Abstract
Purpose
To determine whether multivariate machine learning models of algorithmically assessed magnetic resonance imaging (MRI) features from breast cancer patients are associated with Oncotype DX (ODX) test recurrence scores.
Methods
A set of 261 female patients with invasive breast cancer, pre-operative dynamic contrast enhanced magnetic resonance (DCE-MR) images and available ODX score at our institution was identified. A computer algorithm extracted a comprehensive set of 529 features from the DCE-MR images of these patients. The set of patients was divided into a training set and a test set. Using the training set we developed two machine learning-based models to discriminate (1) high ODX scores from intermediate and low ODX scores, and (2) high and intermediate ODX scores from low ODX scores. The performance of these models was evaluated on the independent test set.
Results
High against low and intermediate ODX scores were predicted by the multivariate model with AUC 0.77 (95% CI 0.56–0.98, p < 0.003). Low against intermediate and high ODX score was predicted with AUC 0.51 (95% CI 0.41–0.61, p = 0.75).
Conclusion
A moderate association between imaging and ODX score was identified. The evaluated models currently do not warrant replacement of ODX with imaging alone.
Keywords: Breast cancer MRI, Oncotype DX, Imaging features, Radiomics, Feature selection, Logistic regression
Introduction
In breast cancer related literature, prior studies (Paik et al. 2004, 2006; Nielsen et al. 2010; Dowsett et al. 2013; Wittner et al. 2008) have shown that multi-assay gene expression profiling is associated with patient outcomes and response to specific systemic therapies. Of the variety of multi-gene assays, the 21-gene expression assay Oncotype DX (ODX) (Genomic Health, Redwood City, CA, USA) test is recommended by the current guidelines (Paik et al. 2004, 2006; Coates et al. 2015) for a specific group of female patients (early-stage, hormone receptor-positive, node-negative, and HER2-negative disease with tumor size equal or greater than 1 cm) and is often used in clinical practice for assessing chemotherapy benefit and the risk of distant recurrence. The 10-year risk of distant recurrence is estimated by the ODX score on a scale of 0 to 100 and considered low if the ODX score is less than 18, and high if ODX score is greater than or equal than 31. Any ODX score between low and high is considered intermediate (Paik et al. 2004). However, there are limitations to obtaining ODX scores (from tumor specimens) as discussed in (Harowicz et al. 2017), since it is an expensive and time-consuming procedure. Therefore, several surrogate measures of ODX were explored in previous studies (Klein et al. 2013; Gage et al. 2015; Tang et al. 2010). These surrogate measures require the immunohistochemical (IHC) analysis of the tumor sample.
Recently, an increased research focus has been placed on using dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) to predict patient outcomes and tumor genomics. These analyses are referred to as radiomics (Gillies et al. 2016) and radiogenomics (Mazurowski 2015). Specifically several of DCE-MRI-based features have been shown to be associated with recurrence-free survival in breast cancer (Mazurowski et al. 2015; Kim et al. 2017). Also, multiple studies (Uematsu et al. 2009; Li et al. 2016a, b; Ashraf et al. 2014; Blaschke and Abe 2015; Wan et al. 2016; Fan et al. 2017; Wu et al. 2017; Sutton et al. 2015), most of which focused on tumor intrinsic molecular subtype, are present in the literature. Some of the radiogenomic studies (Wan et al. 2016; Ashraf et al. 2014; Li et al. 2016a; Sutton et al. 2015) showed initial data on the association of imaging features with ODX, illustrating a potential avenue of a surrogate noninvasive test. In (Ashraf et al. 2014), features from the time-intensity enhancement curve of tumor were delineated on a central representative slice of 56 breast cancer patients from a single institution and were shown to distinguish between high versus low and intermediate risk patients. In Li et al. (2016b), computer-extracted features quantifying size, shape, margin properties, enhancement texture and kinetic assessment from 3D tumor volume from 84 patients (from four institutions) were considered to discriminate between patients with low to medium risk versus high risk. Sutton et al. (2015) explored morphological and texture features from a central slice of tumor in addition to clinical features (age, menopausal status, histologic and nuclear grades, pathological tumor size and lymph node status) and developed a regression model to compute a surrogate ODX for 95 invasive ductal carcinoma patients having ODX scores in the range (0–45). In a recent study by Wan et al. (2016), 96 patients with low and high risk ODX scores were included and features of textural kinetics were explored to differentiate between high and low risk groups. While these studies showed promise for this radiogenomic association, the patient population in each of these studies was restricted to under 100 subjects, a limited number of imaging characteristics were explored, and no independent test set was used to validate the findings.
The purpose of our study is to validate the association of imaging features with ODX in an independent cohort. Our approach includes analysis of a comprehensive set of 529 imaging features from each of the patients, development and evaluation of a multivariate model for prediction of the ODX score.
Materials and methods
Patient population
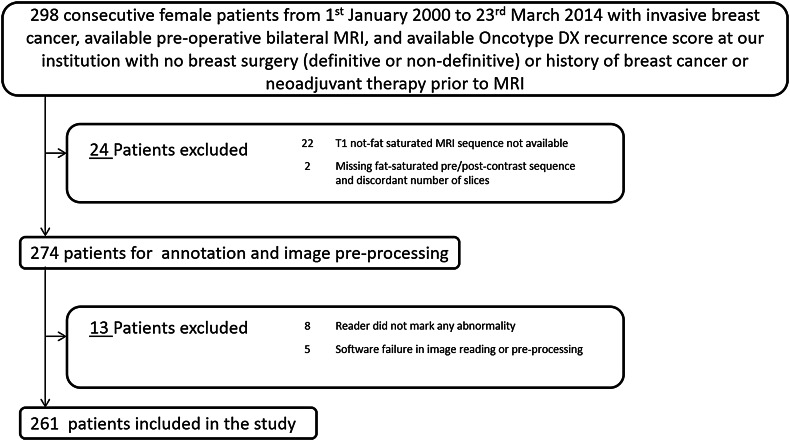
We secured an Institutional Review Board approval for this study. For our analysis, we identified 298 consecutive female patients with an available ODX recurrence score from January 1st, 2000 to March 23rd, 2014 with invasive breast cancer and available pre-operative MRI at our institution, without having any breast surgery, history of breast cancer or neo-adjuvant therapy prior to the MRI acquisition. From these 298 patients, 37 patients were excluded from the study for reasons shown in Fig. 1. We divided the remaining 261 patients into training and test sets having 131 and 130 patients respectively.
Fig. 1.
Flowchart explaining the cohort used in our study
Imaging data
Axial breast DCE-MR images that were acquired by 1.5 or 3 T scanners in the prone position were collected for all patients. The following MR sequences were available: a non-fat saturated T1-weighted sequence and a fat-saturated gradient echo T1-weighted pre-contrast sequence, typically with four post-contrast T1-weighted sequences. The postcontrast sequences were acquired using a weight based protocol of 0.2 mL/kg of the contrast agent (IV administration).
The tumors were annotated by eight fellowship-trained breast radiologists (1–22 years of post-fellowship experience). The following MR sequences were displayed to the reader: (a) the pre-contrast, (b) the first post-contrast, and (c) the subtracted sequence (obtained by subtracting the pre-contrast sequence from the first post-contrast) using an internal GUI (graphical user interface) developed in our laboratory. A three-dimensional bounding box, containing the tumor, was indicated by a reader. Each tumor was annotated by one reader.
Image segmentation
Using a reader’s bounding box, we obtained the tumor mask by a fuzzy C-means automatic segmentation. Using the N4 corrected (Tustison et al. 2010) T1-non-fat saturated (T1-NFS) images and first post-contrast sequences, the breast and fibroglandular tissue masks were extracted automatically. Thus, apart from the semi-automatic tumor mask, we had three more masks extracted automatically for each patient: (a) breast mask (b) FGT mask from T1-NFS (c) FGT mask from post-contrast sequence as described in (Saha et al. 2017).
Imaging feature extraction and organization
We extracted 529 imaging features from DCE-MR image sequences. The feature implementation and extraction where guided mostly by the imaging features existing in the literature and shown to have an association with diagnosis, prognosis and molecular subtypes or considered by different research groups as a potential descriptor of breast cancer characteristics. We selected features that describe the breast, FGT (from T1-NFS or first post contrast sequences), and tumor. The exact list of all features along with references aiding their implementation can be found in the Supplementary Material A of our previous work (Saha et al. 2017). A summary of the feature categories is listed below:
Features of breast and FGT volume (5): The sources of these features are breast mask and FGT mask. DCE-MR voxel values are not required after the corresponding mask has been extracted.
Features of tumor size and morphology (10): The source of these features is the tumor mask, however, DCE-MR voxel values are not required after the corresponding mask has been extracted.
Features of FGT enhancement (82): The sources of these features are the FGT mask and DCE-MR voxel intensities corresponding to the mask, but the spatial relationship between the voxels or variation of feature values within voxels is not used.
Features of tumor enhancement (30): The sources of these features are tumor mask and DCE-MR voxel intensities corresponding to the mask, but the spatial relationship between the voxels or variation of feature values within voxels is not used.
Combined FGT and tumor enhancement (18): The sources of these features are tumor mask, FGT mask and DCE-MR voxel intensities corresponding to the mask, but the spatial relationship between the voxels or variation of feature values within voxels is not used.
Features of FGT enhancement texture (176): The sources of these features are FGT mask and DCE-MR voxel intensities corresponding to the mask along with the spatial relationship of the voxels.
Features of tumor enhancement texture (135): The sources of these features are tumor mask and DCE-MR voxel intensities corresponding to the mask along with the spatial relationship of the voxels.
Features of tumor enhancement spatial heterogeneity (4): The sources of these features are tumor mask and DCE-MR voxel intensities, and these features extract information about the spatial relationship of the clusters formed by tumor enhancement features.
Features of FGT enhancement variation (34): The sources of these features are FGT mask and DCE-MR voxel intensities, dispersion or variation of FGT enhancement feature values only or feature values over timepoints are considered, without considering the spatial relationship between the voxels.
Features of tumor enhancement variation (35): The sources of these features are tumor mask and DCE-MR voxel intensities, dispersion or variation of FGT enhancement feature values only or feature values over timepoints are considered, without considering the spatial relationship between the voxels.
Multivariate predictive models
We considered two prediction tasks (a) prediction of high (i.e., ODX scores greater than or equal to 31 are high) versus low and intermediate ODX scores (b) prediction of high and intermediate (i.e., ODX scores less than 18 are low) versus low. The cut-off values are same as followed in Paik et al. (2004).
To develop the models, we first selected features from the set of 529 features using our training set. First, we computed the area-under the receiver operating characteristic (ROC) curve (AUC) for each of the two prediction tasks in the training set. Then, we sorted the features in descending order according to AUC and iteratively selected low correlated features ( ≤ 0.8) to form sets of top 5 or 10 features for each of the two predictive tasks. Based on the selected features we generated two multivariate logistic regression models for each of the two discrimination tasks. For each one of the tasks, we constructed 2 models: one based on 5 selected features and one based on 10 selected features.
Model evaluation and statistical analysis
The models were evaluated using the test set. We used the AUC (Delong et al. 1988) as the performance metric. To test if the trained models show a statistically significant association (for each of the two predictive tasks), we fitted a logistic regression between the values predicted by a model in the test set and corresponding labels (depending on the predictive task) in the test set. As an exploratory analysis to identify which individual features had the highest association with ODX in test set, we graphically demonstrated the relationship of the top five features in the test set for each of the two tasks. Since in this study we evaluated four models (two models for two classification tasks), we considered p < 0.0125 (0.05/4) as statistically significant.
Results
The clinicopathologic characteristics for these patients are shown in Table 1.
Table 1.
Clinicopathologic characteristics of the patients included in our study
| Patient characteristics | All patients | Patients in training set | Patients in test set |
|---|---|---|---|
| Number of patients | 261 | 131 | 130 |
| Median age (years) | 54 | 53 | 56 |
| Age range (years) | 28–83 | 32–83 | 28–77 |
| Race | |||
| White | 223 | 108 | 115 |
| Black | 25 | 14 | 11 |
| Asian | 3 | 2 | 1 |
| Native | 0 | 0 | 0 |
| Hispanic | 3 | 3 | 0 |
| Multi | 2 | 0 | 2 |
| Hawaiian | 0 | 0 | 0 |
| American Indian | 0 | 0 | 0 |
| Not available | 5 | 4 | 1 |
| Menopausal status | |||
| Pre | 102 | 49 | 53 |
| Post | 156 | 80 | 76 |
| Not available | 3 | 2 | 1 |
| Oncotype DX score | |||
| Low | 145 | 77 | 68 |
| Intermediate | 93 | 38 | 55 |
| High | 23 | 16 | 7 |
| Median ODX | 16 | 16 | 17 |
The variety of scanning equipment and contrast agents used for the patient population are shown in Table 2.
Table 2.
Information about the scanner parameters and contrast agents used
| Characteristics | Technical details | Manufacturer details | Patient count |
|---|---|---|---|
| Magnetic field strength | 1.5 T | Optima MR450w, GE Healthcare, Little Chalfont, UK | 35 |
| Signa HDx, GE Healthcare, Little Chalfont, UK | 9 | ||
| Signa HDxt, GE Healthcare, Little Chalfont, UK | 51 | ||
| Avanto, Siemens, Munich, Germany | 47 | ||
| 3 T | Signa Excite, GE Healthcare, Little Chalfont, UK | 1 | |
| Signa HDx, GE Healthcare, Little Chalfont, UK | 53 | ||
| Signa HDxt, GE Healthcare, Little Chalfont, UK | 31 | ||
| Skyra, Siemens Healthcare, Little Chalfont, UK | 15 | ||
| Trio, Siemens, Munich, Germany | 1 | ||
| Trio Tim, Siemens, Munich, Germany | 18 | ||
| Contrast agent | Gadobutrol | Gadavist, Bayer Healthcare, Berlin, Germany | 1 |
| Gadopentetate dimeglumine | Magnevist, Bayer Healthcare, Berlin, Germany | 145 | |
| Gadobenate dimeglumine | Multihance, Bracco, Milan, Italy | 82 | |
| Unknown | − | 33 |
The details of different MR image acquisition parameters are presented in Table 3.
Table 3.
Values of different MRI acquisition parameters in the dataset
| MRI acquisition parameters | Value details | Values |
|---|---|---|
| Slice thickness (mm) | Range | 1.1–2.5 |
| Median | 2 | |
| Repetition time (ms) | Range | 3.54–7.11 |
| Median | 5.25 | |
| Echo time (ms) | Range | 1.25–2.76 |
| Median | 2.4 | |
| Acquisition matrix | Minimum array size | 320 × 320 |
| Maximum array size | 448 × 448 | |
| Flip angle (°) | Range | 7–12 |
| Median | 10 | |
| FOV (mm) | Range | 250–400 |
| Median | 340 |
The performance of the models for prediction of the ODX category in the test set is shown in Table 4. The models trained to distinguish between high versus intermediate and low recurrence risk achieved an AUC of 0.77 and 0.74. However, the model for discriminating high and intermediate versus low failed to separate patients into these two groups. The performances of the models with five features were slightly better than those with ten features.
Table 4.
Performance of the trained models in the test set
| Name of the task | Model details | AUC in test set with 95% confidence interval | p value for the model in the test set |
|---|---|---|---|
| High versus intermediate and low ODX | Multivariate model using 5 selected features | 0.77 (0.56–0.98) | < 0.003 |
| Multivariate model using 10 selected features | 0.74 (0.51–0.96) | < 0.004 | |
| High and intermediate versus low ODX | Multivariate model using 5 selected features | 0.51 (0.41–0.61) | 0.75 |
| Multivariate model using 10 selected features | 0.50 (0.40–0.60) | 0.74 |
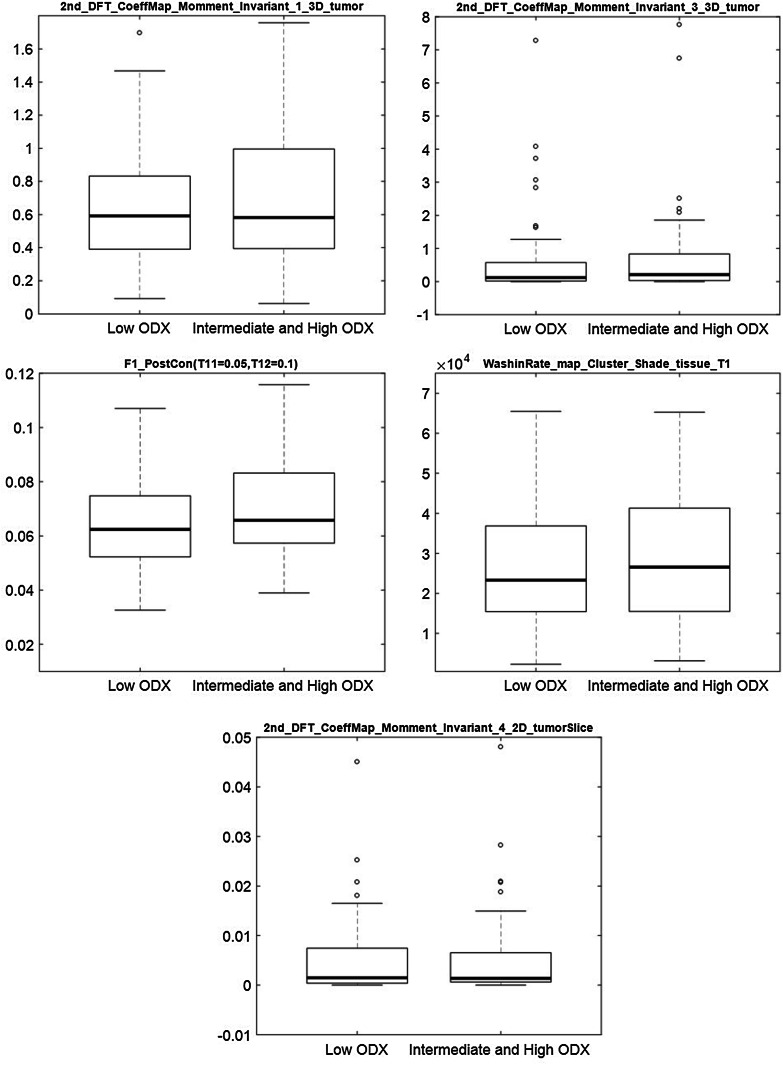
The results for the exploratory univariate analysis of individual features potentially associated with specific ODX categories are shown in Fig. 2 (high versus intermediate and low risk) and Fig. 3 (high and intermediate versus low risk). The features that were selected as most predictive of recurrence score (high versus intermediate and low) were as follows: signal enhancement ratio based washout tumor volume, mean and variance of the washin slope of tumor voxels that had maximum intensity (across all time points) in the first postcontrast sequence, proportion of tumor voxels that enhanced at their maximum intensity in the first postcontrast sequence, and proportion of tumor voxels that enhanced at a particular intensity level when the proportion of FGT voxels reached a certain value of the mean enhancement of the tumor. Consistent with the results of the multivariate analysis, individual features show notably higher discriminative power when distinguishing high risk from intermediate and low risk scores.
Fig. 2.
Boxplot of individual top 5 selected features for discriminating high versus intermediate and low ODX
Fig. 3.
Boxplot of individual top 5 selected features for discriminating high and intermediate versus low ODX
Discussion
We conducted a study to evaluate a comprehensive set of imaging features from DCE-MRI in terms of their ability to predict the distant recurrence risk using ODX scores. Our cohort of 261 patients, showed variability in terms of MRI scanner parameters, MRI acquisition parameters, and contrast agents used. This cohort was divided into training and test sets of approximately equal size and multivariate models were developed to predict the ODX risk categories. Our multivariate models were able to discriminate between high versus intermediate and low ODX scores. However, we found no discriminative power of the examined features for distinguishing high and intermediate ODX score from the low score.
The choice of classification tasks considered in this study was mostly motivated by the literature, where more work has been concentrated on discriminating high risk versus low risk (Wan et al. 2016) or high risk versus intermediate and low risk patients (Li et al. 2016a; Ashraf et al. 2014). However, as discussed in Harowicz et al. (2017), low ODX scores are important for decision making in chemotherapy by potentially sparing patients from chemotherapy because of their low risk of recurrence. Therefore, we considered discrimination of low versus intermediate and high to be an important classification task. Our results for discriminating high versus low and intermediate ODX in the test set were very close to the AUC values reported in studies (Ashraf et al. 2014; Li et al. 2016a). For ODX score prediction, these two studies differed between themselves in the following aspects (a) multi-institutional versus single-institutional (b) use of 3D tumor volume versus a central tumor slice, and (c) types of features considered. We have included features from both of these studies in our set. In the trained model including 10 features, features from both of these studies were selected. Other features selected included signal enhancement ratio (SER) based features (Arasu et al. 2011) extracted from the tumor. An enhancement feature extracted using both tumor and tissue enhancement (Mazurowski et al. 2014) was also selected. A direct comparison of our results with Sutton et al. (2015) and Wan et al. (2016) cannot be discussed, as the entire range (0–100) of ODX scores was not considered for both of these studies though features from both of these studies were included in our set of 529 features.
A strength of our study is the use of an independent test set used to validate the multivariate model developed in training set. This is of high importance given the large number of features evaluated in this and similar studies. A lack of a validation set could result in obtaining high values of correlation between imaging features and genomic features by chance and lack of generalizability of the conclusions.
Our study had some limitations. The proportion of high risk ODX patients in our set is lower compared to the other studies. The proportion of high ODX patients is lower in the test set than in the training set. This results in a wide confidence interval for performance calculated in our study. However, the fact that the multivariate models could learn from small proportion high risk patients shows high robustness of the imaging features found predictive in our study.
An important aspect of our study is the heterogeneity of the cohort in terms of imaging. The acquired images spanned 14 years and varied in terms of the MR scanners used, the MR acquisition parameters and the types of contrast agent used. On one hand, this allowed us to show that some imaging variables are associated with outcomes despite a considerable variability in imaging protocols making the result robust to such changes and more likely generalizable to other data. On the other hand, it is possible that prognostic values of some imaging variables were obscured by this variability and not found in this study. Accounting for variability in imaging is largely an unsolved problem with increasing research attention devoted to it (Saha et al. 2016). Future studies, could repeat this analysis in more controlled imaging conditions with uniform scanning and contrast protocol.
The highest obtained performance of AUC = 0.77, even if thoroughly validated, is likely not sufficient for replacing the ODX tissue-based biomarker. However, it shows promise for potential use as a selection criterion for which patients should undergo this expensive test. Furthermore, the imaging surrogate should be used in combination with the pathology-based surrogate (Klein et al. 2013; Gage et al. 2015; Tang et al. 2010) to obtain a more accurate prediction of the ODX recurrence score.
The negative finding of this study is that despite a comprehensive set of imaging features and a considerable sample size, we were not able to identify imaging predictors of which patients will have a low ODX recurrence score (vs. intermediate and high) in the univariate and multivariate settings. This is a limitation of using imaging in this context since distinguishing these categories is of high importance when deciding on usage of chemotherapy in estrogen receptor positive patients.
In conclusion, we found that there is an association between recurrence risk obtained using multi-gene assay with computer extracted features. However, this association strictly depends on the stratification of ODX scores considered. Therefore, the imaging-based models evaluated in this study are not an appropriate replacement for the ODX scores.
Abbreviations
- Oncotype DX
ODX
- MRI
Magnetic resonance imaging
- DCE-MR
Dynamic contrast enhanced magnetic resonance
- IHC
Immunohistochemical
- NFS
Non-fat-saturated
- FGT
Fibroglandular tissue
- ROC
Receiver operating characteristics
- AUC
Area under receiver operating characteristics
Funding
This study received funding from North Carolina Biotechnology Center (2016-BIG-6520) and National Institutes of Health (R01EB021360).
Compliance with ethical standards
Conflict of interest
Authors have no conflicts of interest to declare.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors.
Informed consent
The requirement for informed consent was waived by the institutional review board.
References
- (2006) NSABP study confirms oncotype DX predicts chemotherapy benefit in breast cancer patients. Oncology 20:789–790 [PubMed]
- Arasu VA, Chen RCY, Newitt DN, Chang CB, Tso H, Hylton NM, Joe BN (2011) Can signal enhancement ratio (SER) reduce the number of recommended biopsies without affecting cancer yield in occult MRI-detected lesions? Acad Radiol 18:716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf AB, Daye D, Gavenonis S, Mies C, Feldman M, Rosen M, Kontos D (2014) Identification of intrinsic imaging phenotypes for breast cancer tumors: preliminary associations with gene expression profiles. Radiology 272:374–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke E, Abe H (2015) MRI phenotype of breast cancer: kinetic assessment for molecular subtypes. J Magn Reson Imaging 42:920–924 [DOI] [PubMed] [Google Scholar]
- Coates AS, Winer EP, Goldhirsch A, Gelber RD, Gnant M, Piccart-Gebhart M, Thürlimann B, Senn HJ, Panel M, André F (2015) Tailoring therapies—improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol 26:1533–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delong ER, Delong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3):837–845 [PubMed]
- Dowsett M, Sestak I, Lopez-Knowles E, Sidhu K, Dunbier AK, Cowens JW, Ferree S, Storhoff J, Schaper C, Cuzick J (2013) Comparison of PAM50 risk of recurrence score with onco type DX and IHC4 for predicting risk of distant recurrence after endocrine therapy. J Clin Oncol 31:2783–2790 [DOI] [PubMed] [Google Scholar]
- Fan M, Li H, Wang S, Zheng B, Zhang J, Li L 2017. Radiomic analysis reveals DCE-MRI features for prediction of molecular subtypes of breast cancer. PLOS One 12:e0171683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage MM, Rosman M, Mylander WC, Giblin E, Kim H-S, Cope L, Umbricht C, Wolff AC, Tafra L (2015) A validated model for identifying patients unlikely to benefit from the 21-gene recurrence score assay. Clin Breast Cancer 15:467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harowicz MR, Robinson TJ, Dinan MA, Saha A, Marks JR, Marcom PK, Mazurowski MA (2017) Algorithms for prediction of the Oncotype DX recurrence score using clinicopathologic data: a review and comparison using an independent dataset. Breast Cancer Res Treat 162:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JJ, Kim JY, Kang HJ, Shin JK, Kang T, Lee SW, Bae YT (2017) Computer-aided diagnosis-generated kinetic features of breast cancer at preoperative MR imaging: association with disease-free survival of patients with primary operable invasive breast cancer. Radiology 284:45–54 [DOI] [PubMed] [Google Scholar]
- Klein ME, Dabbs DJ, Shuai Y, Brufsky AM, Jankowitz R, Puhalla SL, Bhargava R (2013) Prediction of the Oncotype DX recurrence score: use of pathology-generated equations derived by linear regression analysis. Mod Pathol 26:658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijenaar RT, Carvalho S, Velazquez ER, Van Elmpt WJC, Parmar C, Hoekstra OS, Hoekstra CJ, Boellaard R, Dekker ALAJ, Gillies RJ, Aerts HJWL, Lambin P (2013) Stability of FDG-PET Radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol 52(7):1391–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu Y, Burnside ES, Drukker K, Hoadley KA, Fan C, Conzen SD, Whitman GJ, Sutton EJ, Net JM, Ganott M, Huang E, Morris EA, Perou CM, Ji Y, Giger ML (2016a) MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of mammaprint, oncotype DX, and PAM50 gene assays. Radiology 281:382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhu Y, Burnside ES, Huang E, Drukker K, Hoadley KA, Fan C, Conzen SD, Zuley M, Net JM, Sutton E, Whitman GJ, Morris E, Perou CM, Ji Y, Giger ML (2016b) Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2:16012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurowski MA (2015) Radiogenomics: what it is and why it is important. J Am Coll Radiol 12:862–866 [DOI] [PubMed] [Google Scholar]
- Mazurowski MA, Zhang J, Grimm LJ, Yoon SC, Silber JI (2014) Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology 273:365–372 [DOI] [PubMed] [Google Scholar]
- Mazurowski MA, Grimm LJ, Zhang J, Macrom PK, Yoon S, Kim C, Ghate S, Johnson K (2015) Recurrence-free survival in breast cancer is associated with MRI tumor enhancement dynamics quantified using computer algorithms. Eur J Radiol 84(11):2117–2122 [DOI] [PubMed]
- Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J (2010) A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor–positive breast cancer. Clin Cancer Res:1078–0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A Multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351(27):2817–2826 [DOI] [PubMed] [Google Scholar]
- Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, Cronin M, Baehner FL, Watson D, Bryant J (2006) Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor–positive breast cancer. J Clin Oncol 24:3726–3734 [DOI] [PubMed] [Google Scholar]
- Saha A, Grimm LJ, Harowicz M, Ghate SV, Kim C, Walsh R, Mazurowski MA (2016) Interobserver variability in identification of breast tumors in MRI and its implications for prognostic biomarkers and radiogenomics. Med Phys 43:4558–4564 [DOI] [PubMed] [Google Scholar]
- Saha A, Yu X, Sahoo D, Mazurowski MA (2017) Effects of MRI scanner parameters on breast cancer radiomics. Expert Syst Appl 87:384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton EJ, Oh JH, Dashevsky BZ, Veeraraghavan H, Apte AP, Thakur SB, Deasy JO, Morris EA (2015) Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J Magn Reson Imaging 42:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang P, Wang J, Hicks DG, Wang X, Schiffhauer L, Mcmahon L, Yang Q, Shayne M, Huston A, Skinner KA (2010) A lower Allred score for progesterone receptor is strongly associated with a higher recurrence score of 21-gene assay in breast cancer. Cancer Investig 28:978–982 [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu T, Kasami M, Yuen S (2009) Triple-negative breast cancer: correlation between MR imaging and pathologic findings. Radiology 250:638–647 [DOI] [PubMed] [Google Scholar]
- Wan T, Bloch BN, Plecha D, Thompson CL, Gilmore H, Jaffe C, Harris L, Madabhushi A (2016) A radio-genomics approach for identifying high risk estrogen receptor-positive breast cancers on DCE-MRI: preliminary results in predicting OncotypeDX risk scores. Sci Rep 6:21394. 10.1038/srep21394 [DOI] [PMC free article] [PubMed]
- Wittner BS, Sgroi DC, Ryan PD, Bruinsma TJ, Glas AM, Male A, Dahiya S, Habin K, Bernards R, Haber DA (2008) Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res 14:2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Sun X, Wang J, Cui Y, Kato F, Shirato H, Ikeda DM, Li R (2017) Identifying relations between imaging phenotypes and molecular subtypes of breast cancer: model discovery and external validation. J Magn Reson Imaging 46(4):1017–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]