Abstract
Visual information is delivered to the brain by >40 types of retinal ganglion cells (RGCs). Diversity in this representation arises within the inner plexiform layer (IPL), where dendrites of each RGC type are restricted to specific sublaminae, limiting the interneuronal types that can innervate them. How such dendritic restriction arises is unclear. We show that the transcription factor Tbr1 is expressed by four mouse RGC types with dendrites in the outer IPL, and is required for their laminar specification. Loss of Tbr1 results in elaboration of dendrites within the inner IPL, while mis-expression in other cells re-targets their neurites to the outer IPL. Two transmembrane molecules, Sorcs3 and Cdh8, act as effectors of the Tbr1-controlled lamination program. However, they are expressed in just one Tbr1-expressing RGC type, supporting a model in which a single transcription factor implements similar laminar choices in distinct cell types by recruiting partially non-overlapping effectors.
INTRODUCTION
Many regions of the nervous system are arranged into parallel laminae. Neurons that synapse in these areas often confine their axons and dendrites to one or a few of these laminae, restricting their choice of synaptic partners. Laminar specificity is so widespread that it appears to be a major determinant of specific connectivity1,2.
A particularly striking instance of laminar specificity occurs in the vertebrate retina. In mouse, dendrites of >40 retinal ganglion cell (RGC) types arborize in a synaptic layer called the inner plexiform layer (IPL), with dendrites of each type restricted to one or a few of at least 10 sublaminae. There, they receive synapses from ≥70 types of interneurons (amacrine and bipolar cells) whose processes also arborize in specific IPL sublaminae. This stereotyped, stratified arrangement restricts the interneuronal types that can innervate each RGC type, thereby tuning the latter to specific visual features3. The best studied example is a ON/OFF bipartite division of the IPL; RGCs that project dendrites to the outer half of the IPL receive inputs from bipolar cells that are excited by decrements in illumination levels (OFF responses), whereas those that project to the inner half receive inputs from bipolar cells that are excited by increments (ON responses). Consequently these two groups of RGCs are termed OFF- and ON-type respectively4. Further dendritic restrictions within these zones are associated with additional distinctions in RGC response type5.
Multiple cell surface molecules have been shown to mediate intercellular interactions in the IPL, leading in some cases to targeting of neurites to specific sublaminae. They include members of the immunoglobulin6–9 and cadherin superfamilies10, semaphorins and plexins11,12. In contrast, little is known about how the expression of these cell-surface molecules is coordinated to specify laminar targeting of dendrites. Here, we identify the transcription factor T-box brain 1 (Tbr1) as one such regulator. We show that Tbr1 is selectively expressed by four RGC types, all of which bear dendrites that arborize in the outer third of the IPL. Intrigued by this commonality, we used loss- and gain-of-function approaches to ask whether Tbr1 is involved in dendritic targeting. We found that it is required for laminar patterning of Tbr1-expressing RGCs and can re-target dendrites of other neuronal types to the outer IPL when ectopically expressed. We then identified two cell-surface molecules, Cadherin 8 (Cdh8) and Sortilin-related VPS10 domain containing receptor 3 (Sorcs3), which are regulated by Tbr1 and act as two of its downstream effectors to restrict dendrites of one Tbr1-expressing RGC type, the J-RGC13, to the outer IPL. Strikingly, however, Cdh8 and Sorcs3 are not expressed by the other three Tbr1-expressing RGC types. These results suggest that Tbr1 recruits at least partially distinct sets of downstream effectors to specify laminar identity in the different RGC types that express it.
RESULTS
Four RGC types express Tbr1
To identify markers and potential regulators of specific RGC types, we analyzed the expression of transcription factors in mouse retina14. Tbr1 was expressed by ~15% of RGCs but by no other retinal cells (Figure 1a–b). To date, no single RGC type in mouse accounts for more than 10% of total RGCs3. We therefore suspected that Tbr1 labeled multiple RGC types.
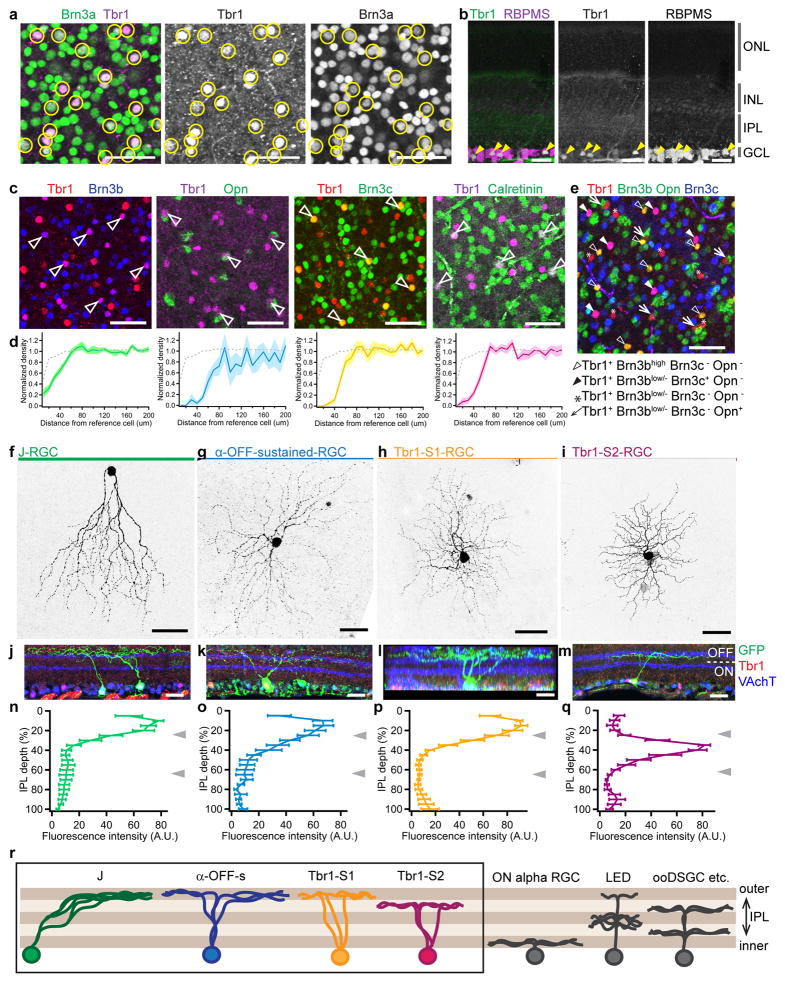
Figure 1. Expression of Tbr1 in four types of OFF-laminating RGCs.
(a) P21 retinal whole mount stained with antibodies to Tbr1 and Brn3a, an RGC marker. A subset of RGCs is Tbr1+. Yellow circles mark Tbr1+ soma. Scale=50μm. (b) Cross-section of P12 retinas showing Tbr1 expression exclusively in RGCs, marked with RBPMS. Arrowheads mark Tbr1+ Rbpms+ RGCs. Scale=25μm. (c) Whole mounts showing that subsets of Tbr1-RGCs express Brn3b, Opn, Brn3c or calretinin (arrowheads). Brn3b and Brn3c are nuclear, Opn is perinuclear and calretinin is cytosolic. Scale=50μm. (d) Density recovery profiles (DRP) for soma co-expressing each marker pair in c. Solid line represents mean and shaded bounds indicate standard error. Dotted gray line indicates normalized density of a heterogenous population consisting of multiple cell types (in this case, the entire Tbr1+ population). n=4 fields per retina, 3 retinas per marker pair, each retina from a different animal. (e) Whole mount of retina stained with a combination of Tbr1, Brn3b, Opn and Brn3c, showing four non-overlapping populations of Tbr1-RGCs. Each population is marked by an open triangle, closed triangle, asterisk or arrow as indicated below the image. Scale=50μm. (f–i) Tbr1-RGCs labeled in whole mounts showing dendritic morphologies of each type. Scale = 50μm. (j–m) Cross-sections (j, k, m) or rotated view (l) of each Tbr1-RGC type from JAM-B-CreER;Thy1-STOP-YFP (j), W7 (k), YFP-H (l) and Cdh4-CreER;Thy1-STOP-YFP (m). Scale = 25μm. (n–q) Quantification of the dendritic stratification for each Tbr1-RGC type. n=7 J-RGCs, 8 α-OFF-s RGCs, 6 Tbr1-S1 RGCs and 6 Tbr1-S2 RGCs from 3 animals each. Line plot and brackets indicate average and standard error. Grey arrowheads mark positions of S2 and S4 as indicated by VAchT immunostaining. (r) Schematic summarizing dendritic stratification of Tbr1-RGCs in comparison to other RGC types. All Tbr1-RGCs laminate dendrites within the outer third of the IPL. Each experiment in a–c, e and f–m was repeated independently in three animals with similar results.
To assess the number and identity of what we will call “Tbr1-RGC” types, we co-stained retinas with Tbr1 plus molecular markers expressed by RGC subsets, including other transcription factors. Tbr1-RGCs did not appreciably express FoxP2, which marks four F-RGC types14; Satb1, which is enriched in four ON-OFF direction-selective RGC (ooDSGC) types9,15; or Tbr2, which marks five intrinsically photosensitive RGC types16,17 (Supplementary Figure 1a). Instead, we found that subsets of Tbr1-RGCs expressed robust levels of Brn3b, Brn3c, osteopontin (Opn; gene symbol Spp1) or calretinin (Figure 1c).
To determine whether these marker pairs labeled distinct RGC types, we made use of a feature of retinal neurons called mosaic spacing: neurons of the same type are less likely to be near neighbors than would be expected by chance, whereas they are randomly distributed with respect to cells of other types18,19. When viewed in whole mounts, somata co-labeled by each marker pair formed a uniformly-spaced mosaic as judged by the density recovery profile (DRP) statistic20, and is therefore likely to represent a single type (Figure 1c–d). Exclusion zone size and soma density varied with each marker pair, indicating that they define four different RGC types (Supplementary Figure 1b). They populated the entire retina, making up 1.8% (Tbr1+ Opn+) to 6.5% (Tbr1+ Brn3b+) of RGCs and totaling ~15% of all RGCs (Supplementary Figure 1c–g). Co-immunostaining for Tbr1 with combinations of Brn3b, Brn3c, Opn and calretinin confirmed that the four types are non-overlapping (Figure 1e; Supplementary Figure 1h). Together, these four RGC types account for most if not all Tbr1-expressing retinal cells.
Tbr1-RGCs laminate in the outer strata of the IPL
To assess the morphology of Tbr1-RGCs, we screened transgenic lines in which RGC subsets are labeled with a fluorescent protein. No labeled RGCs were Tbr1+ in lines that selectively marked ooDSGCs21 or W3-RGCs22 (Supplementary Figure 1m–n). However, all J-RGCs labeled in JAM-B-CreER;Thy1-STOP-YFP13 were Tbr1+ Brn3b+ (Figure 1f,j, Supplementary Figures 1i) and all alpha-OFF-sustained RGCs (α-OFF-s-RGCs) labeled in TYW722–24 were Tbr1+ Opn+ (Figure 1g,k, Supplementary Figure 1j), identifying two previously characterized types as Tbr1+.
The other two Tbr1-RGC types appeared novel. We therefore sought their identities in mouse lines that label multiple RGC types sparsely14,25. Tbr1 plus Brn3c marked RGCs with radial dendrites that laminated within sublamina (S)1 of the IPL; we follow the convention of dividing the IPL into five equal strata, S1–5 (Figure 1h,l, Supplementary Figure 1k). Tbr1 plus calretinin marked another RGC type; its dendrites abutted those of OFF starburst amacrine cells in S2 and bore spine-like protrusions that arose perpendicularly from their parent branches (Figure 1i,m, Supplementary Figure 1l). We will refer to these two types as Tbr1-S1-RGCs and Tbr1-S2-RGCs respectively.
The ability to label Tbr1-RGCs allowed us to assess their topographic distribution, dendritic field area and coverage factor. α-OFF-s-RGCs displayed a temporal-high-nasal-low gradient as previously described26; J-RGCs displayed a dorsal-low-ventral-high gradient and the two novel RGC types displayed shallow center-to-periphery gradients (Supplementary Figure 1c–f). J-RGCs had the highest coverage factor while α-OFF-s-RGCs had the largest dendritic area (Supplementary Figure 1o–p). Remarkably, however, dendrites of all four Tbr1-RGC types were restricted to the outer third of the IPL, with three restricted to the outermost sublamina or S1, and the fourth type to S2 (Figure 1j–r).
Tbr1 is required for laminar specification of RGC dendrites
Before assessing the role of Tbr1 in the retina, we determined its expression pattern through development. Tbr1 immunoreactivity was undetectable at embryonic day(E) 13.5, a time at which RGC production is reaching its peak27. Over the next few days, Tbr1 appeared in post-mitotic cells; by E17.5, it was confined to a subset of RGCs that were already non-overlapping with FoxP2- and Tbr2-positive cells (Supplementary Figure 2a–c). This suggests that Tbr1 could regulate aspects of RGC development, including dendritic morphogenesis, which begins around birth (postnatal day[P] 0)28.
Because constitutive Tbr1 mutant mice die perinatally29, we generated conditional Tbr1 mutants to test this possibility (Supplementary Figure 3a). We first deleted Tbr1 throughout the retina using a line that expresses Cre in embryonic retinal progenitors (Tbr1flox;Six3Cre, henceforth Tbr1ret). Pan-retinal deletion of Tbr1 had no detectable effect on retinal architecture or RGC numbers (Supplementary Figure 3b).
We then deleted Tbr1 selectively from J-RGCs using the JAM-B-CreER line (Tbr1flox;JAM-B-CreER;Thy1-stop-YFP, henceforth Tbr1J; tamoxifen delivered at E14.5 or P0) and verified by immunostaining that ~95% of YFP-labeled J-RGCs lost Tbr1. Remarkably, ~65% of these Tbr1-deficient J-RGCs developed ectopic dendrites. Although they retained dendritic branches in S1, they also extended dendrites in S4, within the inner (ON) region of the IPL (p=1.2×10−6, Cochran-Armitage test) (Figure 2a–c). Other aspects of J-RGCs were not detectably affected: Dendritic field area, total length and ventral asymmetry of the dendritic arbor did not differ significantly between controls and mutants (two-tailed Student’s t-test, p= 0.62, 0.40, 0.39 respectively) (Figure 2d, Supplementary Figure 4a–c). The projection of axons to a primary central target, the superior colliculus, was similarly restricted to the superficial retinorecipient lamina in mutants and controls30 (Supplementary Figure 4d). Finally, Tbr1 mutant J-RGCs neither expressed markers of other RGC types such as FoxP2, Satb1, or Tbr29,14–17, nor did they lose markers expressed by wild-type J-RGCs, such as Brn3b and Rbpms (Supplementary Figure 4e, see also Supplementary Figure 9b). Thus, Tbr1 plays a selective role in specifying the laminar position of J-RGC dendrites.
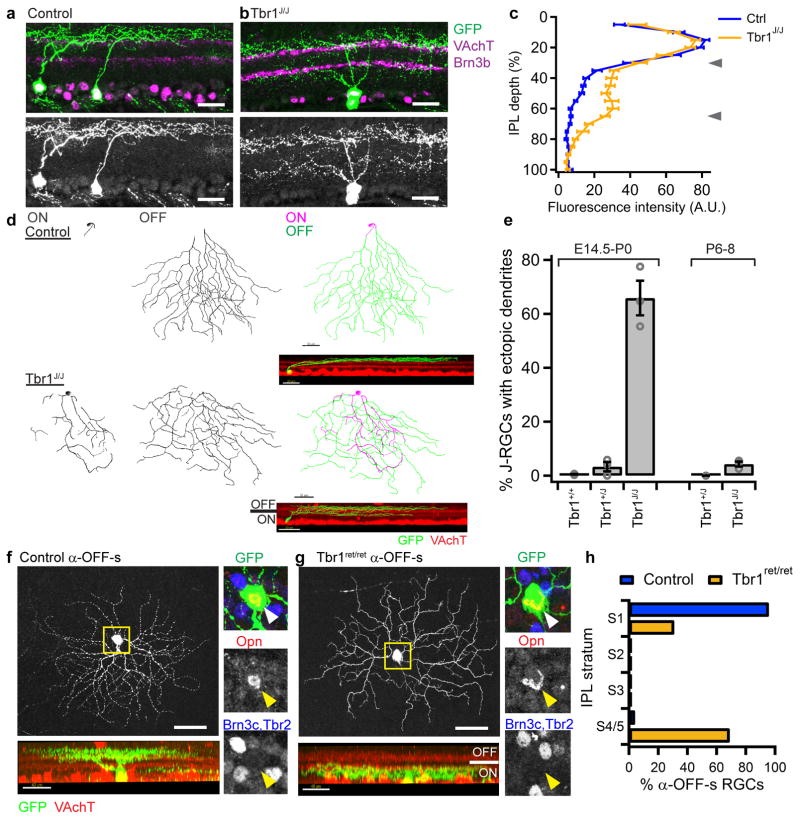
Figure 2. Tbr1 deletion results in ectopic dendritic lamination.
(a–b) Cross-sections of the IPL showing dendritic stratification of P20 J-RGCs in control and Tbr1J/J retina. Scale=25μm. Experiment was repeated independently in 4 animals per genotype with similar results. (c) Distribution of GFP intensities from J-RGC dendrites through the depth of IPL in controls versus Tbr1J/J at P20. n= 20 and 39 sections for control and Tbr1J/J respectively, 4 animals per genotype. Line plots and brackets indicate averages and standard error. p=1.2x10−6, Cochran-Armitage test. Arrowheads indicate peaks of VAchT signals at S2 and S4. (d) Traces (scale=30μm) and rotated views (inset, scale=20μm) of P20 control and Tbr1J/J J-RGCs. Experiment was repeated independently in 3 animals per genotype with similar results. (e) Proportions of J-RGCs with ectopic dendrites when Tbr1 deletion is induced at E14.5-P0 or P6–8. Each bar represents average proportions± standard error. Circles indicate individual retinas. n= 3 retinas per genotype, each retina from a different animal. For E14.5-P0, 144-517, 34-146 and 35-100 cells for Tbr1+/+, Tbr1+/J and Tbr1J/J respectively. For P6–8, n=184–303 and 40–188 cells for Tbr1+/J and Tbr1J/J respectively. (f,g) En face (scale=50μm) and side (scale=40μm) views of α-OFF-s-RGCs labeled in YFP-H control (f) and YFP-H;Tbr1ret/ret (g) retinas. Insets show how cells were identified as α-OFF-s-RGCs (Opn+, Brn3c-,Tbr2-). Loss of Tbr1 re-targeted their dendrites to ON sublaminae of IPL. Experiment was repeated independently in 6 control and 9 Tbr1ret/ret animals with similar results. (h) Proportions of α-OFF-s-RGCs that laminate within each sublamina in the IPL. n= 24 and 29 cells from 6 control and 9 Tbr1ret/ret retinas respectively, each retina from a different animal. p=1.6 x 10−6, Pearson’s chi-square test.
The ability to control the timing of Tbr1 deletion in Tbr1J mice allowed us to determine when it is required for patterning J-RGC dendrites. J-RGC dendrites begin to extend around P0, are concentrated in the outer half of the IPL by P3, and become restricted to S1 by P628 (Supplementary Figure 5a). Deletion at E14.5 and P0 perturbed dendritic lamination to a similar extent (compare Figure 2b and Supplementary Figure 4e), indicating that Tbr1 acts during rather than prior to dendritogenesis. Moreover, mutant J-RGCs extended ectopic dendritic branches within the inner half of the IPL by P4, indicating that Tbr1 acts during the period of laminar restriction (p=0.00018, Cochran-Armitage test) (Supplementary Figure 5b–c). In contrast, deleting Tbr1 at P6 had no detectable effect on dendritic morphology (p=0.096, Cochran-Armitage test) (Figure 2e, Supplementary Figure 5d–e). Thus, Tbr1 is required to direct dendritic stratification rather than to maintain it.
To find out if Tbr1 serves a similar role in other RGC types, we extended the analysis to α-OFF-s-RGCs. We could not use the TYW7 line for this purpose because Cre deletes its YFP cassette22. We therefore used the YFP-H line25 to reveal dendritic morphology and identified α-OFF-s-RGCs as Opn+ RGCs that lacked Brn3c and Tbr2, which are expressed by other alpha types23. As expected, dendrites of α-OFF-s-RGCs marked in this way arborized within S1 in controls (Figure 2f,h). In contrast 70% of these RGCs sent dendritic branches to S4 or S5 in Tbr1ret/ret; YFP-H mice (p=1.6 × 10−6, Pearson’s chi-square test) (Figure 2g–h). As was the case for J-RGCs, dendritic field area and length were unaffected (p=0.75 and 0.18 respectively, two-tailed Student’s t-test) (Supplementary Figure 4f–g). Although we were unable to assay Tbr1-S1- and Tbr1-S2-RGCs, this result suggests a common role for Tbr1 in patterning dendritic lamination for all Tbr1-RGC types.
Loss of visual responses in Tbr1 mutant RGCs
Assessing the role of Tbr1 in RGC function required recording from Tbr1-RGCs. We faced two problems. First, in previous studies, we used transgenic expression of YFP to target cells for intracellular recording6,9,10. In this case, however, we were unable to identify Tbr1-RGCs prospectively. We therefore developed a calcium imaging protocol, in which we expressed the calcium indicator GCaMP6f in a large fraction of RGCs, recorded responses to visual stimuli, and then performed immunostaining to identify individual types (Figure 3a and Supplementary Figure 6a–d). Second, identification initially relied on Tbr1 immunostaining, which was not applicable to Tbr1ret/ret. We therefore used alternative markers. This approach was most successful for α-OFF-s-RGCs because, as noted above, these cells express Opn but not Brn3c or Tbr2.
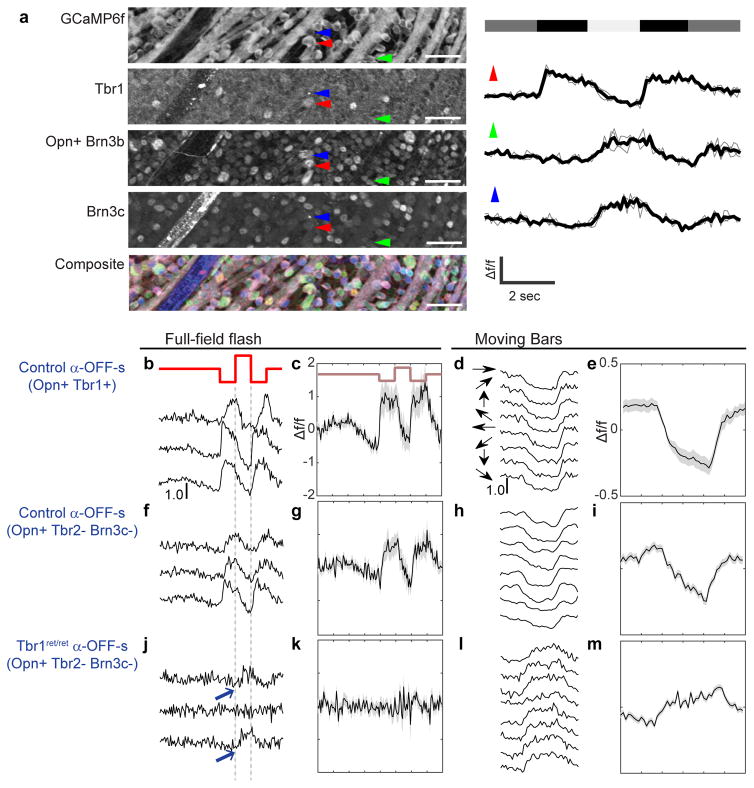
Figure 3. Physiological defects in Tbr1 mutants.
(a) Left, whole mount view of a recorded field from a GCaMP6f-expressing retina that was immunostained for GFP, Tbr1, Opn plus Brn3b and Brn3c. Colors in composite image represent the above markers in gray, red, green and blue respectively. Right, calcium responses to a full-field flash from three cells, marked in a. Thick lines are averages of three repetitions, each represented by a thin line. Red arrow marks α-OFF-s-RGC (Tbr1+Opn+); as expected, it shows sustained activation to decreases in light intensity (top bar). Scale = 50μm. (b–m) Responses of α-OFF-s-RGCs in control and Tbr1ret/ret. Scale denotes 100% Δf/f. Shaded bounds indicate standard error.b,f,j show sample calcium responses evoked by a 2-second full-field flash (in red) from three cells per genotype. Responses are average of three repetitions. c,g,k are averaged responses of all cells to full-field flashes. Black line and shaded bounds represent mean ± SEM. d,h,l are sample calcium responses evoked by a bar moving in eight different directions. Responses are average of three presentations of the bar stimulus. e,i,m are averaged calcium responses to moving bars. Black line and shaded bounds represent mean ± SEM. b–e are control α-OFF-s-RGCs identified by Tbr1 and Opn. n=18 cells from 6 retinas. f–i are control α-OFF-s-RGCs identified by Opn and absence of Tbr2 and Brn3c. n=16 cells from 3 retinas. j–m are α-OFF-s-RGCs from Tbr1ret/ret, similarly identified as in f–i. n=13 cells from 6 retinas.
We first characterized α-OFF-s-RGCs marked with the Tbr1-Opn and Tbr2-Opn-Brn3c combinations. In both cases, RGCs showed the expected properties23,24. They generated a robust increase in GCaMP signal to light offset and a decreased signal to light onset (Figure 3b–c, f–g). They also responded to moving bars, with inhibition when a bright bar entered the receptive field and excitation when it left. However, they showed similar responses to motion in all directions and thus were direction-non-selective (Figure 3d–e, h–i and Supplementary Figure 6e–f).
In contrast, α-OFF-s-RGCs in Tbr1ret/ret responded poorly to both flashes and moving bars (Figure 3j-m, Supplementary Figure 6g). Some non-responsive cells are expected in calcium imaging studies for technical reasons (see Methods), but most control α-OFF-s-RGCs (11/16) responded appropriately whereas only 4 of 13 mutant cells were responsive by criteria described in Methods. Moreover, of the 4 responsive mutant α-OFF-s-RGCs, only one appeared normal; the other 3 generated ON responses (arrows in Figure 3j), a behavior not observed in control α-OFF-s-RGCs. Overall, however, the responsiveness of control and Tbr1 mutant RGCs were similar, suggesting that the defect was specific (Supplementary Figure 6h–i).These results indicate that Tbr1 is required for the visual responsiveness of RGCs that express it. We speculate that this phenotype is a consequence of dendritic displacement, a possibility supported by the acquisition of ON responses in conjunction with the formation of ON arbors. It is also possible, however, that Tbr1 plays additional roles in responsiveness or synaptogenesis (see Discussion).
Tbr1 is sufficient for laminar specification of dendrites
To ask whether Tbr1 expression is sufficient to direct RGC arbors to the outer portion of the IPL, we ectopically expressed it by electroporation in neonatal retinas, along with a plasmid encoding a fluorescent protein (XFP) to mark transfected cells. This technique transfects multiple retinal types including photoreceptors, interneurons and RGCs depending on the site of DNA delivery (see Methods). To transfect RGCs, we delivered DNA intraretinally. Nearly all RGCs transfected with Tbr1+XFP elaborated dendrites within the OFF part of the IPL, whereas dendrites of control RGCs, transfected with XFP only, were equally likely to be found in OFF or ON regions (p=8.0×10−7, Pearson’s chi-square test) (Figure4a–c). Since only 15% of RGCs are endogenously Tbr1+, it seemed likely that forced expression of Tbr1 redirected dendrites of RGCs from other sublaminae to S1. In support of this idea, dendrites of Tbr1-misexpressing RGCs were diverse in arborization patterns and sizes, as well as levels of Brn3b (Supplementary Figure 7a). Moreover, we found that dendrites of CART+ ooDSGCs, which normally stratify in S2 and S431, were re-targeted to S1 (Figure 4b). Thus, Tbr1 plays an instructive role in dendritic lamination.
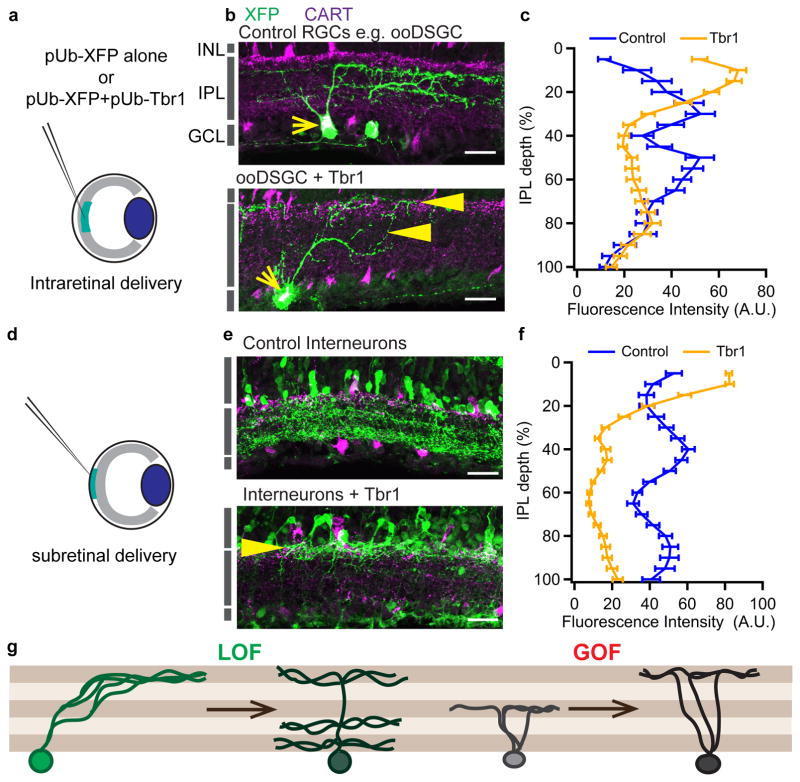
Figure 4. Ectopic expression of Tbr1 re-targets neurites to the outer IPL.
(a) Intraretinal delivery of DNA for overexpression in RGCs. (b) Expression of XFP alone (control) or XFP with Tbr1 into ON-OFF direction selective RGCs (ooDSGCs), identified by CART immunoreactivity (yellow arrows). (c) Quantification of dendrite lamination of control and Tbr1 mis-expressing RGCs. n = 39 control cells from 6 retinas and 62 Tbr1-misexpressing cells from 7 retinas, p=7.95x10−6, Pearson’s chi-square test. (d) Subretinal delivery of DNA construct for overexpression in interneurons. (e) Expression of XFP alone (control) or XFP with Tbr1 in interneurons. In b and e, yellow arrowheads mark neurites re-targeted to the outer IPL. Scale =25μm. (f) Quantification of neurite stratification within the IPL by control and Tbr1-misexpressing interneurons. Line plots and brackets in c and f indicate averages and standard errors. n = 63 and 65 sections from 8 control and 6 Tbr1-electroporated retinas respectively, p=1.0x10−10, Cochran-Armitage test. (g) Schematic summarizing loss- and gain-of-function outcomes for Tbr1 within the IPL.
We also ectopically expressed Tbr1 in interneurons, using subretinal delivery of DNA. Tbr1-misexpressing interneurons similarly re-targeted neurites to the outer IPL (p=1.0×10−10, Cochran-Armitage test) (Figure 4d–f). The effect was specific in that transfected somata remained in the inner nuclear layer, and neither expressed RGC markers (e.g., RBPMS) nor extended axons (Supplementary Figure 7b–c). Amacrine cells that mis-expressed Tbr1 also retained the characteristic marker, AP2 (Supplementary Figure 7d). Together, our loss- and gain-of-function approaches establish Tbr1 as a transcriptional determinant of laminar identity for RGC dendrites (Figure 4g).
Tbr1 regulates Cdh8 and Sorcs3 expression in J-RGCs
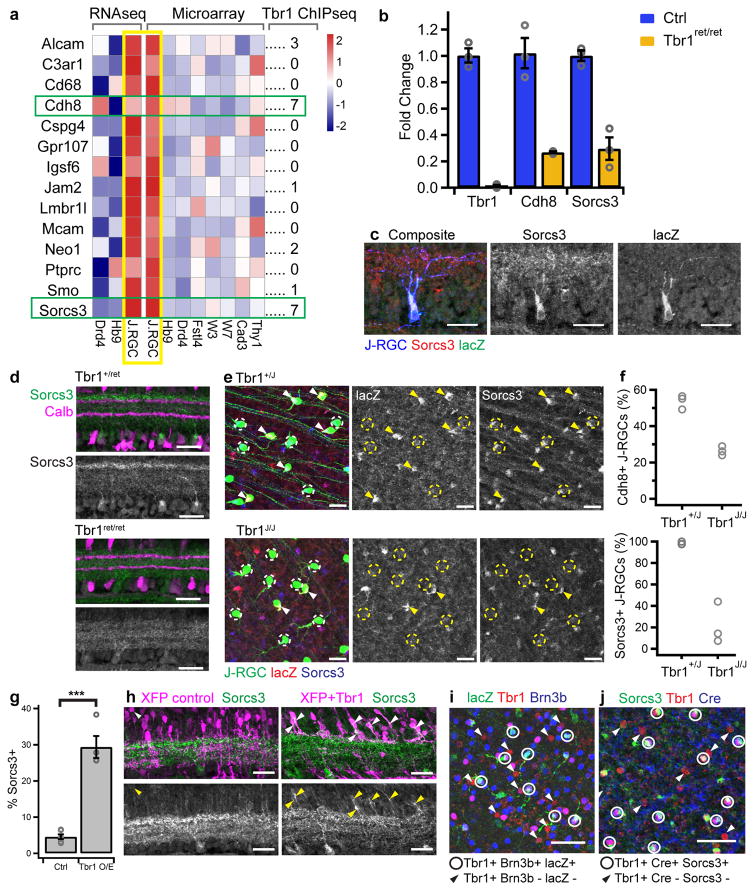
To identify downstream effectors of Tbr1, we focused on J-RGCs, using five criteria to select promising candidates. First, we profiled J-RGCs by RNAseq at P6, when dendritic restriction is nearing completion, and compared them to profiles from two sets of ooDSGCs9. Second, we analyzed microarray data from 7 groups of RGCs, including J-RGCs18. Third, from genes selectively expressed by J-RGCs in both comparisons, we chose those encoding cell-surface proteins, which are the most likely mediators of cell-cell interactions critical for patterning dendrites7–10. Fourth, of the 14 genes that fulfilled these criteria, we identified 6 that contained Tbr1-binding sites in their genomic loci as determined by ChIP-seq of embryonic cortex32: Alcam, Cdh8, Jam2, Neo1, Smo and Sorcs3 (Figure 5a). Of these 6 genes, Cdh8, a classical cadherin and, Sorcs3, a neuronal type I transmembrane receptor33, have the most Tbr1-binding sites (Figure 5a and Supplementary Figure 8a). Finally, we isolated wildtype and Tbr1 mutant J-RGCs and compared them by RNAseq. Thirteen cell surface molecules were significantly downregulated in Tbr1 mutant J-RGCs (p<0.001, two-tailed Fisher’s Exact test), including Sorcs3, Jam2 and Cdh8 but not Alcam, Neo1, or Smo (Supplementary Figure 9a, see also Supplementary Figure 8b–c). None of the remaining 10 candidates were selectively expressed by J-RGCs (Supplementary Figure 9c) and analysis of Jam2 null mutants revealed subtle defects in dendritic morphology but no alterations in lamination28. Based on these considerations, we analyzed Sorcs3 and Cdh8 further.
Figure 5. Cdh8 and Sorcs3 are Tbr1 targets in J-RGCs.
(a) Heatmaps showing expression of J-RGC-enriched cell-surface molecules from RNAseq and microarray data, and number of Tbr1-ChIPseq peaks associated with each gene32. n=2 pup litters per RGC type. (b) Expression of J-RGC candidate genes in P4 RGCs from Tbr1ret/ret relative to controls by RT-qPCR. n=3 animals per genotype; bars indicate mean ± standard error, circles indicate values from individual animals. (c) Retinal cross-section showing expression of Sorcs3 and lacZ from the Cdh8lacZ allele by a P5 J-RGC. (d) Cross-section of control and Tbr1ret/ret IPL stained for Sorcs3. In controls, immunoreactivity is concentrated in a subset of RGC somata and dendrites in outer IPL. Levels are decreased in Tbr1ret/ret. (e) en face views of P4 J-RGC soma in Tbr1J immunostained for Sorcs3 and lacZ. Arrowheads mark J-RGCs that express both markers. Dotted circles mark J-RGCs with no detectable lacZ: these cells still express Sorcs3 in Tbr1+/J but not in Tbr1J/J. (f) Proportions of P4 J-RGCs that express lacZ or Sorcs3 in Tbr1J retinas. n=3 retinas per genotype, 65-683 and 152-520 control and Tbr1J/J cells respectively,. p=0.00050 and 0.0025 for lacZ and Sorcs3 respectively, two-tailed Student’s t-test. Circles represent individual retinas. (g) Proportion of control (Ctrl) or Tbr1-misexpressing (Tbr1 O/E) interneurons that express Sorcs3. n= 6 control and 4 Tbr1-misexpressing retinas, 38-603 and 60-230 cells respectively, *** p<9.9 x 10−6, two-tailed Student’s t-test. Bars and brackets indicate mean ± standard error, circles represent individual retinas. (h) Retinal cross-sections showing Sorcs3 expression in control and Tbr1-misexpressing interneurons. Arrowheads mark Sorcs3+ soma. Each experiment was repeated independently in 6 control and 4 Tbr1-misexpressing animals with similar results. (i) Whole mount of P3 Cdh8lacZ retina stained for lacZ, Tbr1 and Brn3b. Circles mark J-RGCs that are triple-positive. Arrowheads mark the other Tbr1-RGC types, which are Brn3b-. These cells lack lacZ immunoreactivity. (j) Whole mount of P5 JAM-B Cre knock-in retina stained with Sorcs3, Tbr1 and Cre. Dotted circles mark J-RGCs that are triple-positive. Arrowheads mark the other Tbr1- RGC types that are Cre-. These cells do not express Sorcs3. Scale in c–e, h=25μm, in i–j = 50μm. Each experiment in c- e and i–j was repeated independently in three animals with similar results.
We first validated that Sorcs3 and Cdh8 expression in the retina are Tbr1-dependent by performing RT-qPCR on total RGCs isolated from P4 control and Tbr1ret/ret mutants: mRNA levels of both Sorcs3 and Cdh8 decreased by ~80% in mutants (Figure 5b). We also immunostained control and Tbr1 mutant retinas for Sorcs3 and, lacking appropriate antibodies to Cdh8, visualized E. coli beta-galactosidase (lacZ) driven from a Cdh8lacZ knock-in allele10. Control J-RGCs at P4–5 expressed both Sorcs3 and Cdh8 (Figure 5c). However, while we detected Sorcs3 protein in all J-RGCs, we detected lacZ in only about half of them (Figure 5d–f). In Tbr1J/J J-RGCs, levels of both Sorcs3 and lacZ protein were strongly reduced by P4–5 (p=0.0025 and 0.00050 respectively, two-tailed Student’s t-test) (Figure 5d–f). In the converse experiment, overexpression of Tbr1 by electroporation resulted in the up-regulation of Sorcs3 protein in ~30% of Tbr1-misexpressing cells (p<9.9×10−6, two-tailed Student’s t-test) (Figure 5g–h). Together, these results confirm that Sorcs3 and Cdh8 expression in J-RGCs are regulated by Tbr1.
During the first postnatal week, neither Cdh8 nor Sorcs3 was detectably expressed by Tbr1-RGCs other than J-RGCs (Figure 5i, j Supplementary Figure 8g). During the second postnatal week, however, their expression patterns diverged. Cdh8 expression declined in RGCs (Supplementary Figure 8d) but was up-regulated in OFF cone bipolar cells as described previously10. Sorcs3, in contrast, was up-regulated in rod bipolar cells and other RGCs (Supplementary Figure 8e–f). Notably, Sorcs3 was concentrated within the dendrites of both RGCs and rod bipolar cells (Figure 5d, Supplementary Figure 8e).
Cdh8 and Sorcs3 pattern J-RGC dendrites
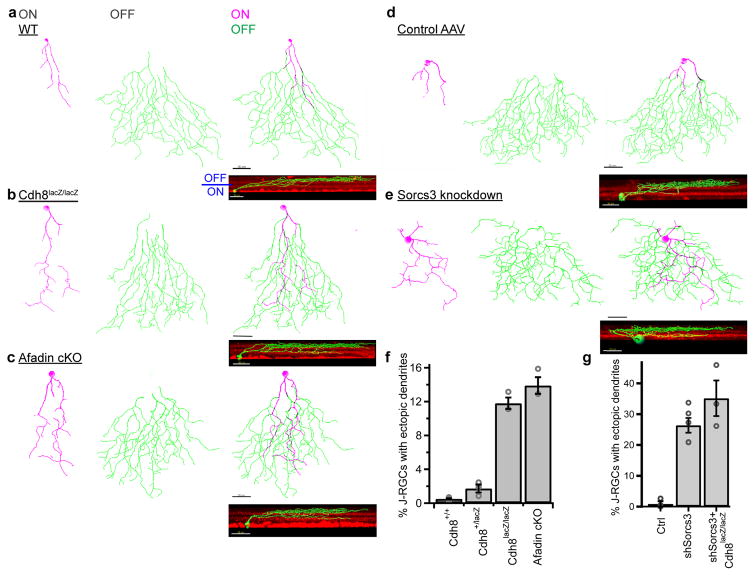
We used loss- and gain-of-function strategies to ask whether Cdh8 and/or Sorcs3 affect dendritic targeting of J-RGCs. To delete Cdh8, we used the Cdh8lacZ line, which carries a null allele. 12% of J-RGCs in Cdh8lacZ/lacZ retinas displayed ectopic ON dendrites that resembled those in Tbr1 mutant J-RGCs while <1% of wildtype J-RGCs and <2% of Cdh8lacZ/+ RGCs had ectopic dendrites (p=1.4×10−7 and 0.38 for wildtype vs Cdh8lacZ/lacZ and wildtype vs Cdh8+/lacZ respectively, Tukey-Kramer test) (Figure 6a–b, f). Given that Cdh8 was lost from all cells, we determined, in two ways, whether the ectopic dendrites were due to the specific loss from J-RGCs, or a secondary consequence of defects in other cells. First, we confirmed that other OFF-type RGCs, including α-OFF-s-RGCs, which express Tbr1 but not Cdh8, were not significantly affected in Cdh8 mutants (p=0.12, Cochran-Armitage test) (Supplementary Figure 10a–b). Second, we used the JAM-B-CreER line to selectively delete afadin, an actin-filament binding protein that interacts with cadherins34, from J-RGCs; afadin mutants phenocopy cadherin mutants in some cases (35, Duan et al. submitted). J-RGCs still formed ectopic dendrites, at a similar frequency to Cdh8 mutants (p=0.11 for Cdh8lacZ/lacZ vs afadin cKO, Tukey-Kramer test) (Figure 6c,f). This result also suggests that the limited penetrance of the Cdh8 mutant is not a result of redundancy with or compensation by other cadherins.
Figure 6. Requirement of Cdh8 and Sorcs3 for laminar restriction in J-RGCs.
(a–c) Traces of J-RGC dendrites in wild-type (WT, a), Cdh8lacZ/lacZ (b) and afadin cKO (c) retinas. Arbors in ON (magenta) and OFF (green) halves of the IPL are shown separately. Insets show rotated views. (d–e) En face and rotated views of single-cell trace of a P12 J-RGC infected at P0 with a TdT control AAV (d) or shSorcs3 AAV (e). (f) Proportion of J-RGCs with ectopic dendrites in each genotype. n= 5 Cdh8+/+, 3 Cdh8+/−, 3 Cdh8−/− and 3 afadin cKO retinas. p=8.7x10−9, F(3,10)=163, one-way ANOVA; p=0.38 and 0.11 for Cdh8+/+ vs Cdh8+/lacZ and Cdh8lacZ/lacZ vs afadin cKO respectively, p=1.4x10−7, 2.8x10−8, 1.2x10−6 and 2.0x10−7 for Cdh8+/+ vs Cdh8lacZ/lacZ, Cdh8+/+ vs afadin cKO, Cdh8+/lacZ vs Cdh8lacZ/lacZ and Cdh8+/lacZ vs afadin cKO respectively, Tukey-Kramer test. (g) Proportion of J-RGCs with ectopic dendrites in retinas infected with AAV encoding TdT (Ctrl) or shSorcs3. n = 3, 5 and 3 retinas respectively for control, shSorcs3 and shSorcs3+Cdh8lacZ/lacZ. p=0.0013, F(2,8)=17.2, one-way ANOVA; p=0.0048, 0.0013 and 0.25 for ctrl vs shSorcs3, ctrl vs shSorcs3+Cdh8lacZ/lacZ, and shSorcs3 vs shSorcs3+Cdh8lacZ/lacZ respectively, Tukey Kramer test. In f and g, circles indicate data from individual retinas of different animals, bar and bracket represent mean and standard error. Scale in a–e = 30μm for en face view, and 20μm for rotated view.
Lacking a germ-line Sorcs3 mutant, we attenuated Sorcs3 expression using RNA interference, injecting intravitreally an adeno-associated virus (AAV) encoding a short hairpin RNA directed against Sorcs3 at P0 (Supplementary Figure 10c–e). Dendrites of J-RGCs infected with a control AAV laminated appropriately (Figure 6d,g), but ~30% of J-RGCs developed ectopic dendrites upon Sorcs3 knockdown, phenocopying the loss of Tbr1 (p=0.0048, Tukey-Kramer test) (Figure 6e,g). Knocking down Sorcs3 in Cdh8lacZ/lacZ mice had comparable effects, though not significantly more effective than in controls (p=0.25, Tukey-Kramer test) (Figure 6g).
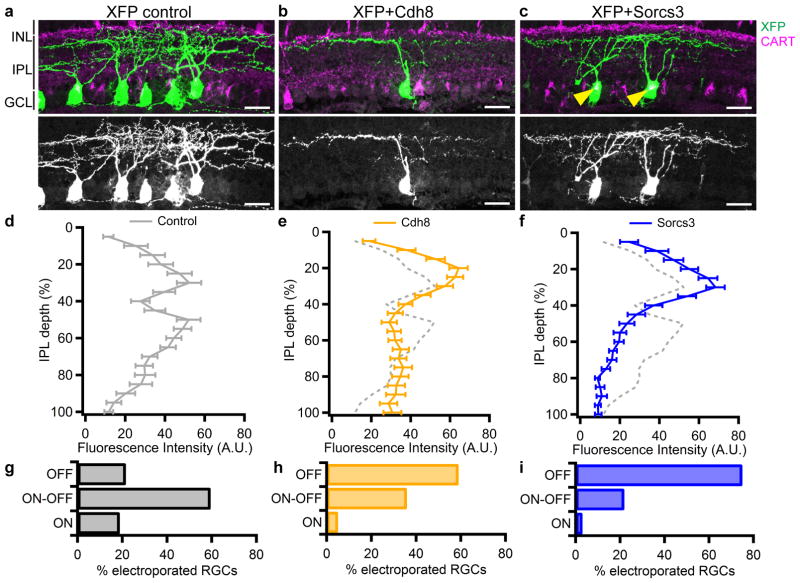
For gain-of-function analyses, we mis-expressed Cdh8 or Sorcs3 in RGCs by neonatal electroporation. Control RGCs showed a similar preference for either the ON or OFF half of the IPL as shown in the average of dendritic distributions from all electroporated RGCs (Figure 7a, d), and they project equally to ON and OFF sublamina (Figure 7g). In contrast, Cdh8-misexpressing RGCs are on average biased to the OFF sublamina (Figure 7b,e).~60% and 35% of Cdh8-misexpressing RGCs developed exclusively OFF dendrites or ON-OFF dendrites respectively (p=0.0028, Pearson’s chi-square test) (Figure 7h). Sorcs3-misexpressing RGCs showed an even stronger bias for the OFF sublamina (Figure 7c,f). 75% and 22% of RGCs that overexpressed Sorcs3 extended OFF and ON-OFF dendrites respectively (p=4.5x 10−5, Pearson’s chi-square test) (Figure 7i). Affected RGCs included immunohistochemically labeled ooDSGCs (Figure 7c). Therefore, both Cdh8 and Sorcs3 bias laminar targeting to the OFF half of the IPL.
Figure 7. Ectopic expression of Cdh8 or Sorcs3 re-targets RGC dendrites to the outer IPL.
(a–c) Retinal cross-section showing RGCs electroporated with control XFP (a), XFP+Cdh8 (b) or XFP+Sorcs3 (c). Arrowheads mark electroporated ooDSGCs, identified by their CART immunoreactivity. Scale=25μm. (d–f) Distribution of fluorescence intensities from dendrites of control RGCs (d, gray, re-plotted from Figure 4c), Cdh8-overexpressing RGCs (e, orange) or Sorcs3-overexpressing RGCs (f, blue). Line plots and brackets indicate average and standard error. Similar results were observed in retinas from 6 control, 4 Cdh8-misexpressing and 5 Sorcs3-misexpressing retinas, each processed independently. (g–h) Proportions of electroporated cells that extended OFF, ON-OFF or ON arbors in each condition. n=39, 32 and 35 RGCs from 6 control, 4 Cdh8-misexpressing and 5 Sorcs3-misexpressing retinas.
Cdh8 and Sorcs3 act downstream of Tbr1
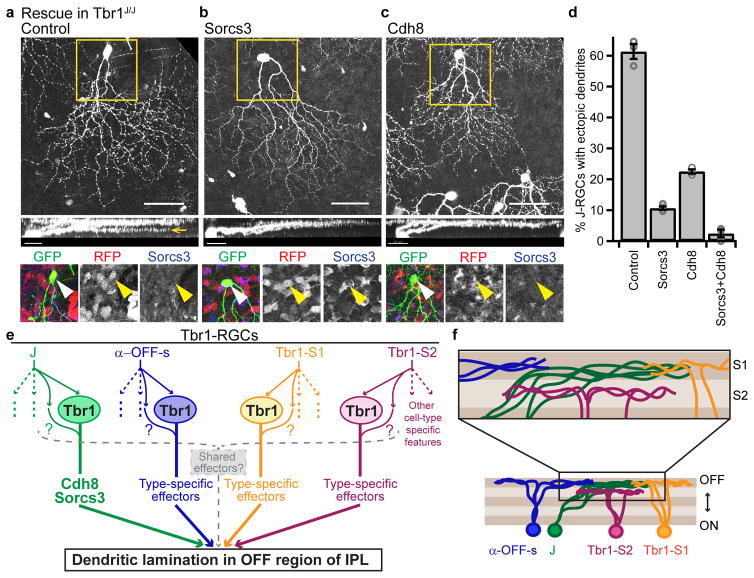
Finally, we asked whether restoration of Cdh8 or Sorcs3 in Tbr1 mutant J-RGCs would rescue laminar defects of their dendrites. We generated AAV vectors for this experiment because insufficient numbers of RGCs were transfected by electroporation. AAVs expressing Sorcs3, a Cdh8-RFP fusion or RFP alone were injected intravitreally into Tbr1J/J animals at P0, and tamoxifen was delivered (also at P0) to delete Tbr1 in J-RGCs and simultaneously label them with YFP. We then scored dendritic lamination of YFP-positive J-RGCs at P10. As in experiments presented above, ~60% of Tbr1 mutant J-RGCs labeled by the control AAV developed ectopic dendrites (Figure 8a,d). In contrast, significantly fewer J-RGCs infected with Sorcs3- or Cdh8-expressing AAVs bore ectopic dendrites (11% for Sorcs3, 23% for Cdh8; p<0.0001 for control vs Sorcs3/Cdh8, Tukey-Kramer test) (Figures 8b–d). Co-delivery of both Sorcs3 and Cdh8 almost completely rescued the Tbr1 phenotype (3%; p<0.0001 for control vs Sorcs3+Cdh8, Tukey-Kramer test) (Figure 8d). Together, these data indicate that Cdh8 and Sorcs3 act downstream of Tbr1 to implement laminar identity of J-RGC dendrites.
Figure 8. Regulation of dendrite laminar identity by Tbr1.
(a–c) En face (scale=50μm) and rotated (scale=25μm) views of Tbr1 mutant J-RGCs that had been infected with a control AAV (a), a Sorcs3-expressing AAV (b) or a Cdh8-expressing AAV (c). Sorcs3 was co-injected with RFP while Cdh8 was RFP-tagged. Yellow boxes mark insets, which show expression of AAVs and Sorcs3 protein in J-RGCs. As expected, Sorcs3 protein is lost from Tbr1 mutant J-RGCs infected with control or Cdh8 AAVs; it is restored in Sorcs3-infected J-RGCs (yellow arrowheads). Dendritic defects in Tbr1-deficient J-RGCs (a, yellow arrow) were rescued by either Sorcs3 (b) or Cdh8 (c). (d) Proportions of J-RGCs with ectopic dendrites in Tbr1J/J retinas. n = 3 control, 4 Sorcs3, 3 Cdh8 and 3 Sorcs3+Cdh8 infected animals. p<0.0001, F(3,9)=376, one-way ANOVA; p<0.0001 for control versus Sorcs3, Cdh8 or Sorcs3+Cdh8. Tukey-Kramer test. (e) Model of Tbr1-regulated laminar patterning. Solid, colored arrows indicate molecular pathways that specify lamination. Dotted, colored arrows indicate pathways for other cellular features. Arrows emanating from ovals marked ‘Tbr1’ indicate cell-type specific effectors that bring dendrites to the OFF strata of the IPL, such as Cdh8 and Sorcs3 in the case of J-RGCs. Colored question marks post the hypothesis that Tbr1-independent mechanisms may act in parallel to Tbr1 to specify lamination. Dotted grey bracket and arrow indicate the possible presence of shared effectors for dendritic lamination that are commonly expressed by all four types. (f) IPL schematic showing that all four Tbr1-RGC types laminate within the OFF half of the IPL (bottom), but differ in their precise lamination.
DISCUSSION
The ability of different RGC types to respond selectively to certain visual features is a consequence of their synaptic inputs, key determinants of which are the IPL sublaminae in which their dendrites arborize. We demonstrate that a single transcriptional regulator can implement a common feature of dendritic laminar patterning by different RGC types. Tbr1 is expressed by four OFF-laminating RGC types, and it is both necessary and sufficient for laminar targeting of their dendrites within the IPL. We also identify two cell-surface molecules, Cdh8 and Sorcs3, as downstream effectors of Tbr1 in just one of the four types. Therefore, Tbr1 may instruct both a common laminar identity and subtle differences within that identity in part by recruiting non-overlapping sets of effectors depending on cell type.
Tbr1 in neural development
Tbr1 belongs to a family of 17 related transcription factors (in mice) that share a conserved T-box DNA binding domain36. It is expressed in various neuronal populations in the vertebrate brain, including cerebral cortex and olfactory bulb29,36–40, where it has been implicated in neuronal differentiation,.
Tbr1 had not been studied in retina previously, but much is known about its roles and expression in cortex. Our results document both similarities and differences between the two structures. Tbr1 is expressed exclusively by layer 6 pyramidal cells in cortex and RGCs in retina. Expression in both populations is initiated early but postmitotically, consistent with roles in neuronal development36. In cortical neurons, loss of Tbr1 disrupts migration, differentiation and axonal targeting, and may result in a partial fate switch in which layer 6 pyramidal neurons assume layer 5 identity39. In contrast, Tbr1 has a remarkably selective effect on dendritic lamination in retina, with no detectable role in fate determination, overall differentiation, or axonal projection.
Tbr2, the closest relative of Tbr1, is also expressed in both brain and retina. In cortex, expression of the two factors is sequential, with Tbr2 expressed in cortical progenitors and Tbr1 in layer 6 projection neurons41. In the retina, expression of both Tbr1 and Tbr2 persist into adulthood and, at least by late gestation (E17.5), is limited to non-overlapping groups of cells. Tbr2 is expressed by intrinsically photosensitive RGCs, which are distinct from Tbr1-RGCs, and is essential for their differentiation and/or survival16,17. Thus, despite their high homology, Tbr1 and Tbr2 appear to regulate distinct genes and processes depending on the brain region.
Laminar targeting by Cdh8 and Sorcs3
Using RNAseq, microarray and ChIP-seq, we chose Cdh8 and Sorcs3 as candidate mediators of Tbr1 effects in J-RGCs and confirmed that their expression is Tbr1-dependent. We do not know whether Tbr1 acts directly on these genes, but loss- and gain-of-function studies provided evidence that both act downstream of Tbr1.
Cdh8 is a member of the cadherin superfamily of adhesion molecules. Several type II cadherins have been implicated in shaping dendritic arbors of ooDSGCs and the axonal arbors of bipolar cells that innervate them (Duan et al. submitted,10). Its role in J-RGCs was therefore not unexpected, but our demonstration that it acts downstream of Tbr1 provides the first clue as to how cadherin expression in the retina is regulated. Cdh8 is expressed transiently in J-RGCs, dissipating after P6, at which time dendritic restriction is complete (Supplementary Figure 5a). Cdh8 is also expressed by Type 2 bipolar cells, which arborize in S110, but it appears in these cells only after J-RGCs dendrites have become restricted. A homophilic interaction between arbors of these two cell types is therefore unlikely to explain the phenotype we observed. Instead, Cdh8 on J-RGCs is likely to interact with a heterophilic partner; one attractive possibility is Cdh11, to which it is known to bind42.
Sorcs3 is a type I transmembrane protein that belongs to a family of vacuolar protein sorting 10 domain-containing receptors. Like other family members, Sorcs3 is expressed by a variety of neuronal populations43,44. Although other family members have been implicated in intracellular trafficking, much of the Sorcs3 protein is present on the cell surface33. Most relevant here, Sorcs3 protein appears to be preferentially localized to neuronal dendrites. In hippocampus, Sorcs3 is present at dendritic spines, where it participates in the modulation of glutamate receptor function44–46. Similarly, it is concentrated in dendrites of J-RGCs. The identity of the ligand for Sorcs3 on J-RGC dendrites is unknown, but it binds nerve growth factor, raising the possibility that it engages neurotrophin signaling in retina.
It is likely that other transcriptional regulators act in parallel with Tbr1 and that other cell surface molecules act in parallel with Cdh8 and Sorcs3 to sculpt J-RGC dendrites. Indeed, the limited penetrance of the Tbr1, Cdh8 and Sorcs3 loss-of-function phenotypes supports this idea, although it is also possible that J-RGCs scored as “normal” by our stringent criteria may have harbored subtle defects.
Regulators of dendrite targeting
Tbr1 expression is shared by four RGC types with dendrites that arborize in the outer third of the retina. Our loss- and gain-of-function methods confirm its role in laminar patterning of dendrites in at least two of the types and possibly in all four (Figure 8e). It is possible that other cell types with a common lamination pattern may also express common transcription factors that specify their patterns. Indeed several including Tbr2, Satb1 and FoxP19,14,16,17 are expressed in multiple cell types that project to one or two common sublaminae in the IPL.
The approaches we have taken here suggest that lamination programs may converge upon single regulators like Tbr1 but diverge at the level of cell types. We identified Cdh8 and Sorcs3 as downstream targets of Tbr1, yet, of the four Tbr1-RGC types, only J-RGCs express these genes at detectable levels during laminar patterning of dendrites. Since Tbr1 confers laminar identity on at least one other Tbr1-RGC type, the α-OFF-s-RGCs, Tbr1 must act through other effectors in those cells. By analogy to a scheme proposed to explain neuronal diversification and differentiation in C. elegans47, factors that define overall identity of a RGC type may cooperate with Tbr1 to regulate unique sets of cell surface molecules in each Tbr1-RGC type (Figure 8e).
The engagement of at least partially non-overlapping sets of effectors in the Tbr1 types provides an explanation for the observation that the precise dendritic lamination within the outer third of the IPL differ across the four types, as though they interact with different cues (Figure 8f). Alternatively or in addition, Tbr1-regulated cell surface effectors may participate in the distinct synaptic choices each Tbr1-RGC type makes. The loss of visual responses from Tbr1 mutant α-OFF-s-RGCs is consistent with this idea.
Finally, Cdh8 and Sorcs3 are also Tbr1 targets in the cortex32 and all three genes have been implicated as risk factors in behavioral disorders such as autism48,49. This conserved pathway is consistent with the speculation that dendritic defects contribute to the pathogenesis of diseases that result from neuronal miswiring.
ONLINE METHODS
Mice
A conditional Tbr1 mutant allele was generated at inGenious Targeting Laboratory (Ronkonkoma, NY). LoxP sites were inserted into introns 1 and 3, flanking Tbr1 exons 2 and 3 (Figure 2b). To enable selection of homologous recombinants, the LoxP site in intron 3 was embedded in a neo cassette that was flanked by Frt sites. The neo cassette was removed by mating to Flp-expressing mice50 to generate the Tbr1flox allele. Cre excision removed exons 2 and 3, including the T-box DNA binding region, similar to the Tbr1 constitutive null allele29.
The following mouse lines were generated in our laboratory and described previously: The JAM-B-CreER transgene was generated from a bacterial artificial chromosome by replacing the translational start site of JAM-B with a cDNA encoding a ligand-activated Cre recombinase, thereby placing the expression of CreER under the control of regulatory elements of Jam213. Thy1-STOP-YFP expresses YFP in many neurons, including all RGCs, following Cre-mediated excision of sequences that terminate transcription and translation51. Thus, in JAM-B-CreER; Thy1-STOP-YFP double transgenics, the administration of tamoxifen results in labeling of J-RGCs. We used homologous recombination to generate a JAM-B Cre knock-in mouse in which the first exon of the endogenous Jam-2 gene has been replaced with the gene encoding Cre recombinase28. Thy1-YFP-H mice label a sparse, nearly random subset of RGCs25. In TYW7, Thy1 regulatory elements drive the expression of lox-flanked YFP; Cre deletes the YFP22. The Cdh4-CreER knock-in line was generated by targeted insertion of a frt-neo-frt cassette, a 6x myc-tagged CreER-T2 and poly-adenylation signal at the translational start site of the Cdh4 coding sequence14.
A Vglut2-Ires-Cre knock-in line52 was obtained from International Mouse Strain Resource (IMSR). Rosa-CAG-lox-STOP-lox-GCaMP6f::deltaNeo53 (floxed GCaMP6f) and HB9-GFP, which label ooDSGCs that prefer ventral motion21,54 were obtained from the Jackson Laboratory. Dopamine receptor D4-GFP (DRD4-GFP) mice, which label ooDSGCs that prefer nasal motion31,55, were obtained from MMRRC-UNC. Six3-cre transgenic mice were provided by William Klein (M.D. Anderson Cancer Center)56. Cdh8lacZ were provided by M. Takeichi (Riken-CDB, Kobe, Japan)10,57. A conditional afadin mutant (afadin cKO) was obtained from Lou Reichardt35.
Tamoxifen (150μg, Sigma) was injected subcutaneously into P0–1 pups or intraperitoneally into P6–8 pups and E14.5 pregnant females. Animals were sacrificed with intraperitoneal injections of sodium pentobarbital followed by cervical dislocation. Animals below P10 were sacrificed by cervical dislocation. Animals of either sex were analyzed. All mice were maintained on a C57BL/6 and CD1 mixed background. The number and age of animals per experiment are indicated in figure legends. Animals were used in accordance with NIH guidelines and protocols approved by Institutional Animal Use and Care Committee at Harvard University.
Histology
Retinas were fixed in cold 4% PFA in PBS for 1.5 hours. For sections, retinas were incubated with 30% sucrose in PBS for at least 2 hours, frozen and cryosectioned at 20μm. Sections were then blocked with 5% normal donkey serum/0.3% Triton-X-100/PBS for 30 minutes, and incubated with primary antibodies in 3% normal donkey serum/0.3% Triton-X-100/PBS overnight. After two PBS washes, the sections were incubated with secondary antibodies for 2 hours, washed and mounted in Fluoromount. For whole mounts, retinas were dissected free of sclera, blocked for at least an hour, incubated in primary antibodies for 5–7 days, followed by secondary antibodies overnight. The retinas were then washed with PBS for at least 3–4 hours and mounted in Vectashield. Following calcium imaging experiments, retinas were fixed in fresh cold 4% PFA for 75 minutes and 0.1% Triton-X-100 used for all blocking and antibody solutions.
For analysis of central projections, animals were anesthetized with intraperitoneal injection of 60mg/kg ketamine plus 7.5mg/kg xylazine and transcardially perfused. Brains were fixed overnight in 4% PFA at 4°C, washed with 1x PBS and sectioned sagitally at 70μm on a vibratome. Sections were blocked for 2 hours, incubated in primary antibodies for 48 hours at 4°C, washed three times with PBS at room temperature over an hour, and re-incubated in secondary antibodies overnight. Sections were washed thrice in PBS at room temperature over 1 hour and mounted in Vectashield.
Antibodies used were as follows: chicken anti-GFP (1:1000, Abcam ab13970), rabbit anti-Tbr1 (1:1000, Abcam ab31940, McKenna et al. 2011), rabbit anti-Tbr2 (1:500, Abcam ab23345), goat FoxP2 (1:1000, Abcam), guinea pig FoxP1 (1:5000, Ben Novitch), goat Satb1 (1:1000, Santa Cruz Biotechnology sc-5989X), goat Pcsk2 (1:1000, R&D Systems AF6018, Supplementary Figure 4e) rabbit anti-mCherry (1:5000, Cai et al. 2012), mouse anti-Cre (1:500, Millipore MAB3120), 1:1000, goat anti-VAchT (1:1000, Millipore ABN100), mouse anti-Brn3a (1:500, Millipore Mab1585), guinea pig anti-VAchT (1:500, Promega G4481), guinea pig Rbpms (1:500, PhosphoSolutions 1832-RBPMS), goat anti-Brn3b (1:500, Santa Cruz Biotechnology sc-6026), mouse anti-Brn3c (1:250, Santa Cruz Biotechnology sc-81980), rabbit anti-Calbindin (1:10000, Swant CB38a), rabbit anti-CART (1:2000, Phoenix Pharmaceuticals H-003-62), Syt2 (1:250, ZIRC Znp-1), goat anti-Opn (1:500, R&D Systems AF-808), goat Sorcs3 (1:1000, R&D systems AF3067, Figure 5d), mouse PKCa (1:500, Abcam ab31), goat anti-Alcam (1:1000, R&D systems AF117258), goat anti-Neo1 (1:1000, R&D systems AF1079, Supplementary Figure 8c) and rabbit β-galactosidase (1:5000 Duan et al. 201410). Dylight405-, Alexa488-, Cy3- and Alexa647-conjugated secondary antibodies (1:1000) were obtained from Jackson Immunoresearch. Unless stated otherwise, these antibodies have been previously validated in Rousso et al. 2016.
In vivo electroporation
Tbr1 and Sorcs3 expression constructs were generated by PCR-amplifying the open reading frames from cDNA isolated from wildtype retinas and cloning them into an expression vector bearing the ubiquitin promoter. A Cdh8-mCherry fusion was obtained by restriction digest from the vector described in ref.10. A fluorescence expression plasmid (GFP or mCherry) was either electroporated alone, as a control, or co-electroporated with Tbr1 or Sorcs3 into neonatal retinas (P0–1) as previously described59,60. The Cdh8-mCherry plasmid was electroporated alone. Expression plasmids (~3mg/ml) were injected intraretinally or into the subretinal space of P0 mice and current pulses (80V, 50ms) were applied across the head using paddle electrodes (Harvard Apparatus, size 9). Intraretinal injection was used to label RGCs60; only P0 pups were used and the glass needle was angled acutely such that it traverses as great a distance as possible within the retina. Subretinal injection labeled photoreceptors, interneurons and Muller glia but not RGCs59. Retinas were analyzed at P12–14 unless otherwise stated. Retinas were taken at ≤P12 for analysis of Sorcs3 expression. For quantification of Sorcs3 expression in Tbr1-mis-expressing cells, discrete cell somata with cytosolic Sorcs3 staining (perinuclear and/or in neurites) were counted as Sorcs3-positive.
RNAseq and transcriptomic analysis
Fluorescently labeled J-RGCs were isolated from JAM-B-CreER x Thy1-STOP-YFP retinas by fluorescence-assisted cell sorting (FACS) at P6. Libraries were prepared using Ovation Ultralow System V2 1–16 (Nugen) and sequenced using Illumina NextSeq (75 cycles, single-end). Hb9-GFP and Drd4-GFP RNAseq were generated similarly9. Two biological replicates were generated for each RGC type. Data were analyzed using Tuxedo suite61 and edgeR62. Reads were trimmed by Trimmomatic63 and mapped onto the mouse genome (mm9 or mm10) by Tophat61. For analysis by Tuxedo suite, transcripts were counted using Cufflinks and differentially expressed genes were detected with Cuffdiff. For analysis by edgeR, mapped reads were counted by HTSeq and counts per million total reads (CPM) generated on edgeR for differential analysis. Microarray data were obtained from a published dataset18.
J-RGC enriched genes with log2 fold change >1 relative to Hb9-GFP and Drd4-GFP were compiled. Genes with log2CPM<2.32 were discarded. Genes with ontology terms associated with cell-surface membrane, excluding ion channels and enzymes were shortlisted. The Genomic Regions Enrichment of Annotation Tool (GREAT)64 was used to analyze a published Tbr1 ChIP-Seq dataset32. Tbr1 ChIP-seq peaks were visualized on Integrative Genomics Viewer (IGV, Broad Institute).
P4–5 Tbr1 wildtype and mutant J-RGCs were isolated by FACS in pools of 100 cells. We collected 11 samples: 5 samples from 4 wildtype mice and 6 samples from 4 Tbr1J/J mice. Smartseq2 and Nextera XT were used to prepare cDNA libraries65. Libraries were sequenced on Nextseq (75 cycles, single-end) and analyzed as described above.
Image and statistical analyses
Imaging was performed on Olympus FV1000 or Zeiss LSM-710 confocal microscopes using 405, 488, 568 and 647nm lasers. Images were acquired at x-y-z resolution of 0.31μm × 0.31μm × 0.5–2μm. Images were analyzed on imageJ. Gamma and contrast settings of the images were adjusted to make dim features visible, without eliminating background signal. To quantify the location of J-RGC dendrites or laminar distributions of neurites from electroporated cells within the IPL, line scans were drawn across the entire IPL and YFP intensity values across IPL depth obtained using the ImageJ/Analyze/Plot Profile function. Depth values were normalized such that 0% indicates the INL-IPL border and 100%, the IPL-GCL border. Intensity values were normalized to the maximum intensity within each line scan. Intensity values, in arbitrary units, were binned for every 5% IPL depth and averaged across samples. Dendritic field areas were measured by drawing convex hulls around dendrites. Coverage factor was calculated as the product of dendritic field area and spatial density. Mosaic analysis was done on WinDRP software18,19. Dendrites were traced using Imaris 7.4.0. Dendrite length was obtained using Imaris or Simple Neurite Tracer on ImageJ 1.49u. Statistical analyses were performed using Pearson’s chi-square tests, Cochran-Armitage tests, one-way ANOVA followed by Tukey-Kramer test or two-tailed Student’s t-tests, as described in figure legends, on Microsoft Excel or GraphPad Prism 7.03. Cochran-Armitage tests were used to assess the distribution of neurites across the IPL between two conditions, where the IPL was divided into equal bins and the order of bins fixed to reflect the arrangement of the IPL. Pearson’s chi-square tests were used when observations such as cell counts were categorized into discrete classes and the order of the classes is arbitrary. Average values are represented as mean ± standard error unless otherwise stated. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications (Krishaswamy et al. 2015, Duan et al. 2014, Liu and Sanes 2017). Data distribution was assumed to be normal but this was not formally tested. Samples were allocated by genotype or treatment (e.g. different electroporation constructs or AAV injections). For experiments involving animals of the same genotype (e.g. electroporation of wildtype pups), animals were randomly assigned to one treatment or another. Data collection and analysis were not performed blind to the conditions of the experiments. Exclusion criteria were pre-established for initial characterization of calcium responses as detailed below. No other data or animals were excluded from analysis.
Reverse transcription-quantitative PCR (RT-qPCR)
RGCs were isolated from P4 control and Tbr1ret/ret retinas by live-staining with microbeads conjugated to monoclonal anti-mouse CD90.2, followed by magnetic column purification (MACS Miltenyi Biotec). RNA was extracted using DirectZol RNA extraction kit (Zymo Research) and assessed on Bioanalyzer. First-strand cDNA synthesis was performed on equal amounts of RNA using Superscript III reagents (Invitrogen). qPCR was performed using KAPA Sybr FAST qPCR kit Master Mix (Kapa Biosystems) on ABI 7900. cDNA levels across samples were normalized using primers against Hprt. Fold change expression relative to controls was calculated by the ΔΔCt method. Primers used were as follows: Cdh8, 5′-AACCAGATTTGCAGTTTATGCCA-3′ and 5′-TTGCCCATATCCACACGGTC-3′; Hprt66, 5′-CAAACTTTGCTTTCCCTGGT-3′ and 5′-CAAGGGCATATCCAACAACA-3′; Sorcs3, 5′-CTCTCGGTGGTATTCGTCGG-3′ and 5′-CAATGCTTCCTATGACCCGC-3′; Tbr140, 5′-CAAGGGAGCATCAAACAACA-3′ and 5′-GTCCTCTGTGCCATCCTCAT-3′.
Intravitreal injections
The AAV construct encoding a shRNA against Sorcs3 was generated by replacing the shPTEN sequence in pAAV9-U6-shPTEN-CMV-mCherry67 with shSorcs3. The efficacy of shSorcs3 was tested in vitro on HEK293T cells that were transfected with a Sorcs3 expression plasmid. AAV9-cag-Td-tomato was used as a control. Animals were euthanized and retinas collected at least 2 weeks post-injection or, in the case of Sorcs3, by P12.
For the generation of Sorcs3- and Cdh8-expressing AAVs, an AAV backbone with an optimized expression cassette, containing a truncated WPRE and SV40 late polyadenylation signal68, was used to accommodate the large sizes of these cDNAs. The GFP sequence in pAAV-CW3SL-EGFP (Addgene plasmid #61463) was first replaced with the sequence of Sorcs3 or RFP-tagged Cdh8. Then, the CamKIIa promoter was replaced with a synuclein promoter. AAVs were delivered intravitreally at P0 and retinas collected 10 days later for whole mount processing.
AAVs to alter Cdh8 or Sorcs3 levels were generated by Boston Children’s Hospital viral core. AAV9.Syn.GCaMP6f.WPRE.SV40 was purchased from the Penn Vector Core. ~0.5–1ul of each AAV was delivered intravitreally. For adults, ophthalmic ointment was applied to the eye post-injection.
For visualizing axonal projections, 1μl of fluorescently tagged recombinant cholera toxin subunit B (CTB) was injected into each eye using a 30.5G Hamilton syringe. The contralateral superior colliculus and uninjected retina were collected 2 days after injection and processed for histology, as described above.
Calcium Imaging
Mice were dark adapted overnight prior to euthanasia. The retina was rapidly dissected under infrared illumination into oxygenated (95% O2; 5% CO2) Ames solution (Sigma). Three relaxing cuts were made and the retina was then placed in a recording chamber, ganglion cells facing up on the stage of a custom-built two-photon microscope6. 5–10μl of 0.2mg/mL sulphorhodamine 101(Sigma) was added to the recording chamber to label blood vessels and the retina was left to rest for 5–10 minutes. GCaMP-expressing neurons were imaged under two photon illumination (wavelength = 960nm) and stimulated with patterned visual stimuli delivered through the objective. Movies (700×100pixels; 420×60 μm) were collected and then regions of interest (ROIs) drawn around individual cells to extract neural responses (see analysis below). To measure stimulus noise, retinas were presented with stimuli without laser activation. Stimulus-generated noise in our calcium imaging movies was worst at the edges of the scan pattern and was all but absent in the center of our movies.
Light stimuli were delivered through the objective from a modified DLP projector suspended above the microscope stage using a custom made lens subsystem, as described previously69. Monochrome light was used (wavelength = 405nm, width 10nm), and the background intensity set to 4.2206 × 104R*/rod/s. The LED was triggered on the edges of every linescan in order to restrict stimulus contamination to the edges of our calcium imaging movies. Visual stimuli were written in Matlab and displayed on the projector using the psychophysics toolbox. Patterns were binary, and grey was achieved by inverting a single pixel checkerboard pattern on every frame. Moving bars were presented as a long bar moving along its long axis; the short axis of the bar corresponded roughly to the receptive field width of the recorded neuron; Bars moved with a velocity of 1000 μm/sec; their length was adjusted to give good separation between the leading and trailing edges.
Following recording, retinas were fixed, immunostained and imaged as described above. Regions of the confocal images that corresponded to the recorded field were located by reference to the pattern of blood vessels. Confocal images were aligned to recorded fields using the affine transform function on ImageJ. ROIs for cell bodies in the transformed image were drawn and molecular signatures were assigned to each cell.
Code written in Matlab R2015b was used to extract Ca2+ traces for each ROI. Traces were first de-trended using Matlab’s moving mean function. The ROI for each cell was applied to the noise movie and the noise trace was subtracted from the response of that cell; this procedure allowed us to account for varying noise across our imaging field. Quality index and z-score were calculated as described previously 70. The quality index provides a measure of consistency across trials. It is calculated as the variance of the mean response for all trials (generally 3), divided by the mean of the variance over trials. Thus, the index spans from 1/3, if all trials are completely random with respect to each other (but have the same variance), to 1 if all responses are identical. Nonresponsive cells tend to have low quality indices, because they are dominated by noise. The z-score of responses to stimuli, a more direct measure of responsiveness, was calculated with respect to the mean and standard deviation of signals recorded during steady gray illumination that preceded the stimulus (8 seconds for full field flashes and 6 seconds for moving bars). For initial characterization (Supplementary Figure 6a–d), we counted only cells with quality index >0.45 and a z-score that was >1.0 for at least two consecutive time-points. For the analysis of alpha-OFF-s-RGCs in control versus Tbr1ret/ret, these selection criteria were omitted and all immunohistochemically identified cells were analyzed so that we could detect changes in responsiveness in Tbr1 mutants.
Life Sciences Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request. No custom code was used in this study.
Supplementary Material
Acknowledgments
This work was supported by NIH grants NS29169 and EY022073 to J.R.S, NS34661 to J.L.R. and MH094589 to B.C. J.L. was funded by an Agency for Science, Technology and Research (A*STAR) fellowship from Singapore. We thank Zhigang He and Chen Wang at Boston Children’s Hospital Viral Core for their generous support (5P30EY012196).
Footnotes
ACCESSION CODES
J-RGC RNAseq data has been deposited at NCBI GEO under accession number GSE102888.
Tbr1 wildtype and mutant J-RGC RNAseq has been deposited at NCBI GEO under accession number GSE108789.
Published gene sets used in this study are available at: Kay et al. 2012: https://www-ncbi-nlm-nih-gov.ezp-prod1.hul.harvard.edu/geo/query/acc.cgi?acc=GSE35077; Notwell et al. 2016: https://www-ncbi-nlm-nih-gov.ezp-prod1.hul.harvard.edu/geo/query/acc.cgi?acc=GSE71384; Peng et al. 2017: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90673.
AUTHOR CONTRIBUTIONS
J.L. and J.R.S. conceived the study, planned experiments, analyzed data and wrote the paper. J.L. performed all experiments unless otherwise stated. J.D.S.R. performed and analyzed calcium imaging experiments. A.K. built instrumentation, wrote stimuli and analyzed calcium imaging experiments. M.A.L. performed experiments on afadin, Cdh8 and axonal projections. S.P. generated cDNA libraries for Tbr1 wildtype and mutant J-RGCs. J.L.R. and B.C. generated conditional Tbr1 mutant mice. The authors declare no competing interests.
COMPETING FINANCIAL INTERESTS
The authors declare no competing interests.
References
- 1.Sanes JR, Yamagata M. Formation of lamina-specific synaptic connections. Curr Opin Neurobiol. 1999;9:79–87. doi: 10.1016/s0959-4388(99)80010-5. [DOI] [PubMed] [Google Scholar]
- 2.Baier H. Synaptic laminae in the visual system: molecular mechanisms forming layers of perception. Annu Rev Cell Dev Biol. 2013;29:385–416. doi: 10.1146/annurev-cellbio-101011-155748. [DOI] [PubMed] [Google Scholar]
- 3.Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- 4.Famiglietti EV, Jr, Kolb H. Structural basis for ON-and OFF-center responses in retinal ganglion cells. Science. 1976;194:193–195. doi: 10.1126/science.959847. [DOI] [PubMed] [Google Scholar]
- 5.Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature. 2001;410:583–587. doi: 10.1038/35069068. [DOI] [PubMed] [Google Scholar]
- 6.Krishnaswamy A, Yamagata M, Duan X, Hong YK, Sanes JR. Sidekick 2 directs formation of a retinal circuit that detects differential motion. Nature. 2015;524:466–470. doi: 10.1038/nature14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamagata M, Sanes JR. Dscam and Sidekick proteins direct lamina-specific synaptic connections in vertebrate retina. Nature. 2008;451:465–469. doi: 10.1038/nature06469. [DOI] [PubMed] [Google Scholar]
- 8.Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. J Neurosci. 2012;32:14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng YR, et al. Satb1 Regulates Contactin 5 to Pattern Dendrites of a Mammalian Retinal Ganglion Cell. Neuron. 2017 doi: 10.1016/j.neuron.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan X, Krishnaswamy A, De la Huerta I, Sanes JR. Type II cadherins guide assembly of a direction-selective retinal circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka RL, et al. Transmembrane semaphorin signalling controls laminar stratification in the mammalian retina. Nature. 2011;470:259–263. doi: 10.1038/nature09675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun LO, et al. On and off retinal circuit assembly by divergent molecular mechanisms. Science. 2013;342:1241974. doi: 10.1126/science.1241974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature. 2008;452:478–482. doi: 10.1038/nature06739. [DOI] [PubMed] [Google Scholar]
- 14.Rousso DL, et al. Two Pairs of ON and OFF Retinal Ganglion Cells Are Defined by Intersectional Patterns of Transcription Factor Expression. Cell Rep. 2016;15:1930–1944. doi: 10.1016/j.celrep.2016.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sweeney NT, James KN, Nistorica A, Lorig-Roach RM, Feldheim DA. Expression of transcription factors divides retinal ganglion cells into distinct classes. J Comp Neurol. 2017 doi: 10.1002/cne.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao CA, et al. T-box transcription regulator Tbr2 is essential for the formation and maintenance of Opn4/melanopsin-expressing intrinsically photosensitive retinal ganglion cells. J Neurosci. 2014;34:13083–13095. doi: 10.1523/JNEUROSCI.1027-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweeney NT, Tierney H, Feldheim DA. Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J Neurosci. 2014;34:5447–5453. doi: 10.1523/JNEUROSCI.0035-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kay JN, Chu MW, Sanes JR. MEGF10 and MEGF11 mediate homotypic interactions required for mosaic spacing of retinal neurons. Nature. 2012;483:465–469. doi: 10.1038/nature10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockhill RL, Euler T, Masland RH. Spatial order within but not between types of retinal neurons. Proc Natl Acad Sci U S A. 2000;97:2303–2307. doi: 10.1073/pnas.030413497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodieck RW. The density recovery profile: a method for the analysis of points in the plane applicable to retinal studies. Vis Neurosci. 1991;6:95–111. doi: 10.1017/s095252380001049x. [DOI] [PubMed] [Google Scholar]
- 21.Trenholm S, Johnson K, Li X, Smith RG, Awatramani GB. Parallel mechanisms encode direction in the retina. Neuron. 2011;71:683–694. doi: 10.1016/j.neuron.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim IJ, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30:1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krieger B, Qiao M, Rousso DL, Sanes JR, Meister M. Four alpha ganglion cell types in mouse retina: Function, structure, and molecular signatures. PLoS One. 2017;12:e0180091. doi: 10.1371/journal.pone.0180091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang JJ, Gao F, Wu SM. Light-evoked excitatory and inhibitory synaptic inputs to ON and OFF alpha ganglion cells in the mouse retina. J Neurosci. 2003;23:6063–6073. doi: 10.1523/JNEUROSCI.23-14-06063.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng G, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 26.Bleckert A, Schwartz GW, Turner MH, Rieke F, Wong RO. Visual space is represented by nonmatching topographies of distinct mouse retinal ganglion cell types. Curr Biol. 2014;24:310–315. doi: 10.1016/j.cub.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassett EA, Wallace VA. Cell fate determination in the vertebrate retina. Trends Neurosci. 2012;35:565–573. doi: 10.1016/j.tins.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Sanes JR. Cellular and Molecular Analysis of Dendritic Morphogenesis in a Retinal Cell Type That Senses Color Contrast and Ventral Motion. J Neurosci. 2017;37:12247–12262. doi: 10.1523/JNEUROSCI.2098-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulfone A, et al. An olfactory sensory map develops in the absence of normal projection neurons or GABAergic interneurons. Neuron. 1998;21:1273–1282. doi: 10.1016/s0896-6273(00)80647-9. [DOI] [PubMed] [Google Scholar]
- 30.Hong YK, Kim IJ, Sanes JR. Stereotyped axonal arbors of retinal ganglion cell subsets in the mouse superior colliculus. J Comp Neurol. 2011;519:1691–1711. doi: 10.1002/cne.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kay JN, et al. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci. 2011;31:7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notwell JH, et al. TBR1 regulates autism risk genes in the developing neocortex. Genome Res. 2016;26:1013–1022. doi: 10.1101/gr.203612.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willnow TE, Petersen CM, Nykjaer A. VPS10P-domain receptors - regulators of neuronal viability and function. Nat Rev Neurosci. 2008;9:899–909. doi: 10.1038/nrn2516. [DOI] [PubMed] [Google Scholar]
- 34.Takai Y, Ikeda W, Ogita H, Rikitake Y. The immunoglobulin-like cell adhesion molecule nectin and its associated protein afadin. Annu Rev Cell Dev Biol. 2008;24:309–342. doi: 10.1146/annurev.cellbio.24.110707.175339. [DOI] [PubMed] [Google Scholar]
- 35.Beaudoin GM, 3rd, et al. Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus. J Neurosci. 2012;32:99–110. doi: 10.1523/JNEUROSCI.4565-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihalas AB, Hevner RF. Control of Neuronal Development by T-Box Genes in the Brain. Curr Top Dev Biol. 2017;122:279–312. doi: 10.1016/bs.ctdb.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 37.Hevner RF, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 38.Han W, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna WL, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang TN, et al. Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat Neurosci. 2014;17:240–247. doi: 10.1038/nn.3626. [DOI] [PubMed] [Google Scholar]
- 41.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimoyama Y, Tsujimoto G, Kitajima M, Natori M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem J. 2000;349:159–167. doi: 10.1042/0264-6021:3490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hermey G, et al. The three sorCS genes are differentially expressed and regulated by synaptic activity. J Neurochem. 2004;88:1470–1476. doi: 10.1046/j.1471-4159.2004.02286.x. [DOI] [PubMed] [Google Scholar]
- 44.Oetjen S, Mahlke C, Hermans-Borgmeyer I, Hermey G. Spatiotemporal expression analysis of the growth factor receptor SorCS3. J Comp Neurol. 2014;522:3386–3402. doi: 10.1002/cne.23606. [DOI] [PubMed] [Google Scholar]
- 45.Breiderhoff T, et al. Sortilin-related receptor SORCS3 is a postsynaptic modulator of synaptic depression and fear extinction. PLoS One. 2013;8:e75006. doi: 10.1371/journal.pone.0075006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christiansen GB, et al. The sorting receptor SorCS3 is a stronger regulator of glutamate receptor functions compared to GABAergic mechanisms in the hippocampus. Hippocampus. 2017;27:235–248. doi: 10.1002/hipo.22689. [DOI] [PubMed] [Google Scholar]
- 47.Hobert O. Terminal Selectors of Neuronal Identity. Curr Top Dev Biol. 2016;116:455–475. doi: 10.1016/bs.ctdb.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Huang TN, Hsueh YP. Brain-specific transcriptional regulator T-brain-1 controls brain wiring and neuronal activity in autism spectrum disorders. Front Neurosci. 2015;9:406. doi: 10.3389/fnins.2015.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 51.Buffelli M, et al. Genetic evidence that relative synaptic efficacy biases the outcome of synaptic competition. Nature. 2003;424:430–434. doi: 10.1038/nature01844. [DOI] [PubMed] [Google Scholar]
- 52.Vong L, et al. Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron. 2011;71:142–154. doi: 10.1016/j.neuron.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madisen L, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron. 2015;85:942–958. doi: 10.1016/j.neuron.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110:385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 55.Huberman AD, et al. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis. 2000;26:130–132. [PubMed] [Google Scholar]
- 57.Suzuki SC, et al. Cadherin-8 is required for the first relay synapses to receive functional inputs from primary sensory afferents for cold sensation. J Neurosci. 2007;27:3466–3476. doi: 10.1523/JNEUROSCI.0243-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buhusi M, et al. ALCAM regulates mediolateral retinotopic mapping in the superior colliculus. J Neurosci. 2009;29:15630–15641. doi: 10.1523/JNEUROSCI.2215-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci U S A. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dhande OS, Crair MC. Transfection of mouse retinal ganglion cells by in vivo electroporation. J Vis Exp. 2011 doi: 10.3791/2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 66.Adachi H, et al. Stage-specific reference genes significant for quantitative PCR during mouse retinal development. Genes Cells. 2015;20:625–635. doi: 10.1111/gtc.12254. [DOI] [PubMed] [Google Scholar]
- 67.Duan X, et al. Subtype-specific regeneration of retinal ganglion cells following axotomy: effects of osteopontin and mTOR signaling. Neuron. 2015;85:1244–1256. doi: 10.1016/j.neuron.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Choi JH, et al. Optimization of AAV expression cassettes to improve packaging capacity and transgene expression in neurons. Mol Brain. 2014;7:17. doi: 10.1186/1756-6606-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Euler T, et al. Eyecup scope--optical recordings of light stimulus-evoked fluorescence signals in the retina. Pflugers Arch. 2009;457:1393–1414. doi: 10.1007/s00424-008-0603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baden T, et al. The functional diversity of retinal ganglion cells in the mouse. Nature. 2016;529:345–350. doi: 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. No custom code was used in this study.