliceiri KW, Berthold MR, Goldberg IG, Ibanez L, Manjunath BS, Martone ME, Murphy RF, Peng H, Plant AL, Roysam B, Stuurman N, Swedlow JR, Tomancak P, Carpenter AE. Biological imaging software tools. Nat. Methods. 2012;9:697–710. doi: 10.1038/nmeth.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston J, Iser WB, Chow DK, Goldberg IG, Wolkow CA. Quantitative image analysis reveals distinct structural transitions during aging in caenorhabditis elegans tissues. PLoS One. 2008;3:e2821. doi: 10.1371/journal.pone.0002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamir L, Orlov N, Eckley DM, Macura T, Johnston J, Goldberg IG. Wndchrm - an open source utility for biological image analysis. Source Code Biol. Med. 2008;3:13. doi: 10.1186/1751-0473-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van de Nobelen S, Rosa-Garrido M, Leers J, Heath H, Soochit W, Joosen L, Jonkers I, Demmers J, van der Reijden M, Torrano V, Grosveld F, Delgado MD, Renkawitz R, Galjart N, Sleutels F. CTCF regulates the local epigenetic state of ribosomal DNA repeats. Epigenetics Chromatin. 2010;3:19. doi: 10.1186/1756-8935-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- 32.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 33.Sloan KE, Bohnsack MT, Watkins NJ. The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell. Rep. 2013;5:237–247. doi: 10.1016/j.celrep.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 35.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsai RY, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 2014;28:3290–3296. doi: 10.1096/fj.14-254680. [DOI] [PubMed] [Google Scholar]
- 37.Hetman M. Role of the nucleolus in human diseases. Preface. Biochim. Biophys. Acta. 2014;1842:757. doi: 10.1016/j.bbadis.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 38.Hetman M, Pietrzak M. Emerging roles of the neuronal nucleolus. Trends Neurosci. 2012;35:305–314. doi: 10.1016/j.tins.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.