Abstract
Precise regulation of the amplitude and duration of receptor tyrosine kinase (RTK) signaling is critical for the execution of cellular programs and behaviors. Understanding these control mechanisms has important implications for the field of developmental biology, and in recent years, the question of how augmentation or attenuation of RTK signaling via feedback loops modulates development has become of increasing interest. RTK feedback regulation is also important for human disease research; for example, germline mutations in genes that encode RTK signaling pathway components cause numerous human congenital syndromes, and somatic alterations contribute to the pathogenesis of diseases such as cancers. In this review, we survey regulators of RTK signaling that tune receptor activity and intracellular transduction cascades, with a focus on the roles of these genes in the developing embryo. We detail the diverse inhibitory mechanisms utilized by negative feedback regulators that, when lost or perturbed, lead to aberrant increases in RTK signaling. We also discuss recent biochemical and genetic insights into positive regulators of RTK signaling and how these proteins function in tandem with negative regulators to guide embryonic development.
Keywords: Receptor tyrosine kinases, signaling pathways, feedback regulation, developmental control, RASopathies, Sprouty
INTRODUCTION
Receptor tyrosine kinases (RTKs) regulate virtually all aspects of embryonic development from early patterning to organogenesis (Lemmon and Schlessinger, 2010; Li and Hristova, 2006). The RTK superfamily encompasses 58 known receptors in humans which are classified into several multi-member subfamilies including, among others, fibroblast growth factor receptors (FGFRs), insulin and insulin-like growth factor receptors (IR and IGF-1R), platelet-derived growth factor receptors (PDGFRs), vascular endothelial growth factor receptors (VEGFRs), and epidermal growth factor receptors (EGFR/HER/ERBBs) (Lemmon and Schlessinger, 2010). Together, these receptors are involved in the entire spectrum of developmental processes. The intracellular signals initiated by RTK activation play pivotal roles in cell fate determination and morphogenesis, and many are highly conserved in evolution from the nematode Caenorhabditis elegans to humans (Pires-daSilva and Sommer, 2003). Furthermore, numerous diseases result from germline or somatic genetic changes that alter the activity, abundance, or cellular distribution of RTKs. Mutations in RTKs or proteins that facilitate their downstream signaling have been implicated in the onset and progression of a wide-range of diseases such as diabetes, inflammation, bone disorders, atherosclerosis, angiogenesis, and various cancers (Lemmon and Schlessinger, 2010).
RTK activation is triggered by binding of extracellular ligands, which leads to receptor oligomerization and auto-phosphorylation on tyrosine residues within the cytoplasmic domains. These phosphorylated residues create docking sites for phosphotyrosine-binding domain-containing proteins that couple RTK activation to downstream signaling pathways (Hubbard, 2004; Hubbard and Miller, 2007; Schlessinger, 2000). Interestingly, a large number of RTKs induce a similar set of downstream effectors, in particular those coupled to activation of the RAS/MAP kinase (MAPK) and phosphatidylinositide-3 kinase (PI3K)/AKT pathways (Blume-Jensen and Hunter, 2001; Ledda and Paratcha, 2007). What distinguishes the signaling outputs between distinct RTKs is often the duration and extent of pathway activation. Feedback regulators play a major role in fine-tuning these variables by attenuating or amplifying the signaling output. They can be already present and act prior to or immediately after receptor activation (early attenuators) (Haglund et al., 2003; Thien and Langdon, 2001) or can be transcriptionally induced by the pathways they eventually inhibit (late attenuators) (Table 1) (Casci and Freeman, 1999; Fiorini et al., 2002; Ghiglione et al., 1999; Golembo et al., 1996; Korsensky and Ron, 2016; Tsang and Dawid, 2004).
Table 1.
Classification of RTK signaling modulators according to their spatiotemporal feedback
| Feedback | Modulator | Target/Pathway | Signaling Output |
Mechanism of Action |
|---|---|---|---|---|
| Late, reversible | Anosmin 1 | FGFR | Amplification | FGF-FGFR signaling complex assembly and stabilization |
| DUSP6 | FGF-MAPK | Attenuation | Dephosphorylation of ERK | |
| FLRT family | FGFR | Amplification | FGF-FGFR signaling complex activation | |
| MIG6 | ErbB receptor family | Attenuation | Inhibition of ErbB receptor family dimerization | |
| SEF | FGF-MAPK/ERK & FGF-PI3K/AKT | Attenuation | FGF-FGFR signaling complex assembly and activation | |
| SPRED | FGF-RAS/RAF | Attenuation | Inhibition of RAF activation | |
| Sprouty family | RAS/MAPK & RAS/PI3K | Attenuation | Inhibition of downstream effector signaling | |
| Early, reversible | PTEN | PI3K/AKT | Attenuation | Dephosphorylation of PIP3 |
| RKIP | RAF/MEK | Attenuation | Inhibition of RAF1 and MEK interaction | |
| SHP2 | EGFR, FGF-MAPK/ERK, IR, & RET | Amplification | Dephosphorylation of inhibitory modulators of downstream effectors | |
| Late, irreversible | CNPY1 | FGFR | Amplification | FGFR maturation |
| LRIG family | ErbB receptor family, MET, & RET | Attenuation | Receptor ubiquitination and degradation | |
| SOCS family | c-KIT, EGFR, FLT3, IGF-1R, & IR | Attenuation | Receptor ubiquitination and degradation | |
| Early, irreversible | CBL family | EGFR, MET, PDGFR, & RET | Attenuation | Receptor ubiquitination and degradation |
| NEDD4 & NEDD4L | ErbB receptor family, FGFR, IGF-1R, IR, NRTK1, & VEGFR | Attenuation | Receptor ubiquitination and degradation | |
| NRDP1 | ERBB3 | Attenuation | ERBB3 trafficking and ubiquitination | |
| SHISA2 | FGFR | Attenuation | Receptor maturation and ubiquitination |
Feedback regulators have also been shown to further control RTK-mediated signaling by modulating the localization of the ligands necessary for RTK activation. One of the characteristic features of RTK signaling in development is the gradation of signal output (Ashe and Briscoe, 2006). This gradation is traditionally thought to arise from a corresponding gradient in ligand concentration or receptor expression. However, it has recently been shown that the presence of regulators significantly contributes to this signal gradient. Indeed, the heparin sulfate proteoglycans (HSPGs), which serve as an amplifier of many RTK signaling pathways, most notably FGFRs, can control morphogen gradient formation by regulating the diffusion rates of ligands (Yan and Lin, 2009). Another example of such a mechanism is the action of CBL, a well-characterized negative feedback regulator of multiple RTK pathways such as EGFR, MET, and RET. This protein regulates the distribution of the Egfr ligand Gurken during Drosophila melanogaster embryogenesis by mediating endocytosis and subsequent degradation of the Egfr-Gurken complex (Chang et al., 2008). In this manner, feedback regulators are essential for not only controlling the level of signal output but also for tuning their spatiotemporal localization.
Here, we review several modulators of RTK signaling with an emphasis on those with known roles in development (Supplemental Table 1) and contributions to human congenital disorders (Figure 5). Our discussion highlights the insights gained from in vivo work in model organisms that can be used to further our biochemical understanding of RTK regulation through feedback pathways. Several other excellent reviews cover feedback regulators in more molecular detail (Avraham and Yarden, 2011; Lemmon et al., 2016; Mohapatra et al., 2013).
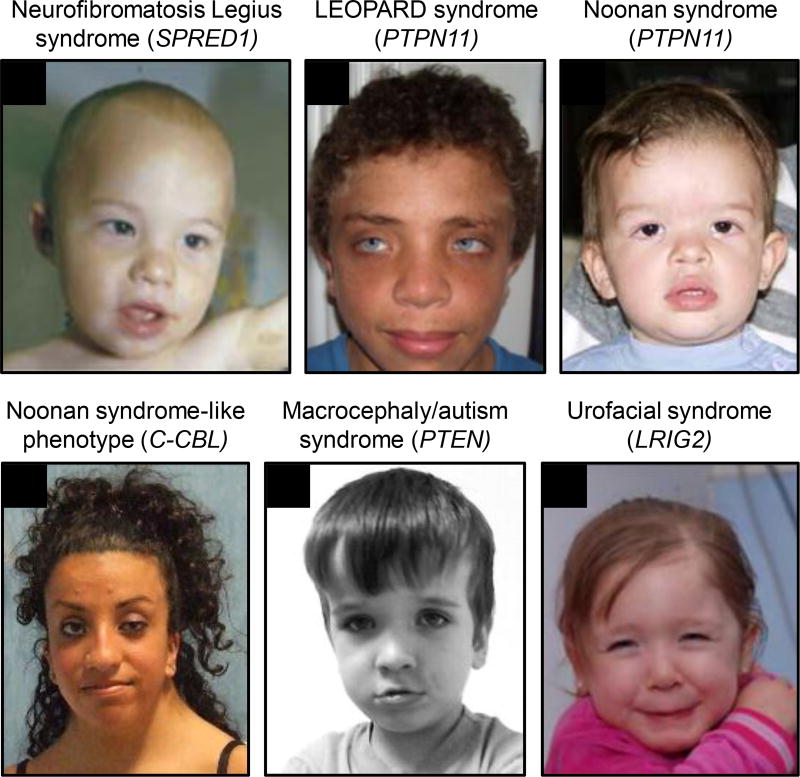
Figure 5. Characteristic features of craniofacial disorders associated with regulators of RTK signaling.
(A) Mild hypertelorism, epicanthic folds, broad nasal tip, full lips and café-au-lait spot on the left upper arm of a child who has Neurofibromatosis Legius syndrome with a SPRED1 mutation (Brems et al., 2007). (B) Dysmorphic features including hypertelorism, downslanting palpebral fissures, epicanthus, coarse facial features, and large, thick, low-set ears of an adolescent boy who has LEOPARD syndrome with a PTPN11 mutation (Santoro et al., 2014). (C) Characteristic craniofacial features including hypertelorism with downslanting palpebral fissures, full or ptotic upper eyelids, and low-set, posteriorly rotated ears with a thickened helix of a young boy who has Noonan syndrome with a PTPN11 mutation (Allanson et al., 2010). (D) Distinctive facial features including hypertelorism, ptosis, downslanting palpebral fissures, epicanthal folds, and low-set, posteriorly rotated ears of a woman who has Noonan syndrome-like phenotype with a C-CBL mutation (Martinelli et al., 2010). (E) Bilateral plantar creases and a flat appearing mid-face with a prominent forehead of a boy who has Macrocephaly/autism syndrome with a PTEN mutation (Butler et al., 2005). (F) Inversion of facial expression when smiling in a young girl who has Urofacial syndrome with a LRIG2 mutation (Stuart et al., 2013). Reprinted or adapted with permission.
Regulation of biosynthesis and maturation of RTKs
Recent studies have revealed that modulation of RTKs’ signaling outputs starts even prior to their arrival at the cell surface. A key aspect of this type of regulation occurs via quality control checks in the endoplasmic reticulum (ER), where newly synthesized receptors undergo folding and maturation by post-translational modification before being trafficked to the cell membrane. RTKs that are not properly synthesized, folded, or modified are degraded through a proteasome-dependent pathway. Several proteins, including the Canopy family, the Shisa family, and NRDP1, have recently been identified to regulate the strength of RTK-mediated signaling through interaction with and modification of receptors in the ER, consequently controlling the number of functional receptors at the cell surface.
The Canopy (CNPY) genes encode four putative ER-resident proteins hypothesized to be positive-feedback regulators of receptor maturation and trafficking (Do et al., 2012; Hart and Tapping, 2012; Hirate and Okamoto, 2006; Matsui et al., 2011), however, only a few studies to date have addressed their structure or function. In Danio rerio (zebrafish), cnpy1 expression is restricted to the midbrain-hindbrain boundary and can be induced by exogenous Fgf8. Knockdown of cnpy1 resulted in midbrain-hindbrain boundary defects with the appearance of an airplane “canopy” and impaired Fgf signaling in a cell-autonomous manner, indicating a positive-feedback relationship between cnpy1 and fgf8 (Hirate and Okamoto, 2006). Cnpy1 was also shown to positively regulate Fgf signaling for proper formation of Kupffer’s vesicle, which orchestrates left–right asymmetric body plan in zebrafish (Matsui et al., 2011). The closely related CNPY2 protein has been linked to FGF signaling in vitro: in mouse macrophages and human hepatocytes, FGF21 enhanced expression of Cnpy2, which resulted in stabilized expression of low-density lipoprotein receptors (Do et al., 2012). More recent work has identified CNPY2 as a HIF-1α-regulated angiogenic secreted factor that stimulates cell proliferation, migration, and angiogenesis in mouse models of cardiovascular pathologies and cancer (Guo et al., 2015b; Guo et al., 2015c; Ito et al., 2014; Taniguchi et al., 2017; Yan et al., 2016). Although these processes are known to be mediated by FGF signaling, no genetic interactions between CNPY2 and FGFs have been reported in vivo. While the involvement of Canopy proteins in FGFR signaling remains poorly understood, CNPY3 and CNPY4 were shown to regulate the toll-like receptors (TLRs), another class of single-pass transmembrane receptors. Co-expression of CNPY3 increased trafficking of exogenously expressed TLRs to the cell membrane via chaperone gp96, leading to elevated TLR-mediated signaling in vitro (Hart and Tapping, 2012). Interestingly, CNPY4 seems to exert an opposite effect and led to the downregulation of TLRs at the cell membrane, subsequently attenuating TLR-mediated signaling. Canopy proteins may play a similar regulatory role on RTKs to control trafficking, as CNPY1 directly interacts with FGFR1 and modulates the extent of mature N-linked glycosylation of the receptor (Matsui et al., 2011), however, further studies will be necessary to understand how Canopy proteins interact with and modulate RTK-mediated signaling.
In a manner potentially similar to the Canopy proteins, the nine Shisa proteins in vertebrates represent a novel class of ER-associated proteins that antagonize FGF-mediated signaling in a cell-autonomous manner by regulating receptor maturation. The founding member of the Shisa family, shisa1, was named based on its expression in the prospective head ectoderm and organizer in Xenopus laevis and in reference to a form of Japanese sculpture with a large head (Yamamoto et al., 2005). Misexpression of shisa1 in X. laevis resulted in enlarged cement glands and anterior head structures due to expansion of otx2 expression, which marks prospective forebrain and midbrain. Accordingly, morpholino knockdown of shisa1 reduced Fgf-mediated xbra expression at the mid-gastrula, and embryos exhibited small eyes and cement glands, suggesting that Shisa1 directs anterior-posterior axis formation through Fgf activity (Yamamoto et al., 2005). Subsequent studies of X. laevis, Gallus gallus (chick), and Mus musculus (mouse) showed that Shisa2 expression along the anteroposterior axis exerts negative regulatory effects on FGF signaling, suggesting that SHISA2 also plays a key role in the proper establishment of segmental patterning of the head (Supplemental Table 1) (Filipe et al., 2006; Furushima et al., 2007; Hedge and Mason, 2008; Nagano et al., 2006). Although the mechanism of action remains unclear, it has been suggested that the Shisa family members bind immature forms of receptors and utilize a conserved PY motif to interact with WW-domain-containing proteins such as the E3 ubiquitin ligase family of NEDD4 proteins, which are discussed below. In doing so, the Shisa proteins aid in bringing these proteins into proximity with immature forms of FGFR in the ER, resulting in ubiquitination of the receptor for retention and degradation (Pei and Grishin, 2012; Yamamoto et al., 2005).
Another example of a modulator of trafficking is the ubiquitin ligase NRDP1 which regulates ERBB3, the catalytically inactive (pseudokinase) member of the ErbB receptor family (Qiu and Goldberg, 2002). In zebrafish, nrdp1 is expressed in the neural crest, nervous system, and muscle during embryogenesis and significantly overlaps with expression of ERBB3, suggesting functional cooperation (Britsch et al., 1998; Lyons et al., 2005; Maddirevula et al., 2011). Knockdown of nrdp1 resulted in decreased expression of melanoblast markers and caused a significant reduction in pigmentation of embryos, a process driven by ERRB3 signaling (Maddirevula et al., 2011). As a RING finger-type ubiquitin ligase, NRDP1 regulates ERBB3 by controlling the abundance of receptor trafficked to the cell surface through constitutive ubiquitination of newly synthesized ERBB3 in the ER (Fry et al., 2011). The mechanism by which the cell regulates NRDP1 activity to fine-tune the precise level of receptor at the membrane was recently found to involve RTNA4. This member of the reticulon family of proteins, which control curvature of ER membranes, counteracts the NRDP1-dependent degradation of ERBB3 by sequestering NRDP1 into ER tubules. As a result, more ERBB3 can be trafficked to the cell surface, where it may engage growth factors and its co-receptors to initiate downstream signaling (Hatakeyama et al., 2016).
Regulation of ligand-receptor signaling complex formation
A single RTK can bind multiple different ligands, and a single ligand can bind to multiple receptors. The specificity of these interactions is primarily driven by relative ligand/receptor affinities and effective concentration of both the receptor and the ligand. While the abundance of the receptor is controlled primarily at the level of biosynthesis and internalization, the pool of available ligands can be significantly influenced by extracellular regulators. For example, HSPGs tightly bind growth factors to limit diffusion in the extracellular matrix and therefore increase their local concentration to drive paracrine signaling by FGF, EGF, MET, VEGF, and PDGF (Abramsson et al., 2007; Cecchi et al., 2012; Fager et al., 1992; Forsten and Schneider, 2005; Gengrinovitch et al., 1999; Rapraeger et al., 1991; Yayon et al., 1991). Other regulators operate intracellularly at the level of the receptor but also modulate the extent of productive ligand/receptor interactions. Recent studies have increased our understanding of such modulators and expanded our knowledge of similar types of feedback regulators beyond HSPGs. Here we discuss Anosmin 1, FLRT3, and SEF; and MIG6, which are FGFR and EGFR pathway-specific protein modulators, respectively, that interact with ligand-receptor signaling complexes to mediate assembly and activation.
The Anosmin 1 gene encodes an extracellular matrix-associated protein that is largely conserved from invertebrates to primates (de Castro et al., 2016), however, no ANOS1 ortholog has been identified in mouse and rat (de Castro et al., 2014). Therefore, the biological functions of Anosmin 1 have primarily been probed by overexpression of human Anosmin 1 in mouse and rat neurons, which led to effects on cell adhesion and migration and neurite outgrowth and branching (Bribian et al., 2008; Garcia-Gonzalez et al., 2016; Soussi-Yanicostas et al., 2002; Soussi-Yanicostas et al., 1998). In development, these processes contribute to cranial neural crest formation and several aspects of neurogenesis, as shown by in vitro and in vivo studies in C. elegans, D. melanogaster, chick, and zebrafish (Supplemental Table 1) (Di Schiavi and Andrenacci, 2013; Endo et al., 2012; Gianola et al., 2009; Murcia-Belmonte et al., 2016). Anosmin 1 enhances FGF2 signaling specifically through FGFR1 in a heparin sulfate (HS)-dependent manner (Bribian et al., 2006; Gonzalez-Martinez et al., 2004). Heparin-bound Anosmin 1 binds to a pre-formed FGF2/FGFR1 complex via extracellular FnIII domains to stabilize the complex, resulting in receptor activation (Figure 1A) (Cariboni et al., 2004; Hu et al., 2009). In both C. elegans and D. melanogaster, perturbation of the FnIII domains ablated biological activity of Anosmin 1 (Andrenacci et al., 2006; Bulow and Hobert, 2004).
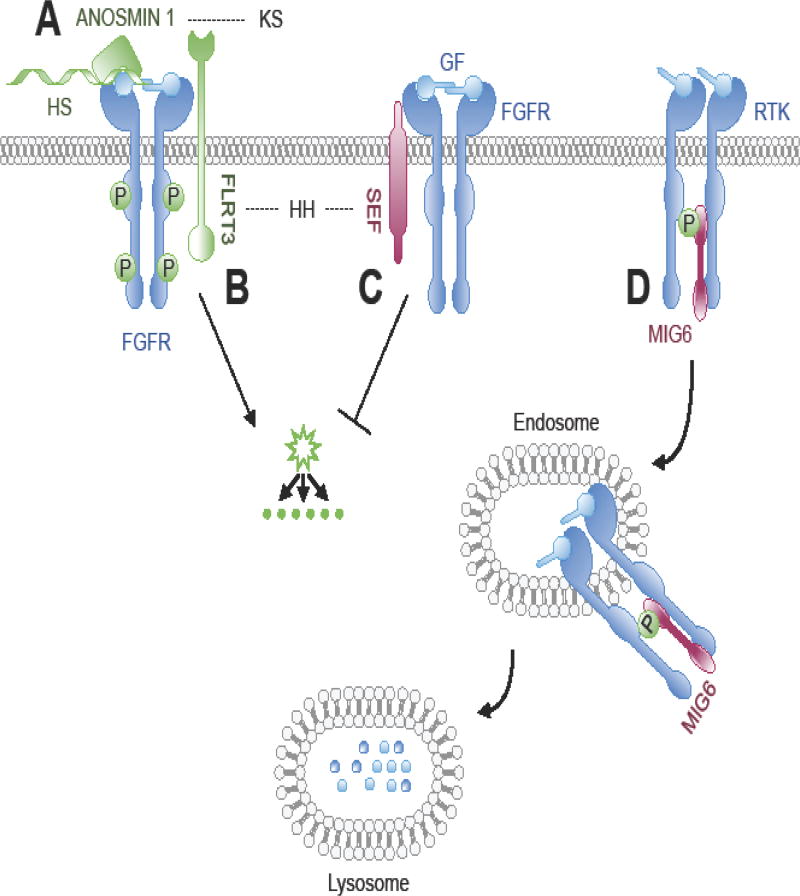
Figure 1. Modulation of RTK signaling by regulation of the ligand-receptor signaling complex formation.
(A) Heparin-bound Anosmin 1 binds to a pre-formed FGF2/FGFR1 complex, promoting its assembly and resulting receptor activation. FGFR signaling induces expression of FLRT3 and SEF via transcriptional activation and translation. (B) FLRT3 complexes with FGFR to promote downstream signaling of the MAPK/ERK pathway via its intracellular domain. (C) SEF complexes with FGFR and blocks receptor phosphorylation and activation of the RAS/MAPK and PI3K/AKT signaling cascades. (D) EGFR signaling induces MIG6 expression via transcriptional activation and translation. MIG6 accumulates in the cytoplasm where it binds directly with the ligand-activated ErbB kinase domain to inhibit auto-phosphorylation. This interaction can direct trafficking of the MIG-bound EGFR from the plasma membrane to late endosomes, targeting the receptor for lysosomal degradation. Dashed lines connecting the human congenital disorder with the protein in the pathway encoded by the causative mutated gene. Syndromes noted in the text and/or Supplemental Table 1. HH, hypogonadotropic hypogonadism with or without anosmia; KS, Kallmann syndrome; FGFR, Fibroblast Growth Factor Receptor; GF, growth factor; HS, heparin-sulfate; P, phosphorylation; RTK, receptor tyrosine kinase.
The three Fibronectin-like domain-containing Leucine-rich Transmembrane (FLRT) genes encode a highly conserved family of glycosylated proteins that mediate cell recognition and FGF signaling in vertebrates in a manner that is distinct from HSPGs and Anosmin 1. Flrt3 was originally identified in X. laevis as a gene with a similar expression pattern to Fgf signaling molecules, particularly at the midbrain/hindbrain boundary (Bottcher et al., 2004). Gain- and loss-of-function of flrt3 or flrt2 phenocopy experiments that perturb Fgf signaling, including effects on gastrulation, microcephaly, anterior truncations, and induction of ectopic tail-like structures (Bottcher et al., 2004; Cho et al., 2013). In chick, flrt3 is necessary but not sufficient for proper formation of the limb organizer called the apical epidermal ridge (AER) and co-localized with fgf8 expression and Erk activity (Tomas et al., 2011). Flrt3 knockout mice are embryonic lethal due to fusion defects and impaired definitive endoderm migration, phenotypes attributed to FLRT3’s function as a cell-adhesion molecule (Egea et al., 2008; Karaulanov et al., 2006; Maretto et al., 2008; Tsuji et al., 2004). X. laevis biochemical analyses in vivo and in vitro revealed that FLRT proteins complex with FGFRs to promote downstream signaling of the MAPK/ERK pathway via their intracellular domain (Figure 1B) (Bottcher et al., 2004). Although rodent FLRT3 similarly physically interacts with FGFR1 (Haines et al., 2006), deletion of Flrt3 in mice had no effect on Fgf8 expression or the expression of known Fgf targets, despite expression of Flrt3 in well-known Fgf signaling centers such as the AER, the midbrain-hindbrain boundary, and the anterior visceral endoderm (Egea et al., 2008; Haines et al., 2006; Maretto et al., 2008). Taken together, these studies suggest that the degree of conservation of the FGF/FLRT3 positive feedback loop varies among species (Supplemental Table 1). Since Flrt3 null mice die at early stages of development, it will be worthwhile to investigate whether FLRT3 modulates FGF signaling at later stages using conditional knockout mice.
Similar to flrt3 in X. laevis, sef (similar expression to fgf gene) was originally identified in zebrafish as a gene whose expression domains overlapped with known signaling centers of Fgfs (Furthauer et al., 2002; Tsang et al., 2002). Loss- or gain-of-function of sef in zebrafish led to various developmental defects, including cephalic malformations, cyclopia, expansion of ventrally derived domains, and reduction of the dorsal-most mesoderm (Furthauer et al., 2002; Tsang et al., 2002). In X. laevis, misexpression of zebrafish sef in the ventral marginal region at the 4-cell stage resulted in posterior truncations and gastrulation defects and was accompanied by suppression of Fgf target genes (Tsang et al., 2002). SEF transcripts have since been detected in zebrafish, chick, and mouse in numerous structures, including somites, the developing brain, limbs, and fin buds (Supplemental Table 1) (Boros et al., 2006; Furthauer et al., 2002; Harduf et al., 2005; Lin et al., 2002; Tsang et al., 2002). Surprisingly, Sef null mice are viable and fertile and do not show any obvious morphological phenotype during embryonic development (Abraira et al., 2007; Lin et al., 2005; Mellett et al., 2015). Lack of severe defects in the Sef mutant mice may be due to compensatory effects by other feedback antagonists. Indeed, Sef and the similarly FGF-induced Sprouty genes, discussed below, are expressed in overlapping regions along the anterior–posterior axis of the mouse embryo (Furthauer et al., 2002; Lin et al., 2002; Minowada et al., 1999).
The prototypic sef in zebrafish encodes a transmembrane receptor-like glycoprotein that blocks phosphorylation of Fgfr and subsequent activation of the Ras/Mapk and PI3K/Akt signaling cascades (Figure 1C) (Furthauer et al., 2002; Harduf et al., 2005; Kovalenko et al., 2006; Kovalenko et al., 2003; Preger et al., 2004; Tsang et al., 2002; Xiong et al., 2003; Yang et al., 2003). In vitro studies with mammalian SEF not only replicated the FGFR-induced antagonism seen in other species but also revealed that SEF can inhibit signaling activated by other growth factors, including EGF, PDGF, and nerve growth factor (NGF) (Kovalenko et al., 2003; Preger et al., 2004; Ren et al., 2008; Torii et al., 2004; Ziv et al., 2006). Interestingly, alternative spliced isoforms of SEF have been identified in humans (Preger et al., 2004; Rong et al., 2007; Ziv et al., 2006). SEF-a is similar to the prototypic SEF reported in zebrafish and mice (Furthauer et al., 2002; Lin et al., 2002; Tsang et al., 2002; Yang et al., 2003), whereas SEF-b, which lacks a signal peptide for secretion, is localized to the cytoplasm and acts at the level of, or downstream from, MEK (Figure 2F) (Preger et al., 2004; Yang et al., 2003; Ziv et al., 2006). Although both isoforms interact with FGFR1, the outcome of this association is not identical, as the cell-surface SEF-a inhibits multiple FGF signaling pathways (Preger et al., 2004). Whether these isoforms function cooperatively or in the same developmental processes remains to be determined; to note, the SEF-b isoform exhibits a restricted pattern of expression in human tissues compared with SEF-a (Preger et al., 2004). Since RTKs deliver varied biological responses, it seems likely that SEF can interfere with RTK signaling at different levels to fine-tune signaling in a cell context- and isoform specific-manner.
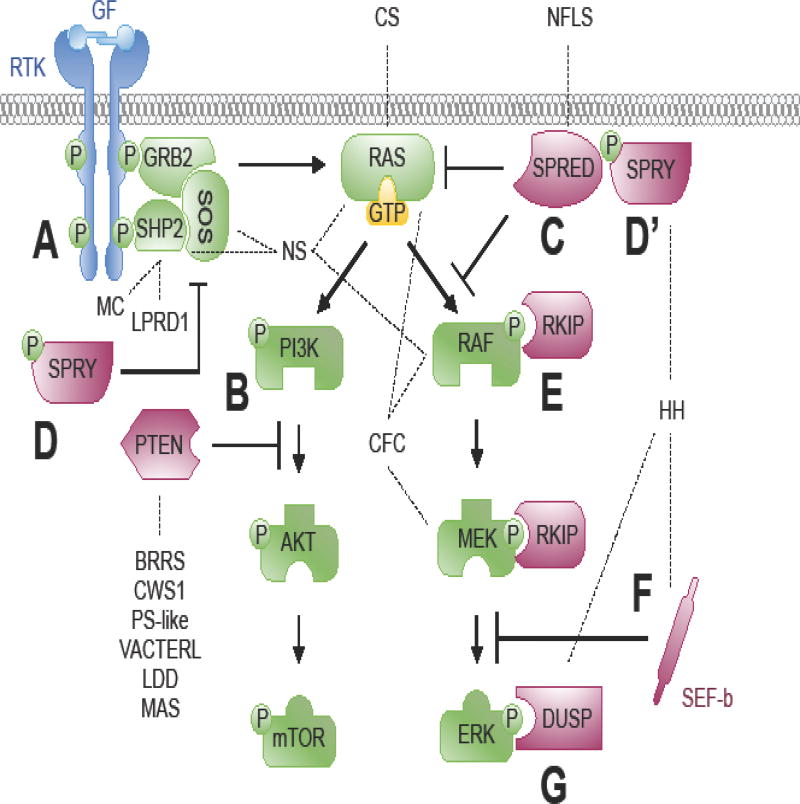
Figure 2. Feedback modulators of intracellular signal transduction cascades.
FGFR signaling induces expression of the SPRED family, the Sprouty family, SEF, and nuclear DUSPs via transcriptional activation and translation to attenuate RAS/MAPK signaling. (A) Growth factor-activated RTKs induce GRB2-mediated recruitment of SHP2 to signaling complexes. GRB2 redirects activated SHP2 to other signaling proteins that normally inhibit RTK signaling, subsequently acting as a positive regulator. (B) Growth factor-activated RTKs recruit and activate PI3K. The PI3K lipid signaling intermediate is dephosphorylated by PTEN, thereby attenuating PI3K/AKT signaling. (C) SPRED proteins increase RAF recruitment to the plasma membrane and prolongs RAS/RAF complexation, withdrawing RAF from activation by phosphorylation. (D, D’) Sprouty proteins translocate to the plasma membrane where they are phosphorylated. This phosphorylation induces a confirmation change that allows Sprouty proteins to bind and disrupt the GRB2/SOS complex, RAS activation, and RAF activation, thereby attenuating RAS/PI3K and RAS/MAPK signaling. (E) RKIP binds to both RAF1 and MEK to prevent their physical interaction and MEK phosphorylation, thereby attenuating RAS/MAPK signaling. (F) SEF-b suppresses activation at the level of, or downstream from, MEK. (G) DUSP6 dephosphorylates ERK. Dashed lines connecting the human congenital disorder with the protein in the pathway encoded by the causative mutated gene. Syndromes noted in the text and/or Supplemental Table 1. BRRS, Bannayan-Ruvalcaba-Riley syndrome; CFC, cardio-facio-cutaneous syndrome; CS, Costello syndrome; CWS1, Cowden syndrome 1; HH, hypogonadotropic hypogonadism with or without anosmia; LDD, Lhermitte-Duclos disease; LPRD1, LEOPARD syndrome 1; MAS, Macrocephaly/autism syndrome; MC, Metachondromatosis; NFLS, Neurofibromatosis Legius syndrome; NS, Noonan syndrome; PS-like, Proteus-like syndrome; VACTERL, vertebral anomalies, anal atresia, congenital cardiac disease, tracheoesophageal fistula, renal anomalies, radial dysplasia, and other limb defects; GF, growth factor; P, phosphorylation; RTK, receptor tyrosine kinase.
Through an evolutionarily conserved modular domain named the ErbB binding region (EBR), the multi-adaptor protein Mitogen-Inducible Gene 6 (MIG6) mediates catalytic repression of ligand-bound ERBB receptors, namely EGFR, ERBB2, and ERBB4 (Anastasi et al., 2007; Hackel et al., 2001). Since ERBB3 signals as an obligate heterodimer with the other members of the ErbB family, MIG6 also inhibits its signaling and thus is a cellular inhibitor of the entire ErbB family. Knockout of Mig6 in mice resulted in aberrant lung development associated with high neonatal mortality (Ferby et al., 2006; Jin et al., 2009; Zhang et al., 2005), and surviving mice developed degenerative joint diseases and spontaneous tumors in organs including the skin, gastrointestinal tract, lung, and endometrium (Supplemental Table 1) (Ferby et al., 2006; Jeong et al., 2009; Jin et al., 2009; Zhang et al., 2005). Importantly, over proliferation and impaired differentiation of epidermal keratinocytes and the resulting skin tumors could be rescued by genetic or pharmacological suppression of EGFR, indicating that unrestrained EGFR activation and sustained signaling through MAPK was a result of loss of Mig6 (Ferby et al., 2006). Tissue-specific deletion of Mig6 in mouse hepatocytes caused hepatomegaly and fatty liver, a phenotype similar to that observed in mice homozygous for a gain-of-function Egfr allele (Ku et al., 2012; Natarajan et al., 2007; Reschke et al., 2010; Scheving et al., 2014). Receptor-induced phosphorylation of the MIG6 ERB domain stabilizes the MIG6/EGFR interaction and prevents activation of EGFR by blocking an allosteric site critical for activation within the receptor dimers (Figure 1D) (Park et al., 2015; Zhang et al., 2007; Zhang et al., 2005). Upon docking onto EGFR, MIG6 is also capable of recruiting components of the endocytic machinery, leading to receptor degradation independent of phosphorylation and ubiquitination (Frosi et al., 2010; Segatto et al., 2011; Walsh and Lazzara, 2014; Ying et al., 2010). This two-tiered mechanism of MIG6-mediated inhibition provides for immediate repression of EGFR signaling (kinase inhibition) coupled to longer term isolation from incoming ErbB receptor ligands (endocytosis) (Anastasi et al., 2016). Whether MIG6 exerts either of these inhibitory functions on other RTKs remains to be determined. MIG6 binding to ErbB receptors is dependent on a protein interface in the kinase domain unique to the ErbB family, so involvement of MIG6 with other RTKs would involve a different mechanism or could imply that these RTKs signal in cooperation with ErbB receptors. In vitro analyses suggest that MET could be a potential target of MIG6, as overexpression of MIG6 was able to inhibit the HGF/MET-induced cell migration and neurite outgrowth (Pante et al., 2005).
Receptor dephosphorylation
The phosphorylation status and signaling output of RTKs is determined by a balance between the intrinsic kinase activity of the receptor and the activities of protein tyrosine phosphatases (PTPs). PTPs have evolved in a number of families that are structurally and mechanistically distinct and control a broad spectrum of RTK signaling pathways (Ostman and Bohmer, 2001; Tonks, 2006). As such, they are arguably one of the most important regulators of the extent and intensity of RTK signaling. Animal studies thus far, however, have yielded only limited insights into specific functions of individual PTPs. While knockout mouse models have been made for all classical PTP genes except Ptpn18, Ptpn20, Ptpn21, and Ptpru (Hendriks et al., 2013), many of these knockout models displayed only mild developmental defects, suggesting significant functional redundancy between PTPs.
Ptpn11 is one of the few exceptions, as Ptpn11 knockout mice died at mid-gestation with multiple defects in mesodermal patterning (Qu et al., 1997; Saxton et al., 1997). Selective deletion of Ptpn11 in developing kidneys of mice caused reduced ureteric bud branching by downregulation of the transcription factors Etv4 and Etv5, which are targets of glial-derived neurotrophic factor (GDNF)/RET signaling and of other RTKs (Willecke et al., 2011). Ptpn11-deficiency in cardiomyocytes resulted in early postnatal lethality and dilated cardiomyopathy associated with increased IR signaling and decreased activation of ERK1/2 and JNK2 (Princen et al., 2009). Numerous other in vivo studies have linked Ptnp11 with FGF-dependent MAPK/ERK signaling and have revealed roles for Ptnp11 in patterning and specification of the optic vesicle, lens and lacrimal gland development, chondrogenesis, intestinal progenitor cell fate, lung branching morphogenesis, and formation of the midbrain-hindbrain boundary, among others (Supplemental Table 1) (Cai et al., 2013; Dee et al., 2016; Heuberger et al., 2014; Pan et al., 2010; Tefft et al., 2005; Yang et al., 2013). PTPN11 encodes the widely expressed non-receptor tyrosine phosphatase Src-homology 2 domain-containing phosphatase 2 (SHP2) (Dance et al., 2008), which in the absence of upstream stimulation, is kept in a low-activity state by an intramolecular interaction between the N-terminal SH2-domain and the catalytic phosphatase domain. Activation of RTKs and/or subsequent activation of scaffolding adaptor proteins leads to recruitment of SHP2 to signaling complexes, where engagement of the SH2-domains induces a conformational change that resolves auto-inhibitory interactions. SHP2-mediated dephosphorylation of FGFRs is controlled by the adaptor protein GRB2, which recruits SHP2 to the activated receptors (Figure 2A). GRB2 additionally redirects activated SHP2 to other signaling proteins, such as Sprouty or STAT proteins, that normally inhibit signaling through ERK1/2, AKT, or STAT5. In this manner, SHP2 can further promote RTK signaling (Ahmed et al., 2010; Ahmed et al., 2013; Hadari et al., 1998; Hanafusa et al., 2004; Tajan et al., 2015; You et al., 1999).
Dephosphorylation of signaling pathway components
The intracellular events downstream of activated RTKs are responsible for transduction and amplification of ligand-induced signaling. This most commonly involves protein phosphorylation, which ultimately results in changes in gene expression and other cellular effects. The PI3K/AKT and RAS/MAPK pathways are principal signaling mechanisms for controlling cell survival, proliferation, differentiation, and migration (Figure 2) (Mendoza et al., 2011), and as such, must be precisely spatially and temporally regulated. Phosphatase and Tensin homolog (PTEN) and Dual-Specificity Phosphatases (DUSPs) represent early and late attenuators, respectively, of RTK-induced intracellular signal transduction cascades. PTEN is the main negative regulator of the PI3K/AKT pathway, whereas DUSPs modulate activation of the RAS/MAPK pathway (Carracedo et al., 2008; Katz et al., 2011) (Figure 2). Multiple mechanisms and modes of crosstalk have been uncovered between these two pathways, further complicating our understanding of their complex roles in development (Mendoza et al., 2011).
The first, and probably still the clearest, indication that PTEN plays an essential role in regulation of cell growth came from early studies in D. melanogaster (Goberdhan and Wilson, 2003). Pten-deficient cells proliferated at a faster rate than their heterozygous counterparts, showed an autonomous increase in cell size, and formed enlarged organs (Gao et al., 2000; Goberdhan et al., 1999; Huang et al., 1999). In vitro and in vivo studies revealed that PTEN controls cell growth and proliferation by antagonizing growth factor-induced activation of the PI3K/AKT pathway. Specifically, PTEN preferentially dephosphorylates membrane-bound PIP3 into PIP2. This prevents PIP3-mediated recruitment of AKT to the plasma membrane and its activation (Figure 2B) (Engelman et al., 2006; Maehama and Dixon, 1998). Numerous subsequent studies in mice and other model organisms have examined the functional role of Pten in various organs and tissues, yielding a diverse spectrum of phenotypes (Supplemental Table 1) (Knobbe et al., 2008). Knockout mouse models of Pten showed that deletion of a single allele resulted in lethal polyclonal autoimmune disorders and various forms of epithelial cancer (Di Cristofano et al., 1999; Di Cristofano et al., 1998). Because of the lethal nature of Pten loss, conditional deletion models have been used to address the roles of PTEN during development. Tissue-specific deletion of Pten in mouse neurons resulted in progressive macrocephaly, seizures, and ataxia, and neurons lacking Pten expressed high levels of phosphorylated AKT (Backman et al., 2001; Groszer et al., 2001; Kwon et al., 2001). In vitro and in vivo analyses revealed that PTEN also regulates cardiac hypertrophy and survival by blocking growth factor signaling through the PI3K/AKT pathway (Crackower et al., 2002; Schwartzbauer and Robbins, 2001). As PI3K pathway signaling is regulated in part by IR signaling and affects downstream proteins involved in metabolism such as mTOR, Pten-deficiency in hepatocytes led to massive hepatomegaly and steatohepatitis with triglyceride accumulation (Horie et al., 2004; Stiles et al., 2004).
DUSPs constitute a large heterogeneous subgroup of the PTP superfamily characterized by their ability to dephosphorylate tyrosine, serine, and threonine residues. Despite a fairly detailed understanding of the biochemical properties and catalytic mechanisms employed by DUSPs (Farooq and Zhou, 2004; Owens and Keyse, 2007), knowledge of their in vivo roles is still expanding (Supplemental Table 1). Of interest here, gene knockdown or overexpression studies in zebrafish, chick, and mouse first identified an in vivo role for DUSP6 in fin/limb bud patterning as a negative feedback regulator of the FGF-RAS/MAPK signaling pathway (Figure 2G) (Kawakami et al., 2003). FGF8 signaling induces expression of DUSP6, which encodes an ERK-specific DUSP, and establishes a negative feedback loop (Bermudez et al., 2010; Groom et al., 1996; Kawakami et al., 2003; Mourey et al., 1996; Muda et al., 1996). Targeted inactivation of Dusp6 in mice led to increased levels of phosphorylated ERK, the phosphorylated ERK target Erm, and transcripts initiated from the Dusp6 promoter itself (Li et al., 2007). Furthermore, Dusp6 knockout mice displayed cardiac hypertrophy and multiple skeletal abnormalities including dwarfism, defects in the middle ear bones and otic capsule, and premature fusion of the cranial sutures (craniosynostosis); histological analysis of the long bones revealed disorganization of chondrocytes in the growth plate (Li et al., 2007; Maillet et al., 2008; Urness et al., 2008). These same skeletal phenotypes are also found in mouse models of human disorders with constitutive activating mutations in FGFRs (Neben and Merrill, 2015; Ornitz and Marie, 2015), highlighting the relationship between FGF signaling and DUSP6. Although many agonists in addition to FGFs activate ERK1/2 during embryonic development, including EGF, NGF, HGF, VEGF, and PDGF, few studies have examined their regulation by DUSP6 in vivo (Bermudez et al., 2010).
Non-catalytic feedback modulators of signaling pathways
Sprouty (SPRY) and SPRED genes encode highly conserved protein families with no apparent enzymatic function that inhibit different steps of the RAS/MAPK signaling pathway and fine-tune RTK signaling in a cell context- and isoform specific-manner. Some evidence suggests that Sprouty proteins indirectly regulate the PI3K/AKT pathway, however, these effects are poorly understood (Castellano and Downward, 2011; Steelman et al., 2011). As late attenuators transcriptionally induced by growth factor activation, Sprouty and SPRED proteins adapt cells to longer term external stimulation, persisting on the timescale of hours (Volinsky and Kholodenko, 2013).
The first member of the Sprouty family identified was found in a screen for genes involved in development of trachea and eyes in Drosophila (Casci et al., 1999; Hacohen et al., 1998). Like Drosophila Sprouty, mammalian Sprouty proteins antagonize FGF signaling in tubular morphogenesis associated with tracheal/lung development (Figure 4A, A’) (Metzger et al., 2008; Shaw et al., 2007; Tefft et al., 1999) and angiogenesis (Taniguchi et al., 2007a; Taniguchi et al., 2009). Since these initial findings, the number of pathways and biological processes regulated by Sprouty proteins continues to expand, including submandibular parasympathetic gangliogenesis (Figure 4B-B’) (Knosp et al., 2015), ureteric branching (Figure 4C, C’) (Basson et al., 2005; Basson et al., 2006; Chi et al., 2004; Gross et al., 2003; Michos et al., 2010), external genitalia development (Figure 4D, D’) (Ching et al., 2014), endochondral bone formation (Figure 4E, E’) (Joo et al., 2016; Minowada et al., 1999), and branchial nerve development (Figure 4F-F”) (Simrick et al., 2011), among others (Supplemental Table 1).
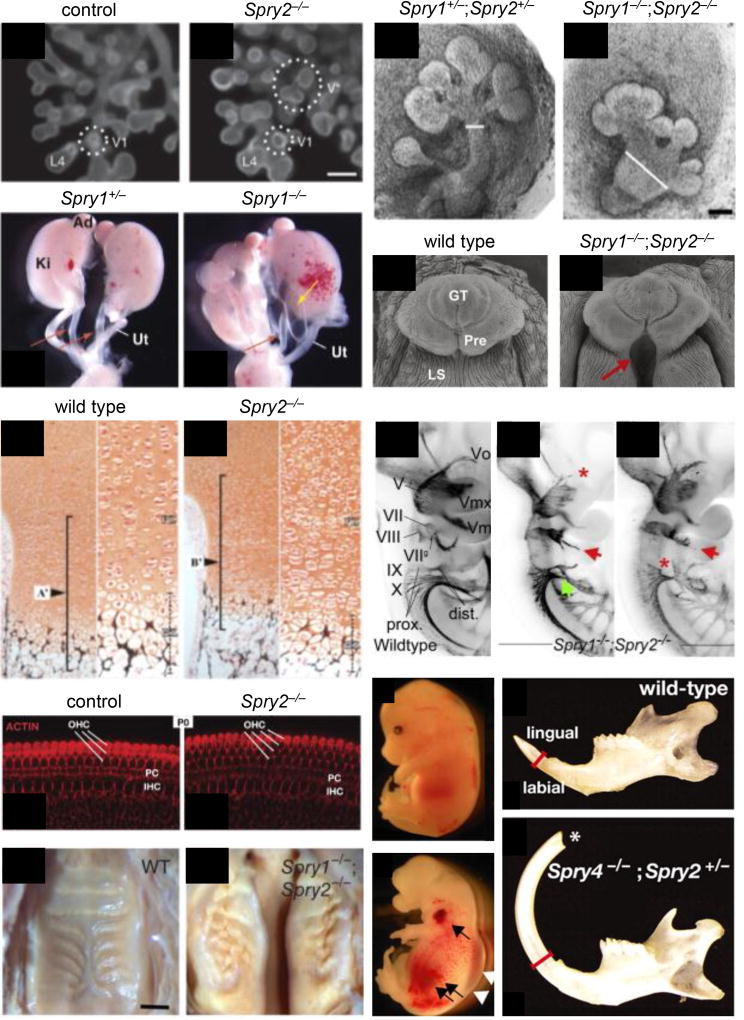
Figure 4. Mouse models are invaluable in decoding the developmental roles of the regulator family of Sprouty proteins.
(A-A’) E12.5 Spry2−/− mouse lung showing the normal ventral secondary branch (V1) and an ectopic branch (V*) that forms earlier and proximal to V1 (Metzger et al., 2008). (B-B’) Genetic deletion of Spry1;Spry2 in mice disrupts submandibular gland epithelial development resulting in a wide primary duct (white lines) and abnormal branching morphogenesis at E13 (Knosp et al., 2015). (C-C’) Kidneys and urogenital tract abnormalities in Spry1−/− newborn pups. Normal ureters and abnormal hydroureters are indicated by red and yellow arrows, respectively. Ad, adrenal; Ki, kidney; Ut, uterus (Basson et al., 2005). (D-D’) Fusion of the preputial (Pre) and labioscrotal (LS) folds along the ventral surface of the genital tubercle (GT) is disrupted in E16.5 male Spry1−/−;Spry2−/− mice, resulting in the absence of an internalized urethra in the proximal GT (red arrow) (Ching et al., 2014). (E-E’) Von Kossa/Safranin-O staining of E18.5 femur sections showed more proliferating chondrocytes in the growth plate of Spry2−/− mice than in that of wild type (Joo et al., 2016). (F-F”) E10.5 Spry1−/−;Spry2−/− mice have trigeminal defects, facial nerve defects, and glossopharyngeal and vagus cranial nerves display incomplete or irregular bridging between proximal and distal ganglia. Arrows highlight abnormal morphology, and asterisks indicate missing portions (Simrick et al., 2011). (G-G’) The region of the P0 Spry2−/− mouse cochlea shown has four rows of outer hair cells (OHCs) instead of the three found in control and elsewhere in the Spry2 null organ of Corti. PC, pillar cells; IHC, inner hair cells (Shim et al., 2005). (H-H’) Increased FGF signaling in Spry1−/−;Spry2−/− mice resulted in disorganized and compacted rugae at P0 (Economou et al., 2012). (I-I’) Gross appearance of wild type and Spry2−/−;Spry4−/− at E12.5. The arrow and arrowhead indicate hemorrhage and edema, respectively (Taniguchi et al., 2009). (J-J’) Abnormal length and thickness of adult Spry2−/−;Spry4+/− incisor as well as the absence of a sharp tip (asterisk) (Klein et al., 2008). Reprinted or adapted with permission.
As the functions of Sprouty proteins in embryonic development have been reviewed previously by others (Cabrita and Christofori, 2008; Horowitz and Simons, 2008; Warburton et al., 2008), we highlight here the specific roles of these proteins in craniofacial and tooth development as an example of the types of effects these genes can have on RTK-mediated signaling. SPRY2 and FGF8/FGFR3 signaling is required for cell fate decisions in the mouse auditory sensory epithelium, as loss of Spry2 resulted in dramatic perturbations in organ of Corti cytoarchitecture (Figure 4G, G’) (Shim et al., 2005). Combined deletion of Spry1 and Spry2 in mice caused highly disorganized palatal rugae including broader and ectopic ruga formation (Figure 4H, H’), indicating that the FGF pathway is activatory in a Turing-type reaction-diffusion system for the striped pattern that establishes and maintains the palatal rugae (Economou et al., 2012). Spry2;Spry4 double knockout mice are embryonic lethal by E12.5 with craniofacial and limb morphogenesis abnormalities (Figure 4I, I’) (Taniguchi et al., 2007a; Taniguchi et al., 2009). The Spry4 loss of function phenotypes, including dwarfism and polysyndactyly, resemble mouse models of human disorders with activating mutations in FGFRs (Neben and Merrill, 2015; Ornitz and Marie, 2015; Taniguchi et al., 2007a), suggesting that loss of Spry4 results in hyperactivation of FGF signaling. Mice carrying single and various combinations of Sprouty mutant alleles also possess supernumerary teeth and display abnormalities in tooth size, shape, and micro-structure (Boran et al., 2009; Charles et al., 2011; Klein et al., 2008; Klein et al., 2006; Lagronova-Churava et al., 2013; Lochovska et al., 2015; Marangoni et al., 2015; Percival et al., 2017). For example, Spry2+/−;Spry4−/− mice develop a ‘tusk’-like incisor in their lower jaws due to the presence of enamel on the lingual surface (Figure 4J, J’) (Boran et al., 2009; Klein et al., 2008). Importantly, the lingual ameloblast phenotype can be rescued in the adult by reducing Fgf gene dosage (Klein et al., 2008), demonstrating the critical role of Sprouty genes in controlling the epithelial-mesenchymal FGF signaling loop.
The four mammalian orthologues of Sprouty proteins share sequence similarity to D. melanogaster Sprouty in the cysteine-rich C-terminus but significantly differ among each other and from the fly ortholog in the N-terminus (de Maximy et al., 1999; Leeksma et al., 2002; Mason et al., 2006). This sequence divergence at the N-terminus could dictate differential functions, potentially by mediating protein-protein interactions. Indeed, Sprouty proteins can interact directly with multiple downstream components of the RTK pathway, including FRS2, GRB2, RAF1, B-RAF, and SHP2. In most cases, however, it remains unclear how these associations modulate signaling. The best studied family members, SPRY1 and SPRY2, have been shown to antagonize RTK signaling at multiple levels, such as binding to the GRB2/SOS complex (Figure 2D) and inhibition of RAF1 activation by RAS (Figure 2D’) depending on the cellular context and/or the identity of the RTK (Mason et al., 2006). The phosphorylation of SPRY1 and SPRY2 at Tyrosine 53 and Tyrosine 55, respectively, induces a conformational change that has been shown to be essential for protein binding and modulation of RAS/MAPK signaling (Alsina et al., 2012; Guy et al., 2009; Hanafusa et al., 2002; Mason et al., 2004; Sasaki et al., 2003; Sasaki et al., 2001). Sprouty proteins may mediate their actions in part by increasing active forms of such phosphatases as PTEN (Edwin et al., 2006; Patel et al., 2013). In cultured cells, SPRY2 increased overall PTEN protein levels while decreasing PTEN phosphorylation, resulting in increased PTEN activity. This was reflected by diminished activation of AKT by EGF signaling and blocked cell proliferation (Edwin et al., 2006). In the context of EGFR signaling, SPRY2 levels are controlled through phosphorylation-dependent complex formation with C-CBL in vitro (Hall et al., 2003; Mason et al., 2004; Rubin et al., 2003). Binding of SPRY2 to C-CBL directs the proteolytic degradation of SPRY2 but also inhibits C-CBL-mediated degradation of EGFR, leading to sustained signaling activity (Egan et al., 2002; Ng et al., 2008; Rubin et al., 2003; Wong et al., 2002). This function may be limited to SPRY2, however, as SPRY4 suppression of MAPK/ERK activation by EGF stimulation did not result in interaction with C-CBL (Mason et al., 2004; Wong et al., 2001).
SPREDs (Sprouty-related PRoteins with an EVH1 Domain) are a family of membrane-associated, negative RAS/MAPK signaling modulators that possess structural and functional similarities to their relatives, the Sprouty proteins. There are four known mammalian SPRED proteins: SPRED1, SPRED2, SPRED3, and EVE-3, the last of which is a splice variant of SPRED3 (Kato et al., 2003; King et al., 2006; Wakioka et al., 2001). Spred1 knockout mice are viable and fertile but exhibit low body weight, a shortened face, and impaired hippocampus-dependent learning capabilities (Brems et al., 2007; Denayer et al., 2008; Inoue et al., 2005; Phoenix and Temple, 2010). Spred2 deficiency in mice suppressed aorta-gonad-mesonephros hematopoiesis and caused defects in bone morphogenesis, with the mice exhibiting a dwarfing phenotype and increase of early hematopoiesis (Bundschu et al., 2005; Nobuhisa et al., 2004). Overlapping expression patterns of different SPRED family members and their possible redundancy might preclude certain phenotypes from being observed in the single null alleles (Supplemental Table 1). Indeed, deletion of both Spred1 and Spred2 in mice, which have overlapping expression patterns in the heart, lung, liver, and bone, resulted in embryonic lethality with subcutaneous hemorrhage, edema, and dilated lymphatic vessels (Engelhardt et al., 2004; Kato et al., 2003; Stowe et al., 2012; Taniguchi et al., 2007b; Tuduce et al., 2010).
Like Sprouty proteins, SPRED proteins inhibit growth factor-mediated MAPK/ERK activation, albeit by different biochemical mechanisms. Overexpression of SPRED1 increases RAF recruitment to the plasma membrane and prolongs RAS/RAF interaction, thus withdrawing RAF from activation by phosphorylation (Figure 2C) (Bundschu et al., 2005; Wakioka et al., 2001). Subsequent studies confirmed that SPRED proteins also inhibit activation of RAS by the small GTPase RAP1 without affecting receptor phosphorylation (King et al., 2006; Nonami et al., 2005; Stowe et al., 2012). Recently, it was proposed that SPRED1-plasma membrane translocation is mediated in a B-RAF- and C-RAF-dependent manner to specifically disturb K-RAS but not H-RAS membrane anchorage (Siljamaki and Abankwa, 2016). This potential mechanism may explain why it has been difficult to pinpoint whether SPRED1 acts at the level of RAS or RAF.
Originally isolated from the bovine brain (Bernier and Jolles, 1984), RAF Kinase Inhibitor Protein (RKIP; also known as PhosphatidylEthanolamine-Binding Protein, PEBP1) was renamed based on its physiologically relevant inhibition of the RAS/MEK/ERK pathway (Yeung et al., 1999; Yeung et al., 2001). Expression of RKIP mRNA has since been detected in all mammalian tissues tested, with high levels in spermatids and brain oligodendrocytes, Purkinje cells, and specific cortical and hippocampal neuronal cell layers (Bernier and Jolles, 1984; Frayne et al., 1999; Theroux et al., 2007). RKIP deficient mice are viable but develop an olfaction deficit, a phenotype that correlates with the expression pattern of the gene in the brain (Theroux et al., 2007). Subsequent studies in model organisms have identified RKIP as critical for neurological functioning, photoreceptor degeneration, myogenesis, reproduction, and spermatogenesis (Supplemental Table 1) (Antoun et al., 2012; Gibbons et al., 2005; Murga-Zamalloa et al., 2011; Nixon et al., 2006; Subramanian et al., 2014; Yamamoto et al., 2012). RKIP inhibits RAF-1 mediated phosphorylation and activation of MEK by competitive physical association which disrupts the interaction between these kinases (Figure 2E). Overexpression of RKIP in vitro reduced cell proliferation and transformation and was accompanied by alterations in MEK-, ERK-, and AP-dependent transcription (Yeung et al., 1999). Interestingly, although RKIP can interact with B-RAF, depletion of RKIP did not affect B-RAF activation, indicating that RKIP may selectively limit the dynamic range of MAPK signaling in response to growth factors (Trakul et al., 2005).
Early attenuation of RTK signaling via receptor ubiquitination and degradation
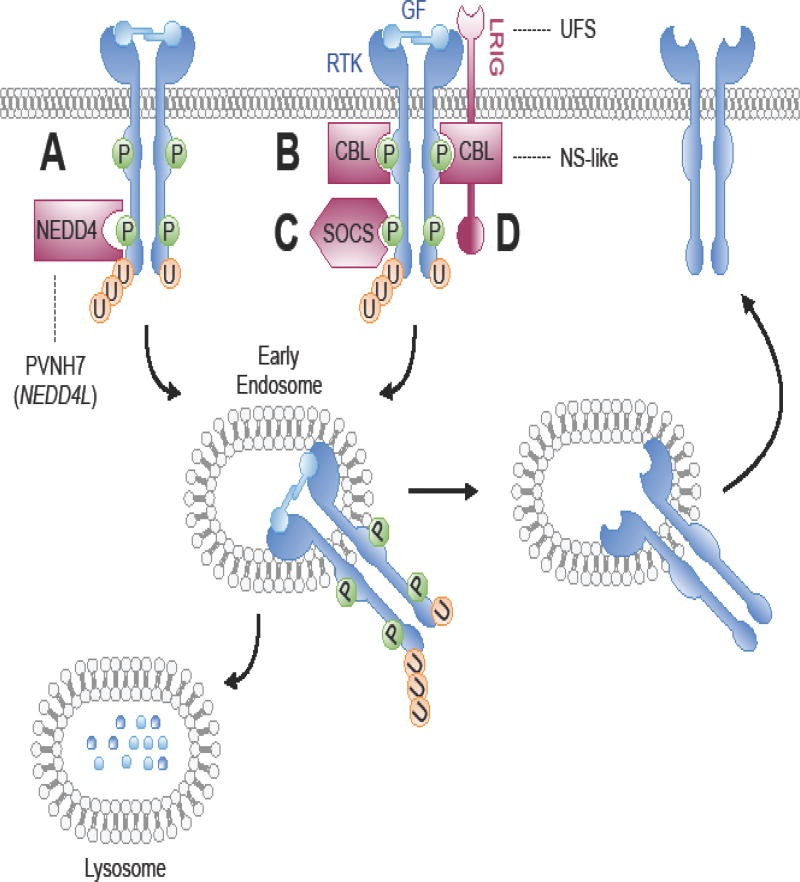
Another common mechanism by which RTK signaling is downregulated is the removal of receptors from the plasma membrane via endocytosis. This can occur either reversibly, when internalized receptors are recycled back to the plasma membrane after a period of time, or irreversibly, when the downregulated receptors are sent for lysosomal degradation. RTK internalization and degradation are regulated upon growth factor induced RTK activation through ubiquitination of the intracellular receptor domains by E3 ubiquitin ligases (Figure 3) (Goh and Sorkin, 2013). The two main E3 ligases involved in RTK ubiquitination during development are the HECT-type ligase NEDD4 and the RING-type ligase CBL. Both NEDD4 and CBL regulate signal duration of multiple RTKs, and their disruption in development results in serious abnormalities.
Figure 3. Attenuation of RTK signaling by receptor ubiquitination and degradation.
(A) Growth factor activation of RTKs leads to recruitment of NEDD4 to the receptor complex. (B) Independent and (C) SOCS- or (D) LRIG-mediated mechanisms recruit CBL to the receptor complex. NEDD4 and CBL direct the ubiquitination of RTKs, resulting in receptor endocytosis and routing to early endosomes. RTKs can then either be recycled to the plasma membrane or targeted for lysosomal degradation, thereby attenuating receptor signaling. Dashed lines connecting the human congenital disorder with the protein in the pathway encoded by the causative mutated gene. Syndromes described in the text and/or Supplemental Table 1. PVNH7, Periventricular nodular heterotopia 7; NS-like, Noonan syndrome-like; UFS, Urofacial syndrome; GF, growth factor; P, phosphorylation; RTK, receptor tyrosine kinase; U, ubiquitination.
NEDD4 (Neuronal precursor cell Expressed and Developmentally Downregulated) proteins are found ubiquitously in eukaryotes and have expanded to nine known family members in mammals, with two of them, NEDD4 and NEDD4L (also known as NEDD4-2) being very closely related (Supplemental Table 1) (Scheffner and Kumar, 2014). Complete loss of Nedd4 in mice resulted in embryonic lethality at mid-gestation with pronounced heart defects, subcutaneous bleeding, and developmental delays (Fouladkou et al., 2010; Kawabe et al., 2010; Liu et al., 2009). Although NEDD4 has several additional substrates beyond RTKs, biochemical analysis suggested that the Nedd4 loss-of-function phenotype can be at least partially attributed to abnormal RTK signaling: the growth retardation in Nedd4 heterozygous mice is associated with reduced cell surface expression and signaling through IR and IGF-1R (Cao et al., 2008). That loss of Nedd4 resulted in loss of IGF-1R signaling contradicts earlier in vitro studies which reported that NEDD4 ubiquitinates and decreases stability of IGF-1R (Vecchione et al., 2003). This suggests that NEDD4 may fine-tune RTK signaling differently in distinct cell types. In agreement with a role for NEDD4 in promoting receptor degradation, Nedd4L-deficient mouse embryos showed increased expression of neurotrophic RTK 1 (NRTK1, also known as TRKA), a possible contributor to the pain sensitivity phenotype in heterozygous adults (Yanpallewar et al., 2016). The binding of NEDD4L to activated NRTK1 leads to receptor ubiquitination and down-regulation and to the modulation of neuronal survival in vitro (Figure 3A) (Arevalo et al., 2006; Yu et al., 2014). NEDD4 may also attenuate RTK signaling by regulating the levels of the tumor suppressor PTEN. In vitro studies demonstrated that NEDD4 was responsible for PTEN ubiquitination (Trotman et al., 2007; Wang et al., 2007), and subsequent studies in X. laevis confirmed that Nedd4-mediated ubiquitination of Pten promoted axonal and dendritic branching by allowing full activation of the PI3K/Akt pathway (Christie et al., 2010; Drinjakovic et al., 2010; Schmeisser et al., 2013). However, it does not appear that aberrant PTEN ubiquitination in mice played a role in impaired axon growth upon deletion of Nedd4 and Nedd4L (Hsia et al., 2014), suggesting that NEDD4 regulation of PTEN may only occur in specific biological contexts.
The first evidence that members of the CBL (Casitas B-lineage Lymphoma proto-oncogene) family of E3 ligases (cbl in D. melanogaster, SLI-1 in C. elegans, and CBL-3, CBL-B, and C-CBL in mammals) act as negative regulators of RTKs was provided by genetic screens in C. elegans and D. melanogaster (Supplemental Table 1). These early studies demonstrated that loss-of-function point mutations in the CBL homologs sli-1 and cbl resulted in aberrant signaling by the EGFR homologs LET-23 and Der, respectively (Jekely et al., 2005; Meisner et al., 1997; Pai et al., 2000; Wang et al., 2008; Yoon et al., 1995). Subsequent studies of the mammalian homologs have shown that c-Cbl- or Cbl-b-deficient mice are viable and fertile with only minor phenotypic differences, but combined deletion results in early embryonic lethality before mid-gestation (Mohapatra et al., 2013; Nakamura et al., 2001). This redundancy is consistent with in vitro work demonstrating that C-CBL and CBL-B work cooperatively to control the duration of EGFR signaling (Pennock and Wang, 2008). Upon ligand-induced receptor activation, phosphorylated CBL proteins complex with EGFR via a highly conserved TKB domain to facilitate receptor ubiquitination and degradation by a catalytic RING finger domain (Figure 3B) (de Melker et al., 2001; Haglund et al., 2003; Levkowitz et al., 1999; Levkowitz et al., 1998; Longva et al., 2002). The c-Cbl knockout phenotype is faithfully recapitulated by a mutation in the RING finger domain that eliminates its E3 ligase activity and resulted in more severe phenotypic changes than a loss-of-function mutation in the c-Cbl TKB domain (Thien and Langdon, 2005; Thien et al., 2003). Thus, TKB domain-mediated interactions with RTKs could not fully explain the spectrum of C-CBL functions. Further confirmation of this hypothesis came from a study analyzing homozygous knock-in mutation of Tyrosine 737, which eliminates the binding site for PI3K in the C-terminal tail of CBL, located outside of the TKB domain (Adapala et al., 2010a; Adapala et al., 2010b). Abrogation of the CBL/PI3K interaction resulted in perturbed RANKL-mediated signaling, leading to increased bone mass due to a cell-autonomous defect in osteoclast function, a phenotype not seen with other Cbl mutations (Adapala et al., 2010a; Adapala et al., 2010b).
Late attenuation of RTK signaling via receptor ubiquitination and degradation
By recruitment of E3 ubiquitin ligases to the receptor complex, members of the Leucine-Rich and Immunoglobulin-like domain (LRIG) and Suppressor of Cytokine Signaling (SOCS) families accelerate receptor ubiquitination and degradation (Figure 3C, D) (Gur et al., 2004; Laederich et al., 2004). Unlike NEDD4 and CBL ubiquitin ligases, the expression of LRIG1 and SOCS genes are induced by growth factor activation of RTKs via transcriptional activation and translation (Segatto et al., 2011). The LRIG1 transmembrane protein and SOCS cytosolic adaptor proteins have been shown to interact with RTKs and attenuate receptor signaling through both ligand-dependent and independent mechanisms.
Genetic approaches in vivo have confirmed the essential biological functions of LRIG proteins and have provided insight into the broad range of their effects on signaling pathways (Supplemental Table 1). Deletion of Lrig1 in mice leads to psoriasis-like epidermal hyperplasia and dramatically increased proliferation of the intestinal crypts and tracheal and bronchial epithelium (Karlsson et al., 2008; Lu et al., 2014; Luetteke et al., 1994; Suzuki et al., 2002). These phenotypes were correlated with a substantial increase in total and phosphorylated protein levels of EGFR, ERBB2, ERBB3, and MET in associated tissues (Suzuki et al., 2002), emphasizing the role of LRIG1 as a negative regulator of RTK signaling in vivo. Importantly, the skin and intestinal phenotypes in Lrig1-deficient mice could be rescued by genetic or chemical inhibition of EGFR phosphorylation, suggesting direct involvement of LRIG1 in controlling the strength of EGFR signaling (Luetteke et al., 1994). Indeed, extensive in vitro studies have demonstrated that LRIG1 attenuates the half-life of all four receptors of the ErbB family and of MET by amplifying C-CBL-mediated ubiquitination (Goldoni et al., 2007; Gur et al., 2004; Laederich et al., 2004; Rafidi et al., 2013; Rondahl et al., 2013; Shattuck et al., 2007; Stutz et al., 2008; Yi et al., 2011). Additional in vitro work suggests that limited proteolysis of the soluble ectodomain of LRIG1 may inhibit EGFR signaling by competing with ligand binding and stabilizing the receptor in the inactive monomeric state (Goldoni et al., 2007). LRIG1 was also shown to restrict RET recruitment to lipid rafts and to inhibit binding of its ligand GDNF preventing receptor activation (Ledda et al., 2008). While the functions of the other members of the LRIG family, LRIG2 and LRIG3, remain poorly understood, studies in X. laevis have demonstrated that lrig3 modulates Fgf-dependent Erk phosphorylation and Wnt signaling during neural crest cell specification and induction. When co-expressed in vitro, Lrig3 co-immunoprecipitated with Fgfr1 via its ectodomain, and this interaction was correlated with reduced levels of Fgfr1 protein (Zhao et al., 2008), suggesting that Lrig3 may attenuate Fgf signaling by the mechanisms similar to those described for EGFR and RET.
Although the biological roles of SOCS proteins have traditionally been considered in the context of cytokine receptor signaling through the JAK/STAT pathway in immunity and hematopoiesis, emerging evidence implicates SOCS proteins in the control of RTK signaling during development (Supplemental Table 1) (Trengrove and Gray, 2013). In vitro and in vivo studies demonstrated that SOCS2 exerts a dual role in the regulation of EGF signaling: Socs2 knockout mice displayed increased intestinal growth due to enhanced responsiveness to EGF (Michaylira et al., 2006), and cortical neurons derived from transgenic Socs2 overexpressing mice had increased neural outgrowth, apparently also due to enhanced EGF signaling (Goldshmit et al., 2004). The gigantism phenotype of Socs2-deficient mice suggests an important role for SOCS2 in the regulation of growth, possibly by modulating growth hormone and IGF-1R signaling (Greenhalgh et al., 2002; Metcalf et al., 2000). These mice exhibited prolonged STAT5B activation, and loss of Stat5b function partially relieved the growth enhancement. In contrast to Socs2 mutants, but similar to Nedd4 heterozygous mice, Socs6 knockout mice displayed a mild growth retardation thought to be due to perturbation of IGF-1R signaling (Krebs et al., 2002). Despite in vitro studies supporting a role for SOCS6 in neural stem differentiation and glucose metabolism (Choi et al., 2010; Gupta et al., 2011; Liu et al., 2008a; Liu et al., 2008b; Vlacich et al., 2010), mice deficient in Socs6 did not display phenotypic alterations consistent with such functions (Krebs et al., 2002). However, transgenic mice overexpressing Socs6 had altered glucose metabolism compared to wild type mice, with enhanced PI3K/AKT activation that was independent of increased IR or IGF-1R phosphorylation (Li et al., 2002). This suggests an additional mechanism by which SOCS6 regulates insulin signaling downstream of the receptor to control glucose metabolism. Similar to the engagement of LRIG1 with many RTKs, in vitro studies suggest that SOCS proteins regulate multiple RTKs including c-KIT, FLT3, IR, IGF-1R, and EGFR by enhancing their degradation via recruitment of E3 ubiquitin ligase complexes (Banks et al., 2005; Kario et al., 2005; Krebs et al., 2002; Nicholson et al., 2005; Trengrove and Gray, 2013). A subset of SOCS proteins – SOCS2, SOCS6, and SOCS7 – protect RTKs from SOCS-mediated degradation by interacting with the domains of other SOCS proteins responsible for the recruitment of E3 ubiquitin ligase complexes (Piessevaux et al., 2006). This suggests a role for these SOCS proteins in restoring cells to a responsive state for subsequent RTK stimulation.
Modulators of RTK signaling associated with human congenital disorders
Given the critical roles of RTK signaling in cell fate determination and morphogenesis, there has been great interest in understanding how RTK regulators are deregulated in human disorders. Indeed, both gain-of-function mutations, which lead to constitutive protein activation, and loss-of-function mutations, which lead to non-functional or dominant negative proteins, have been mapped to regulators of RTK signaling in human disease (Rauen, 2013; Tartaglia and Gelb, 2005). Importantly, mutations in the same gene can cause multiple conditions with wide phenotypic variability, and mutations in different genes can result in disorders with overlapping clinical features, linking these genes into overarching molecular networks. Studying the underlying pathophysiology of these disorders has uncovered novel regulators of RTKs, revealed new biological functions for those already identified, and advanced development of molecular-based therapies for treatment. We highlight here efforts that have provided information regarding human genetic disorders. Several other excellent reviews cover feedback regulators in cancer (Casaletto and McClatchey, 2012; Logue and Morrison, 2012; Regad, 2015)
The key role for Anosmin 1 in neuronal targeting and migration was determined by the identification of missense mutations that result in inactive protein in Kallmann syndrome (KS) (Bick et al., 1992). KS is a disorder characterized by hypogonadotropic hypogonadism (HH), defined as absent or incomplete sexual maturation by the age of 18 years, with or without anosmia. Less frequent symptoms include renal agenesis, cleft palate, mirror movements, and hearing loss (Tsai and Gill, 2006). FGFR1 loss-of-function mutations in an autosomal dominant form of KS first suggested that Anosmin 1 was involved in FGF signaling (Dode et al., 2003). Interestingly, missense mutations in members of the FGF8 set of co-regulated genes, or synexpression group, including DUSP6, SPRY4, FLRT3, and SEF, have also been identified in individuals with HH with or without anosmia (Miraoui et al., 2013). The functional characterization of these mutations may offer new insight into their molecular mechanisms of action and roles of these genes in regulation of FGF signaling in gonadotropin-releasing hormone biology.
The importance of genes that encode protein components or regulators of the RAS/MAPK pathway is elegantly demonstrated by germline mutations associated with a class of developmental disorders known as the RASopathies (Goodwin et al., 2015; Goyal et al., 2017; Jindal et al., 2015; Jindal et al., 2017; Rauen, 2013). In one of these conditions, Costello syndrome (CS), nearly all individuals have a heterozygous de novo germline mutation in HRAS that results in a constitutively active protein (Aoki et al., 2005; Estep et al., 2006), while in cardio-facio-cutaneous syndrome (CFC), patients have heterozygous activating germline mutations in KRAS, BRAF, MEK1, or MEK2, all components of the RAS/MAPK pathway (Niihori et al., 2006; Rodriguez-Viciana et al., 2006). Because of the common underlying pathway dysregulation, RASopathies exhibit numerous overlapping clinical phenotypes. Heterozygous inactivating mutations in SPRED1 cause Neurofibromatosis Legius syndrome (NFLS), a mild form of Neurofibromatosis 1 (NF1), which is characterized by multiple cafe-au-lait skin spots, variable dysmorphic features such as hypertelorism or macrocephaly, lipomas, and mild learning disabilities or attention problems (Figure 5A) (Brems et al., 2007; Brems et al., 2012). The similarities of NFLS and NF1 are explained by the shared underlying molecular mechanism: SPRED1 downregulates the RAS/MAPK pathway through neurofibromin, the NF1 gene product (Stowe et al., 2012). Interaction between these proteins facilities plasma membrane localization of neurofibromin, where it functions as a RAS GTPase-activating protein to negatively regulate RAS signaling (Adapala et al., 2010a; Dunzendorfer-Matt et al., 2016; Hirata et al., 2016; Martin et al., 1990; Stowe et al., 2012; Xu et al., 1990). Association of SPRED2 and SPRED3 with neurofibromin suggests that these isoforms may compensate for loss of Spred1 and thus helps explain the milder phenotype associated with NFLS in comparison with NF1 (Stowe et al., 2012).
The manifestations of another RASopathy, LEOPARD syndrome 1 (LPRD1), are numerous: multiple lentigines, electrocardiographic conduction abnormalities, ocular hypertelorism, pulmonic stenosis, abnormal genitalia, retardation of growth, and sensorineural deafness (Figure 5B) (Legius et al., 2002; Mendez et al., 1985). LPRD1-associated PTPN11 mutations lead to a catalytically defective SHP2 protein that acts in a dominant negative fashion and interferes with MAPK/ERK signaling (Digilio et al., 2002; Kontaridis et al., 2006; Tartaglia et al., 2006). In contrast, heterozygous missense mutations in PTNP11 that result in excessive SHP2 activity are a principal cause of Noonan syndrome (NS) (Fragale et al., 2004; Tartaglia and Gelb, 2005), a relatively common disorder characterized by short stature, facial dysmorphia, and a wide spectrum of congenital heart defects (Figure 5C) (Digilio et al., 2002; Tartaglia et al., 2001). How two mutations with opposite effects on catalytic activity result in syndromes with similar clinical symptoms is a fascinating open question. Genetic and biochemical studies in D. melanogaster and zebrafish successfully demonstrate that ptpn11 mutations associated with LPRD1 and NS result in distinct but similar phenotypes, and in the case of the zebrafish, recapitulate the craniofacial and cardiac defects of human patients (Supplemental Table 1) (Bonetti et al., 2014; Jopling et al., 2007; Oishi et al., 2006; Oishi et al., 2009; Stewart et al., 2010). Mouse models generated for the two most prevalent LPRD1 and NS PTPN11 mutations exhibit developmental defects, including reduced length, craniofacial abnormalities, congenital heart defects, with activation of the PI3K/AKT or RAS/ERK pathways, respectively (Araki et al., 2004; De Rocca Serra-Nedelec et al., 2012; Marin et al., 2011). Importantly, genetic deletion of ERK1/2 prevented cardiac abnormalities in a cardiomyocyte-specific SHP2 gain-of-function mouse model of NS (Nakamura et al., 2007), and injection of the MAPK/ERK kinase inhibitor U0126 in utero prevented craniofacial malformations in newborn pups (Nakamura et al., 2009). Similarly, pharmacological intervention with rapamycin, an inhibitor of mTOR, reversed the hypertrophic cardiomyopathy in a mouse model of LPRD1 (Marin et al., 2011). These studies suggest that some RASopathy-associated PTPN11 mutations can be rescued, opening a new therapeutic avenue for affected individuals. A NS-like phenotype has been associated with several additional genes including C-CBL (Figure 5D) (Martinelli et al., 2010; Niemeyer et al., 2010; Perez et al., 2010). In vitro studies showed that the C-CBL mutations found in patients impaired CBL-mediated degradation of cell-surface receptors in a dominant-negative fashion and caused dysregulation of intracellular signaling through RAS, explaining the overlapping phenotype in NS associated with RAS/MAPK pathway activating mutations (Martinelli et al., 2010; Schubert et al., 2006).
Germline mutations and deletions in PTEN that result in dysregulation of the PI3K/AKT pathway cause Bannayan-Ruvalcaba-Riley syndrome (BRRS) and Cowden syndrome 1 (CWS1) (Liaw et al., 1997; Marsh et al., 1999; Nelen et al., 1997; Zhou et al., 2003). BRRS and CWS1 are rare allelic disorders that share characteristics such as hamartomatous polyps of the gastrointestinal tract, mucocutaneous lesions, and increased risk of developing neoplasms (Blumenthal and Dennis, 2008). It has been suggested that both conditions and several other distinctive phenotypes associated with PTEN mutations be referred to as PTEN hamartoma tumor syndrome (Lachlan et al., 2007; Marsh et al., 1999; Nelen et al., 1997; Sarquis et al., 2006). Also included in this spectrum of disorders are PTEN-related Proteus syndrome (PS) and ‘Proteus-like’ syndrome, complex and highly variable disorders involving vascular malformations and hamartomatous overgrowth of multiple tissues associated with germline and tissue-specific somatic activating mutations in AKT1 or PTEN, respectively (Cohen, 2014; Lindhurst et al., 2011; Smith et al., 2002; Turner et al., 2004; Zhou et al., 2001; Zhou et al., 2000). These correlations demonstrate the critical involvement of PTEN in regulation of the pro-proliferative signals mediated by the PI3K/AKT pathway.
Expanding the array of clinically distinct phenotypes associated with PTEN mutations are VACTERL association and macrocephaly/autism syndrome (Butler et al., 2005; Reardon et al., 2001). VACTERL describes a constellation of congenital anomalies including vertebral anomalies, anal atresia, congenital cardiac disease, tracheoesophageal fistula, renal anomalies, radial dysplasia, and other limb defects (Khoury et al., 1983), whereas macrocephaly/autism syndrome is characterized by increased head circumference, abnormal facial features, and delayed psychomotor development resulting in autistic behavior or intellectual disability (Figure 5E) (Herman et al., 2007; Tsujita et al., 2016). Whether individuals affected with macrocephaly/autism syndrome and VACTERL association develop further clinical manifestations of other PTEN-associated syndromes is unknown. Mouse models with deficient Pten result in macrocephaly and autistic-like behavior with abnormal activation of PI3K/AKT pathway (Supplemental Table 1) (Chen et al., 2015; Clipperton-Allen and Page, 2014; Kwon et al., 2006; Page et al., 2009). Future analyses of specific disease-causing human PTEN mutations will prove useful in understanding the mechanisms underlying these heterogeneous phenotypes.
The recent discovery of autosomal recessive LRIG2 mutations in Urofacial syndrome (UFS) provides additional insight into LRIG2 function as a regulator of RTK signaling. UFS presents with urinary bladder dysfunction associated with abnormal facial expressions (Figure 5F) (Stuart et al., 2013). Interestingly, loss of function mutations in Heparanase-2, which regulates the availability and signaling of growth factors through processing of HSPGs, were also identified as causative for UFS (Daly et al., 2010; Pang et al., 2010). In fact, deletion of Hpse2 but not Lrig2 in X. laevis and mice caused UFS-like urological phenotypes. These observations suggest that HPSE2 might functionally overlap in its mode of RTK inhibition with LRIG2 (Supplemental Table 1) (Guo et al., 2015a; Roberts et al., 2014).
CONCLUDING REMARKS
From the single cell stage, RTKs guide the embryogenesis, development, and postnatal growth of nearly all organisms. Our understanding of the significant contribution that RTKs play has been enabled through extensive work in model organisms and by advances in elucidating the biochemistry, cell biology, and structure of these receptors. Equally important contributions have arisen from studies of human congenital disorders and clinical analyses of RTKs in diseases. These studies reveal that a complex network of proteins is required to guide RTKs during their lifetime in the cell, from their biosynthesis and maturation in the ER, subsequent trafficking to the cell surface, ligand-dependent activation triggering autophosphorylation and downstream signaling, and final desensitization by ubiquitination and endocytosis. Although we have come to appreciate the mechanisms by which many of these regulators contribute to the duration and extent of ligand-dependent RTK activation in development, there are still many exciting discoveries to be made about the individual steps that are necessary to properly regulate RTK signaling. Our knowledge of the specificity of individual regulators for different RTKs and insights into the mechanism of their function remains largely minimalistic. Even more lagging are structural studies on many of these targets, which are necessary to advance development of specific therapeutics for patients in which modulation of RTK regulators could be clinically beneficial. Lastly, it is likely that many feedback regulatory loops still remain to be discovered.
Supplementary Material
Highlights.
RTK signaling plays pivotal roles in cell fate determination and morphogenesis.
Feedback regulators fine-tuning the duration and extent of pathway activation.
Positive regulators function in tandem with negative regulators in development.
Germline mutations in RTK regulators cause human genetic disorders.
Acknowledgments
We thank members of the Klein and Jura laboratories for helpful discussions.
FUNDING
This work was funded by the National Institutes of Health (R35-DE026602 to O.D.K. and R01-GM109176 to N.J.), Program for Breakthrough Biomedical Research Grant to O.D.K and N.J., and Susan G. Komen Foundation Training Grant (CCR14299947 to N.J.). C.L.N. was supported by NIH 1TL1TR001871-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraira VE, Hyun N, Tucker AF, Coling DE, Brown MC, Lu C, Hoffman GR, Goodrich LV. Changes in Sef levels influence auditory brainstem development and function. J Neurosci. 2007;27:4273–4282. doi: 10.1523/JNEUROSCI.3477-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramsson A, Kurup S, Busse M, Yamada S, Lindblom P, Schallmeiner E, Stenzel D, Sauvaget D, Ledin J, Ringvall M, Landegren U, Kjellen L, Bondjers G, Li JP, Lindahl U, Spillmann D, Betsholtz C, Gerhardt H. Defective N-sulfation of heparan sulfate proteoglycans limits PDGF-BB binding and pericyte recruitment in vascular development. Genes Dev. 2007;21:316–331. doi: 10.1101/gad.398207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adapala NS, Barbe MF, Langdon WY, Nakamura MC, Tsygankov AY, Sanjay A. The loss of Cbl-phosphatidylinositol 3-kinase interaction perturbs RANKL-mediated signaling, inhibiting bone resorption and promoting osteoclast survival. J Biol Chem. 2010a;285:36745–36758. doi: 10.1074/jbc.M110.124628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adapala NS, Barbe MF, Langdon WY, Tsygankov AY, Sanjay A. Cbl-phosphatidylinositol 3 kinase interaction differentially regulates macrophage colony-stimulating factor-mediated osteoclast survival and cytoskeletal reorganization. Ann N Y Acad Sci. 2010b;1192:376–384. doi: 10.1111/j.1749-6632.2009.05346.x. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, George R, Lin CC, Suen KM, Levitt JA, Suhling K, Ladbury JE. Direct binding of Grb2 SH3 domain to FGFR2 regulates SHP2 function. Cell Signal. 2010;22:23–33. doi: 10.1016/j.cellsig.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Ahmed Z, Lin CC, Suen KM, Melo FA, Levitt JA, Suhling K, Ladbury JE. Grb2 controls phosphorylation of FGFR2 by inhibiting receptor kinase and Shp2 phosphatase activity. J Cell Biol. 2013;200:493–504. doi: 10.1083/jcb.201204106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanson JE, Bohring A, Dorr HG, Dufke A, Gillessen-Kaesbach G, Horn D, Konig R, Kratz CP, Kutsche K, Pauli S, Raskin S, Rauch A, Turner A, Wieczorek D, Zenker M. The face of Noonan syndrome: Does phenotype predict genotype. Am J Med Genet A. 2010;152A:1960–1966. doi: 10.1002/ajmg.a.33518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsina FC, Irala D, Fontanet PA, Hita FJ, Ledda F, Paratcha G. Sprouty4 is an endogenous negative modulator of TrkA signaling and neuronal differentiation induced by NGF. PLoS One. 2012;7:e32087. doi: 10.1371/journal.pone.0032087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasi S, Baietti MF, Frosi Y, Alema S, Segatto O. The evolutionarily conserved EBR module of RALT/MIG6 mediates suppression of the EGFR catalytic activity. Oncogene. 2007;26:7833–7846. doi: 10.1038/sj.onc.1210590. [DOI] [PubMed] [Google Scholar]
- Anastasi S, Lamberti D, Alema S, Segatto O. Regulation of the ErbB network by the MIG6 feedback loop in physiology, tumor suppression and responses to oncogene-targeted therapeutics. Semin Cell Dev Biol. 2016;50:115–124. doi: 10.1016/j.semcdb.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Andrenacci D, Grimaldi MR, Panetta V, Riano E, Rugarli EI, Graziani F. Functional dissection of the Drosophila Kallmann's syndrome protein DmKal-1. BMC Genet. 2006;7:47. doi: 10.1186/1471-2156-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoun G, Bouchard-Cannon P, Cheng HY. Regulation of MAPK/ERK signaling and photic entrainment of the suprachiasmatic nucleus circadian clock by Raf kinase inhibitor protein. J Neurosci. 2012;32:4867–4877. doi: 10.1523/JNEUROSCI.5650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- Araki T, Mohi MG, Ismat FA, Bronson RT, Williams IR, Kutok JL, Yang W, Pao LI, Gilliland DG, Epstein JA, Neel BG. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- Arevalo JC, Waite J, Rajagopal R, Beyna M, Chen ZY, Lee FS, Chao MV. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–559. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- Ashe HL, Briscoe J. The interpretation of morphogen gradients. Development. 2006;133:385–394. doi: 10.1242/dev.02238. [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Backman SA, Stambolic V, Suzuki A, Haight J, Elia A, Pretorius J, Tsao MS, Shannon P, Bolon B, Ivy GO, Mak TW. Deletion of Pten in mouse brain causes seizures, ataxia and defects in soma size resembling Lhermitte-Duclos disease. Nat Genet. 2001;29:396–403. doi: 10.1038/ng782. [DOI] [PubMed] [Google Scholar]
- Banks AS, Li J, McKeag L, Hribal ML, Kashiwada M, Accili D, Rothman PB. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J Clin Invest. 2005;115:2462–2471. doi: 10.1172/JCI23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson MA, Akbulut S, Watson-Johnson J, Simon R, Carroll TJ, Shakya R, Gross I, Martin GR, Lufkin T, McMahon AP, Wilson PD, Costantini FD, Mason IJ, Licht JD. Sprouty1 is a critical regulator of GDNF/RET-mediated kidney induction. Dev Cell. 2005;8:229–239. doi: 10.1016/j.devcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Basson MA, Watson-Johnson J, Shakya R, Akbulut S, Hyink D, Costantini FD, Wilson PD, Mason IJ, Licht JD. Branching morphogenesis of the ureteric epithelium during kidney development is coordinated by the opposing functions of GDNF and Sprouty1. Dev Biol. 2006;299:466–477. doi: 10.1016/j.ydbio.2006.08.051. [DOI] [PubMed] [Google Scholar]
- Bennasroune A, Gardin A, Aunis D, Cremel G, Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Bermudez O, Pages G, Gimond C. The dual-specificity MAP kinase phosphatases: critical roles in development and cancer. Am J Physiol Cell Physiol. 2010;299:C189–202. doi: 10.1152/ajpcell.00347.2009. [DOI] [PubMed] [Google Scholar]
- Bernier I, Jolles P. Purification and characterization of a basic 23 kDa cytosolic protein from bovine brain. Biochim Biophys Acta. 1984;790:174–181. doi: 10.1016/0167-4838(84)90221-8. [DOI] [PubMed] [Google Scholar]
- Bick D, Franco B, Sherins RJ, Heye B, Pike L, Crawford J, Maddalena A, Incerti B, Pragliola A, Meitinger T, Ballabio A. Brief report: intragenic deletion of the KALIG-1 gene in Kallmann's syndrome. N Engl J Med. 1992;326:1752–1755. doi: 10.1056/NEJM199206253262606. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Blumenthal GM, Dennis PA. PTEN hamartoma tumor syndromes. Eur J Hum Genet. 2008;16:1289–1300. doi: 10.1038/ejhg.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonetti M, Paardekooper Overman J, Tessadori F, Noel E, Bakkers J, den Hertog J. Noonan and LEOPARD syndrome Shp2 variants induce heart displacement defects in zebrafish. Development. 2014;141:1961–1970. doi: 10.1242/dev.106310. [DOI] [PubMed] [Google Scholar]
- Boran T, Peterkova R, Lesot H, Lyons DB, Peterka M, Klein OD. Temporal analysis of ectopic enamel production in incisors from sprouty mutant mice. J Exp Zool B Mol Dev Evol. 2009;312B:473–485. doi: 10.1002/jez.b.21254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boros J, Newitt P, Wang Q, McAvoy JW, Lovicu FJ. Sef and Sprouty expression in the developing ocular lens: implications for regulating lens cell proliferation and differentiation. Semin Cell Dev Biol. 2006;17:741–752. doi: 10.1016/j.semcdb.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher RT, Pollet N, Delius H, Niehrs C. The transmembrane protein XFLRT3 forms a complex with FGF receptors and promotes FGF signalling. Nat Cell Biol. 2004;6:38–44. doi: 10.1038/ncb1082. [DOI] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, Cools J, Marynen P, Thomas G, Yoshimura A, Legius E. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- Brems H, Pasmant E, Van Minkelen R, Wimmer K, Upadhyaya M, Legius E, Messiaen L. Review and update of SPRED1 mutations causing Legius syndrome. Hum Mutat. 2012;33:1538–1546. doi: 10.1002/humu.22152. [DOI] [PubMed] [Google Scholar]
- Bribian A, Barallobre MJ, Soussi-Yanicostas N, de Castro F. Anosmin-1 modulates the FGF-2-dependent migration of oligodendrocyte precursors in the developing optic nerve. Mol Cell Neurosci. 2006;33:2–14. doi: 10.1016/j.mcn.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Bribian A, Esteban PF, Clemente D, Soussi-Yanicostas N, Thomas JL, Zalc B, de Castro F. A novel role for anosmin-1 in the adhesion and migration of oligodendrocyte precursors. Dev Neurobiol. 2008;68:1503–1516. doi: 10.1002/dneu.20678. [DOI] [PubMed] [Google Scholar]
- Britsch S, Li L, Kirchhoff S, Theuring F, Brinkmann V, Birchmeier C, Riethmacher D. The ErbB2 and ErbB3 receptors and their ligand, neuregulin-1, are essential for development of the sympathetic nervous system. Genes Dev. 1998;12:1825–1836. doi: 10.1101/gad.12.12.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulow HE, Hobert O. Differential sulfations and epimerization define heparan sulfate specificity in nervous system development. Neuron. 2004;41:723–736. doi: 10.1016/s0896-6273(04)00084-4. [DOI] [PubMed] [Google Scholar]
- Bundschu K, Knobeloch KP, Ullrich M, Schinke T, Amling M, Engelhardt CM, Renne T, Walter U, Schuh K. Gene disruption of Spred-2 causes dwarfism. J Biol Chem. 2005;280:28572–28580. doi: 10.1074/jbc.M503640200. [DOI] [PubMed] [Google Scholar]
- Butler MG, Dasouki MJ, Zhou XP, Talebizadeh Z, Brown M, Takahashi TN, Miles JH, Wang CH, Stratton R, Pilarski R, Eng C. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. J Med Genet. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrita MA, Christofori G. Sprouty proteins, masterminds of receptor tyrosine kinase signaling. Angiogenesis. 2008;11:53–62. doi: 10.1007/s10456-008-9089-1. [DOI] [PubMed] [Google Scholar]
- Cai Z, Tao C, Li H, Ladher R, Gotoh N, Feng GS, Wang F, Zhang X. Deficient FGF signaling causes optic nerve dysgenesis and ocular coloboma. Development. 2013;140:2711–2723. doi: 10.1242/dev.089987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, Daly RJ, Kumar S, Yang B. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1:ra5. doi: 10.1126/scisignal.1160940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariboni A, Pimpinelli F, Colamarino S, Zaninetti R, Piccolella M, Rumio C, Piva F, Rugarli EI, Maggi R. The product of X-linked Kallmann's syndrome gene (KAL1) affects the migratory activity of gonadotropin-releasing hormone (GnRH)-producing neurons. Hum Mol Genet. 2004;13:2781–2791. doi: 10.1093/hmg/ddh309. [DOI] [PubMed] [Google Scholar]
- Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, Egia A, Sasaki AT, Thomas G, Kozma SC, Papa A, Nardella C, Cantley LC, Baselga J, Pandolfi PP. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Invest. 2008;118:3065–3074. doi: 10.1172/JCI34739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto JB, McClatchey AI. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat Rev Cancer. 2012;12:387–400. doi: 10.1038/nrc3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casci T, Freeman M. Control of EGF receptor signalling: lessons from fruitflies. Cancer Metastasis Rev. 1999;18:181–201. doi: 10.1023/a:1006313122373. [DOI] [PubMed] [Google Scholar]
- Casci T, Vinos J, Freeman M. Sprouty, an intracellular inhibitor of Ras signaling. Cell. 1999;96:655–665. doi: 10.1016/s0092-8674(00)80576-0. [DOI] [PubMed] [Google Scholar]
- Castellano E, Downward J. RAS Interaction with PI3K: More Than Just Another Effector Pathway. Genes Cancer. 2011;2:261–274. doi: 10.1177/1947601911408079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchi F, Pajalunga D, Fowler CA, Uren A, Rabe DC, Peruzzi B, Macdonald NJ, Blackman DK, Stahl SJ, Byrd RA, Bottaro DP. Targeted disruption of heparan sulfate interaction with hepatocyte and vascular endothelial growth factors blocks normal and oncogenic signaling. Cancer Cell. 2012;22:250–262. doi: 10.1016/j.ccr.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WL, Liou W, Pen HC, Chou HY, Chang YW, Li WH, Chiang W, Pai LM. The gradient of Gurken, a long-range morphogen, is directly regulated by Cbl-mediated endocytosis. Development. 2008;135:1923–1933. doi: 10.1242/dev.017103. [DOI] [PubMed] [Google Scholar]
- Charles C, Hovorakova M, Ahn Y, Lyons DB, Marangoni P, Churava S, Biehs B, Jheon A, Lesot H, Balooch G, Krumlauf R, Viriot L, Peterkova R, Klein OD. Regulation of tooth number by fine-tuning levels of receptor-tyrosine kinase signaling. Development. 2011;138:4063–4073. doi: 10.1242/dev.069195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Huang WC, Sejourne J, Clipperton-Allen AE, Page DT. Pten Mutations Alter Brain Growth Trajectory and Allocation of Cell Types through Elevated beta-Catenin Signaling. J Neurosci. 2015;35:10252–10267. doi: 10.1523/JNEUROSCI.5272-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi L, Zhang S, Lin Y, Prunskaite-Hyyrylainen R, Vuolteenaho R, Itaranta P, Vainio S. Sprouty proteins regulate ureteric branching by coordinating reciprocal epithelial Wnt11, mesenchymal Gdnf and stromal Fgf7 signalling during kidney development. Development. 2004;131:3345–3356. doi: 10.1242/dev.01200. [DOI] [PubMed] [Google Scholar]
- Ching ST, Cunha GR, Baskin LS, Basson MA, Klein OD. Coordinated activity of Spry1 and Spry2 is required for normal development of the external genitalia. Dev Biol. 2014;386:1–11. doi: 10.1016/j.ydbio.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho GS, Choi SC, Han JK. BMP signal attenuates FGF pathway in anteroposterior neural patterning. Biochem Biophys Res Commun. 2013;434:509–515. doi: 10.1016/j.bbrc.2013.03.105. [DOI] [PubMed] [Google Scholar]
- Choi YB, Son M, Park M, Shin J, Yun Y. SOCS-6 negatively regulates T cell activation through targeting p56lck to proteasomal degradation. J Biol Chem. 2010;285:7271–7280. doi: 10.1074/jbc.M109.073726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30:9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipperton-Allen AE, Page DT. Pten haploinsufficient mice show broad brain overgrowth but selective impairments in autism-relevant behavioral tests. Hum Mol Genet. 2014;23:3490–3505. doi: 10.1093/hmg/ddu057. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr Proteus syndrome review: molecular, clinical, and pathologic features. Clin Genet. 2014;85:111–119. doi: 10.1111/cge.12266. [DOI] [PubMed] [Google Scholar]
- Crackower MA, Oudit GY, Kozieradzki I, Sarao R, Sun H, Sasaki T, Hirsch E, Suzuki A, Shioi T, Irie-Sasaki J, Sah R, Cheng HY, Rybin VO, Lembo G, Fratta L, Oliveira-dos-Santos AJ, Benovic JL, Kahn CR, Izumo S, Steinberg SF, Wymann MP, Backx PH, Penninger JM. Regulation of myocardial contractility and cell size by distinct PI3K-PTEN signaling pathways. Cell. 2002;110:737–749. doi: 10.1016/s0092-8674(02)00969-8. [DOI] [PubMed] [Google Scholar]
- Daly SB, Urquhart JE, Hilton E, McKenzie EA, Kammerer RA, Lewis M, Kerr B, Stuart H, Donnai D, Long DA, Burgu B, Aydogdu O, Derbent M, Garcia-Minaur S, Reardon W, Gener B, Shalev S, Smith R, Woolf AS, Black GC, Newman WG. Mutations in HPSE2 cause urofacial syndrome. Am J Hum Genet. 2010;86:963–969. doi: 10.1016/j.ajhg.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance M, Montagner A, Salles JP, Yart A, Raynal P. The molecular functions of Shp2 in the Ras/Mitogen-activated protein kinase (ERK1/2) pathway. Cell Signal. 2008;20:453–459. doi: 10.1016/j.cellsig.2007.10.002. [DOI] [PubMed] [Google Scholar]
- de Castro F, Esteban PF, Bribian A, Murcia-Belmonte V, Garcia-Gonzalez D, Clemente D. The adhesion molecule anosmin-1 in neurology: Kallmann syndrome and beyond. Adv Neurobiol. 2014;8:273–292. doi: 10.1007/978-1-4614-8090-7_12. [DOI] [PubMed] [Google Scholar]
- de Castro F, Seal R, Maggi R on behalf of Group of, H.c.f.K.A.L.n. ANOS1: a unified nomenclature for Kallmann syndrome 1 gene (KAL1) and anosmin-1. Brief Funct Genomics. 2016 doi: 10.1093/bfgp/elw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maximy AA, Nakatake Y, Moncada S, Itoh N, Thiery JP, Bellusci S. Cloning and expression pattern of a mouse homologue of drosophila sprouty in the mouse embryo. Mech Dev. 1999;81:213–216. doi: 10.1016/s0925-4773(98)00241-x. [DOI] [PubMed] [Google Scholar]
- de Melker AA, van der Horst G, Calafat J, Jansen H, Borst J. c-Cbl ubiquitinates the EGF receptor at the plasma membrane and remains receptor associated throughout the endocytic route. J Cell Sci. 2001;114:2167–2178. doi: 10.1242/jcs.114.11.2167. [DOI] [PubMed] [Google Scholar]
- De Rocca Serra-Nedelec A, Edouard T, Treguer K, Tajan M, Araki T, Dance M, Mus M, Montagner A, Tauber M, Salles JP, Valet P, Neel BG, Raynal P, Yart A. Noonan syndrome-causing SHP2 mutants inhibit insulin-like growth factor 1 release via growth hormone-induced ERK hyperactivation, which contributes to short stature. Proc Natl Acad Sci U S A. 2012;109:4257–4262. doi: 10.1073/pnas.1119803109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dee A, Li K, Heng X, Guo Q, Li JY. Regulation of self-renewing neural progenitors by FGF/ERK signaling controls formation of the inferior colliculus. Development. 2016;143:3661–3673. doi: 10.1242/dev.138537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denayer E, Ahmed T, Brems H, Van Woerden G, Borgesius NZ, Callaerts-Vegh Z, Yoshimura A, Hartmann D, Elgersma Y, D'Hooge R, Legius E, Balschun D. Spred1 is required for synaptic plasticity and hippocampus-dependent learning. J Neurosci. 2008;28:14443–14449. doi: 10.1523/JNEUROSCI.4698-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Kotsi P, Peng YF, Cordon-Cardo C, Elkon KB, Pandolfi PP. Impaired Fas response and autoimmunity in Pten+/− mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- Di Schiavi E, Andrenacci D. Invertebrate models of kallmann syndrome: molecular pathogenesis and new disease genes. Curr Genomics. 2013;14:2–10. doi: 10.2174/138920213804999174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digilio MC, Conti E, Sarkozy A, Mingarelli R, Dottorini T, Marino B, Pizzuti A, Dallapiccola B. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet. 2002;71:389–394. doi: 10.1086/341528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do HT, Tselykh TV, Makela J, Ho TH, Olkkonen VM, Bornhauser BC, Korhonen L, Zelcer N, Lindholm D. Fibroblast growth factor-21 (FGF21) regulates low-density lipoprotein receptor (LDLR) levels in cells via the E3-ubiquitin ligase Mylip/Idol and the Canopy2 (Cnpy2)/Mylip-interacting saposin-like protein (Msap) J Biol Chem. 2012;287:12602–12611. doi: 10.1074/jbc.M112.341248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dode C, Levilliers J, Dupont JM, De Paepe A, Le Du N, Soussi-Yanicostas N, Coimbra RS, Delmaghani S, Compain-Nouaille S, Baverel F, Pecheux C, Le Tessier D, Cruaud C, Delpech M, Speleman F, Vermeulen S, Amalfitano A, Bachelot Y, Bouchard P, Cabrol S, Carel JC, Delemarre-van de Waal H, Goulet-Salmon B, Kottler ML, Richard O, Sanchez-Franco F, Saura R, Young J, Petit C, Hardelin JP. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–465. doi: 10.1038/ng1122. [DOI] [PubMed] [Google Scholar]
- Drinjakovic J, Jung H, Campbell DS, Strochlic L, Dwivedy A, Holt CE. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunzendorfer-Matt T, Mercado EL, Maly K, McCormick F, Scheffzek K. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc Natl Acad Sci U S A. 2016;113:7497–7502. doi: 10.1073/pnas.1607298113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou AD, Ohazama A, Porntaveetus T, Sharpe PT, Kondo S, Basson MA, Gritli-Linde A, Cobourne MT, Green JB. Periodic stripe formation by a Turing mechanism operating at growth zones in the mammalian palate. Nat Genet. 2012;44:348–351. doi: 10.1038/ng.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwin F, Singh R, Endersby R, Baker SJ, Patel TB. The tumor suppressor PTEN is necessary for human Sprouty 2-mediated inhibition of cell proliferation. J Biol Chem. 2006;281:4816–4822. doi: 10.1074/jbc.M508300200. [DOI] [PubMed] [Google Scholar]
- Egan JE, Hall AB, Yatsula BA, Bar-Sagi D. The bimodal regulation of epidermal growth factor signaling by human Sprouty proteins. Proc Natl Acad Sci U S A. 2002;99:6041–6046. doi: 10.1073/pnas.052090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea J, Erlacher C, Montanez E, Burtscher I, Yamagishi S, Hess M, Hampel F, Sanchez R, Rodriguez-Manzaneque MT, Bosl MR, Fassler R, Lickert H, Klein R. Genetic ablation of FLRT3 reveals a novel morphogenetic function for the anterior visceral endoderm in suppressing mesoderm differentiation. Genes Dev. 2008;22:3349–3362. doi: 10.1101/gad.486708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo Y, Ishiwata-Endo H, Yamada KM. Extracellular matrix protein anosmin promotes neural crest formation and regulates FGF, BMP, and WNT activities. Dev Cell. 2012;23:305–316. doi: 10.1016/j.devcel.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt CM, Bundschu K, Messerschmitt M, Renne T, Walter U, Reinhard M, Schuh K. Expression and subcellular localization of Spred proteins in mouse and human tissues. Histochem Cell Biol. 2004;122:527–538. doi: 10.1007/s00418-004-0725-6. [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am J Med Genet A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- Fager G, Camejo G, Olsson U, Ostergren-Lunden G, Bondjers G. Heparin-like glycosaminoglycans influence growth and phenotype of human arterial smooth muscle cells in vitro. II. The platelet-derived growth factor A-chain contains a sequence that specifically binds heparin. In Vitro Cell Dev Biol. 1992;28A:176–180. doi: 10.1007/BF02631088. [DOI] [PubMed] [Google Scholar]
- Farooq A, Zhou MM. Structure and regulation of MAPK phosphatases. Cell Signal. 2004;16:769–779. doi: 10.1016/j.cellsig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Ferby I, Reschke M, Kudlacek O, Knyazev P, Pante G, Amann K, Sommergruber W, Kraut N, Ullrich A, Fassler R, Klein R. Mig6 is a negative regulator of EGF receptor-mediated skin morphogenesis and tumor formation. Nat Med. 2006;12:568–573. doi: 10.1038/nm1401. [DOI] [PubMed] [Google Scholar]
- Filipe M, Goncalves L, Bento M, Silva AC, Belo JA. Comparative expression of mouse and chicken Shisa homologues during early development. Dev Dyn. 2006;235:2567–2573. doi: 10.1002/dvdy.20862. [DOI] [PubMed] [Google Scholar]
- Fiorini M, Ballaro C, Sala G, Falcone G, Alema S, Segatto O. Expression of RALT, a feedback inhibitor of ErbB receptors, is subjected to an integrated transcriptional and post-translational control. Oncogene. 2002;21:6530–6539. doi: 10.1038/sj.onc.1205823. [DOI] [PubMed] [Google Scholar]
- Forsten R, Schneider B. Treatment of the stress casualty during operation Iraqi freedom one. Psychiatr Q. 2005;76:343–350. doi: 10.1007/s11126-005-4968-9. [DOI] [PubMed] [Google Scholar]
- Fouladkou F, Lu C, Jiang C, Zhou L, She Y, Walls JR, Kawabe H, Brose N, Henkelman RM, Huang A, Bruneau BG, Rotin D. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J Biol Chem. 2010;285:6770–6780. doi: 10.1074/jbc.M109.082347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragale A, Tartaglia M, Wu J, Gelb BD. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum Mutat. 2004;23:267–277. doi: 10.1002/humu.20005. [DOI] [PubMed] [Google Scholar]
- Frayne J, Ingram C, Love S, Hall L. Localisation of phosphatidylethanolamine-binding protein in the brain and other tissues of the rat. Cell Tissue Res. 1999;298:415–423. doi: 10.1007/s004419900113. [DOI] [PubMed] [Google Scholar]
- Frosi Y, Anastasi S, Ballaro C, Varsano G, Castellani L, Maspero E, Polo S, Alema S, Segatto O. A two-tiered mechanism of EGFR inhibition by RALT/MIG6 via kinase suppression and receptor degradation. J Cell Biol. 2010;189:557–571. doi: 10.1083/jcb.201002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry WH, Simion C, Sweeney C, Carraway KL., 3rd Quantity control of the ErbB3 receptor tyrosine kinase at the endoplasmic reticulum. Mol Cell Biol. 2011;31:3009–3018. doi: 10.1128/MCB.05105-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furthauer M, Lin W, Ang SL, Thisse B, Thisse C. Sef is a feedback-induced antagonist of Ras/MAPK-mediated FGF signalling. Nat Cell Biol. 2002;4:170–174. doi: 10.1038/ncb750. [DOI] [PubMed] [Google Scholar]
- Furushima K, Yamamoto A, Nagano T, Shibata M, Miyachi H, Abe T, Ohshima N, Kiyonari H, Aizawa S. Mouse homologues of Shisa antagonistic to Wnt and Fgf signalings. Dev Biol. 2007;306:480–492. doi: 10.1016/j.ydbio.2007.03.028. [DOI] [PubMed] [Google Scholar]
- Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. 2000;221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalez D, Murcia-Belmonte V, Esteban PF, Ortega F, Diaz D, Sanchez-Vera I, Lebron-Galan R, Escobar-Castanondo L, Martinez-Millan L, Weruaga E, Garcia-Verdugo JM, Berninger B, de Castro F. Anosmin-1 over-expression increases adult neurogenesis in the subventricular zone and neuroblast migration to the olfactory bulb. Brain Struct Funct. 2016;221:239–260. doi: 10.1007/s00429-014-0904-8. [DOI] [PubMed] [Google Scholar]
- Gengrinovitch S, Berman B, David G, Witte L, Neufeld G, Ron D. Glypican-1 is a VEGF165 binding proteoglycan that acts as an extracellular chaperone for VEGF165. J Biol Chem. 1999;274:10816–10822. doi: 10.1074/jbc.274.16.10816. [DOI] [PubMed] [Google Scholar]
- Ghiglione C, Carraway KL, 3rd, Amundadottir LT, Boswell RE, Perrimon N, Duffy JB. The transmembrane molecule kekkon 1 acts in a feedback loop to negatively regulate the activity of the Drosophila EGF receptor during oogenesis. Cell. 1999;96:847–856. doi: 10.1016/s0092-8674(00)80594-2. [DOI] [PubMed] [Google Scholar]
- Gianola S, de Castro F, Rossi F. Anosmin-1 stimulates outgrowth and branching of developing Purkinje axons. Neuroscience. 2009;158:570–584. doi: 10.1016/j.neuroscience.2008.10.022. [DOI] [PubMed] [Google Scholar]
- Gibbons R, Adeoya-Osiguwa SA, Fraser LR. A mouse sperm decapacitation factor receptor is phosphatidylethanolamine-binding protein 1. Reproduction. 2005;130:497–508. doi: 10.1530/rep.1.00792. [DOI] [PubMed] [Google Scholar]
- Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 1999;13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goberdhan DC, Wilson C. PTEN: tumour suppressor, multifunctional growth regulator and more. Hum Mol Genet. 2003;12(Spec No 2):R239–248. doi: 10.1093/hmg/ddg288. [DOI] [PubMed] [Google Scholar]
- Goh LK, Sorkin A. Endocytosis of receptor tyrosine kinases. Cold Spring Harb Perspect Biol. 2013;5:a017459. doi: 10.1101/cshperspect.a017459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldoni S, Iozzo RA, Kay P, Campbell S, McQuillan A, Agnew C, Zhu JX, Keene DR, Reed CC, Iozzo RV. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene. 2007;26:368–381. doi: 10.1038/sj.onc.1209803. [DOI] [PubMed] [Google Scholar]
- Goldshmit Y, Walters CE, Scott HJ, Greenhalgh CJ, Turnley AM. SOCS2 induces neurite outgrowth by regulation of epidermal growth factor receptor activation. J Biol Chem. 2004;279:16349–16355. doi: 10.1074/jbc.M312873200. [DOI] [PubMed] [Google Scholar]
- Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996;122:223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Martinez D, Kim SH, Hu Y, Guimond S, Schofield J, Winyard P, Vannelli GB, Turnbull J, Bouloux PM. Anosmin-1 modulates fibroblast growth factor receptor 1 signaling in human gonadotropin-releasing hormone olfactory neuroblasts through a heparan sulfate-dependent mechanism. J Neurosci. 2004;24:10384–10392. doi: 10.1523/JNEUROSCI.3400-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin AF, Kim R, Bush JO, Klein OD. From Bench to Bedside and Back: Improving Diagnosis and Treatment of Craniofacial Malformations Utilizing Animal Models. Curr Top Dev Biol. 2015;115:459–492. doi: 10.1016/bs.ctdb.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal Y, Jindal GA, Pelliccia JL, Yamaya K, Yeung E, Futran AS, Burdine RD, Schupbach T, Shvartsman SY. Divergent effects of intrinsically active MEK variants on developmental Ras signaling. Nat Genet. 2017;49:465–469. doi: 10.1038/ng.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh CJ, Bertolino P, Asa SL, Metcalf D, Corbin JE, Adams TE, Davey HW, Nicola NA, Hilton DJ, Alexander WS. Growth enhancement in suppressor of cytokine signaling 2 (SOCS-2)-deficient mice is dependent on signal transducer and activator of transcription 5b (STAT5b) Mol Endocrinol. 2002;16:1394–1406. doi: 10.1210/mend.16.6.0845. [DOI] [PubMed] [Google Scholar]
- Groom LA, Sneddon AA, Alessi DR, Dowd S, Keyse SM. Differential regulation of the MAP, SAP and RK/p38 kinases by Pyst1, a novel cytosolic dual-specificity phosphatase. EMBO J. 1996;15:3621–3632. [PMC free article] [PubMed] [Google Scholar]
- Gross I, Morrison DJ, Hyink DP, Georgas K, English MA, Mericskay M, Hosono S, Sassoon D, Wilson PD, Little M, Licht JD. The receptor tyrosine kinase regulator Sprouty1 is a target of the tumor suppressor WT1 and important for kidney development. J Biol Chem. 2003;278:41420–41430. doi: 10.1074/jbc.M306425200. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Guo C, Kaneko S, Sun Y, Huang Y, Vlodavsky I, Li X, Li ZR, Li X. A mouse model of urofacial syndrome with dysfunctional urination. Hum Mol Genet. 2015a;24:1991–1999. doi: 10.1093/hmg/ddu613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Mihic A, Wu J, Zhang Y, Singh K, Dhingra S, Weisel RD, Li RK. Canopy 2 attenuates the transition from compensatory hypertrophy to dilated heart failure in hypertrophic cardiomyopathy. Eur Heart J. 2015b;36:2530–2540. doi: 10.1093/eurheartj/ehv294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Zhang Y, Mihic A, Li SH, Sun Z, Shao Z, Wu J, Weisel RD, Li RK. A secreted protein (Canopy 2, CNPY2) enhances angiogenesis and promotes smooth muscle cell migration and proliferation. Cardiovasc Res. 2015c;105:383–393. doi: 10.1093/cvr/cvv010. [DOI] [PubMed] [Google Scholar]
- Gupta S, Mishra K, Surolia A, Banerjee K. Suppressor of cytokine signalling-6 promotes neurite outgrowth via JAK2/STAT5-mediated signalling pathway, involving negative feedback inhibition. PLoS One. 2011;6:e26674. doi: 10.1371/journal.pone.0026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, Amariglio N, Henriksson R, Rechavi G, Hedman H, Wides R, Yarden Y. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J. 2004;23:3270–3281. doi: 10.1038/sj.emboj.7600342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy GR, Jackson RA, Yusoff P, Chow SY. Sprouty proteins: modified modulators, matchmakers or missing links? J Endocrinol. 2009;203:191–202. doi: 10.1677/JOE-09-0110. [DOI] [PubMed] [Google Scholar]
- Hackel PO, Gishizky M, Ullrich A. Mig-6 is a negative regulator of the epidermal growth factor receptor signal. Biol Chem. 2001;382:1649–1662. doi: 10.1515/BC.2001.200. [DOI] [PubMed] [Google Scholar]
- Hacohen N, Kramer S, Sutherland D, Hiromi Y, Krasnow MA. sprouty encodes a novel antagonist of FGF signaling that patterns apical branching of the Drosophila airways. Cell. 1998;92:253–263. doi: 10.1016/s0092-8674(00)80919-8. [DOI] [PubMed] [Google Scholar]
- Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–3973. doi: 10.1128/mcb.18.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–466. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- Haines BP, Wheldon LM, Summerbell D, Heath JK, Rigby PW. Regulated expression of FLRT genes implies a functional role in the regulation of FGF signalling during mouse development. Dev Biol. 2006;297:14–25. doi: 10.1016/j.ydbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Hall AB, Jura N, DaSilva J, Jang YJ, Gong D, Bar-Sagi D. hSpry2 is targeted to the ubiquitin-dependent proteasome pathway by c-Cbl. Curr Biol. 2003;13:308–314. doi: 10.1016/s0960-9822(03)00086-1. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Matsumoto K, Nishida E. Shp2, an SH2-containing protein-tyrosine phosphatase, positively regulates receptor tyrosine kinase signaling by dephosphorylating and inactivating the inhibitor Sprouty. J Biol Chem. 2004;279:22992–22995. doi: 10.1074/jbc.M312498200. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 and Sprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–858. doi: 10.1038/ncb867. [DOI] [PubMed] [Google Scholar]
- Harduf H, Halperin E, Reshef R, Ron D. Sef is synexpressed with FGFs during chick embryogenesis and its expression is differentially regulated by FGFs in the developing limb. Dev Dyn. 2005;233:301–312. doi: 10.1002/dvdy.20364. [DOI] [PubMed] [Google Scholar]
- Hart BE, Tapping RI. Cell surface trafficking of TLR1 is differentially regulated by the chaperones PRAT4A and PRAT4B. J Biol Chem. 2012;287:16550–16562. doi: 10.1074/jbc.M112.342717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J, Wald JH, Rafidi H, Cuevas A, Sweeney C, Carraway KL., 3rd The ER structural protein Rtn4A stabilizes and enhances signaling through the receptor tyrosine kinase ErbB3. Sci Signal. 2016;9:ra65. doi: 10.1126/scisignal.aaf1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedge TA, Mason I. Expression of Shisa2, a modulator of both Wnt and Fgf signaling, in the chick embryo. Int J Dev Biol. 2008;52:81–85. doi: 10.1387/ijdb.072355th. [DOI] [PubMed] [Google Scholar]
- Hendriks WJ, Elson A, Harroch S, Pulido R, Stoker A, den Hertog J. Protein tyrosine phosphatases in health and disease. FEBS J. 2013;280:708–730. doi: 10.1111/febs.12000. [DOI] [PubMed] [Google Scholar]
- Herman GE, Henninger N, Ratliff-Schaub K, Pastore M, Fitzgerald S, McBride KL. Genetic testing in autism: how much is enough? Genet Med. 2007;9:268–274. doi: 10.1097/gim.0b013e31804d683b. [DOI] [PubMed] [Google Scholar]
- Heuberger J, Kosel F, Qi J, Grossmann KS, Rajewsky K, Birchmeier W. Shp2/MAPK signaling controls goblet/paneth cell fate decisions in the intestine. Proc Natl Acad Sci U S A. 2014;111:3472–3477. doi: 10.1073/pnas.1309342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y, Brems H, Suzuki M, Kanamori M, Okada M, Morita R, Llano-Rivas I, Ose T, Messiaen L, Legius E, Yoshimura A. Interaction between a Domain of the Negative Regulator of the Ras-ERK Pathway, SPRED1 Protein, and the GTPase-activating Protein-related Domain of Neurofibromin Is Implicated in Legius Syndrome and Neurofibromatosis Type 1. J Biol Chem. 2016;291:3124–3134. doi: 10.1074/jbc.M115.703710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Okamoto H. Canopy1, a novel regulator of FGF signaling around the midbrain-hindbrain boundary in zebrafish. Curr Biol. 2006;16:421–427. doi: 10.1016/j.cub.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, Mizuno K, Hasegawa G, Kishimoto H, Iizuka M, Naito M, Enomoto K, Watanabe S, Mak TW, Nakano T. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113:1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz A, Simons M. Branching morphogenesis. Circ Res. 2008;103:784–795. doi: 10.1161/CIRCRESAHA.108.181818. [DOI] [PubMed] [Google Scholar]
- Hsia HE, Kumar R, Luca R, Takeda M, Courchet J, Nakashima J, Wu S, Goebbels S, An W, Eickholt BJ, Polleux F, Rotin D, Wu H, Rossner MJ, Bagni C, Rhee JS, Brose N, Kawabe H. Ubiquitin E3 ligase Nedd4-1 acts as a downstream target of PI3K/PTEN-mTORC1 signaling to promote neurite growth. Proc Natl Acad Sci U S A. 2014;111:13205–13210. doi: 10.1073/pnas.1400737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Guimond SE, Travers P, Cadman S, Hohenester E, Turnbull JE, Kim SH, Bouloux PM. Novel mechanisms of fibroblast growth factor receptor 1 regulation by extracellular matrix protein anosmin-1. J Biol Chem. 2009;284:29905–29920. doi: 10.1074/jbc.M109.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Potter CJ, Tao W, Li DM, Brogiolo W, Hafen E, Sun H, Xu T. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. 1999;126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19:117–123. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Kato R, Fukuyama S, Nonami A, Taniguchi K, Matsumoto K, Nakano T, Tsuda M, Matsumura M, Kubo M, Ishikawa F, Moon BG, Takatsu K, Nakanishi Y, Yoshimura A. Spred-1 negatively regulates allergen-induced airway eosinophilia and hyperresponsiveness. J Exp Med. 2005;201:73–82. doi: 10.1084/jem.20040616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Ueda T, Ueno A, Nakagawa H, Taniguchi H, Kayukawa N, Miki T. A genetic screen in Drosophila for regulators of human prostate cancer progression. Biochem Biophys Res Commun. 2014;451:548–555. doi: 10.1016/j.bbrc.2014.08.015. [DOI] [PubMed] [Google Scholar]
- Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Jeong JW, Lee HS, Lee KY, White LD, Broaddus RR, Zhang YW, Vande Woude GF, Giudice LC, Young SL, Lessey BA, Tsai SY, Lydon JP, DeMayo FJ. Mig-6 modulates uterine steroid hormone responsiveness and exhibits altered expression in endometrial disease. Proc Natl Acad Sci U S A. 2009;106:8677–8682. doi: 10.1073/pnas.0903632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin N, Cho SN, Raso MG, Wistuba I, Smith Y, Yang Y, Kurie JM, Yen R, Evans CM, Ludwig T, Jeong JW, DeMayo FJ. Mig-6 is required for appropriate lung development and to ensure normal adult lung homeostasis. Development. 2009;136:3347–3356. doi: 10.1242/dev.032979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Goyal Y, Burdine RD, Rauen KA, Shvartsman SY. RASopathies: unraveling mechanisms with animal models. Dis Model Mech. 2015;8:1167. doi: 10.1242/dmm.022442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Goyal Y, Yamaya K, Futran AS, Kountouridis I, Balgobin CA, Schupbach T, Burdine RD, Shvartsman SY. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc Natl Acad Sci U S A. 2017;114:510–515. doi: 10.1073/pnas.1615651114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo A, Long R, Cheng Z, Alexander C, Chang W, Klein OD. Sprouty2 regulates endochondral bone formation by modulation of RTK and BMP signaling. Bone. 2016;88:170–179. doi: 10.1016/j.bone.2016.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, van Geemen D, den Hertog J. Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 2007;3:e225. doi: 10.1371/journal.pgen.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaulanov EE, Bottcher RT, Niehrs C. A role for fibronectin-leucine-rich transmembrane cell-surface proteins in homotypic cell adhesion. EMBO Rep. 2006;7:283–290. doi: 10.1038/sj.embor.7400614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kario E, Marmor MD, Adamsky K, Citri A, Amit I, Amariglio N, Rechavi G, Yarden Y. Suppressors of cytokine signaling 4 and 5 regulate epidermal growth factor receptor signaling. J Biol Chem. 2005;280:7038–7048. doi: 10.1074/jbc.M408575200. [DOI] [PubMed] [Google Scholar]
- Karlsson T, Mark EB, Henriksson R, Hedman H. Redistribution of LRIG proteins in psoriasis. J Invest Dermatol. 2008;128:1192–1195. doi: 10.1038/sj.jid.5701175. [DOI] [PubMed] [Google Scholar]
- Kato R, Nonami A, Taketomi T, Wakioka T, Kuroiwa A, Matsuda Y, Yoshimura A. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem Biophys Res Commun. 2003;302:767–772. doi: 10.1016/s0006-291x(03)00259-6. [DOI] [PubMed] [Google Scholar]
- Katz S, Ayala V, Santillan G, Boland R. Activation of the PI3K/Akt signaling pathway through P2Y(2) receptors by extracellular ATP is involved in osteoblastic cell proliferation. Arch Biochem Biophys. 2011;513:144–152. doi: 10.1016/j.abb.2011.06.013. [DOI] [PubMed] [Google Scholar]
- Kawabe H, Neeb A, Dimova K, Young SM, Jr, Takeda M, Katsurabayashi S, Mitkovski M, Malakhova OA, Zhang DE, Umikawa M, Kariya K, Goebbels S, Nave KA, Rosenmund C, Jahn O, Rhee J, Brose N. Regulation of Rap2A by the ubiquitin ligase Nedd4-1 controls neurite development. Neuron. 2010;65:358–372. doi: 10.1016/j.neuron.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami Y, Rodriguez-Leon J, Koth CM, Buscher D, Itoh T, Raya A, Ng JK, Esteban CR, Takahashi S, Henrique D, Schwarz MF, Asahara H, Izpisua Belmonte JC. MKP3 mediates the cellular response to FGF8 signalling in the vertebrate limb. Nat Cell Biol. 2003;5:513–519. doi: 10.1038/ncb989. [DOI] [PubMed] [Google Scholar]
- Khoury MJ, Cordero JF, Greenberg F, James LM, Erickson JD. A population study of the VACTERL association: evidence for its etiologic heterogeneity. Pediatrics. 1983;71:815–820. [PubMed] [Google Scholar]
- King JA, Corcoran NM, D'Abaco GM, Straffon AF, Smith CT, Poon CL, Buchert M, I S, Hall NE, Lock P, Hovens CM. Eve-3: a liver enriched suppressor of Ras/MAPK signaling. J Hepatol. 2006;44:758–767. doi: 10.1016/j.jhep.2005.10.031. [DOI] [PubMed] [Google Scholar]
- Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An FGF signaling loop sustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–385. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein OD, Minowada G, Peterkova R, Kangas A, Yu BD, Lesot H, Peterka M, Jernvall J, Martin GR. Sprouty genes control diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–190. doi: 10.1016/j.devcel.2006.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knobbe CB, Lapin V, Suzuki A, Mak TW. The roles of PTEN in development, physiology and tumorigenesis in mouse models: a tissue-by-tissue survey. Oncogene. 2008;27:5398–5415. doi: 10.1038/onc.2008.238. [DOI] [PubMed] [Google Scholar]
- Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, Hoffman MP. Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors. Dev Cell. 2015;32:667–677. doi: 10.1016/j.devcel.2015.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- Korsensky L, Ron D. Regulation of FGF signaling: Recent insights from studying positive and negative modulators. Semin Cell Dev Biol. 2016;53:101–114. doi: 10.1016/j.semcdb.2016.01.023. [DOI] [PubMed] [Google Scholar]
- Kovalenko D, Yang X, Chen PY, Nadeau RJ, Zubanova O, Pigeon K, Friesel R. A role for extracellular and transmembrane domains of Sef in Sef-mediated inhibition of FGF signaling. Cell Signal. 2006;18:1958–1966. doi: 10.1016/j.cellsig.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Kovalenko D, Yang X, Nadeau RJ, Harkins LK, Friesel R. Sef inhibits fibroblast growth factor signaling by inhibiting FGFR1 tyrosine phosphorylation and subsequent ERK activation. J Biol Chem. 2003;278:14087–14091. doi: 10.1074/jbc.C200606200. [DOI] [PubMed] [Google Scholar]
- Krebs DL, Uren RT, Metcalf D, Rakar S, Zhang JG, Starr R, De Souza DP, Hanzinikolas K, Eyles J, Connolly LM, Simpson RJ, Nicola NA, Nicholson SE, Baca M, Hilton DJ, Alexander WS. SOCS-6 binds to insulin receptor substrate 4, and mice lacking the SOCS-6 gene exhibit mild growth retardation. Mol Cell Biol. 2002;22:4567–4578. doi: 10.1128/MCB.22.13.4567-4578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku BJ, Kim TH, Lee JH, Buras ED, White LD, Stevens RD, Ilkayeva OR, Bain JR, Newgard CB, DeMayo FJ, Jeong JW. Mig-6 plays a critical role in the regulation of cholesterol homeostasis and bile acid synthesis. PLoS One. 2012;7:e42915. doi: 10.1371/journal.pone.0042915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, Li Y, Baker SJ, Parada LF. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Zhu X, Zhang J, Knoop LL, Tharp R, Smeyne RJ, Eberhart CG, Burger PC, Baker SJ. Pten regulates neuronal soma size: a mouse model of Lhermitte-Duclos disease. Nat Genet. 2001;29:404–411. doi: 10.1038/ng781. [DOI] [PubMed] [Google Scholar]
- Lachlan KL, Lucassen AM, Bunyan D, Temple IK. Cowden syndrome and Bannayan Riley Ruvalcaba syndrome represent one condition with variable expression and age-related penetrance: results of a clinical study of PTEN mutation carriers. J Med Genet. 2007;44:579–585. doi: 10.1136/jmg.2007.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, Sweeney C. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- Lagronova-Churava S, Spoutil F, Vojtechova S, Lesot H, Peterka M, Klein OD, Peterkova R. The dynamics of supernumerary tooth development are differentially regulated by Sprouty genes. J Exp Zool B Mol Dev Evol. 2013;320:307–320. doi: 10.1002/jez.b.22502. [DOI] [PubMed] [Google Scholar]
- Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci. 2008;28:39–49. doi: 10.1523/JNEUROSCI.2196-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledda F, Paratcha G. Negative Regulation of Receptor Tyrosine Kinase (RTK) Signaling: A Developing Field. Biomark Insights. 2007;2:45–58. [PMC free article] [PubMed] [Google Scholar]
- Leeksma OC, Van Achterberg TA, Tsumura Y, Toshima J, Eldering E, Kroes WG, Mellink C, Spaargaren M, Mizuno K, Pannekoek H, de Vries CJ. Human sprouty 4, a new ras antagonist on 5q31, interacts with the dual specificity kinase TESK1. Eur J Biochem. 2002;269:2546–2556. doi: 10.1046/j.1432-1033.2002.02921.x. [DOI] [PubMed] [Google Scholar]
- Legius E, Schrander-Stumpel C, Schollen E, Pulles-Heintzberger C, Gewillig M, Fryns JP. PTPN11 mutations in LEOPARD syndrome. J Med Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Freed DM, Schlessinger J, Kiyatkin A. The Dark Side of Cell Signaling: Positive Roles for Negative Regulators. Cell. 2016;164:1172–1184. doi: 10.1016/j.cell.2016.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Ettenberg SA, Katz M, Tsygankov AY, Alroy I, Lavi S, Iwai K, Reiss Y, Ciechanover A, Lipkowitz S, Yarden Y. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, Waterman H, Zamir E, Kam Z, Oved S, Langdon WY, Beguinot L, Geiger B, Yarden Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 1998;12:3663–3674. doi: 10.1101/gad.12.23.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Scott DA, Hatch E, Tian X, Mansour SL. Dusp6 (Mkp3) is a negative feedback regulator of FGF-stimulated ERK signaling during mouse development. Development. 2007;134:167–176. doi: 10.1242/dev.02701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Massa PE, Hanidu A, Peet GW, Aro P, Savitt A, Mische S, Li J, Marcu KB. IKKalpha, IKKbeta, and NEMO/IKKgamma are each required for the NF-kappa B-mediated inflammatory response program. J Biol Chem. 2002;277:45129–45140. doi: 10.1074/jbc.M205165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- Lin W, Furthauer M, Thisse B, Thisse C, Jing N, Ang SL. Cloning of the mouse Sef gene and comparative analysis of its expression with Fgf8 and Spry2 during embryogenesis. Mech Dev. 2002;113:163–168. doi: 10.1016/s0925-4773(02)00018-7. [DOI] [PubMed] [Google Scholar]
- Lin W, Jing N, Basson MA, Dierich A, Licht J, Ang SL. Synergistic activity of Sef and Sprouty proteins in regulating the expression of Gbx2 in the mid-hindbrain region. Genesis. 2005;41:110–115. doi: 10.1002/gene.20103. [DOI] [PubMed] [Google Scholar]
- Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, Turner J, Cannons JL, Bick D, Blakemore L, Blumhorst C, Brockmann K, Calder P, Cherman N, Deardorff MA, Everman DB, Golas G, Greenstein RM, Kato BM, Keppler-Noreuil KM, Kuznetsov SA, Miyamoto RT, Newman K, Ng D, O'Brien K, Rothenberg S, Schwartzentruber DJ, Singhal V, Tirabosco R, Upton J, Wientroub S, Zackai EH, Hoag K, Whitewood-Neal T, Robey PG, Schwartzberg PL, Darling TN, Tosi LL, Mullikin JC, Biesecker LG. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Yang T, Wang B, Zhang M, Gu N, Qiu J, Fan HQ, Zhang CM, Fei L, Pan XQ, Guo M, Chen RH, Guo XR. Resistin induces insulin resistance, but does not affect glucose output in rat-derived hepatocytes. Acta Pharmacol Sin. 2008a;29:98–104. doi: 10.1111/j.1745-7254.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- Liu X, Mameza MG, Lee YS, Eseonu CI, Yu CR, Kang Derwent JJ, Egwuagu CE. Suppressors of cytokine-signaling proteins induce insulin resistance in the retina and promote survival of retinal cells. Diabetes. 2008b;57:1651–1658. doi: 10.2337/db07-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Oppenheim RW, Sugiura Y, Lin W. Abnormal development of the neuromuscular junction in Nedd4-deficient mice. Dev Biol. 2009;330:153–166. doi: 10.1016/j.ydbio.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochovska K, Peterkova R, Pavlikova Z, Hovorakova M. Sprouty gene dosage influences temporal-spatial dynamics of primary enamel knot formation. BMC Dev Biol. 2015;15:21. doi: 10.1186/s12861-015-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue JS, Morrison DK. Complexity in the signaling network: insights from the use of targeted inhibitors in cancer therapy. Genes Dev. 2012;26:641–650. doi: 10.1101/gad.186965.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu RL, Hu CP, Yang HP, Li YY, Gu QH, Wu L. Biological characteristics and epidermal growth factor receptor tyrosine kinase inhibitors efficacy of EGFR mutation and its subtypes in lung adenocarcinoma. Pathol Oncol Res. 2014;20:445–451. doi: 10.1007/s12253-013-9715-0. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- Lyons DA, Pogoda HM, Voas MG, Woods IG, Diamond B, Nix R, Arana N, Jacobs J, Talbot WS. erbb3 and erbb2 are essential for schwann cell migration and myelination in zebrafish. Curr Biol. 2005;15:513–524. doi: 10.1016/j.cub.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Maddirevula S, Anuppalle M, Huh TL, Kim SH, Rhee M. Nrdp1 governs differentiation of the melanocyte lineage via Erbb3b signaling in the zebrafish embryogenesis. Biochem Biophys Res Commun. 2011;409:454–458. doi: 10.1016/j.bbrc.2011.05.025. [DOI] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- Maillet M, Purcell NH, Sargent MA, York AJ, Bueno OF, Molkentin JD. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J Biol Chem. 2008;283:31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marangoni P, Charles C, Tafforeau P, Laugel-Haushalter V, Joo A, Bloch-Zupan A, Klein OD, Viriot L. Phenotypic and evolutionary implications of modulating the ERK-MAPK cascade using the dentition as a model. Sci Rep. 2015;5:11658. doi: 10.1038/srep11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretto S, Muller PS, Aricescu AR, Cho KW, Bikoff EK, Robertson EJ. Ventral closure, headfold fusion and definitive endoderm migration defects in mouse embryos lacking the fibronectin leucine-rich transmembrane protein FLRT3. Dev Biol. 2008;318:184–193. doi: 10.1016/j.ydbio.2008.03.021. [DOI] [PubMed] [Google Scholar]
- Marin TM, Keith K, Davies B, Conner DA, Guha P, Kalaitzidis D, Wu X, Lauriol J, Wang B, Bauer M, Bronson R, Franchini KG, Neel BG, Kontaridis MI. Rapamycin reverses hypertrophic cardiomyopathy in a mouse model of LEOPARD syndrome-associated PTPN11 mutation. J Clin Invest. 2011;121:1026–1043. doi: 10.1172/JCI44972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh DJ, Kum JB, Lunetta KL, Bennett MJ, Gorlin RJ, Ahmed SF, Bodurtha J, Crowe C, Curtis MA, Dasouki M, Dunn T, Feit H, Geraghty MT, Graham JM, Jr, Hodgson SV, Hunter A, Korf BR, Manchester D, Miesfeldt S, Murday VA, Nathanson KL, Parisi M, Pober B, Romano C, Eng C, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63:843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- Martinelli S, De Luca A, Stellacci E, Rossi C, Checquolo S, Lepri F, Caputo V, Silvano M, Buscherini F, Consoli F, Ferrara G, Digilio MC, Cavaliere ML, van Hagen JM, Zampino G, van der Burgt I, Ferrero GB, Mazzanti L, Screpanti I, Yntema HG, Nillesen WM, Savarirayan R, Zenker M, Dallapiccola B, Gelb BD, Tartaglia M. Heterozygous germline mutations in the CBL tumor-suppressor gene cause a Noonan syndrome-like phenotype. Am J Hum Genet. 2010;87:250–257. doi: 10.1016/j.ajhg.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JM, Morrison DJ, Bassit B, Dimri M, Band H, Licht JD, Gross I. Tyrosine phosphorylation of Sprouty proteins regulates their ability to inhibit growth factor signaling: a dual feedback loop. Mol Biol Cell. 2004;15:2176–2188. doi: 10.1091/mbc.E03-07-0503. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mason JM, Morrison DJ, Basson MA, Licht JD. Sprouty proteins: multifaceted negative-feedback regulators of receptor tyrosine kinase signaling. Trends Cell Biol. 2006;16:45–54. doi: 10.1016/j.tcb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Matsui T, Thitamadee S, Murata T, Kakinuma H, Nabetani T, Hirabayashi Y, Hirate Y, Okamoto H, Bessho Y. Canopy1, a positive feedback regulator of FGF signaling, controls progenitor cell clustering during Kupffer's vesicle organogenesis. Proc Natl Acad Sci U S A. 2011;108:9881–9886. doi: 10.1073/pnas.1017248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisner H, Daga A, Buxton J, Fernandez B, Chawla A, Banerjee U, Czech MP. Interactions of Drosophila Cbl with epidermal growth factor receptors and role of Cbl in R7 photoreceptor cell development. Mol Cell Biol. 1997;17:2217–2225. doi: 10.1128/mcb.17.4.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellett M, Atzei P, Bergin R, Horgan A, Floss T, Wurst W, Callanan JJ, Moynagh PN. Orphan receptor IL-17RD regulates Toll-like receptor signalling via SEFIR/TIR interactions. Nat Commun. 2015;6:6669. doi: 10.1038/ncomms7669. [DOI] [PubMed] [Google Scholar]
- Mendez HM, Paskulin GA, Vallandro C. The syndrome of retinal pigmentary degeneration, microcephaly, and severe mental retardation (Mirhosseini-Holmes-Walton syndrome): report of two patients. Am J Med Genet. 1985;22:223–228. doi: 10.1002/ajmg.1320220202. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Zhang W, Ballif BA, Elliott HL, Danuser G, Blenis J. ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol Cell. 2011;41:661–671. doi: 10.1016/j.molcel.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453:745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaylira CZ, Simmons JG, Ramocki NM, Scull BP, McNaughton KK, Fuller CR, Lund PK. Suppressor of cytokine signaling-2 limits intestinal growth and enterotrophic actions of IGF-I in vivo. Am J Physiol Gastrointest Liver Physiol. 2006;291:G472–481. doi: 10.1152/ajpgi.00218.2005. [DOI] [PubMed] [Google Scholar]
- Michos O, Cebrian C, Hyink D, Grieshammer U, Williams L, D'Agati V, Licht JD, Martin GR, Costantini F. Kidney development in the absence of Gdnf and Spry1 requires Fgf10. PLoS Genet. 2010;6:e1000809. doi: 10.1371/journal.pgen.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minowada G, Jarvis LA, Chi CL, Neubuser A, Sun X, Hacohen N, Krasnow MA, Martin GR. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126:4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- Miraoui H, Dwyer AA, Sykiotis GP, Plummer L, Chung W, Feng B, Beenken A, Clarke J, Pers TH, Dworzynski P, Keefe K, Niedziela M, Raivio T, Crowley WF, Jr, Seminara SB, Quinton R, Hughes VA, Kumanov P, Young J, Yialamas MA, Hall JE, Van Vliet G, Chanoine JP, Rubenstein J, Mohammadi M, Tsai PS, Sidis Y, Lage K, Pitteloud N. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet. 2013;92:725–743. doi: 10.1016/j.ajhg.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohapatra B, Ahmad G, Nadeau S, Zutshi N, An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, Bailey TA, Palermo N, Borgstahl GE, Natarajan A, Raja SM, Naramura M, Band V, Band H. Protein tyrosine kinase regulation by ubiquitination: critical roles of Cbl-family ubiquitin ligases. Biochim Biophys Acta. 2013;1833:122–139. doi: 10.1016/j.bbamcr.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourey RJ, Vega QC, Campbell JS, Wenderoth MP, Hauschka SD, Krebs EG, Dixon JE. A novel cytoplasmic dual specificity protein tyrosine phosphatase implicated in muscle and neuronal differentiation. J Biol Chem. 1996;271:3795–3802. doi: 10.1074/jbc.271.7.3795. [DOI] [PubMed] [Google Scholar]
- Muda M, Theodosiou A, Rodrigues N, Boschert U, Camps M, Gillieron C, Davies K, Ashworth A, Arkinstall S. The dual specificity phosphatases M3/6 and MKP-3 are highly selective for inactivation of distinct mitogen-activated protein kinases. J Biol Chem. 1996;271:27205–27208. doi: 10.1074/jbc.271.44.27205. [DOI] [PubMed] [Google Scholar]
- Murcia-Belmonte V, Esteban PF, Martinez-Hernandez J, Gruart A, Lujan R, Delgado-Garcia JM, de Castro F. Anosmin-1 over-expression regulates oligodendrocyte precursor cell proliferation, migration and myelin sheath thickness. Brain Struct Funct. 2016;221:1365–1385. doi: 10.1007/s00429-014-0977-4. [DOI] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Ghosh AK, Patil SB, Reed NA, Chan LS, Davuluri S, Peranen J, Hurd TW, Rachel RA, Khanna H. Accumulation of the Raf-1 kinase inhibitory protein (Rkip) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies. J Biol Chem. 2011;286:28276–28286. doi: 10.1074/jbc.M111.237560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Takehara S, Takahashi M, Aizawa S, Yamamoto A. Shisa2 promotes the maturation of somitic precursors and transition to the segmental fate in Xenopus embryos. Development. 2006;133:4643–4654. doi: 10.1242/dev.02657. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Zarycki J, Sullivan JL, Jung JU. Abnormal T cell receptor signal transduction of CD4 Th cells in X-linked lymphoproliferative syndrome. J Immunol. 2001;167:2657–2665. doi: 10.4049/jimmunol.167.5.2657. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Colbert M, Krenz M, Molkentin JD, Hahn HS, Dorn GW, 2nd, Robbins J. Mediating ERK 1/2 signaling rescues congenital heart defects in a mouse model of Noonan syndrome. J Clin Invest. 2007;117:2123–2132. doi: 10.1172/JCI30756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Gulick J, Pratt R, Robbins J. Noonan syndrome is associated with enhanced pERK activity, the repression of which can prevent craniofacial malformations. Proc Natl Acad Sci U S A. 2009;106:15436–15441. doi: 10.1073/pnas.0903302106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proc Natl Acad Sci U S A. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neben CL, Merrill AE. Signaling Pathways in Craniofacial Development: Insights from Rare Skeletal Disorders. Curr Top Dev Biol. 2015;115:493–542. doi: 10.1016/bs.ctdb.2015.09.005. [DOI] [PubMed] [Google Scholar]
- Nelen MR, van Staveren WC, Peeters EA, Hassel MB, Gorlin RJ, Hamm H, Lindboe CF, Fryns JP, Sijmons RH, Woods DG, Mariman EC, Padberg GW, Kremer H. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- Ng C, Jackson RA, Buschdorf JP, Sun Q, Guy GR, Sivaraman J. Structural basis for a novel intrapeptidyl H-bond and reverse binding of c-Cbl-TKB domain substrates. EMBO J. 2008;27:804–816. doi: 10.1038/emboj.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson SE, Metcalf D, Sprigg NS, Columbus R, Walker F, Silva A, Cary D, Willson TA, Zhang JG, Hilton DJ, Alexander WS, Nicola NA. Suppressor of cytokine signaling (SOCS)-5 is a potential negative regulator of epidermal growth factor signaling. Proc Natl Acad Sci U S A. 2005;102:2328–2333. doi: 10.1073/pnas.0409675102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemeyer CM, Kang MW, Shin DH, Furlan I, Erlacher M, Bunin NJ, Bunda S, Finklestein JZ, Sakamoto KM, Gorr TA, Mehta P, Schmid I, Kropshofer G, Corbacioglu S, Lang PJ, Klein C, Schlegel PG, Heinzmann A, Schneider M, Stary J, van den Heuvel-Eibrink MM, Hasle H, Locatelli F, Sakai D, Archambeault S, Chen L, Russell RC, Sybingco SS, Ohh M, Braun BS, Flotho C, Loh ML. Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia. Nat Genet. 2010;42:794–800. doi: 10.1038/ng.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, Kavamura MI, Kurosawa K, Ohashi H, Wilson L, Heron D, Bonneau D, Corona G, Kaname T, Naritomi K, Baumann C, Matsumoto N, Kato K, Kure S, Matsubara Y. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. [DOI] [PubMed] [Google Scholar]
- Nixon B, MacIntyre DA, Mitchell LA, Gibbs GM, O'Bryan M, Aitken RJ. The identification of mouse sperm-surface-associated proteins and characterization of their ability to act as decapacitation factors. Biol Reprod. 2006;74:275–287. doi: 10.1095/biolreprod.105.044644. [DOI] [PubMed] [Google Scholar]
- Nobuhisa I, Kato R, Inoue H, Takizawa M, Okita K, Yoshimura A, Taga T. Spred-2 suppresses aorta-gonad-mesonephros hematopoiesis by inhibiting MAP kinase activation. J Exp Med. 2004;199:737–742. doi: 10.1084/jem.20030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonami A, Taketomi T, Kimura A, Saeki K, Takaki H, Sanada T, Taniguchi K, Harada M, Kato R, Yoshimura A. The Sprouty-related protein, Spred-1, localizes in a lipid raft/caveola and inhibits ERK activation in collaboration with caveolin-1. Genes Cells. 2005;10:887–895. doi: 10.1111/j.1365-2443.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- Oishi K, Gaengel K, Krishnamoorthy S, Kamiya K, Kim IK, Ying H, Weber U, Perkins LA, Tartaglia M, Mlodzik M, Pick L, Gelb BD. Transgenic Drosophila models of Noonan syndrome causing PTPN11 gain-of-function mutations. Hum Mol Genet. 2006;15:543–553. doi: 10.1093/hmg/ddi471. [DOI] [PubMed] [Google Scholar]
- Oishi K, Zhang H, Gault WJ, Wang CJ, Tan CC, Kim IK, Ying H, Rahman T, Pica N, Tartaglia M, Mlodzik M, Gelb BD. Phosphatase-defective LEOPARD syndrome mutations in PTPN11 gene have gain-of-function effects during Drosophila development. Hum Mol Genet. 2009;18:193–201. doi: 10.1093/hmg/ddn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Marie PJ. Fibroblast growth factor signaling in skeletal development and disease. Genes Dev. 2015;29:1463–1486. doi: 10.1101/gad.266551.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostman A, Bohmer FD. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatases. Trends Cell Biol. 2001;11:258–266. doi: 10.1016/s0962-8924(01)01990-0. [DOI] [PubMed] [Google Scholar]
- Owens DM, Keyse SM. Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene. 2007;26:3203–3213. doi: 10.1038/sj.onc.1210412. [DOI] [PubMed] [Google Scholar]
- Page DT, Kuti OJ, Prestia C, Sur M. Haploinsufficiency for Pten and Serotonin transporter cooperatively influences brain size and social behavior. Proc Natl Acad Sci U S A. 2009;106:1989–1994. doi: 10.1073/pnas.0804428106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai LM, Barcelo G, Schupbach T. D-cbl, a negative regulator of the Egfr pathway, is required for dorsoventral patterning in Drosophila oogenesis. Cell. 2000;103:51–61. doi: 10.1016/s0092-8674(00)00104-5. [DOI] [PubMed] [Google Scholar]
- Pan Y, Carbe C, Powers A, Feng GS, Zhang X. Sprouty2-modulated Kras signaling rescues Shp2 deficiency during lens and lacrimal gland development. Development. 2010;137:1085–1093. doi: 10.1242/dev.042820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang J, Zhang S, Yang P, Hawkins-Lee B, Zhong J, Zhang Y, Ochoa B, Agundez JA, Voelckel MA, Fisher RB, Gu W, Xiong WC, Mei L, She JX, Wang CY. Loss-of-function mutations in HPSE2 cause the autosomal recessive urofacial syndrome. Am J Hum Genet. 2010;86:957–962. doi: 10.1016/j.ajhg.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante G, Thompson J, Lamballe F, Iwata T, Ferby I, Barr FA, Davies AM, Maina F, Klein R. Mitogen-inducible gene 6 is an endogenous inhibitor of HGF/Met-induced cell migration and neurite growth. J Cell Biol. 2005;171:337–348. doi: 10.1083/jcb.200502013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Choi HK, Seo JS, Yoo JY, Jeong JW, Choi Y, Choi KC, Yoon HG. DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim Biophys Acta. 2015;1853:2722–2730. doi: 10.1016/j.bbamcr.2015.07.024. [DOI] [PubMed] [Google Scholar]
- Patel R, Gao M, Ahmad I, Fleming J, Singh LB, Rai TS, McKie AB, Seywright M, Barnetson RJ, Edwards J, Sansom OJ, Leung HY. Sprouty2, PTEN, and PP2A interact to regulate prostate cancer progression. J Clin Invest. 2013;123:1157–1175. doi: 10.1172/JCI63672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. Unexpected diversity in Shisa-like proteins suggests the importance of their roles as transmembrane adaptors. Cell Signal. 2012;24:758–769. doi: 10.1016/j.cellsig.2011.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennock S, Wang Z. A tale of two Cbls: interplay of c-Cbl and Cbl-b in epidermal growth factor receptor downregulation. Mol Cell Biol. 2008;28:3020–3037. doi: 10.1128/MCB.01809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival CJ, Marangoni P, Tapaltsyan V, Klein O, Hallgrimsson B. The Interaction of Genetic Background and Mutational Effects in Regulation of Mouse Craniofacial Shape. G3 (Bethesda) 2017;7:1439–1450. doi: 10.1534/g3.117.040659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez B, Kosmider O, Cassinat B, Renneville A, Lachenaud J, Kaltenbach S, Bertrand Y, Baruchel A, Chomienne C, Fontenay M, Preudhomme C, Cave H. Genetic typing of CBL, ASXL1, RUNX1, TET2 and JAK2 in juvenile myelomonocytic leukaemia reveals a genetic profile distinct from chronic myelomonocytic leukaemia. Br J Haematol. 2010;151:460–468. doi: 10.1111/j.1365-2141.2010.08393.x. [DOI] [PubMed] [Google Scholar]
- Phoenix TN, Temple S. Spred1, a negative regulator of Ras-MAPK-ERK, is enriched in CNS germinal zones, dampens NSC proliferation, and maintains ventricular zone structure. Genes Dev. 2010;24:45–56. doi: 10.1101/gad.1839510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piessevaux J, Lavens D, Montoye T, Wauman J, Catteeuw D, Vandekerckhove J, Belsham D, Peelman F, Tavernier J. Functional cross-modulation between SOCS proteins can stimulate cytokine signaling. J Biol Chem. 2006;281:32953–32966. doi: 10.1074/jbc.M600776200. [DOI] [PubMed] [Google Scholar]
- Pires-daSilva A, Sommer RJ. The evolution of signalling pathways in animal development. Nat Rev Genet. 2003;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- Preger E, Ziv I, Shabtay A, Sher I, Tsang M, Dawid IB, Altuvia Y, Ron D. Alternative splicing generates an isoform of the human Sef gene with altered subcellular localization and specificity. Proc Natl Acad Sci U S A. 2004;101:1229–1234. doi: 10.1073/pnas.0307952100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Princen F, Bard E, Sheikh F, Zhang SS, Wang J, Zago WM, Wu D, Trelles RD, Bailly-Maitre B, Kahn CR, Chen Y, Reed JC, Tong GG, Mercola M, Chen J, Feng GS. Deletion of Shp2 tyrosine phosphatase in muscle leads to dilated cardiomyopathy, insulin resistance, and premature death. Mol Cell Biol. 2009;29:378–388. doi: 10.1128/MCB.01661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu XB, Goldberg AL. Nrdp1/FLRF is a ubiquitin ligase promoting ubiquitination and degradation of the epidermal growth factor receptor family member, ErbB3. Proc Natl Acad Sci U S A. 2002;99:14843–14848. doi: 10.1073/pnas.232580999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu CK, Shi ZQ, Shen R, Tsai FY, Orkin SH, Feng GS. A deletion mutation in the SH2-N domain of Shp-2 severely suppresses hematopoietic cell development. Mol Cell Biol. 1997;17:5499–5507. doi: 10.1128/mcb.17.9.5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafidi H, Mercado F, 3rd, Astudillo M, Fry WH, Saldana M, Carraway KL, 3rd, Sweeney C. Leucine-rich repeat and immunoglobulin domain-containing protein-1 (Lrig1) negative regulatory action toward ErbB receptor tyrosine kinases is opposed by leucine-rich repeat and immunoglobulin domain-containing protein 3 (Lrig3) J Biol Chem. 2013;288:21593–21605. doi: 10.1074/jbc.M113.486050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapraeger AC, Krufka A, Olwin BB. Requirement of heparan sulfate for bFGF-mediated fibroblast growth and myoblast differentiation. Science. 1991;252:1705–1708. doi: 10.1126/science.1646484. [DOI] [PubMed] [Google Scholar]
- Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Zhou XP, Eng C. A novel germline mutation of the PTEN gene in a patient with macrocephaly, ventricular dilatation, and features of VATER association. J Med Genet. 2001;38:820–823. doi: 10.1136/jmg.38.12.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regad T. Targeting RTK Signaling Pathways in Cancer. Cancers (Basel) 2015;7:1758–1784. doi: 10.3390/cancers7030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Cheng L, Rong Z, Li Z, Li Y, Zhang X, Xiong S, Hu J, Fu XY, Chang Z. hSef potentiates EGF-mediated MAPK signaling through affecting EGFR trafficking and degradation. Cell Signal. 2008;20:518–533. doi: 10.1016/j.cellsig.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschke M, Ferby I, Stepniak E, Seitzer N, Horst D, Wagner EF, Ullrich A. Mitogen-inducible gene-6 is a negative regulator of epidermal growth factor receptor signaling in hepatocytes and human hepatocellular carcinoma. Hepatology. 2010;51:1383–1390. doi: 10.1002/hep.23428. [DOI] [PubMed] [Google Scholar]
- Roberts NA, Woolf AS, Stuart HM, Thuret R, McKenzie EA, Newman WG, Hilton EN. Heparanase 2, mutated in urofacial syndrome, mediates peripheral neural development in Xenopus. Hum Mol Genet. 2014;23:4302–4314. doi: 10.1093/hmg/ddu147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- Rondahl V, Holmlund C, Karlsson T, Wang B, Faraz M, Henriksson R, Hedman H. Lrig2-deficient mice are protected against PDGFB-induced glioma. PLoS One. 2013;8:e73635. doi: 10.1371/journal.pone.0073635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Z, Ren Y, Cheng L, Li Z, Li Y, Sun Y, Li H, Xiong S, Chang Z. Sef-S, an alternative splice isoform of sef gene, inhibits NIH3T3 cell proliferation via a mitogen-activated protein kinases p42 and p44 (ERK1/2)-independent mechanism. Cell Signal. 2007;19:93–102. doi: 10.1016/j.cellsig.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Rubin C, Litvak V, Medvedovsky H, Zwang Y, Lev S, Yarden Y. Sprouty fine-tunes EGF signaling through interlinked positive and negative feedback loops. Curr Biol. 2003;13:297–307. doi: 10.1016/s0960-9822(03)00053-8. [DOI] [PubMed] [Google Scholar]
- Santoro C, Pacileo G, Limongelli G, Scianguetta S, Giugliano T, Piluso G, Ragione FD, Cirillo M, Mirone G, Perrotta S. LEOPARD syndrome: clinical dilemmas in differential diagnosis of RASopathies. BMC Med Genet. 2014;15:44. doi: 10.1186/1471-2350-15-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarquis MS, Agrawal S, Shen L, Pilarski R, Zhou XP, Eng C. Distinct expression profiles for PTEN transcript and its splice variants in Cowden syndrome and Bannayan-Riley-Ruvalcaba syndrome. Am J Hum Genet. 2006;79:23–30. doi: 10.1086/504392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Taketomi T, Kato R, Saeki K, Nonami A, Sasaki M, Kuriyama M, Saito N, Shibuya M, Yoshimura A. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat Cell Biol. 2003;5:427–432. doi: 10.1038/ncb978. [DOI] [PubMed] [Google Scholar]
- Sasaki A, Taketomi T, Wakioka T, Kato R, Yoshimura A. Identification of a dominant negative mutant of Sprouty that potentiates fibroblast growth factor- but not epidermal growth factor-induced ERK activation. J Biol Chem. 2001;276:36804–36808. doi: 10.1074/jbc.C100386200. [DOI] [PubMed] [Google Scholar]
- Saxton TM, Henkemeyer M, Gasca S, Shen R, Rossi DJ, Shalaby F, Feng GS, Pawson T. Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J. 1997;16:2352–2364. doi: 10.1093/emboj/16.9.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M, Kumar S. Mammalian HECT ubiquitin-protein ligases: biological and pathophysiological aspects. Biochim Biophys Acta. 2014;1843:61–74. doi: 10.1016/j.bbamcr.2013.03.024. [DOI] [PubMed] [Google Scholar]
- Scheving LA, Zhang X, Garcia OA, Wang RF, Stevenson MC, Threadgill DW, Russell WE. Epidermal growth factor receptor plays a role in the regulation of liver and plasma lipid levels in adult male mice. Am J Physiol Gastrointest Liver Physiol. 2014;306:G370–381. doi: 10.1152/ajpgi.00116.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- Schmeisser MJ, Kuhl SJ, Schoen M, Beth NH, Weis TM, Grabrucker AM, Kuhl M, Boeckers TM. The Nedd4-binding protein 3 (N4BP3) is crucial for axonal and dendritic branching in developing neurons. Neural Dev. 2013;8:18. doi: 10.1186/1749-8104-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Raudaskoski M, Knabe N, Kothe E. Ras GTPase-activating protein gap1 of the homobasidiomycete Schizophyllum commune regulates hyphal growth orientation and sexual development. Eukaryot Cell. 2006;5:683–695. doi: 10.1128/EC.5.4.683-695.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzbauer G, Robbins J. The tumor suppressor gene PTEN can regulate cardiac hypertrophy and survival. J Biol Chem. 2001;276:35786–35793. doi: 10.1074/jbc.M102479200. [DOI] [PubMed] [Google Scholar]
- Segatto O, Anastasi S, Alema S. Regulation of epidermal growth factor receptor signalling by inducible feedback inhibitors. J Cell Sci. 2011;124:1785–1793. doi: 10.1242/jcs.083303. [DOI] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, Sweeney C. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AT, Meissner A, Dowdle JA, Crowley D, Magendantz M, Ouyang C, Parisi T, Rajagopal J, Blank LJ, Bronson RT, Stone JR, Tuveson DA, Jaenisch R, Jacks T. Sprouty-2 regulates oncogenic K-ras in lung development and tumorigenesis. Genes Dev. 2007;21:694–707. doi: 10.1101/gad.1526207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim K, Minowada G, Coling DE, Martin GR. Sprouty2, a mouse deafness gene, regulates cell fate decisions in the auditory sensory epithelium by antagonizing FGF signaling. Dev Cell. 2005;8:553–564. doi: 10.1016/j.devcel.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Siljamaki E, Abankwa D. SPRED1 Interferes with K-ras but Not H-ras Membrane Anchorage and Signaling. Mol Cell Biol. 2016;36:2612–2625. doi: 10.1128/MCB.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simrick S, Lickert H, Basson MA. Sprouty genes are essential for the normal development of epibranchial ganglia in the mouse embryo. Dev Biol. 2011;358:147–155. doi: 10.1016/j.ydbio.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Kirk EP, Theodosopoulos G, Marshall GM, Walker J, Rogers M, Field M, Brereton JJ, Marsh DJ. Germline mutation of the tumour suppressor PTEN in Proteus syndrome. J Med Genet. 2002;39:937–940. doi: 10.1136/jmg.39.12.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi-Yanicostas N, de Castro F, Julliard AK, Perfettini I, Chedotal A, Petit C. Anosmin-1, defective in the X-linked form of Kallmann syndrome, promotes axonal branch formation from olfactory bulb output neurons. Cell. 2002;109:217–228. doi: 10.1016/s0092-8674(02)00713-4. [DOI] [PubMed] [Google Scholar]
- Soussi-Yanicostas N, Faivre-Sarrailh C, Hardelin JP, Levilliers J, Rougon G, Petit C. Anosmin-1 underlying the X chromosome-linked Kallmann syndrome is an adhesion molecule that can modulate neurite growth in a cell-type specific manner. J Cell Sci. 1998;111(Pt 19):2953–2965. doi: 10.1242/jcs.111.19.2953. [DOI] [PubMed] [Google Scholar]
- Steelman LS, Chappell WH, Abrams SL, Kempf RC, Long J, Laidler P, Mijatovic S, Maksimovic-Ivanic D, Stivala F, Mazzarino MC, Donia M, Fagone P, Malaponte G, Nicoletti F, Libra M, Milella M, Tafuri A, Bonati A, Basecke J, Cocco L, Evangelisti C, Martelli AM, Montalto G, Cervello M, McCubrey JA. Roles of the Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR pathways in controlling growth and sensitivity to therapy-implications for cancer and aging. Aging (Albany NY) 2011;3:192–222. doi: 10.18632/aging.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RA, Sanda T, Widlund HR, Zhu S, Swanson KD, Hurley AD, Bentires-Alj M, Fisher DE, Kontaridis MI, Look AT, Neel BG. Phosphatase-dependent and - independent functions of Shp2 in neural crest cells underlie LEOPARD syndrome pathogenesis. Dev Cell. 2010;18:750–762. doi: 10.1016/j.devcel.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim YJ, Sherwin R, Devaskar S, Lesche R, Magnuson MA, Wu H. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected] Proc Natl Acad Sci U S A. 2004;101:2082–2087. doi: 10.1073/pnas.0308617100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowe IB, Mercado EL, Stowe TR, Bell EL, Oses-Prieto JA, Hernandez H, Burlingame AL, McCormick F. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart HM, Roberts NA, Burgu B, Daly SB, Urquhart JE, Bhaskar S, Dickerson JE, Mermerkaya M, Silay MS, Lewis MA, Olondriz MB, Gener B, Beetz C, Varga RE, Gulpinar O, Suer E, Soygur T, Ozcakar ZB, Yalcinkaya F, Kavaz A, Bulum B, Gucuk A, Yue WW, Erdogan F, Berry A, Hanley NA, McKenzie EA, Hilton EN, Woolf AS, Newman WG. LRIG2 mutations cause urofacial syndrome. Am J Hum Genet. 2013;92:259–264. doi: 10.1016/j.ajhg.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz MA, Shattuck DL, Laederich MB, Carraway KL, 3rd, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene. 2008;27:5741–5752. doi: 10.1038/onc.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian B, Anand M, Khan NW, Khanna H. Loss of Raf-1 kinase inhibitory protein delays early-onset severe retinal ciliopathy in Cep290rd16 mouse. Invest Ophthalmol Vis Sci. 2014;55:5788–5794. doi: 10.1167/iovs.14-14954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Miura H, Tanemura A, Kobayashi K, Kondoh G, Sano S, Ozawa K, Inui S, Nakata A, Takagi T, Tohyama M, Yoshikawa K, Itami S. Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia. FEBS Lett. 2002;521:67–71. doi: 10.1016/s0014-5793(02)02824-7. [DOI] [PubMed] [Google Scholar]
- Tajan M, de Rocca Serra A, Valet P, Edouard T, Yart A. SHP2 sails from physiology to pathology. Eur J Med Genet. 2015;58:509–525. doi: 10.1016/j.ejmg.2015.08.005. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, Ito S, Ueda T, Morioka Y, Kayukawa N, Ueno A, Nakagawa H, Fujihara A, Ushijima S, Kanazawa M, Hongo F, Ukimura O. CNPY2 promoted the proliferation of renal cell carcinoma cells and increased the expression of TP53. Biochem Biophys Res Commun. 2017;485:267–271. doi: 10.1016/j.bbrc.2017.02.095. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Ayada T, Ichiyama K, Kohno R, Yonemitsu Y, Minami Y, Kikuchi A, Maehara Y, Yoshimura A. Sprouty2 and Sprouty4 are essential for embryonic morphogenesis and regulation of FGF signaling. Biochem Biophys Res Commun. 2007a;352:896–902. doi: 10.1016/j.bbrc.2006.11.107. [DOI] [PubMed] [Google Scholar]
- Taniguchi K, Kohno R, Ayada T, Kato R, Ichiyama K, Morisada T, Oike Y, Yonemitsu Y, Maehara Y, Yoshimura A. Spreds are essential for embryonic lymphangiogenesis by regulating vascular endothelial growth factor receptor 3 signaling. Mol Cell Biol. 2007b;27:4541–4550. doi: 10.1128/MCB.01600-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi K, Sasaki K, Watari K, Yasukawa H, Imaizumi T, Ayada T, Okamoto F, Ishizaki T, Kato R, Kohno R, Kimura H, Sato Y, Ono M, Yonemitsu Y, Yoshimura A. Suppression of Sproutys has a therapeutic effect for a mouse model of ischemia by enhancing angiogenesis. PLoS One. 2009;4:e5467. doi: 10.1371/journal.pone.0005467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Gelb BD. Germ-line and somatic PTPN11 mutations in human disease. Eur J Med Genet. 2005;48:81–96. doi: 10.1016/j.ejmg.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Martinelli S, Stella L, Bocchinfuso G, Flex E, Cordeddu V, Zampino G, Burgt I, Palleschi A, Petrucci TC, Sorcini M, Schoch C, Foa R, Emanuel PD, Gelb BD. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- Tefft D, De Langhe SP, Del Moral PM, Sala F, Shi W, Bellusci S, Warburton D. A novel function for the protein tyrosine phosphatase Shp2 during lung branching morphogenesis. Dev Biol. 2005;282:422–431. doi: 10.1016/j.ydbio.2005.03.022. [DOI] [PubMed] [Google Scholar]
- Tefft JD, Lee M, Smith S, Leinwand M, Zhao J, Bringas P, Jr, Crowe DL, Warburton D. Conserved function of mSpry-2, a murine homolog of Drosophila sprouty, which negatively modulates respiratory organogenesis. Curr Biol. 1999;9:219–222. doi: 10.1016/s0960-9822(99)80094-3. [DOI] [PubMed] [Google Scholar]
- Theroux S, Pereira M, Casten KS, Burwell RD, Yeung KC, Sedivy JM, Klysik J. Raf kinase inhibitory protein knockout mice: expression in the brain and olfaction deficit. Brain Res Bull. 2007;71:559–567. doi: 10.1016/j.brainresbull.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien CB, Langdon WY. Cbl: many adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- Thien CB, Langdon WY. c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J. 2005;391:153–166. doi: 10.1042/BJ20050892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thien CB, Scaife RM, Papadimitriou JM, Murphy MA, Bowtell DD, Langdon WY. A mouse with a loss-of-function mutation in the c-Cbl TKB domain shows perturbed thymocyte signaling without enhancing the activity of the ZAP-70 tyrosine kinase. J Exp Med. 2003;197:503–513. doi: 10.1084/jem.20021498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas AR, Certal AC, Rodriguez-Leon J. FLRT3 as a key player on chick limb development. Dev Biol. 2011;355:324–333. doi: 10.1016/j.ydbio.2011.04.031. [DOI] [PubMed] [Google Scholar]
- Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- Torii S, Kusakabe M, Yamamoto T, Maekawa M, Nishida E. Sef is a spatial regulator for Ras/MAP kinase signaling. Dev Cell. 2004;7:33–44. doi: 10.1016/j.devcel.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem. 2005;280:24931–24940. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]
- Trengrove HG, Gray A. The role of military dental capabilities in mass fatality situations. Mil Med. 2013;178:523–528. doi: 10.7205/MILMED-D-12-00399. [DOI] [PubMed] [Google Scholar]
- Trotman LC, Wang X, Alimonti A, Chen Z, Teruya-Feldstein J, Yang H, Pavletich NP, Carver BS, Cordon-Cardo C, Erdjument-Bromage H, Tempst P, Chi SG, Kim HJ, Misteli T, Jiang X, Pandolfi PP. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell. 2007;128:141–156. doi: 10.1016/j.cell.2006.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PS, Gill JC. Mechanisms of disease: Insights into X-linked and autosomal-dominant Kallmann syndrome. Nat Clin Pract Endocrinol Metab. 2006;2:160–171. doi: 10.1038/ncpendmet0119. [DOI] [PubMed] [Google Scholar]
- Tsang M, Dawid IB. Promotion and attenuation of FGF signaling through the Ras-MAPK pathway. Sci STKE. 2004;2004:pe17. doi: 10.1126/stke.2282004pe17. [DOI] [PubMed] [Google Scholar]
- Tsang M, Friesel R, Kudoh T, Dawid IB. Identification of Sef, a novel modulator of FGF signalling. Nat Cell Biol. 2002;4:165–169. doi: 10.1038/ncb749. [DOI] [PubMed] [Google Scholar]
- Tsuji L, Yamashita T, Kubo T, Madura T, Tanaka H, Hosokawa K, Tohyama M. FLRT3, a cell surface molecule containing LRR repeats and a FNIII domain, promotes neurite outgrowth. Biochem Biophys Res Commun. 2004;313:1086–1091. doi: 10.1016/j.bbrc.2003.12.047. [DOI] [PubMed] [Google Scholar]
- Tsujita Y, Mitsui-Sekinaka K, Imai K, Yeh TW, Mitsuiki N, Asano T, Ohnishi H, Kato Z, Sekinaka Y, Zaha K, Kato T, Okano T, Takashima T, Kobayashi K, Kimura M, Kunitsu T, Maruo Y, Kanegane H, Takagi M, Yoshida K, Okuno Y, Muramatsu H, Shiraishi Y, Chiba K, Tanaka H, Miyano S, Kojima S, Ogawa S, Ohara O, Okada S, Kobayashi M, Morio T, Nonoyama S. Phosphatase and tensin homolog (PTEN) mutation can cause activated phosphatidylinositol 3-kinase delta syndrome-like immunodeficiency. J Allergy Clin Immunol. 2016;138:1672–1680. e1610. doi: 10.1016/j.jaci.2016.03.055. [DOI] [PubMed] [Google Scholar]
- Tuduce IL, Schuh K, Bundschu K. Spred2 expression during mouse development. Dev Dyn. 2010;239:3072–3085. doi: 10.1002/dvdy.22432. [DOI] [PubMed] [Google Scholar]
- Turner JT, Cohen MM, Jr, Biesecker LG. Reassessment of the Proteus syndrome literature: application of diagnostic criteria to published cases. Am J Med Genet A. 2004;130A:111–122. doi: 10.1002/ajmg.a.30327. [DOI] [PubMed] [Google Scholar]
- Urness LD, Li C, Wang X, Mansour SL. Expression of ERK signaling inhibitors Dusp6, Dusp7, and Dusp9 during mouse ear development. Dev Dyn. 2008;237:163–169. doi: 10.1002/dvdy.21380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlacich G, Nawijn MC, Webb GC, Steiner DF. Pim3 negatively regulates glucose-stimulated insulin secretion. Islets. 2010;2:308–317. doi: 10.4161/isl.2.5.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinsky N, Kholodenko BN. Complexity of receptor tyrosine kinase signal processing. Cold Spring Harb Perspect Biol. 2013;5:a009043. doi: 10.1101/cshperspect.a009043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- Walsh AM, Lazzara MJ. Differential parsing of EGFR endocytic flux among parallel internalization pathways in lung cancer cells with EGFR-activating mutations. Integr Biol (Camb) 2014;6:312–323. doi: 10.1039/c3ib40176f. [DOI] [PubMed] [Google Scholar]
- Wang X, Trotman LC, Koppie T, Alimonti A, Chen Z, Gao Z, Wang J, Erdjument-Bromage H, Tempst P, Cordon-Cardo C, Pandolfi PP, Jiang X. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell. 2007;128:129–139. doi: 10.1016/j.cell.2006.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Werz C, Xu D, Chen Z, Li Y, Hafen E, Bergmann A. Drosophila cbl is essential for control of cell death and cell differentiation during eye development. PLoS One. 2008;3:e1447. doi: 10.1371/journal.pone.0001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton D, Perin L, Defilippo R, Bellusci S, Shi W, Driscoll B. Stem/progenitor cells in lung development, injury repair, and regeneration. Proc Am Thorac Soc. 2008;5:703–706. doi: 10.1513/pats.200801-012AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke R, Heuberger J, Grossmann K, Michos O, Schmidt-Ott K, Walentin K, Costantini F, Birchmeier W. The tyrosine phosphatase Shp2 acts downstream of GDNF/Ret in branching morphogenesis of the developing mouse kidney. Dev Biol. 2011;360:310–317. doi: 10.1016/j.ydbio.2011.09.029. [DOI] [PubMed] [Google Scholar]
- Wong ES, Fong CW, Lim J, Yusoff P, Low BC, Langdon WY, Guy GR. Sprouty2 attenuates epidermal growth factor receptor ubiquitylation and endocytosis, and consequently enhances Ras/ERK signalling. EMBO J. 2002;21:4796–4808. doi: 10.1093/emboj/cdf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong ES, Lim J, Low BC, Chen Q, Guy GR. Evidence for direct interaction between Sprouty and Cbl. J Biol Chem. 2001;276:5866–5875. doi: 10.1074/jbc.M006945200. [DOI] [PubMed] [Google Scholar]
- Xiong S, Zhao Q, Rong Z, Huang G, Huang Y, Chen P, Zhang S, Liu L, Chang Z. hSef inhibits PC-12 cell differentiation by interfering with Ras-mitogen-activated protein kinase MAPK signaling. J Biol Chem. 2003;278:50273–50282. doi: 10.1074/jbc.M306936200. [DOI] [PubMed] [Google Scholar]
- Xu GF, O'Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell. 1990;62:599–608. doi: 10.1016/0092-8674(90)90024-9. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, Nagano T, Takehara S, Hibi M, Aizawa S. Shisa promotes head formation through the inhibition of receptor protein maturation for the caudalizing factors, Wnt and FGF. Cell. 2005;120:223–235. doi: 10.1016/j.cell.2004.11.051. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Lee D, Kim Y, Lee B, Seo C, Kawasaki H, Kuroda S, Tanaka-Yamamoto K. Raf kinase inhibitory protein is required for cerebellar long-term synaptic depression by mediating PKC-dependent MAPK activation. J Neurosci. 2012;32:14254–14264. doi: 10.1523/JNEUROSCI.2812-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harb Perspect Biol. 2009;1:a002493. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan P, Gong H, Zhai X, Feng Y, Wu J, He S, Guo J, Wang X, Guo R, Xie J, Li RK. Decreasing CNPY2 Expression Diminishes Colorectal Tumor Growth and Development through Activation of p53 Pathway. Am J Pathol. 2016;186:1015–1024. doi: 10.1016/j.ajpath.2015.11.012. [DOI] [PubMed] [Google Scholar]
- Yang RB, Ng CK, Wasserman SM, Komuves LG, Gerritsen ME, Topper JN. A novel interleukin-17 receptor-like protein identified in human umbilical vein endothelial cells antagonizes basic fibroblast growth factor-induced signaling. J Biol Chem. 2003;278:33232–33238. doi: 10.1074/jbc.M305022200. [DOI] [PubMed] [Google Scholar]
- Yang W, Wang J, Moore DC, Liang H, Dooner M, Wu Q, Terek R, Chen Q, Ehrlich MG, Quesenberry PJ, Neel BG. Ptpn11 deletion in a novel progenitor causes metachondromatosis by inducing hedgehog signalling. Nature. 2013;499:491–495. doi: 10.1038/nature12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanpallewar S, Wang T, Koh DC, Quarta E, Fulgenzi G, Tessarollo L. Nedd4-2 haploinsufficiency causes hyperactivity and increased sensitivity to inflammatory stimuli. Sci Rep. 2016;6:32957. doi: 10.1038/srep32957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayon A, Klagsbrun M, Esko JD, Leder P, Ornitz DM. Cell surface, heparin-like molecules are required for binding of basic fibroblast growth factor to its high affinity receptor. Cell. 1991;64:841–848. doi: 10.1016/0092-8674(91)90512-w. [DOI] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- Yeung KC, Rose DW, Dhillon AS, Yaros D, Gustafsson M, Chatterjee D, McFerran B, Wyche J, Kolch W, Sedivy JM. Raf kinase inhibitor protein interacts with NF-kappaB-inducing kinase and TAK1 and inhibits NF-kappaB activation. Mol Cell Biol. 2001;21:7207–7217. doi: 10.1128/MCB.21.21.7207-7217.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Holmlund C, Nilsson J, Inui S, Lei T, Itami S, Henriksson R, Hedman H. Paracrine regulation of growth factor signaling by shed leucine-rich repeats and immunoglobulin-like domains 1. Exp Cell Res. 2011;317:504–512. doi: 10.1016/j.yexcr.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Ying H, Zheng H, Scott K, Wiedemeyer R, Yan H, Lim C, Huang J, Dhakal S, Ivanova E, Xiao Y, Zhang H, Hu J, Stommel JM, Lee MA, Chen AJ, Paik JH, Segatto O, Brennan C, Elferink LA, Wang YA, Chin L, DePinho RA. Mig-6 controls EGFR trafficking and suppresses gliomagenesis. Proc Natl Acad Sci U S A. 2010;107:6912–6917. doi: 10.1073/pnas.0914930107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon CH, Lee J, Jongeward GD, Sternberg PW. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science. 1995;269:1102–1105. doi: 10.1126/science.7652556. [DOI] [PubMed] [Google Scholar]
- You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Mol Cell Biol. 1999;19:2416–2424. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Calvo L, Anta B, Lopez-Benito S, Lopez-Bellido R, Vicente-Garcia C, Tessarollo L, Rodriguez RE, Arevalo JC. In vivo regulation of NGF-mediated functions by Nedd4-2 ubiquitination of TrkA. J Neurosci. 2014;34:6098–6106. doi: 10.1523/JNEUROSCI.4271-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J. Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature. 2007;450:741–744. doi: 10.1038/nature05998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Su Y, Lanning N, Swiatek PJ, Bronson RT, Sigler R, Martin RW, Vande Woude GF. Targeted disruption of Mig-6 in the mouse genome leads to early onset degenerative joint disease. Proc Natl Acad Sci U S A. 2005;102:11740–11745. doi: 10.1073/pnas.0505171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development. 2008;135:1283–1293. doi: 10.1242/dev.015073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Hampel H, Thiele H, Gorlin RJ, Hennekam RC, Parisi M, Winter RM, Eng C. Association of germline mutation in the PTEN tumour suppressor gene and Proteus and Proteus-like syndromes. Lancet. 2001;358:210–211. doi: 10.1016/s0140-6736(01)05412-5. [DOI] [PubMed] [Google Scholar]
- Zhou XP, Marsh DJ, Hampel H, Mulliken JB, Gimm O, Eng C. Germline and germline mosaic PTEN mutations associated with a Proteus-like syndrome of hemihypertrophy, lower limb asymmetry, arteriovenous malformations and lipomatosis. Hum Mol Genet. 2000;9:765–768. doi: 10.1093/hmg/9.5.765. [DOI] [PubMed] [Google Scholar]
- Zhou XP, Waite KA, Pilarski R, Hampel H, Fernandez MJ, Bos C, Dasouki M, Feldman GL, Greenberg LA, Ivanovich J, Matloff E, Patterson A, Pierpont ME, Russo D, Nassif NT, Eng C. Germline PTEN promoter mutations and deletions in Cowden/Bannayan-Riley-Ruvalcaba syndrome result in aberrant PTEN protein and dysregulation of the phosphoinositol-3-kinase/Akt pathway. Am J Hum Genet. 2003;73:404–411. doi: 10.1086/377109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv I, Fuchs Y, Preger E, Shabtay A, Harduf H, Zilpa T, Dym N, Ron D. The human sef-a isoform utilizes different mechanisms to regulate receptor tyrosine kinase signaling pathways and subsequent cell fate. J Biol Chem. 2006;281:39225–39235. doi: 10.1074/jbc.M607327200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.