Abstract
The hormone ethylene regulates many aspects of plant growth and development, including fruit ripening. In transgenic tomato (Lycopersicon esculentum) plants, antisense inhibition of ethylene biosynthetic genes results in inhibited or delayed ripening. The dominant tomato mutant, Never-ripe (Nr), is insensitive to ethylene and fruit fail to ripen. The Nr phenotype results from mutation of the ethylene receptor encoded by the NR gene, such that it can no longer bind the hormone. NR has homology to the Arabidopsis ethylene receptors. Studies on ethylene perception in Arabidopsis have demonstrated that receptors operate by a “receptor inhibition” mode of action, in which they actively repress ethylene responses in the absence of the hormone, and are inactive when bound to ethylene. In ripening tomato fruit, expression of NR is highly regulated, increasing in expression at the onset of ripening, coincident with increased ethylene production. This expression suggests a requirement for the NR gene product during the ripening process, and implies that ethylene signaling via the tomato NR receptor might not operate by receptor inhibition. We used antisense inhibition to investigate the role of NR in ripening tomato fruit and determine its mode of action. We demonstrate restoration of normal ripening in Nr fruit by inhibition of the mutant Nr gene, indicating that this receptor is not required for normal ripening, and confirming receptor inhibition as the mode of action of the NR protein.
The plant hormone ethylene controls a number of developmental processes including seedling growth and morphology, fruit ripening, organ senescence, and abscission. Ethylene is synthesized from S-adenosyl-l-Met through the activity of the enzymes 1-aminocyclopropane-1-carboxylic acid (ACC) synthase and ACC oxidase. Ethylene perception affects the expression of a number of genes that are important for the biological response. The signal transduction pathway(s) that allows cells to perceive and respond to ethylene has not yet been fully elucidated, although a number of signaling components have recently been identified through the use of Arabidopsis mutants.
Genes for putative ethylene receptors were isolated from Arabidopsis following identification of the ethylene-insensitive mutant etr1, which failed to show the classical seedling “triple response” to ethylene (Bleecker et al., 1988). ETR1 encodes a protein with homology to bacterial two-component receptors (Chang et al., 1993). These receptors allow bacteria to respond to environmental stimuli, and consist of a sensor and transmitter protein and separate response regulator (Chang and Stewart, 1998). ETR1 shares sequence identity with the His kinase domain of the bacterial transmitter region, and with the response regulator, which is situated at the carboxyl terminus of ETR1 rather than on a separate peptide (Chang et al., 1993). The bacterial receptors bind ligands through the N termini of their sensor modules. The N terminus of ETR1 has no homology to the bacterial proteins, but contains three hydrophobic regions and has been shown through expression studies in yeast to be membrane associated and to bind ethylene (Schaller and Bleecker, 1995). ETR1 is one of a five-member gene family in Arabidopsis. The other members include ETR2 and EIN4 (Hua et al., 1998; Sakai et al., 1998), which have similar structures to ETR1, and ERS1 and ERS2 (Hua et al., 1995, 1998), which encode receptors lacking the carboxy-terminal response-regulator-like domain present in the other three proteins. Loss of the ability to bind ethylene by any of the five proteins results in dominant insensitivity to ethylene. etr2 and ein4 mutants (Roman et al., 1995; Sakai et al., 1998) were identified in genetic screens in which disruption of their ethylene-binding ability resulted in insensitivity to ethylene. Transgenic Arabidopsis plants expressing in vitro mutated ers1 and ers2 genes, whose products could not bind ethylene, were also insensitive to the hormone (Hua et al., 1995, 1998). Loss of function of any one receptor, however, had no effect on ethylene sensitivity (Hua and Meyerowitz, 1998), indicating functional redundancy among receptors, whereas quadruple mutants in which ETR1, ETR2, EIN4, and ERS2 were knocked out showed a constitutive ethylene response phenotype (Hua and Meyerowitz, 1998). These observations are consistent with the “receptor inhibition” model of ethylene action (Bleecker et al., 1998) in which absence of ethylene results in active receptors and repression of ethylene responses (Hua and Meyerowitz, 1998). According to this model, in the presence of ethylene, receptors switch to an inactive state, and responses such as the triple response are observed. Disruption of ethylene binding in any one receptor thus leads to active repression of the response pathway and insensitivity to ethylene.
Tomato (Lycopersicon esculentum) has five genes encoding proteins with similarity to ETR1, including NR (Wilkinson et al., 1995), LeETR1 (Zhou et al., 1996a; Lashbrook et al., 1998), LeETR2 (Zhou et al., 1996b; Lashbrook et al., 1998), LeETR4 (Tieman and Klee, 1999), and LeETR5 (Tieman and Klee, 1999). They show different patterns of expression: NR and LeETR4 transcripts increase in abundance during fruit ripening (Payton et al., 1996; Lashbrook et al., 1998; Tieman and Klee, 1999), whereas LeETR1 and LeETR2 show a more or less constitutive pattern of expression (Lashbrook et al., 1998). LeETR5 expression increases in flower tissue (Tieman and Klee, 1999). The NR gene was identified through its homology to ETR1, and it was shown that the tomato-ripening mutant Never-ripe (Nr) (Rick and Butler, 1956), which bears fruit that are impaired in color change and softening, has a mutation in the ethylene-binding domain of the NR receptor (Wilkinson et al., 1995) and is unable to bind the hormone. The effect of the Nr mutation, together with the increase in expression of NR observed at onset of ripening in wild-type plants, indicated a specific role for this receptor during ripening. The Arabidopsis model, however, suggests that ethylene response is dependent upon receptor inactivation by ligand binding. If this model is true for tomato, it is difficult to explain why there should be an increase in expression of NR at the same time as increased ethylene evolution and ripening response to ethylene. In a receptor inhibition model, no change in NR gene expression would be expected. To address this question, we down-regulated expression of the mutant Nr gene by antisense inhibition. We anticipated that if the receptor inhibition model is correct, down-regulation of the mutant gene should lead to restoration of normal ripening. This would not be the case, however, if NR is specifically required for fruit ripening. The results presented confirm receptor inhibition as the mode of action of the NR receptor and indicate that NR is not required for normal ripening.
RESULTS
Transformation of Nr Mutants
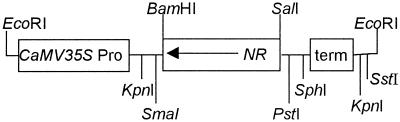
A partial cDNA clone designated tETR was isolated previously (Payton et al., 1996), and found to be identical to the reported NR cDNA sequence. A full-length NR cDNA was subsequently isolated and a fragment of the cDNA, from nucleotides +34 to +748, was inserted downstream of the cauliflower mosaic virus (CaMV) 35S promoter in the antisense orientation in pBin 19 (Bevan, 1984) (Fig. 1). The transgene was introduced into cells of 3-week-old Nr cotyledons by Agrobacterium tumefaciens-mediated transformation. Four primary transformants were initially regenerated on selective media containing 100 mg L−1 kanamycin and grown to maturity. Southern analysis indicated that transformants contained either one or two copies of the transgene.
Figure 1.
Structure of the NR antisense gene inserted into the EcoRI site of pBin 19 (Bevan, 1984). The tomato NR gene (0.714 kb) was placed in the antisense orientation relative to the CaMV 35S promoter (CaMV 35S Pro), upstream of the CaMV 35S terminator sequence (term). Relative positions of additional restriction endonuclease recognition sites are shown.
Analysis of Ripening in NR Antisense Transformants
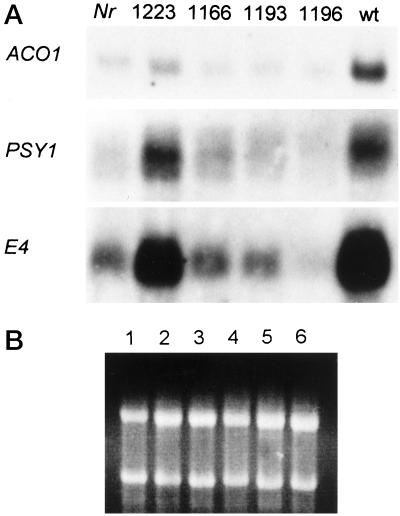
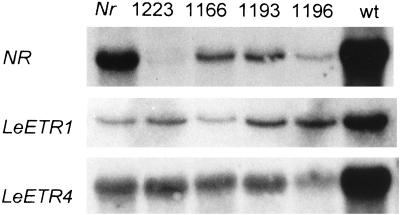
Visual examination of the four primary transformants obtained (1166, 1193, 1196, and 1223) showed that fruit of one plant, 1223, appeared to ripen normally and turned red as rapidly as wild-type fruit. This plant contained two copies of the NR antisense transgene. Fruit of the remaining three plants were yellow throughout ripening, and were visually similar to those of the Nr mutant, although studies on the progeny of these plants (described below) showed an inherited antisense gene dose-dependent restoration of ripening. Expression of several ripening genes including ACC oxidase 1 (ACO1; Hamilton et al., 1991), phytoene synthase 1 (PSY1; Ray et al., 1992), and E4 (Lincoln et al., 1987), was studied in the fruit of each of the primary transformants. Total RNA was extracted from fruit at the onset of ripening (breaker) for use in northern analysis. The results showed that for each of the three genes studied, transcripts were more abundant in the wild type than Nr mutant fruit (Fig. 2). However, in the NR antisense transformants, expression of ACO1, E4, and PSY1 was higher in plant 1223 when compared to the other transformants (Fig. 2). Fruit of this transformant therefore had a wild-type phenotype and showed levels of expression of ripening and ethylene-responsive genes that were similar to wild type. Northern analysis was carried out to determine whether expression of the mutant Nr gene was altered in any of the antisense plants. Abundance of Nr transcripts was found to be reduced in three of the four antisense plants compared to non-transgenic Nr fruit, but the degree of reduction varied (Fig. 3). In plant 1223, which ripened normally, Nr transcripts were virtually undetectable (Fig. 3). Expression of two further ethylene receptor genes, LeETR1 and LeETR4, was also analyzed in breaker fruit of Nr transformants (Fig. 3). LeETR1 and LeETR4 transcripts appeared more abundant in wild type compared to non-transgenic Nr fruit. There was some variation in the level of LeETR1 transcripts in transformants 1166 and 1196, and a reduction of LeETR4 transcripts in transformant 1196, compared to Nr fruit. There was, however, no effect on fruit phenotype in these transformants.
Figure 2.
Expression of ACO1, PSY1, and E4 in the fruit of transgenic and non-transformed plants (A). Total RNA (20 μg) was isolated from fruit at breaker stage from (lanes 1–6) non-transformed Nr, transformant 1223, transformant 1166, transformant 1193, transformant 1196, and non-transformed wild type (wt). Ethidium staining of RNA prior to blotting confirmed RNA loading (B).
Figure 3.
Expression of tomato ethylene receptor genes in the fruit of transgenic and non-transformed plants at breaker stage. Forty micrograms of total RNA from non-transformed Nr, transformant 1223, transformant 1166, transformant 1193, transformant 1196, and non-transformed wild type (wt) were blotted and hybridized with probes corresponding to NR, LeETR1, and LeETR4.
Effect of Transgene Inheritance on Progeny Phenotype
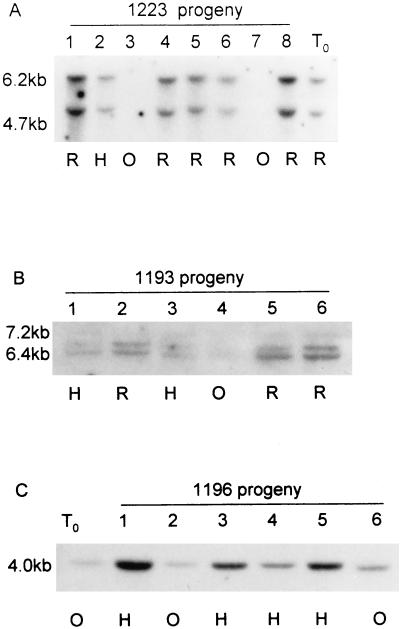
Inheritance of the antisense gene was studied in the progeny (T1 generation) of transformants germinated from self-seed, and the fruit phenotypes of azygous, hemizygous, and homozygous progeny were observed. Seed from transformant 1223, which had a wild-type fruit phenotype, was sown directly into compost in the absence of selection for the transgene. Eight progeny plants (1223.1–1223.8) were studied. Of these, two had yellow fruit and were phenotypically identical to Nr, whereas the remaining six had red wild-type fruit. Southern analysis of progeny plants using the NPTII gene as a probe showed that plants with yellow Nr-type fruit (1223.3 and 1223.7) did not contain a transgene (Fig. 4A), whereas the plants bearing red wild-type fruit (1223.1, 1223.2, 1223.4, 1223.5, 1223.6, and 1223.8) contained two copies of the transgene in either the homozygous or hemizygous state (Figs. 4A and 5).
Figure 4.
Transgene inheritance and phenotype of progeny of Nr transformants. The sizes of DNA fragments produced following Southern analysis are indicated. The color of fruit produced by individual plants is shown below each lane. R, Red; O, orange; H, half-red. A, Transformant 1223 (T0), and (in lanes 1–8) progeny plants 1223.1, 1223.2, 1223.3, 1223.4, 1223.5, 1223.6, 1223.7, and 1223.8. B, Progeny of transformant 1193 (in lanes 1–6): 1193.1, 1193.2, 1193.3, 1193.4, 1193.5, and 1193.6. C, Transformant 1196 (T0), and (in lanes 1–6) progeny plants 1196.1, 1196.2, 1196.3, 1196.4, 1196.5, and 1196.6.
Figure 5.
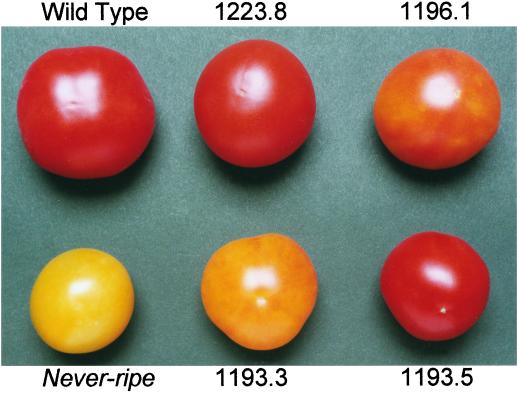
The phenotype of fruit from T1 progeny of Nr transformants. Top row, left to right: non-transformed wild type, T1 plant 1223.8 (red fruit), and T1 plant 1196.1 (half-red fruit); lower row, left to right: non-transformed Nr, T1 plant 1193.3 (half-red fruit), and T1 plant 1193.5 (red fruit). Fruit were harvested at 7 d post-breaker and allowed to ripen for a further 16 d before photographing.
Seedlings from the remaining three transformants, all of which had produced Nr-type fruit, were selected on kanamycin-containing media before being transferred to compost and grown to maturity. Progeny of transformant 1166 produced orange Nr-type fruit, identical to those produced by the transgenic parent. Of the six progeny from transformant 1193 (1193.1–1193.6), however, three produced fully red fruit that were phenotypically indistinguishable from wild-type fruit, whereas a further two progeny produced fruit that appeared to be intermediate in color between Nr and wild-type fruit. These fruit were designated half-red. The sixth T1 plant produced Nr-type orange fruit. The phenotype of fruit was compared to the transgene copy number of the progeny plants. Transformant 1193 contained two copies of the transgene. Southern analysis showed that progeny that inherited both transgenes in the homozygous state (1193.2, 1193.5, and 1193.6) produced red fruit (Figs. 4B and 5). The half-red fruit were produced by plants that contained both transgene copies in a hemizygous state (1193.1 and 1193.3) (Figs. 4B and 5). The single progeny plant producing orange Nr-type fruit appeared to have inherited only one of the transgenes in a hemizygous state (1193.4) (Fig. 4B).
Fruit of progeny of a fourth transformant, 1196, were half-red in four of the T1 plants (1196.1, 1196.3, 1196.4, and 1196.5), and identical to Nr in a further two plants (1196.2 and 1196.6). Plant 1196 contained a single transgene insert, and progeny bearing half-red fruit were shown to have inherited this transgene in the homozygous state (Figs. 4C and 5), whereas Nr-type plants had inherited the transgene in a hemizygous condition (Fig. 4C).
DISCUSSION
We have used antisense inhibition of a mutant tomato ethylene receptor gene to demonstrate that ethylene perception and signaling via the NR receptor is consistent with the receptor-inhibition model proposed to describe ethylene perception in Arabidopsis. According to this model, receptors negatively regulate ethylene responses and this inhibition is released when ethylene is bound to the receptors (Hua and Meyerowitz, 1998). The results presented demonstrate that removal of the mutant Nr receptor by antisense inhibition results in activation of ethylene responses and onset of normal ripening. Thus, the inability of the mutant Nr receptor to bind ethylene prevents its inactivation and in Nr mutant fruit ethylene responses are therefore suppressed.
In a transgenic antisense Nr plant (transformant 1223), inhibition of the mutant receptor gene was sufficient to allow fruit to turn red in color and to achieve wild-type levels of expression of ripening-related (PSY1 and ACO1) and ethylene-responsive (E4) genes. In the other primary transformants generated, the extent of down-regulation of the Nr gene was insufficient to allow fruit to ripen normally, and expression of ACO1, PSY1, and E4 was reduced as in the untransformed Nr fruit. However, some of the progeny of these plants produced fruit that were either wild type in appearance or intermediate between wild type and Nr. The phenotype of the fruit correlated with transgene dosage in the progeny plants: Where a single transgene was present (transformant 1196), inheritance of the transgene in a homozygous state gave fruit of intermediate phenotype, whereas hemizygotes produced Nr-type fruit. Progeny of a plant that contained two copies of the transgene were able to produce wild-type fruit if they were homozygous for both copies, but gave intermediate or Nr-type fruit if they were hemizygous for both copies, or inherited one transgene, respectively. This finding suggests that a threshold level of mutant receptor is required to suppress the ethylene response pathway and prevent normal ripening. It has previously been reported that genetic background affects the severity of the Nr phenotype (Lanahan et al., 1994). Ripening occurs to a greater extent in Nr fruit in tomato cv Ailsa Craig than where the mutation is present in tomato cv Pearson. It was suggested that expression of the Nr gene is incomplete in tomato cv Ailsa Craig, leading to a less severe phenotype in homozygous plants and partial ripening in heterozygotes when compared with tomato cv Pearson Nr plants (Lanahan et al., 1994). This is consistent with our observation that restoration of normal ripening in Nr plants depends on the extent of antisense inhibition of Nr gene expression and the inheritance of antisense genes.
Expression of the NR gene in wild-type plants is up-regulated by ethylene in mature green fruit (Wilkinson et al., 1995), and the increase in NR expression at the onset of ripening reflects the increase in ethylene synthesis in fruit at this stage. It is not clear why a receptor gene that is apparently not required for normal ripening should be up-regulated in mature fruit in a system operating by receptor inhibition. In Arabidopsis, the ethylene receptor genes ERS1, ERS2, and ETR2 are up-regulated in response to ethylene (Hua et al., 1998), and loss-of-function mutants reported for two of these genes, ETR2 and ERS2, showed normal ethylene sensitivity. Our previous observations (H.C.C. Foote and D. Grierson, unpublished data) and those of others (Wilkinson et al., 1997) have indicated that antisense inhibition of the wild-type NR gene has no effect on ripening, raising questions about the normal function of this gene. It has been suggested that in Arabidopsis, the different ethylene receptors allow plants to respond to a range of concentrations of ethylene in different tissues throughout plant development (Hua and Meyerowitz, 1998). Induction of NR during fruit ripening might be required to allow a more subtle subset of ethylene responses to occur than those investigated in the present study, and these responses might become apparent once the complex biochemical and physiological changes that occur in fruit during the ripening process are understood in sufficient detail. The ethylene receptor LeEtr4 has been reported to have a higher level of expression in fruit than NR and is expressed throughout fruit development, not only at ripening. In contrast with a previous report describing expression of LeETR4 in tomato cv Pearson (Tieman and Klee, 1999), our results show that LeETR4 expression is reduced in Nr compared to wild-type tomato cv Ailsa Craig fruit. Furthermore, this reduction in transcript levels is not altered by down-regulation of the NR gene in mutant plants, indicating that a functional NR gene might be required to achieve wild-type levels of LeETR4 expression.
The Nr mutation has been shown to affect ethylene responses in tissues other than ripe fruit (Lanahan et al., 1994), including seedlings. When germinated in the dark on media containing the ethylene precursor ACC, Nr seedlings failed to show a classic triple response (shortened, swollen hypocotyl, exaggerated apical hook, and shortened root). When seeds from homozygous NR antisense T1 plants of lines 1223, 1196, and 1193, all of which produced red or half-red fruit, were germinated on ACC in the dark, no significant differences in hypocotyl length were observed between these seedlings and Nr seedlings similarly treated (data not shown). This finding indicates that although down-regulation of the mutant Nr gene was sufficient to alleviate its effect on fruit ripening in these lines, insensitivity of seedlings to ethylene was not altered. It is possible that the extent of antisense down-regulation of the mutant gene is not as severe in seedling tissue of these lines. The fact that there are multiple ethylene receptors, and some show differential expression, might also be important. The differences in response shown by different tissues might reflect changes in receptor function or interplay at different stages of development; the level of mutant receptor produced in these plants might be sufficient to give an insensitive response in seedling tissue, whereas in fruit it might be below the level required to inhibit ripening. Differences in receptor function throughout development have been reported in Arabidopsis, where a double etr1; ein4 loss-of-function mutant was found to have more severe defects in root and leaf development than the single etr1 mutant, whereas hypocotyl elongation in etiolated seedlings was the same in both the double and single mutants (Hua and Meyerowitz, 1998).
Although the importance of receptor levels and possible receptor interplay at different stages of development remain to be elucidated, the present results show that the wild-type NR gene product is not required for normal ripening. Furthermore, the observation that antisense inhibition of the mutant Nr gene product restores normal ripening provides strong support for a receptor-inhibition model for the ethylene regulation of ripening in tomato.
MATERIALS AND METHODS
Construction of the NR Antisense Gene
All molecular cloning procedures were carried out using standard procedures (Sambrook et al., 1989). A fragment of the NR gene from nucleotide +34 to nucleotide +748 was amplified using the PCR. The fragment was ligated into PCR-cloning vector pTAG (Novagen Inc., Madison, WI), and the nucleotide sequence was verified by sequence analysis. The NR insert in pTAG was excised by digestion with BamHI and SalI and inserted into BamHI/SalI-digested pDH51 (Pietrzak et al., 1986). This insertion resulted in the NR gene fragment being in the antisense orientation with respect to the CaMV 35S promoter in pDH51. The entire pDH51 insert was then excised using EcoRI and inserted into pBin 19 (Bevan, 1984). The resulting vector was designated pNR6.1AS.
Plant Transformation
pNR6.1AS was introduced into competent Agrobacterium tumefaciens LBA4404 cells (Bevan, 1984) and used to transform cotyledon cells of the tomato (Lycopersicon esculentum cv Ailsa Craig) Nr mutant. The transformation was carried out according to a standard procedure (Bird et al., 1988). Plantlets were regenerated on 100 mg/mL−1 kanamycin and transferred to compost. Transformants were grown in the greenhouse under standard conditions used for cultivation of transgenic and non-transgenic wild-type and Nr tomato plants.
RNA Isolation and Northern Analysis
Pericarp tissue from fruit harvested at breaker was frozen in liquid nitrogen and stored at −80°C until further use. RNA was extracted according to the protocol described by Griffiths et al. (1999). RNA was separated on a 1% (w/v) formaldehyde gel and transferred by capillary blotting to a GeneScreen Plus (DuPont, Boston) hybridization membrane. Hybridization using probes to detect NR, LeETR1, and LeETR4 transcripts was carried out for 16 h at 42°C in buffer containing 1% (w/v) SDS, 50% (v/v) deionized formamide, 5× SSC, 50 mm sodium phosphate (pH 6.8), 0.1% (w/v) sodium pyrophosphate, 10% (w/v) dextran sulfate, and 50 μg/mL−1 salmon sperm DNA. Hybridization using probes to detect ACO1, PSY1, and E4 transcripts was carried out for 16 h at 65°C in buffer containing 5× sodium chloride-sodium dihydrogen phosphate-EDTA (SSPE), 5× Denhardt's solution (2% [w/v] bovine serum albumin fraction V, 20% [w/v] Ficoll 400, and 2% [w/v] polyvinylpyrollidone), 0.5% (w/v) SDS, and 100 μg/mL−1 salmon sperm DNA. Radiolabeled probes were prepared using the Rediprime II random prime labeling system (Amersham Pharmacia Biotech, Buckinghamshire, UK). Following hybridization, membranes were washed twice in 2× SSPE and 0.1% (w/v) SDS for 30 min at room temperature, once in 1× SSPE and 0.1% (w/v) SDS for 30 min at room temperature, and finally in 0.1× SSPE and 0.1% (w/v) SDS for 45 min at 60°C. Autoradiography was used to detect signal intensity.
DNA Isolation and Southern Analysis
Genomic DNA was isolated from 3 to 5 g of leaf tissue. Tissue was frozen and ground in liquid nitrogen. Ten milliliters of urea extraction buffer (42% [w/v] urea, 0.31 m NaCL, 50 mm Tris-HCl [pH 8.0], 20 mm EDTA [pH 8.0], and 0.1% [w/v] sodium sarcosine) were added to the powder and ground for a further 1 to 2 min. Ten milliliters of phenol/chloroform was added and the mixture was incubated at 65°C for 15 min before centrifugation at 12,000g for 15 min. An equal volume of isopropanol was added to the supernatant, which was then incubated at −20°C for 30 min. DNA was pelleted by centrifugation at 12,000g for 15 min, washed in 70% (v/v) ethanol, dried, and resuspended in water. RNA was removed by the addition of 25 μg/mL−1 Rnase (Dnase-free; Boehringer Mannheim/Roche, Basel). To detect transgene inserts, 30 μg of DNA was digested with either EcoRI or HindIII. DNA was then separated by electrophoresis through a 0.8% (w/v) agarose gel, and blotted to a GeneScreen Plus membrane (DuPont). Hybridization was then carried out according to the manufacturer's instructions (GeneScreen Plus, DuPont) using a radiolabeled probe prepared using the Rediprime II random prime labeling system (Amersham Pharmacia Biotech).
Footnotes
This work was supported by a grant from the Biotechnology and Biological Sciences Research Council.
LITERATURE CITED
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird CR, Smith CJS, Ray JA, Moureau P, Bevan MW, Bird AS, Hughes S, Morris PC, Grierson D, Schuch W. The tomato polygalacturonase gene and ripening-specific expression in transgenic plants. Plant Mol Biol. 1988;11:651–662. doi: 10.1007/BF00017465. [DOI] [PubMed] [Google Scholar]
- Bleecker AB, Esch JJ, Hall AE, Rodriguez FI, Binder BM. The ethylene-receptor family from Arabidopsis: structure and function. Philos Trans R Soc Lond B. 1998;353:1405–1412. doi: 10.1098/rstb.1998.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: similarity of products to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chang C, Stewart RC. The two-component system. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths A, Barry C, Alpuche-Solis AG, Grierson D. Ethylene and developmental signals regulate expression of lipoxygenase genes during tomato fruit ripening. J Exp Bot. 1999;50:793–798. [Google Scholar]
- Hamilton AJ, Bouzayen M, Grierson D. Identification of a tomato gene for the ethylene-forming enzyme by expression in yeast. Proc Natl Acad Sci USA. 1991;88:7434–7437. doi: 10.1073/pnas.88.16.7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyerowitz E. Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell. 1998;94:261–271. doi: 10.1016/s0092-8674(00)81425-7. [DOI] [PubMed] [Google Scholar]
- Hua J, Sakai S, Nourizadeh S, Chen QC, Bleecker AB, Ecker JR, Meyerowitz EM. EIN4 and ERS2 are members of the putative ethylene receptor gene family. Plant Cell. 1998;10:1321–1332. doi: 10.1105/tpc.10.8.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. The Never-ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lashbrook CC, Tieman DM, Klee HJ. Differential regulation of the tomato ETR gene family throughout plant development. Plant J. 1998;15:243–252. doi: 10.1046/j.1365-313x.1998.00202.x. [DOI] [PubMed] [Google Scholar]
- Lincoln JE, Cordes S, Read E, Fischer RL. Regulation of gene expression by ethylene during Lycopersicon esculentum (tomato) fruit development. Proc Natl Acad Sci USA. 1987;84:2793–2797. doi: 10.1073/pnas.84.9.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton S, Fray RG, Brown S, Grierson D. Ethylene receptor expression is regulated during fruit ripening, flower senescence and abscission. Plant Mol Biol. 1996;31:1227–1231. doi: 10.1007/BF00040839. [DOI] [PubMed] [Google Scholar]
- Pietrzak M, Shillito RD, Hohn T, Potrykus I. Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res. 1986;14:5857–5868. doi: 10.1093/nar/14.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray J, Moreau P, Bird C, Bird A, Grierson D, Maunders M, Truesdale M, Bramley P, Schuch W. Cloning and characterization of phytoene synthase genes from tomato. Plant Mol Biol. 1992;19:401–404. doi: 10.1007/BF00023387. [DOI] [PubMed] [Google Scholar]
- Rick CM, Butler L. Phytogenetics of the tomato. Adv Genet. 1956;8:267–382. [Google Scholar]
- Roman G, Lubarsky B, Kieber JJ, Rothenburg M, Ecker JR. Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: five novel mutant loci integrated into a stress response pathway. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai h, Hua J, Chen GQ, Chang C, Bleecker AB, Meyerowitz EM. ETR2 is an ETR1-like gene involved in ethylene signal transduction in Arabidopsis. Proc Nat Acad Sci USA. 1998;95:5812–5817. doi: 10.1073/pnas.95.10.5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schaller GE, Bleecker AB. Ethylene-binding sites generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Tieman D, Klee HJ. Differential expression of two novel members of the tomato ethylene-receptor family. Plant Physiol. 1999;120:165–172. doi: 10.1104/pp.120.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleecker AB, Chang C, Meyerowitz EM, Klee HJ. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Zhou D, Kalaitzis P, Matoo AK, Tucker ML. The mRNA for an ETR1 homologue in tomato is constitutively expressed in vegetative and reproductive tissues. Plant Mol Biol. 1996a;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- Zhou D, Matoo AK, Tucker ML. Molecular cloning of a tomato cDNA (accession no. U4279) encoding an ethylene receptor. Plant Physiol. 1996b;110:1435. [Google Scholar]