Abstract
Emergence of the field of exosome biology has opened an exciting door to better understanding communication between cells. These tiny nanovesicles act as potent regulators of biological function by delivering proteins, lipids and nucleic acids from the cell of origin to target cells. Recently, several enzymes including membrane-type 1 matrix metalloproteinase (MT1-MMP), insulin-degrading enzyme (IDE), sialidase and heparanase, among others, were localized on the surface of exosomes secreted by various cell types. These exosomal surface enzymes retain their activity and can degrade their natural substrates present within extracellular spaces. To date, enzymes on exosome surfaces have been associated with the mobilization of growth factors, degradation of extracellular matrix macromolecules and destruction of amyloid β plaques. This review focuses on the emerging role of exosomal surface enzymes and how this mechanism of remodeling within the extracellular space may regulate disease progression as related to cancer, inflammation and Alzheimer’s disease.
Keywords: extracellular matrix, cancer, Alzheimer’s, extracellular vesicles, enzyme
Introduction
Exosomes are extracellular vesicles contained within a lipid bilayer that range in size from ~30–150 nm. Although the identification of exosomes is somewhat complicated because there is no single protein marker or set of markers that positively identifies a vesicle as an exosome, by definition they are generated intracellularly within endosomes. This distinguishes exosomes from microvesicles that are extracellular vesicles derived from the cell surface and are generally much larger, ranging in size up to ~1 µm. Exosomes bear cargo that can be transferred to recipient cells and alter cell behaviors thereby impacting both normal and pathological cell functions. These functions, to name a few, include alteration of cell signaling, changes in patterns of cell adhesion and migration, and regulation of cell proliferation and gene expression. A number of excellent comprehensive reviews that discuss the biogenesis, secretion and functions of exosomes are available [1–3].
The cargo of exosomes includes nucleic acids, lipids and proteins. Nucleic acids include mRNAs, miRNAs and other noncoding RNAs and together these have been found to regulate the expression of numerous genes in recipient cells [4]. Proteins found in exosomes include those involved in antigen presentation, adhesion, signal transduction as well as those composing the cytoskeleton [1]. In addition, exosomes are endowed with a host of membrane transport proteins and those involved in the formation and secretion of exosomes including the ESCRT proteins (the endosomal sorting complex required for transport protein family). Also, a number of proteolytic proteins have been found in exosome cargo, many of which are members of the metalloproteinase family including the secreted MMPs, the membrane-anchored MMPs as well as the ADAMs and ADAMTS proteases [5]. Together these can cleave a wide range of substrates within the extracellular matrix (e.g., collagens) as well as cleave and sometimes activate soluble proteins that are thereby released from the cell surface (i.e. act as sheddases). There is also evidence that exosomes can carry glycosidases as cargo. Although this class of enzymes has not been extensively studied in exosomes, evidence is emerging that they are present and can actively degrade substrates on cells or within extracellular matrices [6–8].
Much of exosome function is dependent on their capacity to dock with nearby cells or to travel locally or distally before docking. Once exosomes have docked with a recipient cell, they can directly activate signaling cascades at the cell surface or their membranes can fuse with the recipient cell and the exosomal contents released into the cytoplasm. Alternatively, exosomes can be endocytosed by a recipient cell and the exosomal contents released when the exosomes are lysed within endosomal/lysosomal compartments. In addition to functions resulting directly from exosome docking with cells, biologically active molecules on the surface of exosomes can play important functional roles. For example, Wnts were found on the surface of exosomes secreted by Drosophila and human cells and could activate Wnt signaling on recipient cells [9]. Fibronectin has been found on the surface of exosomes secreted by many types of cells and demonstrated to be important in exosome docking and in promoting cell motility [10, 11].
Enzymes, including proteases and glycosidases, can localize on the exosome surface. One possibility as to how this occurs can be understood by examining the fate of cell surface enzymes during exosome biogenesis (Fig. 1). Endosomes form following invagination of the cell membrane leaving molecules on the cell surface localized to the luminal side of the endosomal membrane. Multiple small invaginations of the endosomal membrane occur resulting in the cell surface molecules now being present on the surface of the forming vesicles. These vesicles collect within the multivesicular body, which eventually fuses with the cell membrane and releases the newly formed exosomes into the extracellular space (Fig. 1). Thus, enzymes having transmembrane domains (e.g., MT1-MMP), or enzymes bound to other cell surface molecules (e.g., heparanase), localize on the surface of exosomes. Another possibility is that soluble enzymes bind to the surface of exosomes within either the endosomal compartment, or following secretion of the exosomes into the extracellular space.
Figure 1.
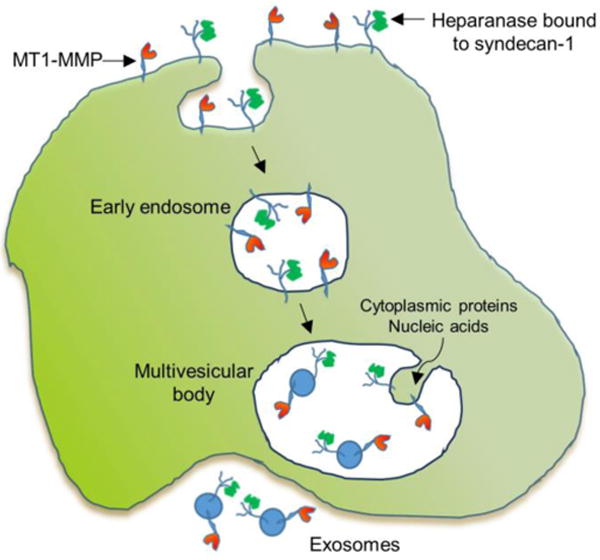
This figure traces the route of MT1-MMP and heparanase through their endocytosis and subsequent transit through early endosome, multivesicular body and eventual secretion as surface components of the secreted exosomes. MT1-MMP localizes to the cell surface via its transmembrane domain; heparanase is retained on the cell surface through binding to the heparan sulfate chains of the transmembrane proteoglycan syndecan-1. Following membrane invagination and formation of the early endosome, the cell surface MT1-MMP and heparanase are present on the interior face of the early endosomal membrane. Within the multivesicular body, vesicles begin to form as the membrane invaginates and the forming vesicle fills with cytoplasmic contents (proteins, nucleic acids). Eventually the invaginations are pinched off to form vesicles followed by merging of the multivesicular body with the cell membrane and release of exosomes having MT1-MMP and heparanase on their surface.
Once localized on the exosome surface, the enzymes are available to act on molecules they encounter either at the cell surface or with soluble or insoluble components of the extracellular matrix. Several examples of exosomal surface enzyme activity have been described and their functions studied. Figure 2 summarizes the activity of four of these enzymes and highlights their potential importance in pathological states. Degradation of extracellular molecules by intact exosomes represents a new area of investigation that will likely expand dramatically as more enzymes on the surface of exosomes are discovered and their functions delineated.
Figure 2.
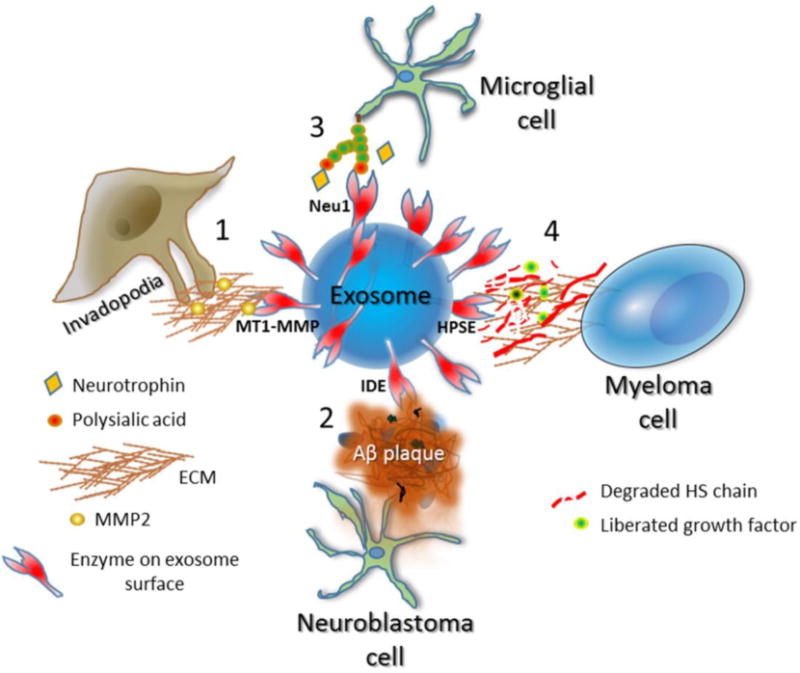
Examples of enzymes on the exosome surface and their known functions. Depicted is an exosome having active enzymes on its surface and the functional impact of those enzymes within the extracellular space. The specific enzyme present depends on the cell that secreted the exosome. 1, MT1-MMP is on the surface of exosomes secreted at the site of invadopodia formation in breast and other cancers. MT1-MMP mediates shedding of cell surface molecules including syndecan-1 and CD44 and degrades fibrillar collagens, a host of other matrix molecules including fibronectin and vitronectin, and soluble molecules within the extracellular space. In addition, MT1-MMP activates MMP-2. Through this range of activities, MT1-MMP on the exosome surface may contribute to tumor migration and invasion as well as angiogenesis. 2, N2a neuroblastoma cells or BV-2 microglial cells secrete exosomes having a high level of insulin-degrading enzyme (IDE) on their surface. This enzyme is capable of degrading Aβ peptides that are prevalent in Alzheimer’s plaques. Statins enhance secretion of these exosomes containing high IDE levels and thus could prove therapeutically useful to attack plaques and diminish Alzheimer’s disease progression. 3, Microglial cells have high levels of polysialic acid on their surface. When stimulated by LPS, the cells rapidly enhance secretion of exosomes having the sialidase neuraminidase 1 (Neu1) on their surface. The sialidase cleaves the polysialic acid thereby liberating neurotrophic factors that are bound to the polysialic acid. The released neurotrophic factors have functional impact by regulating neural cell development and growth. 4, Myeloma cells expressing a high level of heparanase (HPSE) secrete exosomes having HPSE localized to the exosome surface. When added to ECM, the HPSE on the exosomes is capable of degrading heparan sulfate (HS) chains embedded within the ECM. This likely liberates growth factors bound to HS and by degrading ECM promotes tumor invasion and metastasis. Treatment of the myeloma cells with chemotherapeutic drugs enhances the secretion of exosomes loaded with HPSE as cargo leading to enhanced ECM degradation.
Protease activity on the exosome surface
MT1-MMP
MT1-MMP (MMP-14) is a transmembrane protein present on the surface of many cell types including both normal and pathologically altered cells. MT1-MMP is credited with playing a major role in promoting migration of cells, particularly during tumor invasion and metastasis, because it degrades fibrillar collagens (types I, II and III) as well as other matrix components including fibronectin, vitronectin and some members of the laminin family [12]. In addition, MT1-MMP also acts as a sheddase by cleaving, for example, syndecan-1, ICAM-1 and CD44 releasing them from the surface of cells and thereby altering cell behavior. MT1-MMP also cleaves a host of soluble molecules with varying effects including activation of proMMP-2 and proMMP-13. Through these activities MT1-MMP is involved in angiogenesis, epithelial morphogenesis, skeletal development, wound healing and inflammation [12].
Because it is expressed on the surface of cells, it is not surprising that MT1-MMP has also been localized to the exosome surface. Exosomes secreted by cells of the G361 human melanoma cell line displayed MT1-MMP on their surface as determined by immunoelectron microscopy [13]. Incubation of exosomes secreted by HT-1080 human fibrocarcinoma cells stably expressing recombinant MT1-MMP readily degraded its substrate type I collagen into its characteristic peptide fragments [13]. When exosomes from MT1-MMP expressing HT-1080 cells were incubated with MMP-2 proenzyme, the MMP-2 was activated as detected in zymograms [13]. Together these findings indicate that tumor cells can secrete exosomes having surface MT1-MMP that is biologically active and targets the ECM.
Interestingly, MT1-MMP has been credited with playing a key role in the formation and function of invadopodia, membrane protrusions prominent on metastatic tumor cells. These protrusions bear enzymes that degrade extracellular matrix, including MT1-MMP. Inhibition of MT1-MMP protease activity, or siRNA knockdown of MT1-MMP in MDA-MB-231 breast cancer cells resulted in decreased formation of invadopodia and the invadopodia that did form failed to degrade ECM [14]. In related studies with SCC61 carcinoma cells, it was shown that multivesicular bodies, the intracellular sites of exosome genesis, localize to invadopodia [15]. Exosomes from these cells were strongly positive for MT1-MMP. When Rab27a, a docking factor key for formation of exosomes, was knocked down in these cells, there was a dramatic decrease in the rate of invadopodia formation and in the lifetime of invadopodia that did form [15]. Also, after Rab27a knockdown, the percentage of invadopodia that were MT1-MMP positive was greatly diminished compared to control invadopodia that did form. Introduction of exogenous purified exosomes to the Rab27a knockdown cells growing in culture stimulated new invadopodia formation and extended invadopodia lifetime. The authors speculate that induction of invadopodia is a consequence of the tumor cells being simulated by growth factors or signaling molecules associated with the exosomes, or that exosome proteinases, membrane or other cargo may stabilize developing invadopodia. In total, these studies indicate that exosomes play a critical function in regulating invadopodia and that exosomal MT1-MMP-mediated degradation likely is a key factor in this process. Moreover they provide evidence that formation of invadopodia enhances exosome secretion that not only supports invadopodia but likely impacts the local tumor microenvironment and aids in establishment of metastatic niches, events that together drive tumor progression [15].
Notably, exosomes secretion has also been found to be essential for directional and efficient cell migration [11]. Exosomes enhanced the speed of tumor cell migration by delivering ECM constituents present on the exosome surface that served to promote adhesion formation. Fibronectin, attached to the exosome surface via interaction with integrins, was found to be critical for promoting tumor cell motility. Essentially, secreted exosomes provide a fibronectin-rich ECM “carpet” on which the leading edge of the migrating cell can adhere to and travel [11]. Presumably, the ability to migrate on this exosome carpet depends on both making new adhesion interactions as well as breaking those interactions as needed for progressive movement. Again, MT1-MMP may be involved in the latter because it can degrade ECM substrates including fibronectin and vitronectin as well as cell surface syndecan-1, a molecule that via its heparan sulfate chains mediates exosome binding to fibronectin [10].
ADAM-17
ADAM-17, an important sheddase of many cell surface molecules, was also found on the surface of exosomes secreted by lung epithelial A549 cells following their stimulation with phorbol-12-myristate-13-acetate (PMA) [16]. Interestingly cell activation in this manner led to downregulation of cell surface ADAM-17 indicating that cell activation caused a re-routing of ADAM-17 to the extracellular compartment through its association with exosomes. Immunostaining and flow cytometry confirmed the presence of ADAM-17 on the exosome surface. Similarly, monocytes and primary endothelial cells when stimulated with lipopolysaccharide (LPS) induced exosomal ADAM-17 release [16]. Incubation of ADAM-17 exosomes with cells expressing the ADAM-17 substrates TGFα or amphiregulin caused shedding of both substrates. These data indicate that activation of cells can downregulate cell surface ADAM-17 and stimulate release of exosomal ADAM-17 which can then mediate shedding of ADAM-17 substrates at distal sites [16]. This has important implications in numerous disease states whose characteristics can be regulated by the shedding of cell surface molecules, many of which are cleaved by ADAM-17. Beyond its role as a sheddase, ADAM17 has a diverse array of substrates including cytokines and growth factors as well as adhesion molecules that are activated or inactivated by their cleavage with ADAM-17 [17]. It is therefore not surprising that ADAM-17 is implicated in driving numerous human diseases including cancer, heart disease, diabetes, rheumatoid arthritis, kidney fibrosis and Alzheimer’s disease. The finding that ADAM-17 can be delivered and traffic within the extracellular space as cargo on the surface of exosomes provides new insight into the complex biology of this enzyme and its relationship to disease progression.
Insulin-degrading enzyme (IDE)
Accumulation of amyloid β (Aβ) peptides within plaques is thought to play a significant role in the development and progression of Alzheimer’s disease. Thus, mechanisms regulating plaque formation and degradation are of great interest in understanding the disease and in developing interventional therapies. Insulin-degrading enzyme (IDE), also known as insulysin, is a 110 kDa thiol zinc-metalloendopeptidase localizing in the cytosol, peroxisomes, endosomes and the cell surface. Multiple functions have been attributed to IDE including degradation of insulin, amylin and glucagon as well as modulation of the ubiquitin-proteasome system indicating it may regulate protein turnover and homeostasis [18]. IDE is known to cleave the amyloid β-protein and the β amyloid precursor protein and to eliminate the neurotoxic effects of Aβ peptides [19–21]. Moreover, it has been demonstrated that in vivo, IDE regulates the levels of amyloid β-protein and the β-amyloid precursor protein intracellular domain [22].
Interestingly, in addition to its cellular localization, IDE is also found extracellularly, although it lacks a signal sequence necessary for targeting proteins to the secretory pathway. Studies using BV-2 microglial cells or N2a neuroblastoma cells demonstrated that a large proportion of the secreted IDE is associated with exosomes [23, 24]. Studies with the microglial cells revealed that statins enhanced both exosome secretion and the level of IDE, resulting in a dramatic increase in Aβ degradation, a finding confirmed in vivo by treating mice with lovastatin [24]. In N2a cells, exosomal IDE was found to be proteolytically active and treatment of purified exosomes with proteinase K removed exosomal surface proteins including IDE indicating that the enzyme is present on the exosome surface and available to degrade Aβ peptides [23]. Growth of the N2a cells in hypoxic conditions enhanced the exosome secretory rate and increased IDE levels extracellularly. Interfering with formation of exosomes by disrupting the ATPase Vps4 resulted in reduced IDE secretion and subsequent increase of ~43% in the amount of extracellular Aβ peptide compared to cells secreting normal levels of exosomes. These results imply that IDE present on the exosome surface plays a key role in destruction of Aβ peptides within the extracellular matrix of the brain.
It is noteworthy that in a separate study it was demonstrated that the cellular prion protein PrPc is present on the surface of exosomes secreted by N2a cells and that this protein binds to oligmers of Aβ, sequestering them on the exosome surface and preventing their accumulation within the extracellular matrix [25]. Thus exosomes, in addition to acting as carriers of IDE, may have additional functions as negative regulators of Alzheimer’s plaque formation. In patients with early stage Alzheimer’s disease, it has been shown that abnormally large early endosomes are present [26]. Because endosomes are precursors of the multivesicular bodies that contain newly formed exosomes, their abnormal size may signal disruption in exosome formation. Misregulation of exosome secretion or failure of IDE to be properly loaded into exosomes could lead to accumulation of Aβ peptides thereby contributing to the onset or progression of Alzheimer’s. Therapeutics that increase exosome secretion and/or enhance IDE expression (e.g., statins) have potential to diminish the level of extracellular Aβ and disease severity. Statins have been tested in Alzheimer’s patients and while a number of studies indicated that statins can reduce Alzheimer’s severity, this remains controversial and awaits further clinical trials [27].
The case of exosomal IDE and its relationship to degradation of Aβ peptides provides a potential opportunity for an exosome-based therapy. A recurring challenge in delivering drugs to the brain is the necessity to cross the blood-brain barrier. In a recent paper, a methodology was described in which an exosome address signal was engineered into a protein resulting in that protein being specifically targeted to exosomes [28]. When delivered through the nasal route, the exosomes crossed the blood-brain barrier and delivered the biologically active protein to recipient neurons in a number of brain regions. A strategy similar to this might prove useful for delivering exosomes bearing enzymatically active IDE into regions containing aggregates of Aβ peptides.
Other ECM-degrading exosomal proteins
Exosomes secreted by a rat pancreatic adenocarcinoma cell line (BSp73ASML) were shown to bind to and degrade extracellular matrix proteins including collagens, laminins and fibronectin [29]. Binding to ECM was not random; rather it varied depending on ECM composition and adhesion receptor profile of the exosomes. Flow cytometry analysis of purified exosomes bound to latex beads detected the presence of a number of proteases including MMPs -2, -3, -9, -13, -14, ADAM-10, ADAM-17, ADAMTS-5, ADAMTS-8, uPAR and the glycosidase hyaluronidase [29]. As some of these are secreted enzymes, it is not known if or how they become associated with the exosome surface and whether these associations occur within the multivesicular body during exosome formation, or after the exosomes are secreted. Results also suggested that exosome degradation of the ECM could impact tumor and host cell adhesion, motility and invasiveness [29].
Glycosidase activity on the exosome surface
Sialidase
Present in abundance on many cell types are sialic acids, nine-carbon-backbone carboxylated sugars [30]. Sialic acids are usually present as monosialyl residues at non-reducing terminus of glycoproteins and glycolipids where they play important functional roles in a diverse array of ligand-receptor interactions [30]. Sialic acids are occasionally present in α2→8-linked polysialic acid chains. An example of this is on the surface of neuronal cells where a polymer of the Neu5Ac (N-acetylneuraminic acid) sialic acid is present and associated with embryonic brain development and adult brain functions [31, 32]. Polysialic acid mediates neural cell adhesion and migration and binds to neurotrophins including brain-derived neurotrophic factor (BDNF), fibroblast growth factor-2 (FGF2) and neurotransmitters [33, 34]. Neurotrophins are a family of proteins that promote the survival, development and function of neurons. They are essential for development of the vertebrate nervous system and in general act through cell surface receptors including Trk receptor tyrosine kinases and the p75 neurotrophin receptor [35]. Binding of neurotrophins to polysialic acid regulates their availability and concentration in the brain, thus precise control of polysialic acid levels at the cell surface plays an important functional role. This control is mediated by sialyltransferases that synthesize and sialidases that degrade sialic acid. Sialidases fall within the family of enzymes known as glycosidases, enzymes that catalyze the hydrolysis of glycosidic linkages, thereby degrading oligosaccharides and glycoconjugates. Often, hydrolysis of these linkages at the cell surface results in breaking the interaction between proteins and carbohydrates; interactions that mediate a large number of biological functions including cell adhesion and cell signaling.
Treatment of the microglial cell line Ra2 with LPS to simulate inflammatory conditions resulted in loss of polysialic acid from the cell surface [6]. This loss was rapid, with 80% being lost within 10 minutes of exposure of cells to LPS. The effect of LPS could be blocked by the sialidase inhibitor Neu5Ac2en, pointing to an endogenous extracellular sialidase as the mediator of polysialic acid loss from the cell surface. Further analysis revealed the sialidase to be neuraminidase-1 (Neu1). Examination of extracellular vesicles secreted by the microglial cells following stimulation with LPS revealed that upon density gradient centrifugation, Neu1 was present in the low-density membrane fraction (1.13–1.19 g/ml) and co-presented with CD63 and CD9, molecules known to be associated with exosomes [6]. Further analysis confirmed the presence of Neu1 on the vesicle surface. Interestingly, trimming of the cell surface polysialic acid by Neu1 present on the exosomes triggered the release of BDNF [6]. This exosome-facilitated mechanism of neurotrophin regulation is potentially of great importance as microglia are known to be involved in maintenance of brain cells and in development of neurological diseases including Alzheimer’s and multiple sclerosis [36]. Moreover, this control of neurotrophin release may regulate adult neural plasticity and participate in response and repair of damage caused by inflammation or other tissue insults.
This example of an intact exosome degrading a surface carbohydrate to release a factor that can stimulate cell signaling is particularly interesting as it highlights a previously unrecognized function of exosomes. It will be important to assess whether other cell types respond to stimuli by releasing a burst of exosomes and whether those exosomes have a similar impact on regulating the availability of growth factors at the cell surface. If this is the case, it would suggest that exosome release may be used by cells as a means to rapidly respond to changing conditions within tissue microenvironments.
Heparanase
Heparanase is an endoglycosidase that trims heparan sulfate chains thereby altering the cell surface and remodeling the ECM. Heparanase, via both enzymatic and non-enzymatic mechanisms, stimulates a range of biological activities including tumor growth, metastasis, angiogenesis and inflammation [37–40]. Interestingly, syndecan heparan sulfate proteoglycans were found to be essential for formation of at least a significant portion of exosomes expressed by some cell types. This occurs through syndecan interaction with the PDZ domains of the cytoplasmic adaptor syntenin, a protein that also binds ALIX, an auxiliary component of the ESCRT family [41]. The syndecan-syntenin-ALIX complex supports the membrane budding process leading to generation of multivesicular endosomes that eventually fuse with the plasma membrane and release exosomes. The process of membrane budding within endosomes also requires heparan sulfate thus raising the possibility that trimming of heparan sulfate by heparanase regulates this process [41]. In support of this notion, analysis of CAG human myeloma or ARH-77 human lymphoblastoid cells transfected with the cDNA for heparanase revealed a dramatic increase in exosome secretion compared to control cells [7]. Addition of exogenous recombinant heparanase to wild-type CAG myeloma or MDA-MB-231 breast cancer cells also stimulated exosome secretion. Upregulation of exosome secretion by heparanase was dependent on heparanase enzymatic activity, indicating that heparanase degradation of heparan sulfate chains was regulating exosome secretion [7]. Subsequently this was confirmed by demonstration that trimming of syndecan heparan sulfate chains by heparanase modulated the syndecan-syntenin-ALIX pathway resulting in enhanced endosomal intraluminal budding and biogenesis of exosomes [42].
In addition to enhancing exosome secretion, heparanase alters exosome composition in ways that stimulated tumor cell spreading and endothelial cell invasion significantly more than did exosomes from control cells [7]. Included in the changes in exosome composition was the finding that heparanase itself was present as exosome cargo. In another set of experiments it was discovered that exposure of myeloma cells to bortezomib, a proteasome inhibitor widely used to treat myeloma patients, caused increased heparanase expression and a burst of exosome secretion [8, 43]. These exosomes (referred to as chemoexosomes) were loaded with very high levels of heparanase. When chemoexosomes were bound to magnetic beads using anti-CD63 antibodies, heparanase was detected on the exosome surface by flow cytometry [8]. Treatment of the bead-bound exosomes with bacterial Heparinase III released the heparanase from the exosome surface. This demonstrates that heparanase is bound to heparan sulfate chains present on the exosome surface. These heparan sulfate chains are likely from syndecan-1, a heparan sulfate-bearing proteoglycan known to be present in a high level on exosomes secreted by myeloma cells [7]. Moreover, western blotting of the chemoexosomes following removal of surface-bound heparanase revealed that no detectable heparanase remained. This indicates that most, if not all, of the heparanase present in chemoexosomes is associated with the exosome surface. To determine if the heparanase present on the chemoexosome surface could degrade heparan sulfate, intact chemoexosomes were incubated with an ECM deposited by bovine endothelial cells. Following 16 hours of exposure of chemoexosomes to the ECM, heparan sulfate fragments liberated by the exosomal heparanase were detected in the culture medium [8]. Similarly, exosomes secreted by CAG myeloma cells expressing a high level of heparanase also degraded heparan sulfate within the ECM [8]. However, consistent with their having lower levels of exosomal heparanase, the exosomes secreted by heparanase-high cells liberated less heparan sulfate than did the chemoexosomes. Together these findings demonstrate that heparanase present on the surface of exosomes can degrade heparan sulfate present within the ECM. Moreover, the finding that chemoexosomes are loaded with a high level of surface heparanase and that this can degrade ECM reveals a previously unappreciated potentially negative side effect of anti-tumor therapy.
The ability of exosomal heparanase to degrade heparan sulfate as exosomes diffuse through the tumor microenvironment could be important in promoting cell migration through ECM during tumor metastasis and/or inflammation. This may be particularly relevant regarding degradation of basement membranes that are known to be rich in heparan sulfate due to the presence of the perlecan heparan sulfate proteoglycan. Additionally, exosomal heparanase may participate in endothelial cell migration during angiogenesis, wound healing and inflammation. Fibronectin, a common component of exosomes and known to be involved in cell migration, associates with the exosome surface at least in part via interaction with exosomal heparan sulfate chains [10]. Heparanase trimming of exosomal heparan sulfate could possibly release fibronectin into the ECM in ways that modulate cell-ECM adhesion. Thus, heparanase on the surface of exosomes could impact exosome-target cell interactions in either positive or negative ways. In addition, heparan sulfate on the surface of cells binds to fibronectin on exosomes to mediate exosome docking [10, 44]. As such, heparan sulfate seems an appropriate target to block exosome docking as a potential therapeutic intervention of tumor-host crosstalk. Within this context, the heparin mimic Roneparstat was found to block exosome docking with target cells presumably by competing against cell surface heparan sulfate [10].
Summary and future perspectives.
Exosomes have emerged as important regulators of cell behavior in both normal and pathological settings. The recent finding that enzymes important in regulating various aspects of cell behavior can localize to the exosome surface extends the importance of these extracellular vesicles and raises new questions about their function. Although there are at present a limited number of examples of exosomal surface enzymes, within this context it is possible to envision a number of activities in which exosomes may participate where surface enzymes could play a role. This underscores the importance of further assessing the enzymes associated with exosomes, and determining their localization (e.g., exosome surface and/or encapsulated within exosomes). It will also be important in future studies to understand how enzymes are associated with the exosome surface. This could be via transmembrane domains as a component of the enzymes structure (as with MT1-MMP) or by their binding to molecules on the exosome surface as demonstrated in the case of heparanase binding to exosomal surface heparan sulfate. In addition, it is possible that enzymes form complexes with other molecules at the exosome surface, some of which may regulate the activity of the enzyme or protect the enzyme from proteolytic degradation. As described in two of the examples in this review, stimulation of cells by LPS or chemotherapy resulted in a burst in secretion of exosomes having a high level of enzyme on their surface. It will be important to determine if exosomes from these same cells prior to stimulation display the enzyme on their surface, or whether the stimulation event caused relocation of the enzyme to the exosome surface. Exosomes are also being closely investigated for their potential as biomarkers for disease diagnosis and prognosis. Because the exosomal surface enzymes described to date are linked to disease processes, they may prove to be useful biomarkers particularly as related to cancer progression. Also, because they are on the exosome surface they are rendered relatively easy to detect. Lastly, exosomes and their manipulation provide a ripe opportunity for development of new therapeutic approaches. As it is now becoming feasible to load exosomes with specific proteins, enzymes that degrade specific pathological complexes such as Aβ peptides could be used to combat Alzheimer’s disease. Targeted delivery of exosomes bearing sialidases on their surface could release cell-bound neurotrophins that would stimulate growth of injured nerve tissue thereby helping to speed recovery and regain nerve function. In conclusion, the emerging evidence that enzymes can be localized to the exosome surface, retain their enzymatic activity and regulate biological processes within the extracellular space opens exciting new opportunities to probe, understand and treat a myriad of human diseases.
Highlights.
Exosomes regulate cell communication by transferring proteins, lipids and nucleic acids between cells
New evidence demonstrates that enzymatically active proteases and glycosidases are present on the surface of some exosomes
These enzymes on exosome surfaces can degrade the ECM, liberate growth factors and alter cell adhesion and invasion
This newly appreciated mechanism of exosome function has implications in the progression of cancer, inflammation and Alzheimer’s disease
Acknowledgments
Funding information
Original research related to heparanase was supported by grants from the National Institutes of Health CA138340 and CA211752 (to RDS) and the United States – Israel Binational Science Foundation (jointly to RDS and IV). Israel Vlodavsky is a Research Professor of the Israel Cancer Research Fund (ICRF). The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used
- Aβ
amyloid β
- ADAM
a disintegrin and metalloproteinase
- ADAMTS
a disintegrin and metalloproteinase with thrombospondin motifs
- ALIX
ALG-2-interacting protein X
- BDNF
brain-derived neurotropic factor
- ECM
Extracellular matrix
- ESCRT
endosomal sorting complex required for transport
- FGF2
fibroblast growth factor 2
- HPSE
heparanase
- HS
heparan sulfate
- IDE
insulin-degrading enzyme
- LPS
lipopolysaccharide
- MMP
matrix metalloproteinase
- MT1-MMP
membrane-type 1 matrix metalloproteinase
- Neu1
neuraminidase 1
- PMA
phorbol-12-myristate-13-acetate
- TGFα
transforming growth factor alpha
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest
The authors declare no competing financial interests.
References
- 1.Colombo M, Raposo G, Thery C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–89. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 2.Kalluri R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126:1208–15. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4:27066. doi: 10.3402/jev.v4.27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimoda M, Khokha R. Proteolytic factors in exosomes. Proteomics. 2013;13:1624–36. doi: 10.1002/pmic.201200458. [DOI] [PubMed] [Google Scholar]
- 6.Sumida M, Hane M, Yabe U, Shimoda Y, Pearce OM, Kiso M, Miyagi T, Sawada M, Varki A, Kitajima K, Sato C. Rapid trimming of cell surface polysialic acid (PolySia) by exovesicular sialidase triggers release of preexisting surface neurotrophin. J Biol Chem. 2015;290:13202–14. doi: 10.1074/jbc.M115.638759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thompson CA, Purushothaman A, Ramani VC, Vlodavsky I, Sanderson RD. Heparanase regulates secretion, composition, and function of tumor cell-derived exosomes. J Biol Chem. 2013;288:10093–9. doi: 10.1074/jbc.C112.444562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandari SK, Purushothaman A, Ramani VC, Brinkley GJ, Chandrashekar DS, Varambally S, Mobley JA, Zhang Y, Brown EE, Vlodavsky I, Sanderson RD. Chemotherapy induces secretion of exosomes loaded with heparanase that degrades extracellular matrix and impacts tumor and host cell behavior. Matrix Biol. doi: 10.1016/j.matbio.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross JC, Chaudhary V, Bartscherer K, Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14:1036–45. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 10.Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem. 2016;291:1652–63. doi: 10.1074/jbc.M115.686295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sung BH, Ketova T, Hoshino D, Zijlstra A, Weaver AM. Directional cell movement through tissues is controlled by exosome secretion. Nat Commun. 2015;6:7164. doi: 10.1038/ncomms8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Itoh Y. Membrane-type matrix metalloproteinases: Their functions and regulations. Matrix Biol. 2015;44–46:207–23. doi: 10.1016/j.matbio.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Hakulinen J, Sankkila L, Sugiyama N, Lehti K, Keski-Oja J. Secretion of active membrane type 1 matrix metalloproteinase (MMP-14) into extracellular space in microvesicular exosomes. J Cell Biochem. 2008;105:1211–8. doi: 10.1002/jcb.21923. [DOI] [PubMed] [Google Scholar]
- 14.Artym VV, Zhang Y, Seillier-Moiseiwitsch F, Yamada KM, Mueller SC. Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 2006;66:3034–43. doi: 10.1158/0008-5472.CAN-05-2177. [DOI] [PubMed] [Google Scholar]
- 15.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. Exosome secretion is enhanced by invadopodia and drives invasive behavior. Cell Rep. 2013;5:1159–68. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groth E, Pruessmeyer J, Babendreyer A, Schumacher J, Pasqualon T, Dreymueller D, Higashiyama S, Lorenzen I, Grotzinger J, Cataldo D, Ludwig A. Stimulated release and functional activity of surface expressed metalloproteinase ADAM17 in exosomes. Biochim Biophys Acta. 2016;1863:2795–2808. doi: 10.1016/j.bbamcr.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Gooz M. ADAM-17: the enzyme that does it all. Crit Rev Biochem Mol Biol. 2010;45:146–69. doi: 10.3109/10409231003628015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tundo GR, Sbardella D, Ciaccio C, Grasso G, Gioia M, Coletta A, Polticelli F, Di Pierro D, Milardi D, Van Endert P, Marini S, Coletta M. Multiple functions of insulin-degrading enzyme: a metabolic crosslight? Crit Rev Biochem Mol Biol. 2017;52:554–582. doi: 10.1080/10409238.2017.1337707. [DOI] [PubMed] [Google Scholar]
- 19.Vekrellis K, Ye Z, Qiu WQ, Walsh D, Hartley D, Chesneau V, Rosner MR, Selkoe DJ. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci. 2000;20:1657–65. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu WQ, Ye Z, Kholodenko D, Seubert P, Selkoe DJ. Degradation of amyloid beta-protein by a metalloprotease secreted by microglia and other neural and non-neural cells. J Biol Chem. 1997;272:6641–6. doi: 10.1074/jbc.272.10.6641. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee A, Song E, Kihiko-Ehmann M, Goodman JP, Jr, Pyrek JS, Estus S, Hersh LB. Insulysin hydrolyzes amyloid beta peptides to products that are neither neurotoxic nor deposit on amyloid plaques. J Neurosci. 2000;20:8745–9. doi: 10.1523/JNEUROSCI.20-23-08745.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–7. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bulloj A, Leal MC, Xu H, Castano EM, Morelli L. Insulin-degrading enzyme sorting in exosomes: a secretory pathway for a key brain amyloid-beta degrading protease. J Alzheimers Dis. 2010;19:79–95. doi: 10.3233/JAD-2010-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamboli IY, Barth E, Christian L, Siepmann M, Kumar S, Singh S, Tolksdorf K, Heneka MT, Lutjohann D, Wunderlich P, Walter J. Statins promote the degradation of extracellular amyloid {beta}-peptide by microglia via stimulation of exosome-associated insulin-degrading enzyme (IDE) secretion. J Biol Chem. 2010;285:37405–14. doi: 10.1074/jbc.M110.149468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An K, Klyubin I, Kim Y, Jung JH, Mably AJ, O’Dowd ST, Lynch T, Kanmert D, Lemere CA, Finan GM, Park JW, Kim TW, Walsh DM, Rowan MJ, Kim JH. Exosomes neutralize synaptic-plasticity-disrupting activity of Abeta assemblies in vivo. Mol Brain. 2013;6:47. doi: 10.1186/1756-6606-6-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nixon RA. Autophagy, amyloidogenesis and Alzheimer disease. J Cell Sci. 2007;120:4081–91. doi: 10.1242/jcs.019265. [DOI] [PubMed] [Google Scholar]
- 27.Mendoza-Oliva A, Zepeda A, Arias C. The complex actions of statins in brain and their relevance for Alzheimer’s disease treatment: an analytical review. Curr Alzheimer Res. 2014;11:817–33. [PubMed] [Google Scholar]
- 28.Sterzenbach U, Putz U, Low LH, Silke J, Tan SS, Howitt J. Engineered exosomes as vehicles for biologically active proteins. Mol Ther. 2017;25:1269–1278. doi: 10.1016/j.ymthe.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mu W, Rana S, Zoller M. Host matrix modulation by tumor exosomes promotes motility and invasiveness. Neoplasia. 2013;15:875–87. doi: 10.1593/neo.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–69. doi: 10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- 31.Sato C, Kitajima K. Disialic, oligosialic and polysialic acids: distribution, functions and related disease. J Biochem. 2013;154:115–36. doi: 10.1093/jb/mvt057. [DOI] [PubMed] [Google Scholar]
- 32.Colley KJ, Kitajima K, Sato C. Polysialic acid: biosynthesis, novel functions and applications. Crit Rev Biochem Mol Biol. 2014;49:498–532. doi: 10.3109/10409238.2014.976606. [DOI] [PubMed] [Google Scholar]
- 33.Kanato Y, Kitajima K, Sato C. Direct binding of polysialic acid to a brain-derived neurotrophic factor depends on the degree of polymerization. Glycobiology. 2008;18:1044–53. doi: 10.1093/glycob/cwn084. [DOI] [PubMed] [Google Scholar]
- 34.Ono S, Hane M, Kitajima K, Sato C. Novel regulation of fibroblast growth factor 2 (FGF2)-mediated cell growth by polysialic acid. J Biol Chem. 2012;287:3710–22. doi: 10.1074/jbc.M111.276618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao MV. Neurotrophins and their receptors: a convergence point for many signalling pathways. Nat Rev Neurosci. 2003;4:299–309. doi: 10.1038/nrn1078. [DOI] [PubMed] [Google Scholar]
- 36.Graeber MB, Li W, Rodriguez ML. Role of microglia in CNS inflammation. FEBS Lett. 2011;585:3798–805. doi: 10.1016/j.febslet.2011.08.033. [DOI] [PubMed] [Google Scholar]
- 37.Sanderson RD, Elkin M, Rapraeger AC, Ilan N, Vlodavsky I. Heparanase regulation of cancer, autophagy and inflammation: new mechanisms and targets for therapy. FEBS J. 2017;284:42–55. doi: 10.1111/febs.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vlodavsky I, Singh P, Boyango I, Gutter-Kapon L, Elkin M, Sanderson RD, Ilan N. Heparanase: From basic research to therapeutic applications in cancer and inflammation. Drug Resist Updat. 2016;29:54–75. doi: 10.1016/j.drup.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vlodavsky I, Iozzo RV, Sanderson RD. Heparanase: Multiple functions in inflammation, diabetes and atherosclerosis. Matrix Biol. 2013;32:220–22. doi: 10.1016/j.matbio.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 40.Goldberg R, Meirovitz A, Hirshoren N, Bulvik R, Binder A, Rubinstein AM, Elkin M. Versatile role of heparanase in inflammation. Matrix Biol. 2013;32:234–240. doi: 10.1016/j.matbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–85. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 42.Roucourt B, Meeussen S, Bao J, Zimmermann P, David G. Heparanase activates the syndecan-syntenin-ALIX exosome pathway. Cell Res. 2015;25:412–28. doi: 10.1038/cr.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramani VC, Vlodavsky I, Ng M, Zhang Y, Barbieri P, Noseda A, Sanderson RD. Chemotherapy induces expression and release of heparanase leading to changes associated with an aggressive tumor phenotype. Matrix Biol. 2016;55:22–34. doi: 10.1016/j.matbio.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci U S A. 2013;110:17380–5. doi: 10.1073/pnas.1304266110. [DOI] [PMC free article] [PubMed] [Google Scholar]


