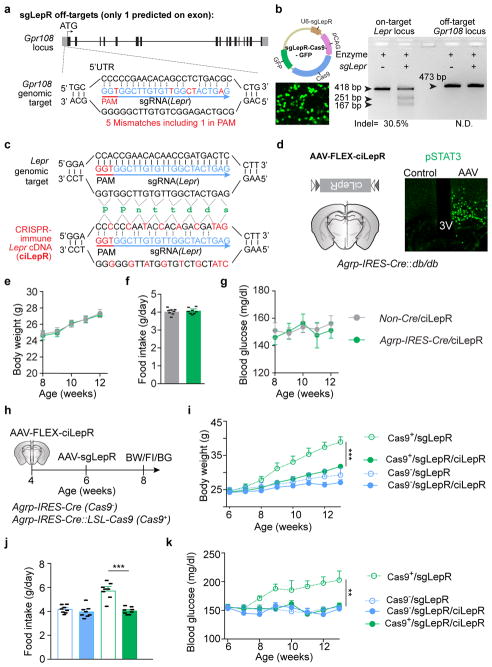
Extended Data Figure 4. Analysis of off-target effects of CRISPR-mediated genome editing; Expression of CRISPR-immune leptin receptors in AgRP neurons occludes alterations in body weight, food intake, or blood glucose levels caused by CRISPR-mediated disruption of Lepr.
a, Diagram of the predicted off-target site of sgLepR on the first exon of Gpr108 locus. b, Diagram of plasmid expressing both sgLepR and Cas9-GFP proteins (upper panel), fluorescent imaging of GFP following transfection of the plasmid into mouse N2a cells (lower panel), and on-target as well as off-target indel detection results demonstrating that sgLepR effectively induces mutations in Lepr locus but not in Gpr108 locus (right panel). c, sgRNA targeting the mouse Lepr locus (upper panel) and of CRISPR-immune Lepr cDNA encoding the long-form leptin receptors (ciLepR, lower panel) with indicated silent mutations to prevent binding of sgRNA. d, Schematic diagram of Cre-dependent AAV pEF1α-FLEX-ciLepr (AAV-FLEX-ciLepR) injected unilaterally into the ARC of Agrp-IRES-Cre::Leprdb/db mice (left) and representative images of leptin-induced pSTAT3 (Tyr705) immunostaining (right), indicating that ciLepR is fully responsive to leptin stimulation. e–g, Body weight (e), food intake (f), and blood glucose (g) of Non-Cre and Agrp-IRES-Cre littermates following bilateral injection of AAV-FLEX-ciLepR into the ARC (n=8 mice per group), suggesting that AAV-mediated expression of ciLepR in AgRP neurons produces no obvious effects on energy or glucose balances. h–k, Diagram of experiment flow (h), body weight (i), daily food intake (j), and blood glucose measurements (k) in virus-transduced ad libitum fed littermates (n=8 mice per group). Since expression of ciLepR in AgRP neurons totally prevented body weight gain, increased food intake, and hyperglycemia induced by CRISPR-mediated deletion, potential contributions from off-site mutagenesis are excluded. Data are mean ± s.e.m. and representative of three independent experiments; **P<0.01, ***P<0.001; Student’s two-tailed, unpaired t-test (f, j) or two-way ANOVA analysis (e, g, i, k).