Abstract
Cancer and other cells residing in the same niche engage various modes of interactions to synchronize and to buffer the negative effects of environmental changes. Extracellular miRNAs have been recently implicated in the intercellular crosstalk. Here we show a mechanistic model involving breast-cancer-secreted, extracellular-vesicle-encapsulated miR-105, which is induced by the oncoprotein MYC in cancer cells and in turn activates MYC signaling in cancer-associated fibroblasts (CAFs) to induce a metabolic program. This results in CAFs’ capacity to display different metabolic features in response to changes in the metabolic environment. When nutrients are sufficient, miR-105-reprogrammed CAFs enhance glucose and glutamine metabolism to fuel adjacent cancer cells. When nutrients are deprived whereas metabolic byproducts are accumulated, these CAFs detoxify metabolic wastes, including lactic acid and ammonium, by converting them into energy-rich metabolites. Thus, the miR-105-mediated metabolic reprogramming of stromal cells contributes to sustained tumour growth by conditioning the shared metabolic environment.
Keywords: Cancer-associated fibroblasts, Tumour microenvironment, Extracellular miRNA, MYC, Cell metabolism
Introduction
Cancer engages a dynamic interplay amongst cells in the cancer-hosting niche1, 2. Across the many levels of intercellular crosstalk, a role for cancer-secreted extracellular vesicles (EVs) has been recognized3, 4. Through travelling in the interstitial spaces and blood, EVs transfer biomolecules between adjacent or distant cells5–7. Cancer-derived EVs are implicated in tissue invasion, angiogenesis, immune evasion, and metastasis7–15, with many of these functions attributed to EV-encapsulated miRNA post-transcriptionally regulating gene expression in niche cells16–18. We previously reported that breast cancer (BC) cells secrete miR-105 to destroy tight junctions in vascular endothelial cells and promote metastasis11, and that BC-secreted miR-122 suppresses glucose utilization in a pre-metastatic niche to outsmart nutrient competition10. Increased levels of circulating miR-105 and miR-122 can be detected at a pre-metastatic stage and correlate with the occurrence of metastasis in BC patients; systemic interventions of these miRNAs suppress BC metastases10, 11, 19. Here, we extend our study of cancer-secreted miRNA to the metabolic reprogramming of cancer-associated fibroblasts (CAFs), a major cellular component of tumour stroma20.
Proliferative cancer cells often acquire enhanced abilities to exploit nutrients, such as glucose and glutamine, to support their energy demand and biomass synthesis21–23. This, along with the generation of metabolic byproducts including lactic acid (LA; from aerobic glycolysis known as the “Warburg effect”) and ammonium (NH4+), influence the metabolic environment of tumour–milieu-residing niche cells, highlighting the necessity to rebalance the metabolic needs of cancer and niche cells. Interestingly, in some tumours CAFs are found to also undergo aerobic glycolysis24, 25, and may secrete energy-rich metabolites such as pyruvate and lactate to fuel neighbouring cancer cells26, 27. Expression of monocarboxylate transporter (MCT) 4 in tumour stroma is associated with progression to invasive BC, suggesting a role for MCT-dependent stroma-to-cancer metabolite transfer28. However, this model requires adequate glucose to suffice both glycolytic cancer cells and CAFs, to enable a pro-cancerous metabolic interplay rather than provoking a competition between the two cell populations. When the environment is deprived of nutrients, which is a common intermittent stress in a growing tumour often concurring with accumulation of metabolic wastes due to insufficient vascular drainage, CAFs may undergo different metabolic interplay with cancer cells that is yet to be elucidated. We therefore set out to unravel the complex metabolic interactions between BC cells and CAFs.
Results
miR-105 induces a MYC-dependent metabolic program
Patient-derived CAFs (29; Supplementary Fig. 1) exhibited high and comparable uptake efficiencies for EVs from MDA-MB-231 BC cells and MCF10A non-cancer cells (Fig. 1a). RNA-seq transcriptome profiling followed by gene set enrichment analysis (GSEA) revealed enrichment of a gene signature related to MYC activation in CAFs treated with MDA-MB-231 EVs compared to those receiving MCF10A EVs or PBS (Fig. 1b). In quest of the potential upstream event leading to MYC activation, we observed >50% downregulation of the gene encoding MXI1 (MAX-interacting protein 1) (Fig. 1c), a protein that antagonizes the MYC–MAX dimeric transcriptional factor through forming heterodimers with MAX30, recruiting transcription repressors31, 32, and directly suppressing transcription from MYC promoter33. Eight miRNAs are predicted by three independent algorithms to recognize the 3’UTR of MXI1; among these, we focused on miR-105 because of its highest cancer-vs-normal secretion with an undetectable level in MCF10A EVs (Supplementary Table 1), its capacity to decrease MXI1 and elevate MYC protein levels in CAFs (Fig. 1d), and its association with BC progression11. Indeed, EVs from MCF10A engineered to overexpress and secrete miR-105 also induced the MYC activation gene signature and downregulated MXI1 in CAFs (Fig. 1b–c). Characterization of EVs by nanoparticle tracking analysis and density gradient fractionation indicated miR-105’s enrichment in exosome-containing fractions (Supplementary Fig. 2).
Figure 1. miR-105 induces a MYC-dependent metabolic program.
(a) CAFs were incubated with DiI-labelled EVs (red) for 24 h before fluorescent and phase contrast images were captured. Bar=100 μm. The experiment was repeated independently three times with similar results. (b) GSEA demonstrating the enrichment of a MYC target gene set in CAFs treated with MDA-MB-231 EVs or MCF10A/miR-105 EVs vs. those treated with PBS or MCF10A EVs. Based on data from two independent replicates, genes were ranked by signed P value score from edgeR (see Methods) and subjected to GSEA interrogation, which generated the indicated P value, q value and normalized enrichment score (NES) for each gene set based on 1,000 random permutations. (c) Heat map showing the normalized counts of MXI1 in all CAF RNA samples (exact test by edgeR, n=2 independent experiments). P value was calculated by edgeR using exact test. (d) Western blots showing indicated protein levels in miRNA-mimic-transfected CAFs. (e) Western blots showing indicated protein levels in MCF10A overexpressing miR-105 or MYC, or both. (f) Relative RNA levels detected by RT-qPCR and compared to the MCF10A/vec cells (one-way ANOVA, n=3 independent experiments). (g) ECAR and OCR assays in MCF10A overexpressing the empty vector, miR-155, miR-105, MYC, or both miR-105 and MYC (one-way ANOVA, n=3 independent experiments). *ECAR P<0.05, ***ECAR P<0.001, †OCR P<0.001. (h) Changes of metabolite levels in the medium within 72 h in indicated cells transfected with MYC siRNA or control siRNA (one-way ANOVA, n=3 independent experiments). (i) Western blots showing indicated protein levels in MCF10A with or without miR-105 overexpression and previously transfected with an expression plasmid of MXI1 cDNA lacking 3’UTR or control vector. (j) RNA and protein levels of MXI1 in MDA-MB-231 cells transfected with anti-miR-105 or control (two-sided t-test, n=3 independent experiments). (k) Changes of metabolite levels in the medium over 72 h by MDA-MB-231 cells treated as indicated (one-way ANOVA, n=3 independent experiments). For the entire figure, data are shown as mean ± SD; *P<0.05, **P<0.01, ***P<0.001. Unprocessed original scans of blots are shown in Supplementary Figure 9. Source data are shown in Supplementary Table 5.
Gene expression associated with miR-105 overexpression in MCF10A revealed enrichment of gene sets related to MYC activation (Supplementary Fig. 3a). In addition, Ingenuity pathway analysis predicted MYC as the top upstream regulator of miR-105-regulated genes, whereas the ENCODE ChIP-Seq analysis identified MYC, MAX, and MXI1 among the potentially involved transcription factors (Supplementary Table 2). When compared to gene expression associated with MYC overexpression, a significant subset of genes, including known MYC targets in glucose metabolism, were regulated in the same direction by miR-105 and MYC (Supplementary Fig. 3b; Fig. 1e–f), suggesting a functional overlap between miR-105 and MYC. This was confirmed by the similar capacities of miR-105 and MYC to enhance glycolysis (evidenced by an increase in ECAR and decrease in OCR; Fig. 1g) and accelerate nutrient use (increases in the consumption of glucose and glutamine and in the production of LA and NH4+; Fig. 1h). Some of these effects were more pronounced when miR-105 and MYC were co-expressed. Knockdown of MYC expression abolished miR-105- and MYC-induced nutrient metabolism (Fig. 1h), whereas ectopic expression of MXI1 lacking 3’UTR abolished the induction of glycolytic genes (Fig. 1i). In contrast to the effects of miR-105 overexpression, antagonization of endogenous miR-105 increased MXI1 expression in MDA-MB-231 (Fig. 1j), and resembled the effects of MYC knockdown by reducing glucose and glutamine metabolism (Fig. 1k).
miR-105 targets MXI1 to activate MYC signaling and is also induced by MYC
We next constructed a series of luciferase reporters containing the wild-type (wt) or mutated (ΔSite I or ΔSite II) full-length MXI1 3’UTR, or the annealed oligonucleotides encompassing each putative miR-105 site in MXI1 3’UTR (wt or mutated; Supplementary Fig. 4a). Mutations of the highly conserved proximal miR-105-binding site (Site I), but not Site II, abolished the reporters’ responsiveness to miR-105 (Fig. 2a). Supporting the hypothesized function of miR-105 to activate MYC through targeting MXI1, miR-105 induced an E-box-driven luciferase reporter, which was abolished by ectopic expression of MXI1 lacking 3’UTR (Fig. 2b).
Figure 2. miR-105 targets MXI1 to activate MYC signaling and is induced by MYC.
(a) Responsiveness of wild-type and mutant MXI1 3’UTR reporters shown in Supplementary Fig. 4a to miR-105 mimic in transfected MCF10A cells (two-sided t-test, n=3 independent experiments). (b) Responsiveness of an E-box reporter and the control reporter to miR-105 mimic in the absence or presence of ectopic expression of MXI1 cDNA lacking 3’UTR (two-sided t-test, n=3 independent experiments). (c) MCF10A- or MDA-MB-231-secreted EVs were fed to CAFs in the presence or absence of DRB (20 μM). After 24 h, RNA extracted from the recipient cells was analyzed for miR-105 level using U6 as an internal reference (one-way ANOVA, n=3 independent experiments). (d) Responsiveness of wild-type and mutant miR-105 promoter reporters to a MYC expression vector in transfected MCF10A cells (two-sided t-test, n=3 independent experiments). (e–f) Levels of miR-105 in cells (normalized to U6) and EVs (normalized to cel-miR-39-3p spike-in control) under MYC overexpression (e) or siRNA knockdown for 24 h (f) (two-sided t-test, n=3 independent experiments). (g) Western blots showing protein levels in indicated non-cancerous breast cells (MCF10A) and BC cells. (h) RT-qPCR-determined levels of miR-105 in equal amounts of EVs from indicated cells (normalized to cel-miR-39-3p spike-in control; n=3 independent experiments). (i) Western blots showing indicated protein levels in CAFs treated with equal amounts of EVs from indicated cells. For the entire figure, data are shown as mean ± SD; **P<0.01, ***P<0.001. Unprocessed original scans of blots are shown in Supplementary Figure 9. Source data are shown in Supplementary Table 5.
In CAFs treated with MDA-MB-231 EVs, we detected an increase in miR-105, which was partially reduced by an RNA polymerase II inhibitor (Fig. 2c). This suggests that EV-transferred miR-105 may further induce the endogenous miR-105 expression in recipient CAFs to augment the effect. A thorough search of the promoter of GABRA3, the hosting gene harboring hsa-mir-105-1/2, revealed the presence of an E-box which responded to MYC to activate reporter gene expression (Supplementary Fig. 4b; Fig. 2d). Ectopic expression of MYC indeed increased miR-105 both inside the cells and in EVs (Fig. 2e), whereas MYC knockdown decreased intracellular and secreted miR-105 in BC cells expressing high levels of endogenous miR-105 (Fig. 2f). We therefore proposed a MYC–miR-105–MYC activation loop, whereby the high MYC activity in cancer cells leads to a high secretion level of miR-105 to activate MYC signaling in stromal cells, phenotypically extending the effects of MYC originating from cancer cells. BC cells expressing higher levels of MYC indeed tend to secrete higher levels of miR-105 (Fig. 2g–h). EVs from these cells, but not cells with low MYC expression and low miR-105 secretion, were able to downregulate MXI1 and induce MYC and target genes in recipient CAFs (Fig. 2i).
miR-105 enhances glycolysis and glutaminolysis in CAFs
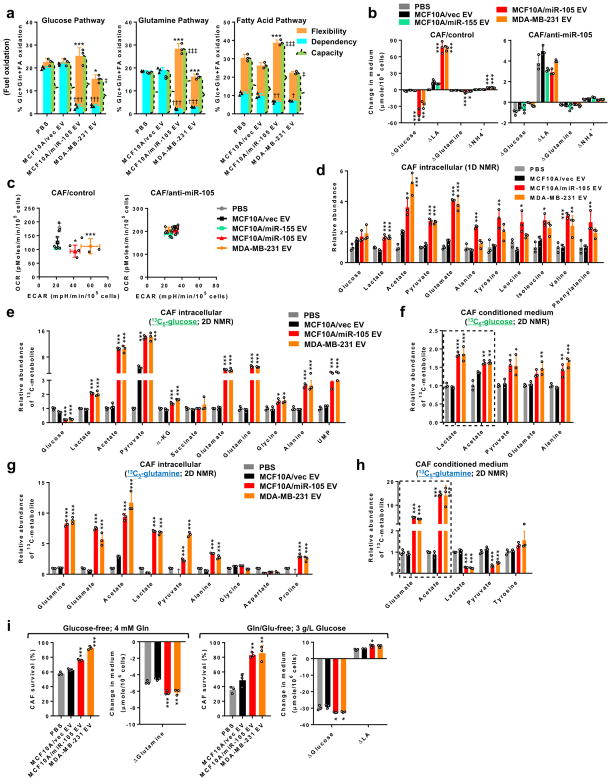
We went on to determine changes in bioenergetics, a major cellular process regulated by MYC23, 34, in CAFs following treatment with high-miR-105 EVs (from MCF10A/miR-105 and MDA-MB-231 cells) compared to low-miR-105 EV (from MCF10A/vec control cells) and PBS treatments. Comparing the use among three mitochondrial fuels, miR-105-reprogrammed CAFs exhibited reduced dependency and enhanced flexibility to oxidize glucose and glutamine, with less significant alterations with the fatty acid pathway (Fig. 3a). In a miR-105-dependent manner, the EV-reprogrammed CAFs as well as NIH3T3 cells both significantly increased the catabolism of glucose and glutamine resulting in accelerated extracellular acidification (Fig. 3b–c; Supplementary Fig. 5a–b). In contrast, miR-155 that is also highly secreted by MDA-MB-231 cells (11; Supplementary Fig. 2e) and used as a control here, did not significantly influence cell metabolism of glucose and glutamine (Fig. 3b–c). Nuclear magnetic resonance (NMR) spectral profiling revealed higher intracellular levels of lactate, acetate, and glutamate in both fibroblast models upon EV transfer of miR-105 (Fig. 3d; Supplementary Fig. 5c).
Figure 3. CAFs are reprogrammed by cancer-secreted miR-105 to enhance secretion of glucose- and glutamine-derived metabolites.
(a) Mitochondrial fuel test to determine the flexibility, dependency, and capacity of glucose, glutamine, and fatty acid pathways in CAFs treated as indicated for 48 h (n=3 independent experiments). The y axis shows fuel oxidation, indicated by the % of target pathway OCR out of the OCR from all three pathways. Flexibility: *P<0.05, ***P<0.001; dependency: †P<0.05, ††P<0.01, †††P<0.001; capacity: ‡P<0.05; ‡‡‡P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups). (b) Changes of metabolite levels in the medium over 72 h by CAFs expressing anti-miR-105 or control and treated as indicated (n=3 independent experiments). (c) ECAR and OCR assays in treated CAFs (n=3 independent experiments). *ECAR P<0.05, ***ECAR P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups). (d) Levels of metabolites in CAFs cultured in regular medium (3 g/L glucose; 4 mM glutamine) measured by 1D NMR. Data was normalized to the PBS group (n=3 independent experiments). (e–f) CAFs pretreated as indicated for 48 h were incubated with 13C6-glucose (3 g/L) for 6 h in 12C-glucose-free medium. Cell extracts (e) and the conditioned media (f) were analysed by 2D NMR for the abundance of 13C-containing metabolites. Data was normalized to the PBS group (n=3 independent experiments). (g–h) Pretreated CAFs were incubated with 13C5-glutamine (4 mM) for 6 h in 12C-glutamine-free medium. Cell extracts (g) and the conditioned media (h) were analysed by 2D NMR. Data was normalized to the PBS group (n=3 independent experiments). †undetectable. (i) Pretreated CAFs were cultured in glucose-free medium or glutamine/glutamate-free medium for 24 h. Cell survival was determined by comparing to cells grown in regular medium. Changes of metabolite levels in the medium were also determined (n=3 independent experiments). For the entire figure, data are shown as mean ± SD, and statistical significance was assessed using one-way ANOVA. *P<0.05, **P<0.01, ***P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups) unless indicated separately. Source data are shown in Supplementary Table 5.
To define nutrient-specific metabolic flows, we incubated EV-pretreated CAFs with 13C-labelled glucose or glutamine and measured labelled metabolites by 2D NMR spectra. Higher levels of 13C-containing lactate, acetate, pyruvate, and several amino acids were detected in miR-105-reprogrammed CAFs following 13C-glucose incubation (Fig. 3e). To our particular interest, these CAFs also secreted higher amounts of labelled metabolites, especially lactate and acetate (Fig. 3f), which could potentially benefit adjacent cancer cells. Incubation with 13C-glutamine resulted in increased levels of labelled glutamate, acetate, lactate, and pyruvate in reprogrammed CAFs (Fig. 3g). Among these metabolites, labelled glutamate and acetate exhibited remarkably increased secretion by CAFs and could be involved in fuelling cancer cells (Fig. 3h). Thus, simultaneous induction of glycolysis and glutaminolysis in CAFs may enhance the flexibility to use either nutrient as the major fuel source. This was confirmed by the enhanced ability of reprogrammed CAFs to survive under single-nutrient deprivation of either glucose or glutamine/glutamate by increasing the use of the alternative nutrients (Fig. 3i).
The altered patterns of glucose and glutamine metabolism could be explained at the gene expression level, where genes coding for key enzymes in glycolysis (HK2, LDHA, LDHB, etc.) and glutaminolysis (GLS) and for metabolite transportation (SLC2A1, SLC16A1, SLC16A3, and SLC1A5) were upregulated in fibroblasts reprogrammed by high-miR-105 EVs (Fig. 4a; Supplementary Fig. 5d–e). These genes could also be induced by a synthetic miR-105 mimic (Fig. 4b) and are known MYC targets. Their regulations by high-miR-105 EVs were abolished by treating CAFs with an EV uptake inhibitor (Fig. 4c), and by expressing MXI1 lacking 3’UTR or carrying miR-105-site-I-mutated 3’UTR but not MXI1 carrying the wild-type 3’UTR (Fig. 4d–e). Furthermore, MDA-MB-231 cells with miR-105 gene knockout, which exhibited a higher MXI1 level and suppressed expression of MYC targets, failed to influence miR-105-regulated genes in CAFs via secreted EVs (Supplementary Fig. 4c; Fig. 4f–g).
Figure 4. miR-105 induces genes in glucose and glutamine metabolism through targeting MXI1.
(a) Relative RNA levels in EV-treated CAFs were detected by RT-qPCR and compared to PBS-treated cells (n=3 independent experiments). (b) Relative RNA levels in CAFs transfected with miR-105 mimic, control miRNA mimic, or PBS (n=3 independent experiments). (c) Western blots showing indicated protein levels in CAFs treated with EVs or PBS in the absence or presence of Dynasore (10 μM). (d) Western blots showing indicated protein levels in EV-treated CAFs previously transfected with an expression plasmid of MXI1 cDNA lacking 3’UTR or control vector for 16 h. (e) Western blots showing indicated protein levels in EV-treated CAFs previously transfected with an expression plasmid of MXI1 cDNA carrying the full-length wild-type or miR-105-site-I-mutated 3’UTR. (f) Western blots showing protein levels in wild-type and miR-105-knockout MDA-MB-231 cells. (g) Western blots showing protein levels in CAFs treated with PBS or EVs. For the entire figure, data are shown as mean ± SD, and statistical significance was assessed using one-way ANOVA. *P<0.05, **P<0.01, ***P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups unless indicated). Unprocessed original scans of blots are shown in Supplementary Figure 9. Source data are shown in Supplementary Table 5.
miR-105 enables detoxification of LA and NH4+ by CAFs
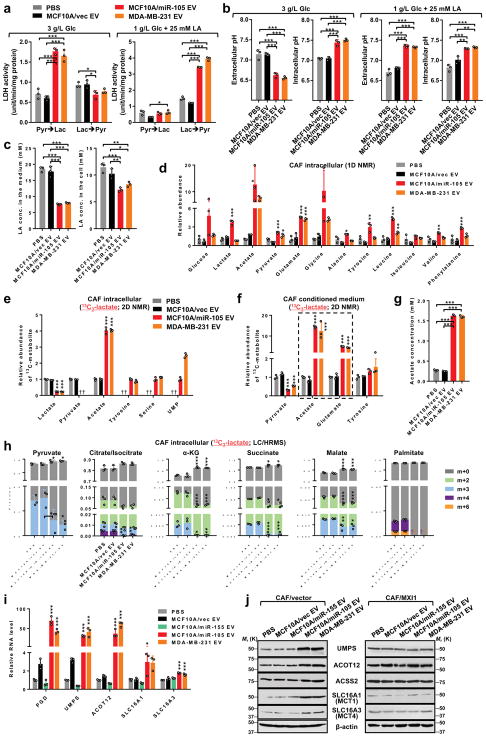
The tetrameric enzyme lactate dehydrogenase (LDH) catalyses the interconversion between pyruvate and lactate in a tissue- and substrate-dependent manner. Elevated expression of both LDHA and LDHB in miR-105-reprogrammed fibroblasts therefore enhanced the reaction in both directions, leading to increased LA production under high glucose and increased pyruvate production under high LA (Fig. 5a; Supplementary Fig. 6a). This was accompanied by glycolysis-associated extracellular acidification but also alleviation of extreme lactic acidosis partially through the consumption of extracellular LA (Fig. 5b–c; Supplementary Fig. 6b). Under a high level of LA, miR-105-reprogrammed fibroblasts contained higher intracellular levels of acetate and glutamate (Fig. 5d), exhibited enhanced growth (Supplementary Fig. 6c), and conditioned the medium to enhance cancer cell survival (Supplementary Fig. 6d). By incubating CAFs with 13C-lactate and profiling metabolites using 2D NMR and LC/HRMS, we detected increased 13C-containing acetate, amino acids, and UMP but decreased TCA intermediates and fatty acids in the cells, along with higher secretion levels of acetate and glutamate (Fig. 5e–h). CAFs reprogrammed by miR-105 increased the expression of ACOT12 (but not ACSS2), SLC16A1/3, PGD, and UMPS in a MXI1-dependent manner (Fig. 5i–j), which may respectively explain increased acetate production and secretion and suggest enhanced flow of lactate-derived gluconeogenesis into the pentose phosphate shunt for UMP biosynthesis.
Figure 5. miR-105-reprogrammed CAFs have enhanced abilities to consume extracellular LA and secrete LA-derived metabolites.
(a) LDH activity assays to measure the interconversion between pyruvate and lactate in EV-treated CAFs under high-glucose (3 g/L glucose; no LA) or high-LA (1 g/L glucose; 25 mM LA) conditions (n=3 independent experiments). (b) Extracellular (in non-buffered conditioned medium) and intracellular pH of treated CAFs following 24-h incubation in medium containing indicated levels of glucose and LA (n=3 independent experiments). (c) LA concentrations in the medium (10 mL medium from 107 cells) and in cells following 24 h incubation in medium containing 1 g/L glucose and 25 mM LA as the starting levels (n=3 independent experiments). (d) Levels of metabolites in CAFs pretreated as indicated and cultured in high-LA medium for 24 h were measured by 1D NMR. Data was normalized to the PBS group (n=3 independent experiments). (e–h) CAFs pretreated with EVs or PBS for 48 h were incubated in glucose-free medium containing 10 mM lactate for 24 h, and then changed to glucose-free medium containing 10 mM 13C3-lactate. After 6 h, cell extracts (e,h) and the conditioned media (f,g; 10 mL medium from 107 cells) were analysed by 2D NMR (e,f; data normalized to the PBS group; n=3 independent experiments), acetate assay kit (g; n=3 independent experiments), or LC/HRMS (h; n=3 independent experiments) for the abundances of 13C-containing metabolites. †undetectable. (i) Relative RNA levels in EV-treated CAFs grown under high-LA condition were detected by RT-qPCR and compared to PBS-treated cells (n=3 independent experiments). (j) Western blots showing indicated protein levels in EV-treated CAFs previously transfected with an expression plasmid of MXI1 cDNA lacking 3’UTR or control vector for 16 h and grown under high-LA condition. For the entire figure, data are shown as mean ± SD, and statistical significance was assessed using one-way ANOVA. *P<0.05, **P<0.01, ***P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups unless indicated). †undetectable. Unprocessed original scans of blots are shown in Supplementary Figure 9. Source data are shown in Supplementary Table 5.
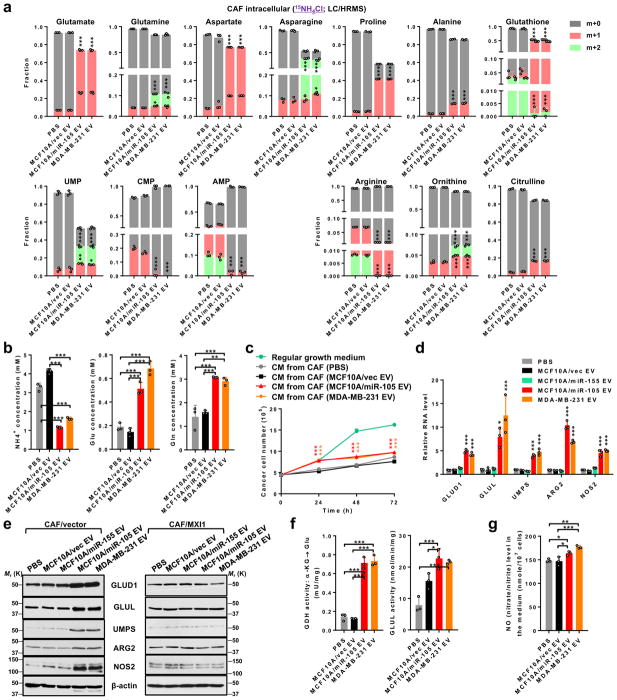
We next examined the ability of EV-treated CAFs to cope with another extreme condition in glutamine/glutamate-depleted, NH4+-containing medium. miR-105-reprogrammed CAFs significantly increased the conversion of inorganic 15NH4+ into several amino acids (especially glutamate and aspartate) and into UMP (but not other nucleotides), along with enhanced arginine hydrolysis evidenced by decreased arginine and increased ornithine and citrulline containing the 15N tracer (Fig. 6a). De novo glutathione synthesis was also altered, with the m+1 form dominating in miR-105-reprogrammed CAFs. This could be related to altered oxidative stress and/or redox regulation in the reprogrammed CAFs, where reduced levels of reactive oxygen species were detected (Supplementary Fig. 7a). Reprogrammed CAFs significantly enhanced the clearance of extracellular NH4+, and increased the secretion of glutamate as well as glutamine which has been previously reported to fuel glutamine-addicted cancer cells35, 36 (Fig. 6b). Indeed, BC cells exhibited enhanced growth and migration in glutamine/glutamate-depleted, NH4+-containing media that have been conditioned by miR-105-reprogramed CAFs (Fig. 6c; Supplementary Fig. 7b). Increased and MYC-mediated expression of GLUD1, GLUL (as previously reported35, 37), UMPS, ARG2, and NOS2 (Fig. 6d–e), along with increased enzymatic activities of GDH and GLUL (Fig. 6f) and increased nitric oxide (NO) production (Fig. 6g) were detected in miR-105-reprogrammed CAFs. These genes respectively control the de novo synthesis of glutamate and glutamine from NH4+, UMP synthesis, as well as arginine hydrolysis and inducible synthesis of NO. It is noted that the upregulation of GLUD1 is specific to the stress condition, as the gene was instead downregulated in miR-105-reprogramed CAFs cultured under growth condition (Supplementary Fig. 7c). Thus, our results collectively indicate a detoxifying function of miR-105-reprogrammed CAFs under extreme metabolic conditions including high levels of extracellular LA and NH4+.
Figure 6. miR-105-reprogrammed CAFs have an enhanced ability to consume extracellular ammonium.
(a) CAFs pretreated with EVs or PBS for 48 h were incubated in glutamine/glutamate-free medium containing 5 mM 15NH4Cl for 16 h. Cell extracts were analysed by LC/HRMS for the abundance of 15N-containing metabolites (n=3 independent experiments). (b) Concentrations of NH4+, glutamate, and glutamine in the medium of pretreated CAFs (10 mL medium from 107 cells) following 24 h incubation in glutamine/glutamate-free medium containing 5 mM NH4Cl (determined by assay kits; n=3 independent experiments). (c) MDA-MB-231 cells were cultured in regular growth medium (3 g/L glucose; 4 mM glutamine; no NH4+), or in NH4+-containing medium (3 g/L glucose; glutamine/glutamate-free; 5 mM NH4Cl) that had been conditioned by CAFs treated as indicated. Growth of MDA-MB-231 cancer cells in the CM was determined every 24 h (n=3 independent experiments). (d) Relative RNA levels in EV-treated CAFs grown in NH4+-containing medium (3 g/L glucose; glutamine/glutamate-free; 5 mM NH4Cl) were detected by RT-qPCR and compared to PBS-treated cells (n=3 independent experiments). (e) Western blots showing indicated protein levels in EV-treated CAFs previously transfected with an expression plasmid of MXI1 cDNA lacking 3’UTR or control vector for 16 h and grown in NH4+-containing medium. (f) Activities of GDH (α-KG converting to glutamate) and GLUL were measured in pretreated CAFs grown for 24 h in glutamine/glutamate-free medium containing 5 mM NH4Cl (n=3 independent experiments). (g) Levels of NO (measured as total nitrate/nitrite by an assay kit) in the medium of pretreated CAFs following 24 h incubation in glutamine/glutamate-free medium containing 5 mM NH4Cl (n=3 independent experiments). For the entire figure, data are shown as mean ± SD, and statistical significance was assessed using one-way ANOVA. *P<0.05, **P<0.01, ***P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups unless indicated). Unprocessed original scans of blots are shown in Supplementary Figure 9. Source data are shown in Supplementary Table 5.
Reprogrammed CAFs assist tumour growth
To determine if miR-105-mediated CAF reprogramming contributes to tumour growth in vivo, we orthotopically transplanted patient-derived BC cells (PDX) that were developed from the same tumour as CAFs29 and that secreted a high level of miR-105, together with CAFs stably expressing anti-miR-105 or a control sequence, into female NOD/SCID/IL2Rγ-null (NSG) mice. Compared to transplanting cancer cells alone, co-transplantation with the control CAFs, but not with those expressing anti-miR-105, significantly enhanced tumour growth (Fig. 7a). Homogenized tumour tissues were analysed by 2D NMR following individual administration of 13C-labelled tracers, including glucose and glutamine that were intravenously injected over a 30-min period and lactate injected directly into the centre of tumours. Comparing to tumours containing wild-type CAFs, those with miR-105-resistant CAFs had lower amounts of labelled acetate and glutamate following 13C-glucose injection (Fig. 7b), broad inhibition of a panel of labelled metabolites including α-KG, acetate, and multiple amino acids following 13C-glutamine injection (Fig. 7c), as well as lower amounts of labelled acetate and amino acids following 13C-lactate injection (Fig. 7d). When cancer cells were purified following tumour dissociation and analysed by LC/HRMS, those from tumours containing wild-type CAFs contained higher amounts of biosynthesis precursors including nucleotides, amino acids, NADH, and glutathione (Fig. 7e).
Figure 7. Cancer-secreted miR-105 enhances cancer–stroma metabolic coupling and promotes tumour growth.
(a) Female NSG mice received mammary fat pad injection of 2×105 PDX BC cells, either alone or mixed with 106 CAFs stably expressing anti-miR-105 or control. Tumour volume was measured (two-way ANOVA, n=7 mice per condition). (b–d) Tumour-bearing mice were infused with 13C6-glucose (b) or 13C5-glutamine (c) for 30 min, or injected with 13C3-lactate (d) into the tumour. Mice were sacrificed after 1 h and tumours collected. Abundances of 13C-containing metabolites were measured by 2D NMR. Data was normalized to the “PDX+CAF/control” group (two-sided t-test, n=3 mice per condition). (e) PDX cancer cells isolated from tumours were analysed by LC/HRMS for the levels of metabolites (data normalized to the “PDX only” group; one-way ANOVA, n=4 mice per condition). (f) Mice were randomized within individual groups and treated with an MCT inhibitor or vehicle starting on day 40. Tumour volume was recorded (two-way ANOVA, n=6 mice per condition). (g) Tumours from (f) were collected on day 46. Intratumoural pH in the edge and core areas of harvested tumours was measured (one-way ANOVA, n=3 mice per condition). (h) Representative IHC images showing Ki67 staining and the overall percentage of Ki67+ tumour cells (one-way ANOVA, n=5 mice per condition). Bar=100 μm. (i) Tumours from (f) were extracted for 1D NMR metabolic analysis (one-way ANOVA, n=3 mice per condition). †P<0.05, ††P<0.01, †††P<0.001 compared to the corresponding MCTi-free group. (j) Female NSG mice received mammary fat pad injection of 105 MCFDCIS cells mixed with 106 CAFs pretreated with EVs or PBS. Mice continued to receive EV or PBS treatment at the tumour site twice a week until tumour collection. Tumour volume was measured (two-way ANOVA, n=6 mice per condition). (k) Representative IHC images showing Ki67 staining and the overall percentage of Ki67+ tumour cells (one-way ANOVA, n=5 mice per condition). Bar=100 μm. (l) CAFs isolated from tumours were analysed by RT-qPCR for gene expression (one-way ANOVA, n=3 mice per condition). For the entire figure, data are shown as mean ± SD; *P<0.05, **P<0.01, ***P<0.001. Source data are shown in Supplementary Table 5.
Because acetate outstood as a common mediator of the metabolic coupling between CAFs and cancer cells in our in vitro and in vivo studies, and because the cross-plasma-membrane transport of acetate among other monocarboxylates depends on MCTs, we further determined the in vivo effect of a MCT1/2 inhibitor that was administered in established tumours. It is important to note that the MCT1/2 inhibitor would block the multi-directional transports of monocarboxylates within the tumour microenvironment, including those establishing the cancer–stroma metabolic coupling studied herein. Inhibition of MCT halted the progression of wild-type tumours within a week and abolished their growing advantages over those tumours with miR-105-resistant CAFs (Fig. 7f). Tumours containing wild-type CAFs also exhibited a less steep pH decline from tumour edge to the core (Fig. 7g), along with increased cancer cell proliferation indicated by immunohistochemistry (IHC) of Ki67 (Fig. 7h) as well as higher levels of metabolites including lactate and acetate (Fig. 7i). These effects were all abolished by MCT inhibition.
Similar to the PDX+CAF tumour model, co-transplantation of MDA-MB-231 and modified NIH3T3 also indicated that specifically blocking miR-105’s effect in fibroblasts suppressed tumour growth and cancer cell proliferation (Supplementary Fig. 8a–b). In a different tumour model we co-transplanted mice with wild-type CAFs and MCFDCIS cells that we have previously shown to produce a very low level of miR-10511. EVs with high or low miR-105 levels, or PBS as a control, were injected at the site of tumour growth to directly monitor the effect of EV-encapsulated miR-105. Compared to the other two groups, high-miR-105 EVs enhanced tumour growth and cancer cell proliferation (Fig. 7j–k), and CAFs purified from this group had lower expression of MXI1 and higher expression of MYC-regulated metabolic genes (Fig. 7l). Collectively, the in vivo data support our hypothesis that miR-105-reprogrammed CAFs nourish cancer cells with energy-rich metabolites and contribute to sustained tumour growth.
A MYC–miR-105–MXI1–MYC loop is observed in primary tumours
Unlike lung fibroblasts and astrocytes, which reduce glucose flux by targeting the glycolytic gene PKM with BC-secreted miR-12210, CAFs reprogrammed by BC-secreted EVs are highly responsive to miR-105-mediated MYC activation, inducing multiple genes involved in glucose and glutamine metabolism. We found that three out of the four cases of primary CAFs tested as well as the NIH3T3 embryonic fibroblasts expressed higher basal levels of MYC proteins compared to normal primary fibroblasts of the mammary, brain, and lung as well as astrocytes, which may affect cell responsiveness to regulators of MYC activity (Fig. 8a). Additionally, all tested mammary-derived fibroblasts had lower basal levels of MXI1, which may also sensitize the effect of an MXI1-targeting miRNA. The herein described gene regulation by EV-transferred miR-105 were also observed in other cases of CAFs from primary BCs (Fig. 8b), suggesting that the miR-105-mediated, MYC-dependent metabolic reprogramming of stromal cells is a relatively general event in BCs.
Figure 8. miR-105 is associated with the MYC pathway in primary tumours.
(a) Western blots showing protein levels of MYC and MXI1 in primary CAFs isolated from triple-negative BCs (CAF265922, the CAFs used in this study, and CAF1) and HER2+ BCs (CAF2 and CAF3) and other indicated primary cells and cell lines. (b) Relative RNA levels in EV-treated CAF1 and CAF3 were detected by RT-qPCR and compared to PBS-treated cells (one-way ANOVA, n=3 independent experiments). Data are shown as mean ± SD; *P<0.05, **P<0.01, ***P<0.001 (when individually compared to both PBS and MCF10A/vec EV control groups). (c) A BC tissue array was analysed for correlations among selected gene expression by IHC/ISH-determined scores. Correlations between nuclear MYC and miR-105 staining in tumour cells, miR-105 and MXI1 staining in the stroma, miR-105 and nuclear MYC staining in the stroma, nuclear MYC staining in the tumour and stroma, nuclear MYC and LDHB staining in the stroma, and between nuclear MYC and GLUL staining in the stroma were shown. The Kendall’s tau, sample size (N; number of independent tissue samples), and P value (two-sided) are indicated. (d) Representative ISH and IHC images showing staining in tumour (T) and stroma (S). The staining was repeated twice independently with similar results. Bar=100 μm. (e) A proposed model of the metabolic reprogramming of CAFs by cancer-secreted EVs through a miR-105-mediated mechanism identified herein. Unprocessed original scans of blots are shown in Supplementary Figure 9. Source data are shown in Supplementary Table 5.
Primary human breast tumours were examined to seek evidence for the herein identified gene regulation loop consisting of the tumour-initiated MYC–miR-105 axis as well as the consequent MXI1–MYC axis extending to the stroma. In tumour cells, levels of miR-105 and nuclear-localized MYC were positively correlated, whereas in the stroma, miR-105 exhibited an inverse correlation with MXI1 and a positive correlation with nuclear MYC (Fig. 8c–d). A strong correlation between MYC levels in the tumour and stroma compartments was detected, supporting the proposed model of “MYC extension” from cancer to stromal cells through EV transfer of miR-105. In addition, LDHB and GLUL indeed exhibited correlations with nuclear MYC in the stroma (Fig. 8c–d).
Discussion
Altered nutrient metabolism in tumour stroma is emerging as an important aspect of cancer-associated metabolic reprogramming. Our work collectively suggest that cancer-secreted miRNAs reprogram niche cell metabolism through various mechanisms: (1) biasing the nutrient “competition” toward cancer cells by suppressing niche cells’ consumption (e.g., miR-122-mediated suppression of glucose uptake in lung fibroblasts and astrocytes10); (2) establishing a “symbiosis” between cancer and niche cells through which niche-produced energy-rich metabolites (acetate, glutamate, etc.) feed anabolic cancer cells (shown in CAFs; Fig. 8e); and (3) engaging “scavengers” to support a rapidly increasing population of cancer cells by accelerating waste elimination through adjacent niche cells, which convert cancer-produced metabolic byproducts into non-toxic metabolites to re-enter cancer bioenergetics (shown in CAFs; Fig. 8e). These metabolic interactions resembling those in an ecosystem may exist in different types, stages, and compartments of tumours, and may depend on the metabolic pattern of cancer cells and the dynamic metabolic conditions in the tumour microenvironment. Latest findings indicate that certain tumours use lactate to fuel TCA cycle38, 39 and to a certain extent can recycle ammonia into central amino acid metabolism40. These functions can be facilitated by CAFs through the herein reported mechanism. As CAF-derived exosomes also contain metabolic intermediates41, we used EV-depleted CAF-conditioned media to measure the direct secretion of metabolites. We also detected glucose, lactate, acetate, and glutamate in EVs derived from CAFs and epithelial cells, but at much lower levels compared to the EV-depleted medium. Beyond direct cancer–stroma metabolic interactions, reprogrammed stromal cells also exert non-metabolic effects on cancer cells, such as by inducing IL-6 production and stromal inflammation42. Our data suggesting altered CAF production of NO may implicate another mechanism in the associated adaptation of tumour microenvironment.
An emerging physiological significance of EVs is to transfer a “prototype” message reflecting genetic and epigenetic alterations in donor cells (viral genome and proteins as an extreme example43, 44) to recipient cells without these features. Some oncoproteins, oncomiRs, and DNA fragments harboring oncogenic mutations are found in cancer-derived EVs45–47, and may participate in transferring a cancer prototype to non-cancer cells. For instance, melanoma-derived exosomes transfer MET to bone marrow progenitors to induce a pro-metastatic phenotype8. EGFR-containing exosomes from gastric cancer cells influence liver stromal cells to promote liver metastasis48. Some glioblastomas secrete vesicles containing the tumour-specific EGFRvIII mRNA4. Medulloblastoma cells with MYC amplification secrete EVs carrying MYC DNA and RNA sequences49, which could serve as an alternative mechanism to activate MYC pathway in recipient cells. The horizontal spreading of oncogenic signals adds non-epithelial niche cells into the scenario of “field cancerization50”, and may underpin another advantage of targeted therapies that would also affect non-cancer cells acquiring a cancer prototype, those that are less proliferative but may serve as a “powerhouse” and even a reservoir of oncogenic signals due to the bidirectional transfer potential of EVs.
Methods
Cells and constructs
Human BC cell lines, fetal lung fibroblast cell line WI-38, and the mouse embryonic fibroblast cell line NIH3T3 were obtained from American Type Culture Collection (Manassas, VA) and cultured in the recommended media. The non-cancerous mammary epithelial cell line MCF10A was constructed to stably overexpress miR-105 (MCF10A/miR-105), miR-155 (MCF10A/miR-155), or the empty vector (MCF10A/vec) as previously described11. MCF10A and derived cells were cultured as reported11, 51. Patient-derived primary fibroblasts CAF265922 (denoted as CAF in the study herein) and BC cells (PDX265922, propagated in NSG mice and denoted as PDX) derived from the same triple-negative breast tumour, as well as other patient-derived primary CAFs (CAF1 from another triple-negative breast tumour; CAF2 and CAF3 from HER2+ breast tumours) were isolated as previously described29 and cultured in Iscove’s modified dulbecco’s media (Thermo Fisher Scientific; Waltham, MA) supplemented with 20% fetal bovine serum. Primary normal human mammary fibroblasts were obtained from Zen-Bio (Research Triangle Park, NC) and cultured in the recommended media. Primary human astrocytes, lung fibroblasts, and brain fibroblasts were obtained from ScienCell (Carlsbad, CA) and cultured in the recommended media. All cells used herein were tested to be free of mycoplasma contamination and authenticated by using the short tandem repeat profiling method. Purity of PDX cancer cells and CAFs was ensured by fluorescence activated cell sorting using human EpCAM as a marker for epithelial cancer cells and PDGFRβ as a marker for fibroblasts. CAFs in culture were frequently checked to confirm they were negative for EpCAM or CD31 and positive for PDGFRβ and Vimentin. A lentiviral construct expressing an miArrest miR-105 inhibitor (anti-miR-105) as well as the scrambled control were purchased from GeneCopoeia (Rockville, MD) to generate CAF and NIH3T3 cells with stable inhibition of miR-105 as well as the control cells. PCR primers 5′-ATCACTCGAGAACCCAGCATGACATAAC and 5′-ATCAGCGGCCGCTTCTTCGTTCACAGTT were used to clone the wild-type 3′UTR of human MXI1. The PCR-amplified fragments were digested with XhoI and NotI and then inserted into the same sites of psiCHECK-2 reporter vector (Promega; Madison, WI) downstream of the Renilla luciferase gene. PCR primers 5′-TTGATGCAGCGGGTGATAATGAT and 5′-ATCATTATCACCCGCTGCATCAA (for mutated miR-105 binding site I; mutated nucleotides underlined), or 5′-GCTGGAAAGCGGGTGTTATAAAC and 5′-GTTTATAACACCCGCTTTCCAGC (for mutated miR-105 binding site II; mutated nucleotides underlined), were used to clone mutants of full-length MXI1 3′UTR. To construct the reporters with individual miR-105 sites, annealed oligonucleotides encompassing the putative sites indicated in Supplementary Fig. 4a, either in wild type or with a scrambled seed sequence, were inserted into psiCHECK-2. The MXI1 cDNA expression plasmid was constructed by PCR cloning the full-length MXI1 cDNA lacking 3′UTR using primers 5′-ACTGGATCCATGGAGCGGGTGAAGATGA and 5′-ACTGAATTCCTATGAAGTGAATGAAAG from MCF10A cells, and inserting the open reading frame (ORF) into the BamHI/EcoRI sites of pcDNA3.1(+) vector (Thermo Fisher Scientific). To clone the expression plasmids of MXI1 that contains an in-frame C-terminal HA tag as well as the full-length MXI1 3′UTR (either wild-type or miR-105-site-I-mutated), the PCR-derived 3′UTR fragments used to construct the psiCHECK-2 reporters were first inserted into the pcDNA3.1-MXI1 plasmid, downstream of the MXI1 cDNA. Next, primers 5′-AACATCGTATGGGTATGAAGTGAATGAAAGTTT and 5′-CCAGATTACGCTTAGGAATTCTGCAGATATCCA were used to insert the cDNA of an in-frame HA tag immediately before the stop codon of MXI1 through PCR with the assistance of Q5 high-fidelity DNA polymerase (New England Biolabs; Ipswich, MA) following the manufacturer’s protocol. The pBABE-MYC plasmid was a kind gift from Dr. Mei Kong. The promoter region (−161 to +33) of GABRA3/hsa-mir-105/mir-767 was cloned from MCF10A cells using the PCR primers 5′-ATATATCTCGAGCAGCACAGCCGCGTGCTAAGCACCA (for E-box-mutated promoter, the underlined “GT” were replaced with “TA”) and 5′-GCGCGCAAGCTTCTGGTTTGCTCGCTAGCTT; PCR-amplified fragment was then ligated into the XhoI/HindIII sites of pGL3-Basic reporter vector (Promega). The E-box luciferase reporter plasmid was constructed by inserting 4 tandem copies of sequence “CACGTG” upstream to the luciferase gene in pGL3-Promoter vector (Promega). To sequentially knock out both hsa-mir-105-1 and hsa-mir-105-2 genes using the CRISPR-Cas9 genomic editing system, sgRNA sequences predicted by the sgRNA Designer (portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design) were synthesized in DNA form and annealed into double strands, treated with T4 polynucleotide kinase, and inserted into the BbsI-digested pSpCas9(BB)-2A-GFP vector52. The constructs were transiently transfected into MDA-MB-231 cells in combinations of two against the same gene; among the 4 sgRNAs designed for each gene, the combination of CAGAACCTGAGTGTGCATCG and GCTACATTGCCGTCTGCTTC was selected for hsa-mir-105-1, and the combination of GGTATCAGCTCATGTTGCAG and GGATGTTTGAGCATGTGCTA was selected for hsa-mir-105-2 based on their highest efficiency to induce genomic deletion of the flanking region in pilot experiments. At 24 h after transfection, GFP+ cells were sorted by FACS and monoclones were screened by genotyping PCR and confirmed by sequencing (Supplementary Fig. 4c). For miR-155 overexpression, the hsa-mir-155 gene was cloned by PCR using primers 5′-ACAAACCAGGAAGGGGAAAT and 5′-GCAGCAATTTGTTCCATGTG, and constructed into the EcoRI/SalI sites of pBABE-GFP retroviral vector. All plasmid constructs were verified by sequencing. miScript miRNA mimics and inhibitors as well as their corresponding negative controls were purchased from Qiagen (Hilden, Germany). The MYC siRNA and control siRNA were purchased from Santa Cruz Biotechnology (Dallas, TX) with detailed information provided in Supplementary Table 3. Cell transfection, reporter assays, production of viruses, infection and selection of transduced cells, flow cytometry for cell characterization, as well as transwell migration assays were carried out as previously described10, 11, 29.
EV purification and characterization
EVs secreted by cultured cells were prepared as previously reported10, 11. Conditioned media (CM) was first prepared by incubating cells grown at sub-confluence in growth media containing 10% EV-depleted FBS (prepared by overnight ultracentrifugation of medium-diluted FBS at 100,000 ×g at 4ºC) for 48 h, and pre-cleared by centrifugation at 500 ×g for 15 min and then at 10,000 ×g for 20 min. EVs were isolated by ultracentrifugation at 110,000 ×g for 70 min, and washed in PBS using the same ultracentrifugation conditions. When indicated, DiI (1,1′-Dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; Sigma-Aldrich; St. Louis, MO) was added into the PBS at 1 μM and incubated for 20 min before the washing spin, followed by an additional wash to remove the excess dye. The pelleted EVs were resuspended in ~100 μl of PBS, and subjected to nanoparticle tracking analysis (NTA) using a NanoSight NS300 (Malvern; Westborough, MA), iodixanol/OptiPrep gradient separation, RNA extraction by TRIZOL LS (Thermo Fisher Scientific), and treatment of cells and animals. For gradient separation we used a protocol modified from53–55. EVs isolated by ultracentrifugation were loaded onto a 12-step OptiPrep (Sigma-Aldrich) gradient consisted of 30, 27.5, 25, 22.5, 20, 17.5, 15, 12.5, 10, 7.5, 5, and 2.5% iodixanol in 20 mM Hepes (pH 7.2), 150 mM NaCl, 1 mM Na3VO4, and 50 mM NaF. After centrifugation in a SW 40 Ti rotor (Beckman Coulter; Indianapolis, IN) at 110,000 ×g at 4°C for 16 h, 12 1-mL fractions were collected and washed in PBS by another spin at 110,000 ×g for 70 min before Western analysis and RNA extraction for RT-qPCR. For cell treatment, 2 μg of EVs (equivalent to those collected from ~5×106 producer cells) based on protein measurement using Pierce™ BCA protein assay kit (Thermo Fisher Scientific) were added to 2×105 recipient cells. Dynasore was obtained from Sigma-Aldrich.
RNA extraction and quantitative reverse transcription PCR
These procedures were performed as described previously11, 29. Primers used in RT-qPCR are indicated in Supplementary Table 3. An annealing temperature of 55°C was used for all primers. U6 primer was used as an internal control for intracellular miRNA levels. As a spike-in control for EV miRNA levels, 20 fmole of synthetic cel-miR-39-3p was added to EVs from equal number of cells during RNA extraction and its levels were subsequently used for data normalization following miScript miRNA RT-qPCR assays (Qiagen).
Western blot analysis
Protein extracts were separated by electrophoresis on a 10% or 12% SDS polyacrylamide gel. Protein detection was performed using antibodies described in Supplementary Table 3.
Cell bioenergetics assays
ECAR and OCR were measured using an XFe24 extracellular flux analyser (Seahorse Bioscience; North Billerica, MA). Briefly, cells were seeded at 4×104 per well in 525 μL of XF assay medium supplemented with 2 mM glutamine (for ECAR) or 2 mM glutamine, 10 mM glucose, and 1 mM pyruvate (for OCR). Cells were incubated in a CO2-free incubator for 1 h at 37°C to allow for temperature and pH equilibration prior to loading into the XFe24 analyser for measurement following the manufacturer’s protocol. Basal glycolysis was calculated by subtracting the third baseline (non-glycolytic acidification) ECAR reading from the third glycolysis ECAR reading following glucose addition. Basal respiration was derived by subtracting the third OCR reading following antimycin A addition from the third basal OCR reading. For Mito Fuel Flex Test, inhibitors of glucose (UK5099), glutamine (BPTES), and fatty acid (etomoxir) pathways were applied following the manufacturer’s protocol. Fuel dependency is tested by first injecting an inhibitor of the target pathway, followed by inhibition of the two alternative pathways. Fuel capacity is tested by first injecting inhibitors of the alternative pathways, followed by inhibition of the target pathway. Fuel flexibility is calculated as the difference between capacity and dependency.
Medium metabolite measurements
Cells seeded at equal number were cultured in growth media containing 3 g/L glucose but no pyruvate for 72 h before CM was collected, cleared by centrifugation, and subjected to metabolite measurement using a BioProfile 100 Plus (Nova Biomedical; Waltham, MA). Media collected from cell-free plates after 72 h incubation was used as the baseline control to calculate the consumption or production of each metabolite, which was further normalized to the cell number in each plate determined at the time of CM collection.
Intracellular and extracellular pH measurements
Cells were incubated with 5 μM of the cell-permeant pH indicator BCECF AM (2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein, acetoxymethyl ester; Thermo Fisher Scientific) in Hank’s buffered salt solution (HBSS) for 25 min at 37°C. Intracellular pH was measured by determining the ratio of fluorescence intensities at 535 nm when excited at 490 nm and 440 nm, and comparing to a calibration curve. Extracellular pH was measured by a pH meter (Thermo Fisher Scientific).
Measurements of LDH, GDH, and GLUL enzymatic activities
LDH activity was assessed using a protocol modified from a previous study56 by measuring the reaction in two directions. The reaction of pyruvate converting to lactate was conducted at 25˚C in a mixture containing 50 mM phosphate buffer (pH 7.5), 0.18 mM NADH, 0.6 mM sodium pyruvate, and cell lysate to obtain a measurable decrease in absorbance at 340 nm using a Beckman Coulter DU70 UV-Vis spectrophotometer (Indianapolis, IN). For the reaction of lactate converting to pyruvate, the enzymatic activity was determined by measuring an increase in absorbance at 340 nm in a reaction mixture containing 50 mM Tris-HCl buffer (pH 9.5), 50 mM lithium lactate, 0.1 mM NAD+, and cell lysate. One unit of LDH activity was defined as the amount that consumed or produced 1 μmole of NADH per min under the assay conditions. GDH aminating activity was assessed by adding cell lysate to a reaction mixture containing 86 mM Tris-HCl buffer (pH 8.3), 7.6 mM α-KG, 0.22 M NH4Cl, and 0.25 mM NADH. The decrease in absorbance at 340 nm was recorded at 30°C in a spectrophotometer. Weight activity (mU/mg) was calculated by ΔOD/min for 0–5 min and normalized to cell pellet weight. One unit of GDH activity was defined as the amount that oxidized one μmole of NADH per min under the conditions. GLUL activity was measured following previously reported method37.
Metabolic profiling by NMR spectroscopy
All NMR analyses were performed at the City of Hope NMR Core facility. Sample preparation, 1D NMR spectroscopy, and data analysis were performed as described10. For experiments with 13C-labelled glucose and glutamine, CAFs pretreated with EVs for 48 h were incubated with 16.6 mM (3 g/L) D-glucose-13C6 (≥99%; Sigma-Aldrich) in medium free of 12C-glucose, or with 4 mM L-glutamine-13C5,15N2 (98%, 98%; Sigma-Aldrich) in medium free of 12C-glutamine. For experiments with 13C-labelled lactate, pretreated CAFs were cultured in glucose-free medium containing 10 mM lactate for 24 h, and then changed to glucose-free medium containing 10 mM sodium L-lactate-13C3 (≥99%; Sigma-Aldrich). Cells were analysed for metabolic profiles after 6 h. For CM analysis, cells were washed after incubating with the tracer for 6 h, and changed to the corresponding medium containing regular unlabelled (12C) metabolites; CM was collected after 24 h. To determine the abundances of 13C-containing metabolites, cells and the EV-depleted CM were extracted to collect the hydrophilic fraction, which was resuspended in 500 μL 100% D2O containing 5 mM sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS; Cambridge Isotope Laboratories; Tewksbury, MA) that serves as an internal chemical shift reference and a concentration standard in 2D spectra. NMR spectra were acquired at 25°C on a Bruker Avance spectrometer equipped with a cryoprobe operating at 600.19 MHz 1H frequency. One-dimensional NOESY (nuclear Overhauser effect spectroscopy) with presaturation spectra57 was collected with spectral width of 10 kHz, 32k data points, 3s relaxation delay, and 1,024 transients. Data was processed using Bruker topspin 3.1, and analysed with Chenomx NMR Suite Processor (version 7.5; Chenomx; Edmonton, Canada). Two-dimensional 1H-13C HSQC (Heteronuclear Single Quantum Coherence) was used to analyse cell samples and constant-time 1H-13C HSQC was used to analyse medium and tumour samples. The spectrum width for 1H and 13C are 16 ppm and 14 ppm, respectively. The acquisition time for 1H and 13C are 3.4 s and 122 ms, respectively. 1H-13C correlation spectra were processed using Bruker topspin 3.1, and analysed with Sparky software (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco).
Liquid chromatography coupled to high-resolution mass spectrometry (LC/HRMS)
CAFs seeded in 6-well plates at 2×105 cells per well were pretreated with EVs for 48 h before metabolic tracing. For lactate tracing, cells were cultured in glucose-free medium containing 10 mM lactate for 24 h, and then changed to glucose-free medium containing 10 mM sodium L-lactate-13C3 (≥99%; Sigma-Aldrich) for 6 h. For ammonium tracing, cells were cultured in glutamine/glutamate-free medium containing 5 mM 15NH4Cl (99%; Cambridge Isotope Laboratories) for 16 h. Cells were extracted and analysed by LC/HRMS for quantification of metabolites and mass isotopomer distributions as previously described58 except the following modifications. Mobile phase A was replaced with water containing 5 mM ammonium acetate (pH 6.8). Scan range (m/z) was slightly modified: 70 to 900 for pos mode (1.31 to 12.5 min) and neg mode (1.31 to 6.6 min) and 100 to 1000 for neg mode (6.61 to 12.5 min). LC/HRMS-detected metabolites are summarized in Supplementary Table 4.
Measurements of metabolite levels by kits
Levels of selected metabolites were measured by commercial kits to confirm the results of metabolic profiling. These include the acetate colourimetric assay kit (BioVision; Milpitas, CA), PicoProbe™ lactate fluorometric assay kit (BioVision), Glutamate-Glo™ assay (Promega), glutamine colourimetric assay kit (BioVision), EnzyChrom™ ammonia/ammonium assay kit (BioAssay Systems; Hayward, CA), Autokit Glucose (Wako Diagnostics; Richmond, VA), and NO colourimetric assay kit (BioVision). The manufacturers’ protocols were followed. CM-H2DCFDA was used as an indicator for reactive oxygen species (ROS) following the manufacturer’s protocol (Thermo Fisher Scientific).
Animals
All animal experiments were approved by the institutional animal care and use committees at the University of California San Diego and the Beckman Research Institute of the City of Hope. The study is compliant with all relevant ethical regulations regarding animal research. Female NOD/SCID/IL2Rγ-null (NSG) mice of 6–8 week old were used in this study. Xenograft tumours were established in NSG mice through mammary fat pad injection of 2×105 patient derived xenograft (PDX) BC cells, either alone or mixed with 1×106 CAFs stably expressing anti-miR-105 or control (CAF/anti-miR-105 or CAF/control), that were combined at 1:1 with Matrigel (BD Biosciences; San Jose, CA). Tumour volume was assessed by caliper measurements using the formula (width2×length)/2. A needle pH microelectrode and reference electrode (Microelectrodes; Bedford, NH) were used to measure intratumoural pH in core and periphery areas. Administration of stable isotope-labelled metabolites was performed when tumours reached ≈250 mm3. D-glucose-13C6 and L-glutamine-13C5,15N2 were individually administered via tail-vein infusion, whereas sodium L-lactate-13C3 (25 mM, 100 μl) was directly injected into the core of tumour. For tail-vein infusion, a 27G catheter was placed in the lateral tail vein under anesthesia. Using a syringe pump (Braintree Scientific; Braintree, MA), solutions of labelled glucose (90 g/L) or glutamine (40 mM) were infused at the speed of 700 μL/h over a total period of 30 min. Mice were sacrificed 1 h after infusion/injection; tumours were collected and metabolites extracted for 2D NMR metabolic profiling. For LC/HRMS, tumours were collected and dissociated as described29, sorted PDX cancer cells (previously labelled with GFP by lentiviral transduction) were analysed for the relative levels of metabolites. For the MCT inhibitor study, PDX/CAF xenograft tumours were established as described above. Mice were randomized within individual groups and started to receive MCT1/2 inhibitor AZD3965 (100 mg/kg; oral twice a day) or vehicle on day 40. On day 46, tumour extracts were analysed for 1D NMR metabolic profiling. In another experiment, 1×106 MDA-MB-231 cells combined with 1×106 NIH3T3/anti-miR-105 or NIH3T3/control cells were injected. For the EV treatment experiment, mice were injected with 1×105 MCFDCIS cells mixed with 1×106 CAFs that had been previously treated with EVs or PBS in vitro for 2 days. The treatment with EVs or PBS continued in mice at 2 μg EVs per injection at the tumour site and twice a week starting one day after cell transplantation until tumour collection. Sorted CAFs (previously labelled with mCherry by lentiviral transduction) were analysed for gene expression.
IHC, in situ hybridization (ISH), and immunofluorescence (IF)
BC tissue arrays were purchased from US Biomax (Cat #BR1505b; Rockville, MD). IHC and ISH were performed as previously described10, 11, 29. Stained slides were scored according to intensity of staining (−: 0; +: 1; ++: 2; and +++: 3) and percentage of the cells of interest staining positive for each antigen (0%: 0; 1~25%: 1; 26~50%: 2; 51~75%: 3; and 76~100%: 4). The intensity score was multiplied by the percentage score to obtain a final score, which was used in the statistical analyses. IF was performed as described11. Antibodies used are described in Supplementary Table 3.
RNA-seq and bioinformatics
RNA sequencing was performed by the City of Hope Integrative Genomics Core using RNA samples from CAFs treated with EVs derived from MDA-MB-231, MCF10A/miR-105, or MCF10A cells or with PBS, and from MCF10A cells stably overexpressing miR-105, MYC, GFP, or the vector (the latter two both as controls). RNA was sonicated and reverse-transcribed into cDNA, followed by end repair, A-tailing, and linker ligation. The ligated material was amplified by PCR and then analysed on a HiSeq2500 (Illumina; San Diego, CA) for parallel sequencing. Sequences were aligned to human genome assembly hg19. Bioconductor package “edgeR” 3.4.2 was used to normalize the data and calculate P value and log2 fold change between different groups59. For gene set enrichment analysis (GSEA), genes were ranked by the signed P value score, which is -log10(P) with sign of the log2 fold change. The preranked data were uploaded to GSEA 3.0 and enrichment of MSigDB C2 CGP (chemical and genetic perturbation) gene sets were interrogated with 1,000 random permutations to obtain the P value, q value and normalized enrichment score (NES). For Ingenuity pathway analysis (content version: 42012434; build: ing_pandora; Qiagen), genes altered by miR-105 overexpression in MCF10A, defined as fold change ≥2 with P<0.05 compared to the controls, were interrogated to predict the potential upstream regulators60. These genes were also interrogated by the ENCODE datasets61 using Enrichr online web server (2017 version)62, 63 to identify the transcription factors that could be involved in gene regulation.
Statistics and reproducibility
All quantitative data are presented as mean ± standard deviation (SD) unless otherwise specified. Statistical tests were performed using GraphPad Prism 7.01 and SPSS 22 unless otherwise specified. Two-sided Student’s t-tests were used for comparisons of the means of data between two groups. For multiple independent groups, one-way or two-way ANOVA with post-hoc Tukey tests were used. The correlations between IHC/ISH-determined gene expression levels were evaluated by Kendall’s tau correlation tests. Samples without tumour or stromal cells were removed from the correlation analysis. Values of P<0.05 were considered significant. Sample size was generally chosen based on preliminary data indicating the variance within each group and the differences between groups. For animal studies, sample size was predetermined to allow an 80% power to detect a difference of 50%. All samples/animals that have received the proper procedures with confidence were included for the analyses. Animals were randomized before treatments. For animal studies, the investigators were blinded to allocation during outcome assessment. All Western blots were repeated independently three times with similar results, and representative images are shown. ISH and IHC staining was repeated twice independently with similar results.
Data availability
All RNA-seq data generated in this study have been deposited into the NCBI Gene Expression Omnibus (GEO) with the accession code GSE106508. Previously published GEO dataset GSE50429 was re-analysed for miRNA levels in the cells and EVs of MDA-MB-231 and MCF10A in Supplementary Table 1. Previously published ENCODE ChIP-Seq data that were re-analysed and shown in Supplementary Table 2 are available under GEO accession codes GSE33213, GSE30399, GSE12782, GSE25416, GSE28286, GSE12781, GSE31363, GSE31477, and GSE32465. NMR spectrometry data for metabolic profiling have been deposited to the EMBL-EBI MetaboLights database64 with the identifier MTBLS626 (https://www.ebi.ac.uk/metabolights/MTBLS626). Source data for Figs. 1c,f–h,j,k, 2a–f,h, 3b,c,i, 4a,b, 5a–c,g,i, 6b–d,f,g, 7a,f,g,h,j–l, 8b, and Supplementary Figs. 2a,b,d,e, 5a,b,d, 6a–d, 7a–c, 8a,b have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH)/National Cancer Institute (NCI) grants R01CA218140 (SEW), R01CA166020 (SEW), R01CA206911 (SEW), and R01CA163586 (SEW), California Breast Cancer Research Program grant 20IB-0118 (SEW), Breast Cancer Research Foundation-AACR grant 12-60-26-WANG (SEW), and by the National Key Technology R&D Program of China Grant 2015BAI12B12 (XR). Research reported in this publication included work performed in Core facilities supported by the NIH/NCI under grant number P30CA23100 (UCSD Cancer Center) and P30CA33572 (City of Hope Cancer Center). We acknowledge the ENCODE Consortium and the ENCODE production laboratories generating the datasets used herein for the analysis.
Footnotes
Author Contributions
S.E.W. and W.Y. conceived ideas, and J.W.L., Y.C., and X.R. contributed to project planning. W.Y. and S.E.W. designed and performed most of the experiments. X.W. performed all bioinformatics analyses. W.Z. performed some experiments with the MCF10A-derived lines. M.Y.F., M.C., J.W., and L.L. assisted with cell line construction and mouse experiments. X.L. and A.R.C. assisted with the nanoparticle tracking analysis and EV gradient separation. J.L., X.L., and J.W.L. performed LC/HRMS and data analysis. C.H.C. and Y.C. assisted with NMR analysis. M.Y.F., D.P.P., and O.F. performed IHC and pathological evaluation. S.E.W. and W.Y. wrote the manuscript.
Competing Interests
The authors declare no competing financial interests.
Additional Information
Supplementary Information is included in the submitted version of the paper.
References
- 1.Barcellos-Hoff MH, Lyden D, Wang TC. The evolution of the cancer niche during multistage carcinogenesis. Nat Rev Cancer. 2013;13:511–518. doi: 10.1038/nrc3536. [DOI] [PubMed] [Google Scholar]
- 2.Chin AR, Wang SE. Cancer Tills the Premetastatic Field: Mechanistic Basis and Clinical Implications. Clin Cancer Res. 2016;22:3725–3733. doi: 10.1158/1078-0432.CCR-16-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valadi H, et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 4.Skog J, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 6.ELA S, Mager I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nature reviews Drug discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 7.Becker A, et al. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016;30:836–848. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Webber J, Steadman R, Mason MD, Tabi Z, Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 10.Fong MY, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183–194. doi: 10.1038/ncb3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W, et al. Cancer-Secreted miR-105 Destroys Vascular Endothelial Barriers to Promote Metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chow A, et al. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-kappaB. Scientific reports. 2014;4:5750. doi: 10.1038/srep05750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redzic JS, Balaj L, van der Vos KE, Breakefield XO. Extracellular RNA mediates and marks cancer progression. Semin Cancer Biol. 2014;28:14–23. doi: 10.1016/j.semcancer.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin AR, Wang SE. Cancer-derived extracellular vesicles: the ‘soil conditioner’ in breast cancer metastasis? Cancer Metastasis Rev. 2016;35:669–676. doi: 10.1007/s10555-016-9639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol. 2015 doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le MT, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J Clin Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang G, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31:3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tominaga N, et al. Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier. Nat Commun. 2015;6:6716. doi: 10.1038/ncomms7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu X, et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J Transl Med. 2012;10:42. doi: 10.1186/1479-5876-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 21.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, Metabolism, and Cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavlides S, et al. The reverse Warburg effect: aerobic glycolysis in cancer associated fibroblasts and the tumor stroma. Cell Cycle. 2009;8:3984–4001. doi: 10.4161/cc.8.23.10238. [DOI] [PubMed] [Google Scholar]
- 25.Zhang D, et al. Metabolic reprogramming of cancer-associated fibroblasts by IDH3alpha downregulation. Cell reports. 2015;10:1335–1348. doi: 10.1016/j.celrep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Lisanti MP, Martinez-Outschoorn UE, Sotgia F. Oncogenes induce the cancer-associated fibroblast phenotype: metabolic symbiosis and “fibroblast addiction” are new therapeutic targets for drug discovery. Cell Cycle. 2013;12:2723–2732. doi: 10.4161/cc.25695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Outschoorn UE, Lisanti MP, Sotgia F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin Cancer Biol. 2014;25:47–60. doi: 10.1016/j.semcancer.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Martins D, et al. Loss of caveolin-1 and gain of MCT4 expression in the tumor stroma: key events in the progression from an in situ to an invasive breast carcinoma. Cell Cycle. 2013;12:2684–2690. doi: 10.4161/cc.25794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsuyada A, et al. CCL2 mediates cross-talk between cancer cells and stromal fibroblasts that regulates breast cancer stem cells. Cancer Res. 2012;72:2768–2779. doi: 10.1158/0008-5472.CAN-11-3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zervos AS, Gyuris J, Brent R. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber-Agus N, et al. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995;80:777–786. doi: 10.1016/0092-8674(95)90356-9. [DOI] [PubMed] [Google Scholar]
- 32.Ayer DE, Lawrence QA, Eisenman RN. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- 33.Lee TC, Ziff EB. Mxi1 is a repressor of the c-Myc promoter and reverses activation by USF. J Biol Chem. 1999;274:595–606. doi: 10.1074/jbc.274.2.595. [DOI] [PubMed] [Google Scholar]
- 34.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harbor perspectives in medicine. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L, et al. Targeting Stromal Glutamine Synthetase in Tumors Disrupts Tumor Microenvironment-Regulated Cancer Cell Growth. Cell Metab. 2016;24:685–700. doi: 10.1016/j.cmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bott AJ, et al. Oncogenic Myc Induces Expression of Glutamine Synthetase through Promoter Demethylation. Cell Metab. 2015;22:1068–1077. doi: 10.1016/j.cmet.2015.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faubert B, et al. Lactate Metabolism in Human Lung Tumors. Cell. 2017;171:358–371. e359. doi: 10.1016/j.cell.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hui S, et al. Glucose feeds the TCA cycle via circulating lactate. Nature. 2017;551:115–118. doi: 10.1038/nature24057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spinelli JB, et al. Metabolic recycling of ammonia via glutamate dehydrogenase supports breast cancer biomass. Science. 2017 doi: 10.1126/science.aam9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao H, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. doi: 10.7554/eLife.10250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valencia T, et al. Metabolic reprogramming of stromal fibroblasts through p62-mTORC1 signaling promotes inflammation and tumorigenesis. Cancer Cell. 2014;26:121–135. doi: 10.1016/j.ccr.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booth AM, et al. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006;172:923–935. doi: 10.1083/jcb.200508014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnaiah V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells. Proc Natl Acad Sci U S A. 2013;110:13109–13113. doi: 10.1073/pnas.1221899110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahlert C, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.San Lucas FA, et al. Minimally invasive genomic and transcriptomic profiling of visceral cancers by next-generation sequencing of circulating exosomes. Ann Oncol. 2016;27:635–641. doi: 10.1093/annonc/mdv604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thakur BK, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016. doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balaj L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 51.Debnath J, et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- 52.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tauro BJ, et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 54.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A. 2016;113:E968–977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X, et al. Na+-H+ exchanger 3 (NHE3) is present in lipid rafts in the rabbit ileal brush border: a role for rafts in trafficking and rapid stimulation of NHE3. J Physiol. 2001;537:537–552. doi: 10.1111/j.1469-7793.2001.00537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karamanos Y. Purification and Characterisation of Lactate Dehydrogenase: An Undergraduate Biochemistry Laboratory Experiment. Advances in Biochemistry. 2014;2:14–23. [Google Scholar]
- 57.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Romero IL, Litchfield LM, Lengyel E, Locasale JW. Metformin Targets Central Carbon Metabolism and Reveals Mitochondrial Requirements in Human Cancers. Cell Metab. 2016;24:728–739. doi: 10.1016/j.cmet.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kramer A, Green J, Pollard J, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. Bmc Bioinformatics. 2013;14 doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haug K, et al. MetaboLights--an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res. 2013;41:D781–786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data generated in this study have been deposited into the NCBI Gene Expression Omnibus (GEO) with the accession code GSE106508. Previously published GEO dataset GSE50429 was re-analysed for miRNA levels in the cells and EVs of MDA-MB-231 and MCF10A in Supplementary Table 1. Previously published ENCODE ChIP-Seq data that were re-analysed and shown in Supplementary Table 2 are available under GEO accession codes GSE33213, GSE30399, GSE12782, GSE25416, GSE28286, GSE12781, GSE31363, GSE31477, and GSE32465. NMR spectrometry data for metabolic profiling have been deposited to the EMBL-EBI MetaboLights database64 with the identifier MTBLS626 (https://www.ebi.ac.uk/metabolights/MTBLS626). Source data for Figs. 1c,f–h,j,k, 2a–f,h, 3b,c,i, 4a,b, 5a–c,g,i, 6b–d,f,g, 7a,f,g,h,j–l, 8b, and Supplementary Figs. 2a,b,d,e, 5a,b,d, 6a–d, 7a–c, 8a,b have been provided as Supplementary Table 5. All other data supporting the findings of this study are available from the corresponding author on reasonable request.