Abstract
Proliferative vitreoretinopathy (PVR) is a common complication of open globe injury and the most common cause of failed retinal detachment surgery. The response by retinal pigment epithelial (RPE) cells liberated into the vitreous includes proliferation and migration; most importantly, epithelial to mesenchymal transition (EMT) of RPE plays a central role in the development and progress of PVR. For the first time, we show that knockdown of BIRC5, a member of the inhibitor of apoptosis family, using either lentiviral vector based CRISPR/Cas9 nickase gene editing or inhibition of survivin using the small-molecule inhibitor YM155, results in the suppression of EMT in RPE cells. Knockdown of survivin or inhibition of survivin significantly reduced TGFβ-induced cell proliferation and migration. We further demonstrated that knockdown or inhibition of survivin attenuated the TGFβ signaling by showing reduced phospho-SMAD2 in BIRC5 knockdown or YM155-treated cells compared to controls. Inhibition of the TGFβ pathway using TGFβ receptor inhibitor also suppressed survivin expression in RPE cells. Our studies demonstrate that survivin contributes to EMT by cross-talking with the TGFβ pathway in RPE cells. Targeting survivin using small-molecule inhibitors may provide a novel approach to treat PVR disease.
Keywords: BIRC5, survivin, lentiviral CRISPR/Cas9 nickase vector, YM155, retinal pigment epithelial cells, epithelial to mesenchymal transition (EMT)
Introduction
Proliferative vitreoretinopathy (PVR) is a common complication of open globe injury and the most common cause of failed retinal detachment surgery. It is characterized by the formation and contraction of epiretinal membranes (ERM) and causes rhegmatogenous retinal detachment (RRD) [1]. PVR occurs in 5%–10% of all RRD, and it is a major risk factor for redetachment after surgery [2]. PVR also occurs in other ocular disorders including large retinal tears or injuries, and endoresection of tumors. Currently, the standard treatment for PVR is surgery by restoring normal anatomy and the ciliary body function, including scleral buckling, vitrectomy, membrane peeling, and retinotomies. However, the success rate is only 40–80% [2]. Therefore, understanding the molecular mechanisms of PVR and developing new therapies including pharmacological intervention are essential to better treat this disease.
There are several cell types identified in ERMs including retinal pigment epithelial (RPE) cells, astrocytes, microglia, macrophages, and Muller cells [3, 4]. RPE cells are primary contributors in forming ERM by transforming into fibroblastic and myofibroblastic phenotypes via epithelial to mesenchymal transition (EMT) [5]. However, the molecular mechanisms underlying EMT in RPE cells remain elusive. Previous studies indicated that multiple signaling pathways were involved in EMT of RPE cells including Wnt/β-catenin[6], TGFβ [7], Notch[8], HIF1α[9], ERK1/2[10], and p38MAPK[11]. Activation of those pathways promoted EMT in RPE cells.
Survivin, a member of the inhibitor of apoptosis protein (IAP) family encoded by the BIRC5 gene, functions by inhibiting the caspase activation and promotes cell growth, which is required for embryonic development [12]. Survivin was extensively studied in various human cancers and highly expressed in cancer cells but rarely expressed in normal corresponding cells, thus making is as a drug target for cancer therapy. Several studies demonstrated that survivin promoted EMT in cancer cells by participating in multiple signaling pathways: TGFβ, ERK1/2, and PI3/AKT [13–15]. Survivin was expressed in RPE cells as a survival factor [16, 17] and induced by Activin A, a member of the TGFβ superfamily [18] and TGFβ in RPE cells [19]. However, the function of survivin and how survivin contributes to EMT in RPE cells is little understood.
In this study, we investigated the role of survivin in ARPE19 cells by using lentiviral CRISPR/Cas9 nickase vector-mediated gene editing and inhibition of survivin expression using a small-molecule survivin inhibitor, YM155. For the first time, we show that survivin regulates EMT by activating the TGFβ pathway in RPE cells, and thus, survivin inhibitors may provide a novel approach to treat PVR.
Materials and Methods
Cell culture
ARPE-19 cells were obtained from the American Type Culture Collection and maintained at 37°C in a 5% CO2 in DMEM/F-12 medium supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA) and antibiotics (Penicillin– Streptomycin solution, Invitrogen; Carlsbad, CA). Cells were cultured to 90% confluence and serum-starved for 12 h before survivin inhibitor treatment. HEK293 FT cells were purchased from Invitrogen and cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, 100 µg/mL streptomycin, 1% glutamine and 1% nonessential amino acids. TGFβ1 was purchased from Sigma (St. Louis, MO).
Lentiviral vector production
The lentiviral CRISPR/Cas9 nickase-mediated BIRC5 gene editing vectors were described previously [13]. Survivin knockdown (KD) stable cell lines were generated by transducing the ARPE-19 cells with the lentiviral CRISPR/Cas9 nickase BIRC5 vector and lentiCas9-blast Cas9 nickase vectors and selected with 5 µg/ml puromycin or 10 µg/ml blasticidin. LentiCas9-blast was used as the control vector without gRNAs.
Immunofluorescent staining
Cells were fixed for 5 min in 4% paraformaldehyde and washed three times with 0.1% Tween 20 in PBS (PBST), and then incubated with blocking buffer (5% normal goat serum, 3% bovine serum albumin, and 0.1% Triton-X 100 in PBS) for 1 h. Cells were incubated with the primary antibodies to cytokeratin-7 (Abcam, Cambridge, MA) and β-catenin (1:200 dilution, Cell Signaling, Danvers, MA) overnight at 4°C and then washed three times with PBST. Secondary antibodies, Alexa 488 or 594 conjugated goat anti-rabbit or mouse (1:200 dilution, Life Technologies), were added and incubated for 1 h at room temperature. Cell nuclei were counterstained with DAPI (Vector Laboratories, Inc.; Burlingame, CA). Images were taken using a Nikon inverted fluorescence microscope.
Cell migration assay
The cell migration assay was performed using a modified transwell chamber (BD Falcon™, San Jose, CA). These chambers were inserted into 24-well cell culture plates. ARPE-19 cells transduced with lentiviral BIRC5 Cas9 nickase gRNAs and control vectors (3 × 104) in 300 µl serum-free DMEM were added to the upper chamber. DMEM containing 6 ng/ml TGFβ in DMEM was added into the lower chamber of each well and incubated for 24 h. The medium and nonmigrated cells in the upper chamber were removed, while the migrated cells on the lower side of the membranes were fixed with methanol and stained with crystal violet. Pictures were taken at 20× magnification, and cells from at least three different fields were counted.
MTT assay
ARPE-19 cells (5,000/well), transduced with lentiviral CRISPR/Cas9 nickase for BIRC5 editing and control vectors, were plated into 96-well plates and cells and were then cultured at 2% DMEM with or without 6 ng/ml TGFβ for different time points (24, 48 and 72 h). OD570 was measured at indicated time points by adding 10 µl of MTT reagent into each well and incubated for ~4 h and then terminated by adding 100 µl detergent reagent to incubate at 22°C in the dark for 2 h. Cell proliferation was compared between control and BIRC5 KD cells with or without TGFβ treatment.
Western blot
ARPE-19 cells were collected and lysed in RIPA buffer (Thermo Scientific; Rockford, IL) containing 1% Halt Proteinase Inhibitor Cocktail (Thermo Scientific; Rockford, IL). An equal amount of protein was loaded onto 10% SDS-PAGE gels and transferred onto nitrocellulose membranes, which were then blocked with 5% non-fat milk for 1 h and incubated with primary antibodies against survivin, N-cadherin, vimentin (Cell Signaling), cytokeratin-7(Abcam) and GAPDH (Sigma; St. Louis, MO) for 24 h at 4°C. The membranes were washed with PBST and incubated with horseradish peroxidase-conjugated secondary antibodies at room temperature for 1 h. Protein bands were visualized using chemiluminescence by exposing on X-ray film.
Statistical analysis
Significant differences were determined from independent experiments performed in triplicate and presented as means ± S.D. using Student's t-test. p < 0.05 was considered to be a significant difference.
Results
Knockdown of BIRC5 using lentiviral CRISPR/Cas9 nickase-mediated editing or inhibition of survivin using the small-molecule survivin inhibitor YM155 leads to suppression of EMT in RPE cells
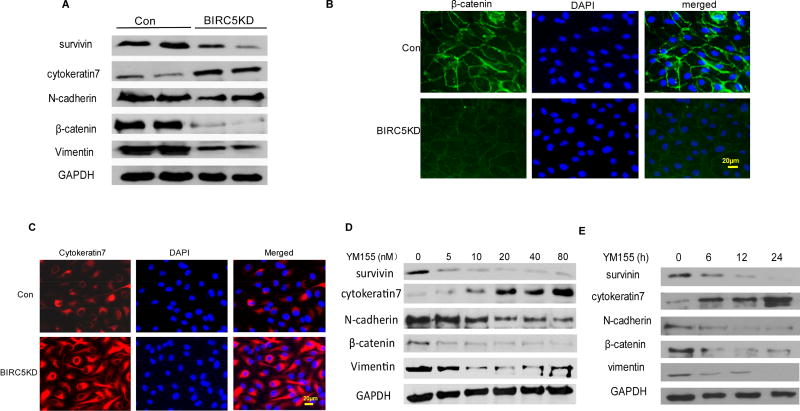
To investigate the role of BIRC5 in RPE cells, we generated BIRC5 KD and control stable cells by transducing ARPE-19 cell line with lentiviral BIRC5 CRISPR/Cas9 nickase and control vectors, respectively. In BIRC5 KD cell line, survivin expression was significantly reduced compared to control cells. EMT markers were also significantly altered in survivin KD compared to control cells, including epithelial cell marker cytokeratin-7 upregulation and mesenchymal markers N-cadherin, β-catenin, and vimentin downregulation, indicating that knockdown of survivin inhibited EMT in RPE cells (Fig. 1A). We also examined EMT markers by performing immunofluorescent staining of mesenchymal marker β-catenin and epithelial marker cytokeratin-7. β-catenin was stained in cell membranes with weak fluorescence, whereas cytokeratin-7 was strongly stained in cytoplasm and cell membrane in BIRC5 KD compared to control cells (Fig. 1B and 1C). Immunofluorescent staining data also verified our finding that knockdown of survivin inhibited EMT in RPE cells. In addition to the CRISPR/Cas9 nickase gene editing approach, we also applied a pharmacological approach by treating APRE19 cells using different doses of the small-molecule survivin inhibitor YM155. EMT markers were altered in a dose-dependent manner including epithelial marker cytokeratin-7 upregulation and mesenchymal markers N-cadherin, β-catenin, and vimentin downregulation (Fig. 1D). We also tested the efficacy of YM155 using a 20 nM dose at different time points. Survivin and mesenchymal markers were significantly inhibited whereas the epithelial marker cytokeratin-7 was upregulated at indicated time points (Fig. 1E). Our data demonstrated that knockdown or inhibition of survivin suppresses EMT in RPE cells.
Figure 1. Knockdown or inhibition of survivin suppresses EMT in ARPE-19 cells.
A. Western blot analysis of survivin and EMT markers in survivin knockdown(KD) and control ARPE-19 cells generated using lentiviral CRISPR/Cas9nickase vector. B. and C. Immunofluorescent staining of mesenchymal marker β-catenin (B) and epithelial marker cytokeratin-7 (C) in survivin KD and control ARPE-19 cells. D. Western blot analysis of EMT markers in wildtype ARPE-19 cells at 48 h following different doses of survivin inhibitor YM155 treatment. E. Western blot analysis of EMT markers at different time points following 20 nM YM155 treatment.
Knockdown or inhibition of survivin suppresses TGFβ-induced cell proliferation in RPE cells
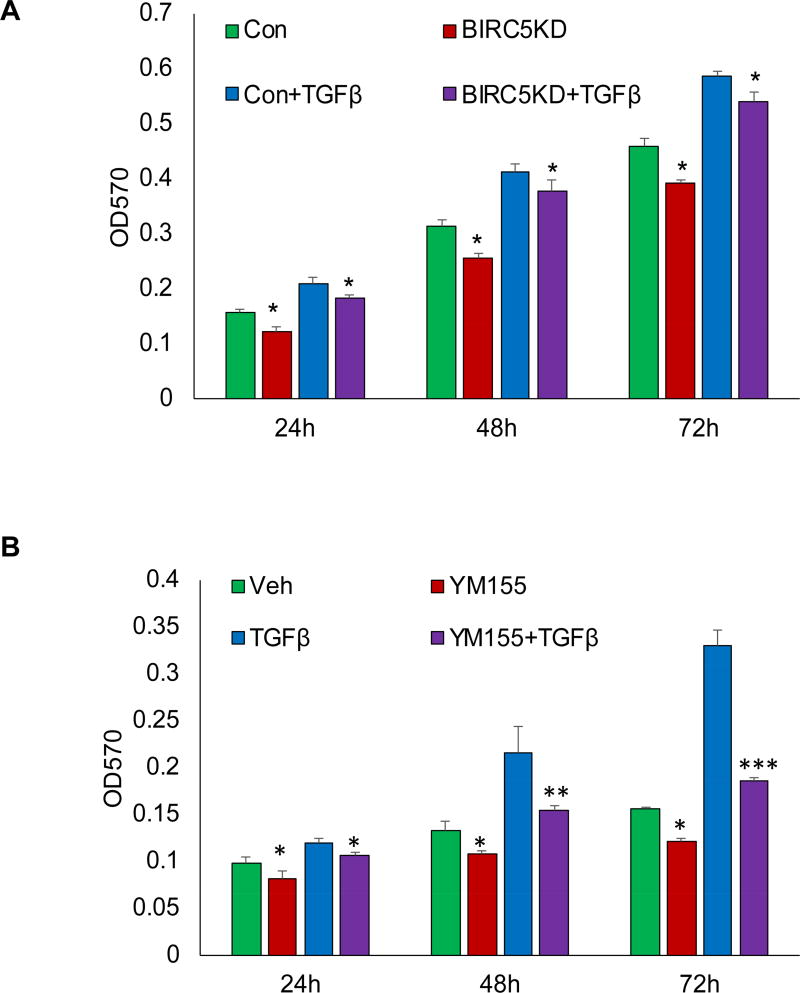
A previous study showed that TGFβ regulates cell cycle progression during EMT by upregulating survivin in ARPE-19 cells [19], suggesting that survivin was regulated by the TGFβ pathway. Our study indicated that knockdown of survivin inhibited EMT in RPE cells, implicating survivin as a contributor to RPE cell proliferation by activating in the TGFβ pathway. To determine whether knockdown of survivin reduces TGFβ-induced cell proliferation, we performed MTT assay in BIRC5 KD and control cells following 6 ng/ml TGFβ treatment at different time points. We found that the knockdown of survivin significantly inhibited cell proliferation at 24, 48, and 72 h (Fig. 2A). In addition, we inhibited survivin expression using its small-molecule inhibitor YM155 in RPE cells, and then measured cell proliferation following 20 nM YM155 treatment at different time points. We found that inhibition of survivin in turn significantly inhibited TGFβ-induced cell proliferation at 24, 48, and 72 h (Fig. 2B).
Figure 2. Knockdown or inhibition of survivin suppresses TGFβ-induced cell proliferation.
A. Cell proliferation in survivin KD and control cells was measured by MTT assay at different time points following 6 ng/ml TGFβ treatment (*p<0.05). B. Cell proliferation was measured using MTT assay following 20 nM YM155 treatment for 4 h and then treated with 6 ng/ml TGFβ(*p<0.05,**p<0.01,***p<0.001).
Knockdown or inhibition of survivin suppresses TGFβ-induced cell migration in RPE cells
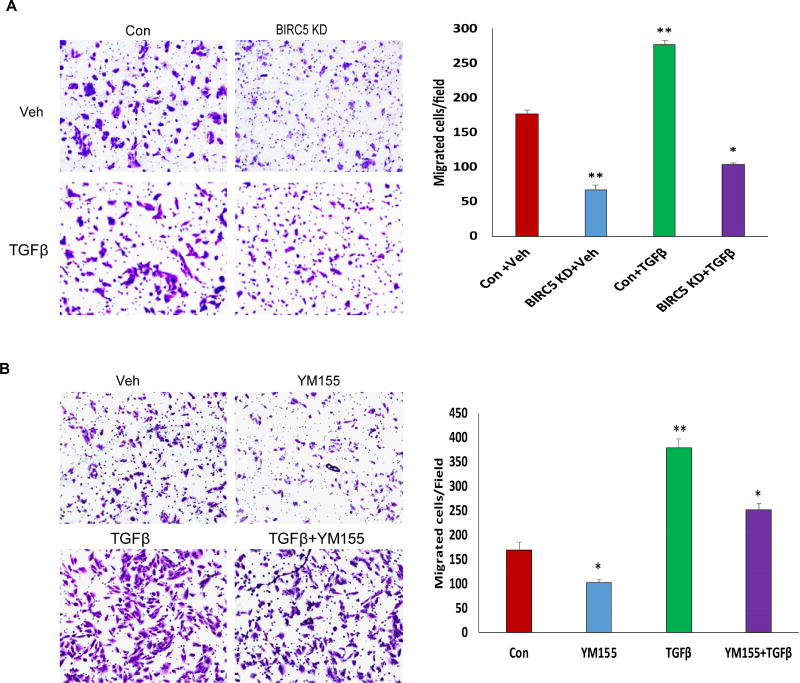
During the EMT process, RPE cells migrate to facilitate the formation of fibrotic phenotype. To understand how survivin contributes to TGFβ-induced cell migration, we performed transwell cell migration assay using BIRC5 KD and control cells. Knockdown of BIRC5 in RPE cells significantly reduced TGFβ-induced cell migration compared to control cells (Fig. 3A). We also treated wildtype ARPE-19 cells using 20 nM YM155 for 24 h and then performed cell migration assay. Similarly, inhibition of survivin using YM155 significantly reduced cell migration (Fig. 3B).
Figure 3. Knockdown or inhibition of survivin suppresses TGFβ-induced cell migration.
A. Cell migration in survivin KD and control cells was examined using transwell plates at different time points following 6 ng/ml TGFβ treatment(*p<0.05;**p<0.01). B. Cell migration was measured using transwell plates following 20 nM YM155 treatment for 4 h and then treated with 6 ng/ml TGFβ (*p<0.05;**p<0.01).
Knockdown or inhibition of survivin attenuated the TGFβ pathway in RPE cells
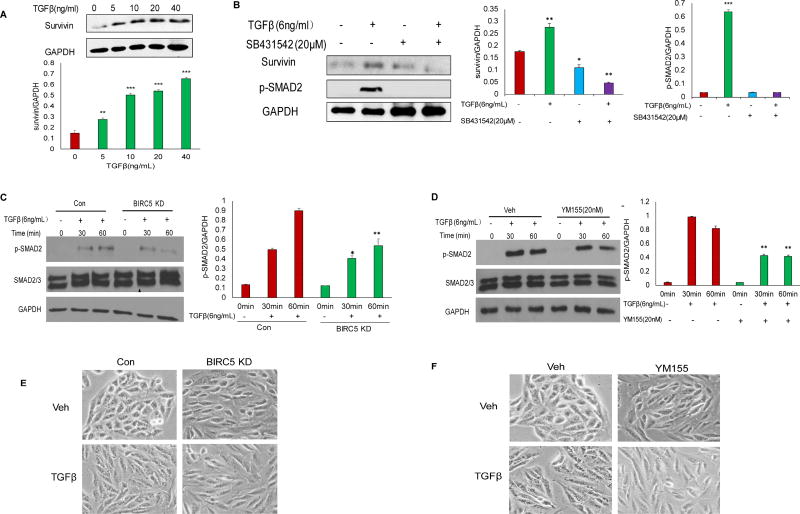
Previous studies reported that TGFβ promoted EMT in RPE cells [20–22]. To understand the molecular mechanisms of survivin in regulating EMT in RPE cells, we treated ARPE-19 cells using different doses (5, 10, 20, 40 ng/mL) of TGFβ for 24 h. Survivin was significantly upregulated among those doses compared to control (0 ng/mL) (Fig. 4A). To examine how survivin interacts with the TGFβ pathway, we also treated wildtype ARPE-19 cells using a TGFβ pathway inhibitor SB431542 at 20 µM for 12 h and then treated the cells with 6 ng/mL TGFβ We observed that TGFβ induced survivin expression, whereas the TGFβ inhibitor inhibited survivin expression and attenuated phospho-SMAD2 (Fig. 4B). We further examined the TGFβ pathway in survivin KD and control cells by detecting phospho-SMAD2 using Western blot, which showed significant attenuation in survivin KD compared to control cells while total SMAD2/3 was not altered (Fig. 4C). We also treated cells with survivin inhibitor 20 nM YM155 for 4h and then treated with 6 ng/ml TGFβ at different time points. Similar to the results shown in Fig. 4C, inhibition of survivin attenuated the TGFβ signaling pathway in this case (Fig. 4D). Our data indicated that survivin cross-talked with the TGFβ pathway in regulating TGFβ-induced EMT. To characterize the EMT phenotypic switch, we also treated survivin KD and control cells with 6 ng/ml TGFβ, and we observed that TGFβ induced typical fibroblast-like, mesenchymal phenotypic APRE19 control cells, but not in the survivin KD cells (Fig. 4E). Similarly, TGFβ could not induce a typical mesenchymal phenotype in YM155-treated cells compared with wildtype control cells (Fig. 4F). Our data thus indicated that survivin contributed to EMT by cross-talking with the TGFβ pathway in RPE cells.
Figure 4. Survivin contributes to EMT by activating TGFβ pathways.
A. Western blot analysis of survivin expression following different doses of TGFβ treatment for 24 h (**p<0.01;***p<0.001). B. Western blot analysis of survivin (*p<0.05) and phospho-SMAD2 (***p<0.001) expression following 20 µM TGFβ inhibitor and 6 ng/ml TGFβ treatment. C. Western blot analysis of phospho-SMAD2 (*p<0.05;**p<0.01) and total SMAD2/3 at different time points in survivin KD and control cells following 6 ng/ml TGFβ treatment. D. Western blot analysis of phospho-SMAD2 (**p<0.01) and total SMAD2/3 in wildtype ARPE-19 cells following 20 nM YM155 treatment for 4 h, then treated with 6 ng/ml TGFβ. E. Survivin KD and control cell morphology following 6 ng/ml TGFβ treatment for 24 h. F. Wildtype ARPE-19 cell morphology at 24 h following 20 nM YM155 treatment and then with 6 ng/ml TGFβ treatment.
Discussion
Proliferative vitreoretinopathy (PVR) is a common, blinding clinical complication of posterior segment ocular trauma and retinal detachment. Epithelial to mesenchymal transition of RPE cells plays a central role in the development of PVR. During EMT, epithelial cells lose intracellular junctions and dissociate from contact cell inhibition to develop mesenchymal-like characteristics. It is a dynamic scarring process during which RPE cells are able to migrate and proliferate in the vitreous, and on the retinal surface, transforming into fibroblastic-like cells to produce fibrotic tissue and tractional retinal detachment.
In this study we demonstrated for the first time that survivin contributes to EMT in RPE cells by using lentiviral CRISRP/Cas9 nickase vector-mediated gene editing and pharmacological inhibitor YM155. Knockdown or inhibition of survivin suppressed EMT (Fig. 1). Functional analysis indicated that knockdown of survivin significantly reduced TGFβ-induced cell proliferation and migration (Figs. 2 & 3). Survivin regulated EMT by cross-talking with the TGFβ signaling pathway in RPE cells (Fig. 4). Although previous studies indicated that several transcriptional factors and multiple signaling pathways were involved in EMT in RPE cells including the TGFβ-mediated pathway [7, 23–26], it is still unknown how survivin contributes to EMT in RPE cells. Our data showed that phospho-SMAD2 was significantly reduced in survivin KD cells or in YM155-treated cells compared to control, suggesting that survivin potentially regulates EMT by interacting with the TGFβ signaling pathway.
Previous studies showed that TGFβ induced survivin expression, thus promoted cell cycle progression, and inhibited cell apoptosis by promoting EMT in RPE cells, while TGFβ inhibited survivin expression and induced cell apoptosis in hepatocellular carcinoma Hep3B cells [19]. We recently reported that TGFβ induced survivin expression and inhibited cell apoptosis by promoting EMT in ovarian cancer epithelial cells [13], which was similar to our finding in RPE cells. Those studies suggested that the TGFβ dual function in inhibiting or inducing cell proliferation was cellular context-dependent by regulating survivin expression. Based on our finding in this study that knockdown or inhibition of survivin attenuates the TGFβ pathway by reducing phospho-SMAD2. We concluded that survivin regulates EMT by cross-talking with the TGFβ pathway.
Although it is not clear how survivin participates in the TGFβ pathway in regulating EMT in RPE cells, it is possible that survivin may activate the TGFβ pathway through interactions with TGFβ receptors. Survivin is the smallest member of IAP (inhibitor of apoptosis protein) family, which contains a single baculovirus IAP repeat (BIR) domain and lacks a C-terminus RING finger domain. In a previous study, XIAP (X-linked inhibitor of apoptosis protein), a member of the IAP family, was shown to directly interact with TGFβ receptor1 (TGFβR1) through the BIR domain [27]. Therefore, survivin may activate the TGFβ pathway through interaction with the BIR domain in RPE cells as XIAP does, which needs to be addressed further in future studies. In this study, we solely focused on the TGFβ/SMAD pathway and showed that survivin was required for TGFβ-induced EMT in RPE cells. However, survivin may also contribute to EMT by participating in other multiple non-SMAD-dependent signaling pathways activated by TGFβ. For example, cellular survival pathways ERK1/2 and AKT were activated by TGFβ in RPE cells [19]. In addition to the TGFβ pathway, survivin may regulate EMT by activating other signaling pathways in RPE cells such as Wnt/β-catenin [18] and NF-kB [16]. Those studies indicate that survivin is a key factor in promoting RPE cell survival by activating multiple signaling pathways.
Currently, vitreous surgery is a standard treatment for PVR. However, the surgical results are often not satisfactory and recurrences are common. Given the relative poor outcome of surgery, pharmacological interventions are needed to improve the efficacy of surgery. Several drugs used for inhibiting cell proliferation, anti-inflammatory and anti-growth factor-mediated pathways were tested in clinical trials, and some improvement was observed [2]. YM155 is a small-molecule inhibitor and approved by FDA for clinical trials in cancer therapy. For the first time, we showed that YM155 inhibited EMT in RPE cells by attenuating the TGFβ pathway, and that it suppressed TGFβ-induced cell proliferation and migration, suggesting that YM155 is a potential drug for the treatment of PVR. Although we showed that YM155 was efficient in inhibiting cell proliferation and migration, it is not clear whether it is effective in vivo. It is essential to evaluate the efficacy in vivo using a PVR animal model.
In conclusion, this study demonstrated that survivin contributed to EMT in RPE cells by activating the TGFβ pathway. Targeting survivin using YM155 or other survivin inhibitors potentially provides a new avenue for PVR therapy.
Supplementary Material
Highlights.
TGFβ activates BIRC5 expression in RPE cells
Lentiviral CRISPR/Cas9 nickase mediated knockdown of BIRC5 results in the suppression of epithelial to mesenchymal transition in RPE cells
Reduction of survivin attenuates TGFβ pathway in RPE cells
Inhibition of TGFβ pathway downregulates survivin expression in RPE cells.
Acknowledgments
This work was partially supported by AHA grant and West Cancer Center to J. Yue, and the National Institute of Health grant 1R01CA193609-01A1 to W. Li. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tamiya S, Kaplan HJ. Role of epithelial-mesenchymal transition in proliferative vitreoretinopathy. Exp Eye Res. 2016;142:26–31. doi: 10.1016/j.exer.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Sadaka A, Giuliari GP. Proliferative vitreoretinopathy: current and emerging treatments. Clin Ophthalmol. 2012;6:1325–1333. doi: 10.2147/OPTH.S27896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iribarne M, Ogawa L, Torbidoni V, Dodds CM, Dodds RA, Suburo AM. Blockade of endothelinergic receptors prevents development of proliferative vitreoretinopathy in mice. Am J Pathol. 2008;172:1030–1042. doi: 10.2353/ajpath.2008.070605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sethi CS, Lewis GP, Fisher SK, Leitner WP, Mann DL, Luthert PJ, Charteris DG. Glial remodeling and neural plasticity in human retinal detachment with proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci. 2005;46:329–342. doi: 10.1167/iovs.03-0518. [DOI] [PubMed] [Google Scholar]
- 5.Zhou X, Kuang X, Long C, Liu W, Tang Y, Liu L, Liu H, He J, Huang Z, Fan Y, Zhang Q, Shen H. Curcumin Inhibits Proliferation and Epithelial-Mesenchymal Transition of Retinal Pigment Epithelial Cells Via Multiple Pathways. Curr Mol Med. 2017;17:312–319. doi: 10.2174/1566524017666171106115655. [DOI] [PubMed] [Google Scholar]
- 6.Han JW, Lyu J, Park YJ, Jang SY, Park TK. Wnt/beta-Catenin Signaling Mediates Regeneration of Retinal Pigment Epithelium After Laser Photocoagulation in Mouse Eye. Invest Ophthalmol Vis Sci. 2015;56:8314–8324. doi: 10.1167/iovs.15-18359. [DOI] [PubMed] [Google Scholar]
- 7.Wang HF, Ma JX, Shang QL, An JB, Chen HT. Crocetin inhibits the proliferation, migration and TGF-beta2-induced epithelial-mesenchymal transition of retinal pigment epithelial cells. Eur J Pharmacol. 2017;815:391–398. doi: 10.1016/j.ejphar.2017.09.041. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Yuan G, Dong M, Zhang T, Hua G, Zhou Q, Shi W. Notch signaling modulates proliferative vitreoretinopathy via regulating retinal pigment epithelial-to-mesenchymal transition. Histochem Cell Biol. 2017;147:367–375. doi: 10.1007/s00418-016-1484-x. [DOI] [PubMed] [Google Scholar]
- 9.Lai K, Luo C, Zhang X, Ye P, Zhang Y, He J, Yao K. Regulation of angiogenin expression and epithelial-mesenchymal transition by HIF-1alpha signaling in hypoxic retinal pigment epithelial cells. Biochim Biophys Acta. 2016;1862:1594–1607. doi: 10.1016/j.bbadis.2016.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Chen X, Xiao W, Wang W, Luo L, Ye S, Liu Y. The complex interplay between ERK1/2, TGFbeta/Smad, and Jagged/Notch signaling pathways in the regulation of epithelial-mesenchymal transition in retinal pigment epithelium cells. PLoS One. 2014;9:e96365. doi: 10.1371/journal.pone.0096365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi E, Haga A, Tanihara H. Merlin Regulates Epithelial-to-Mesenchymal Transition of ARPE-19 Cells via TAK1-p38MAPK-Mediated Activation. Invest Ophthalmol Vis Sci. 2015;56:2449–2458. doi: 10.1167/iovs.14-16300. [DOI] [PubMed] [Google Scholar]
- 12.Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH. Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol. 2000;10:1319–1328. doi: 10.1016/s0960-9822(00)00769-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhao G, Wang Q, Gu Q, Qiang W, Wei JJ, Dong P, Watari H, Li W, Yue J. Lentiviral CRISPR/Cas9 nickase vector mediated BIRC5 editing inhibits epithelial to mesenchymal transition in ovarian cancer cells. Oncotarget. 2017;8:94666–94680. doi: 10.18632/oncotarget.21863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yi XP, Han T, Li YX, Long XY, Li WZ. Simultaneous silencing of XIAP and survivin causes partial mesenchymal-epithelial transition of human pancreatic cancer cells via the PTEN/PI3K/Akt pathway. Mol Med Rep. 2015;12:601–608. doi: 10.3892/mmr.2015.3380. [DOI] [PubMed] [Google Scholar]
- 15.Yang P, Wang G, Huo H, Li Q, Zhao Y, Liu Y. SDF-1/CXCR4 signaling up-regulates survivin to regulate human sacral chondrosarcoma cell cycle and epithelial-mesenchymal transition via ERK and PI3K/AKT pathway. Med Oncol. 2015;32:377. doi: 10.1007/s12032-014-0377-x. [DOI] [PubMed] [Google Scholar]
- 16.Yang P, Wiser JL, Peairs JJ, Ebright JN, Zavodni ZJ, Bowes Rickman C, Jaffe GJ. Human RPE expression of cell survival factors. Invest Ophthalmol Vis Sci. 2005;46:1755–1764. doi: 10.1167/iovs.04-1039. [DOI] [PubMed] [Google Scholar]
- 17.Xu JY, Wu LY, Zheng XQ, Lu JL, Wu MY, Liang YR. Green tea polyphenols attenuating ultraviolet B-induced damage to human retinal pigment epithelial cells in vitro. Invest Ophthalmol Vis Sci. 2010;51:6665–6670. doi: 10.1167/iovs.10-5698. [DOI] [PubMed] [Google Scholar]
- 18.Guo X, Zhu D, Lian R, Han Y, Guo Y, Li Z, Tang S, Chen J. Matrigel and Activin A promote cell-cell contact and anti-apoptotic activity in cultured human retinal pigment epithelium cells. Exp Eye Res. 2016;147:37–49. doi: 10.1016/j.exer.2016.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Lee J, Choi JH, Joo CK. TGF-beta1 regulates cell fate during epithelial-mesenchymal transition by upregulating survivin. Cell Death Dis. 2013;4:e714. doi: 10.1038/cddis.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Ye S, Xiao W, Luo L, Liu Y. Differentially expressed microRNAs in TGFbeta2-induced epithelial-mesenchymal transition in retinal pigment epithelium cells. Int J Mol Med. 2014;33:1195–1200. doi: 10.3892/ijmm.2014.1688. [DOI] [PubMed] [Google Scholar]
- 21.Mony S, Lee SJ, Harper JF, Barwe SP, Langhans SA. Regulation of Na,K-ATPase beta1-subunit in TGF-beta2-mediated epithelial-to-mesenchymal transition in human retinal pigmented epithelial cells. Exp Eye Res. 2013;115:113–122. doi: 10.1016/j.exer.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Wang H, Wang F, Gu Q, Xu X. Snail involves in the transforming growth factor beta1-mediated epithelial-mesenchymal transition of retinal pigment epithelial cells. PLoS One. 2011;6:e23322. doi: 10.1371/journal.pone.0023322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Zhang C, Su L, Song Z. GSK3beta attenuates TGF-beta1 induced epithelial-mesenchymal transition and metabolic alterations in ARPE-19 cells. Biochem Biophys Res Commun. 2017;486:744–751. doi: 10.1016/j.bbrc.2017.03.113. [DOI] [PubMed] [Google Scholar]
- 24.Chen CL, Chen YH, Tai MC, Liang CM, Lu DW, Chen JT. Resveratrol inhibits transforming growth factor-beta2-induced epithelial-to-mesenchymal transition in human retinal pigment epithelial cells by suppressing the Smad pathway. Drug Des Devel Ther. 2017;11:163–173. doi: 10.2147/DDDT.S126743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimsa M, Strzalka-Mrozik B, Kimsa-Dudek M, Kruszniewska-Rajs C, Gola J, Adamska J, Mazurek U. Transforming growth factor beta-related genes in human retinal pigment epithelial cells after tacrolimus treatment. Pharmacol Rep. 2016;68:969–974. doi: 10.1016/j.pharep.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 26.Feng Z, Li R, Shi H, Bi W, Hou W, Zhang X. Combined silencing of TGF-beta2 and Snail genes inhibit epithelial-mesenchymal transition of retinal pigment epithelial cells under hypoxia. Graefes Arch Clin Exp Ophthalmol. 2015;253:875–884. doi: 10.1007/s00417-014-2922-x. [DOI] [PubMed] [Google Scholar]
- 27.Neil JR, Tian M, Schiemann WP. X-linked inhibitor of apoptosis protein and its E3 ligase activity promote transforming growth factor-{beta}-mediated nuclear factor-{kappa}B activation during breast cancer progression. J Biol Chem. 2009;284:21209–21217. doi: 10.1074/jbc.M109.018374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.