Abstract
Increased expression of CBX3/HP1γ, a core component of heterochromatin protein 1, has recently proved to be involved in human tumorigenesis and patient prognosis. The present study aimed to investigate the expression of CBX3/HP1γ and its clinicopathological significance in primary tongue squamous cell carcinoma (TSCC). Gene expression profiles of CBX3/HP1γ in TSCC from Oncomine database were analyzed. The expression of CBX3/HP1γ at protein level was measured using immunohistochemistry (IHC). The potential associations between CBX3/HP1γ expression and multiple clinicopathological parameters were estimated using the Chi square test. In addition, the effect of CBX3/HP1γ expression on patients' survival was further assessed by Kaplan-Meier and Cox regression analyses. The agreement of elevated CBX3/HP1γ expression was indicated in four datasets on the Oncomine database. Aberrant overexpression of CBX3/HP1γ was identified in TSCC tissues compared with cancer-adjacent normal tissue, which was significantly associated with cervical nodes metastasis (P=0.010) and clinical stage (P=0.025). Furthermore, patients with high CBX3/HP1γ expression exhibited a reduced survival compared with those with low expression (Log-rank test, P=0.004). Univariate and multivariate Cox regression analysis suggested that the expression status of CBX3/HP1γ could be regarded as an independent prognostic factor for TSCC patients (HR=2.461; 95% CI=1.128–5.370; P=0.024). The present study indicated that aberrant overexpression of Cbx3/HP1γ was associated with cervical nodes metastasis and unfavorable survival in TSCC. These findings suggest that CBX3/HP1γ may serve an important role in tongue tumorigenesis and may be a valuable candidate diagnostic and prognostic marker for TSCC patients.
Keywords: tongue neoplasms, prognosis, Cbx3/HP1γ
Introduction
Tongue squamous cell carcinoma (TSCC) is the most common malignancy in the oral cavity, accounting for about one-third of all oral cancers (1). The morbidity and typical mortality rates of TSCC has been increasing rapidly during the past five years (2,3). Being an aggressive subtype of oral cancer, TSCC is characterized by high frequency of local invasion and occult lymph node metastasis. Consequently, the long-term survival rate continue to be low, with the overall five-year survival rate at the level of less than 50% (4). Therefore, identification of novel biomarkers and therapeutic targets against TSCC is necessary and urgent to illustrate the clinical behavior of TSCC and to personalize therapy.
The heterochromatin proteins 1 (HP1) family, known as chromatin binding proteins, directly bind to the promoter region of the methylated H3K9 (the methyl groups of histone H3 at lysine 9) to participate in the heterochromatin silence of gene expression (5). Recent studies indicated that H3K9 methylation may play key roles in tongue cancer tumorigenesis (6). HP1γ, encoded by the CBX3, is a paralog of HP1. By binding methylated H3K9, CBX3/HP1γ can recruit lots of cofactors which perform various biological functions including RNA alternative splicing, DNA damage response, transcription elongation, cell growth and differentiation (7). Moreover, CBX3/HP1γ is also located in euchromatin areas, which suggest that it may be also associated wih transcriptional activation (8). Previous studies proved that CBX3/HP1γ was abnormally expressed in multiple cancers such as prostate, colon and lung cancer (9–11), and was always an unfavorable prognostic factor. Thus, it seems that CBX3/HP1γ plays a potential role as a novel cancer biomarker and a putative oncogene. However, the expression level of CBX3/HP1γ in TSCC and its clinical pathologic parameters remains unclear.
Herein, we sought to investigate the expression of CBX3/HP1γ in primary human TSCC specimens and identify potential relationship between its expression and clinicopathological features as well as patients' survival.
Materials and methods
Oncomine database analysis
To obtain the outline of the CBX3/HP1γ expression pattern, we explored the CBX3 mRNA levels of human TSCC in Oncomine database (http://www.oncomine.org), which is publicly available.
Patients and specimen collection
A cohort of 126 patients with primary TSCC treated in Sun Yat-sen University Cancer Center between January 2009 and December 2010 were enrolled retrospectively. The enrollment criteria included: i) primary TSCC without any chemotherapy or radiotherapy before surgery; ii) patients underwent radical tumor resection and neck dissection and iii) complete information available about clinical, pathological and follow-up data. The formalin-fixed paraffin-embedded samples were collected from the archives of the hospital, with adjacent non-tumor tissues from the same patients as a control group. Cancer stage was defined according to the TNM staging based on American Joint Committee on Cancer (AJCC), 7th edition (12). In order to verify the diagnosis, histological grade and stage, all samples were reviewed by a pathologist. This study was under the approval of the Clinical Research Ethics Committee of Sun Yat-sen University Cancer Center. Informed consent was obtained from all patients.
Immunohistochemistry (IHC)
IHC studies were performed on formalin-fixed, paraffin-embedded specimens following routine procedure. In brief, 3 µm-thick tissue sections were deparaffnized in xylene and hydrated in graded alcohol ranging from 100, 95, and 80 to 70%. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 min, and then washed with phosphate-buffered saline (PBS, pH 7.3). Antigen retrival was completed by boiling in citric acid buffer (10 mM, pH 6.0) for 15 min. In order to block nonspecific binding, slides were incubated with 10% normal goat serum for 15 min at room temperature. These sections were in further incubation of a rabbit anti-CBX3 antibody (diluted 1:400 in PBS; Wuhan Sanying Biotechnology, Wuhan, China) overnight at 4°C, following with incubation of secondary anti-rabbit antibody at a concentration of 1:100 at 37°C for 30 min. Subsequently, the sections were washed with PBS, colorized by diaminobenzidine (DAB) solution and counterstained with haematoxylin. PBS was used to replace the primary antibody as a negative control, whereas known IHC-positive CBX3/HP1γ staining slides of colon cancer were used as positive control.
The immunoreactivity was scored independently by two pathologists insensible of relevant clinicopathological data according to the stained intensity of cancer cells and positively-stained proportion. The estimated fraction of positive-stained tumor cells was defined as the proportion score (0, none; 1, 10%; 2, 11–50%; 3, 51–80%; 4, >80%), whereas the estimated staining intensity was demonstrated as the intensity score (0, no staining; 1, weak; 2, moderate; 3, strong) with the aggregate score ranging from 0 to 12, as described elsewhere (13). Overexpression of CBX3/HP1γ was defined as an aggregate score of >4. Therefore, the immunoreactivity of each slide was categorized into three subgroups based on the final score: negative (0); low expression (1–4); high expression (6–12).
Statistical analyses
Comparisons of raw IHC outcomes between groups were performed using the Wilcoxon signed-rank test. The Chi-square test was used to evaluate the associations between clinicpathological parameters of patients and CBX3/HP1γ expression. The overall survival was analyzed using Kaplan-Meier method and compared with Log-rank test. Univariate and multivariate Cox regression models were used to estimate individual clinical and pathological variables with patients' overall survival. P-values <0.05 (two-sided) were considered to indicate a statistically significant difference. All statistical analyses were performed using Graphpad Prism 5 (GraphPad Software, Inc., La Jolla, CA, USA) or SPSS v24.0 (IBM Corp., Armonk, NY, USA).
Results
mRNA expressing pattern of CBX3 on oncomine database
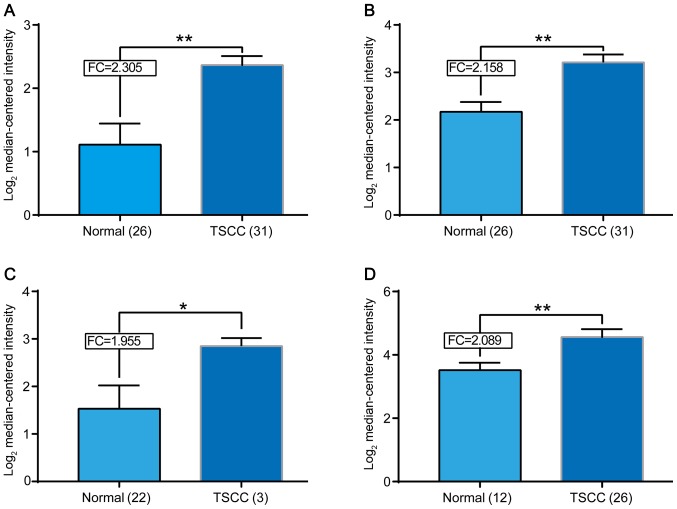
We explored the mRNA expression of CBX3 in 4 datasets on Oncomine Database (Fig. 1). CBX3 mRNA was found significantly elevated in human TSCC tissues compared with normal tissues in datasets from Oncomine Database [Talbot et al (14), Estilo et al (15), Kuriakose et al (16) and Ye et al (17)]. We noticed that the number of cases examined in each group was quite different. For example, in Kuriakose's group, there are only 3 cancer cases against 22 normal controls. However, the data from study came to a consistent result that CBX3 mRNA significantly elevated in human TSCC tissues.
Figure 1.
Search for Cbx3 mRNA expression of TSCC in Oncomine database. The mRNA levels of Cbx3 of TSCC were elevated in different datasets. The different datasets from the researchers (A) Talbot et al (14), (B) Estilo et al (15), (C) Kuriakose et al (16) and (D) Ye et al (17), who provide the data in Oncomine were indicated. The expression level of CBX3 mRNA was indicated in the form of log2 median-centered intensity. *P<0.05, **P<0.01. FC, fold change.
Overexpression of CBX3/HP1γ at protein level in TSCC tissues
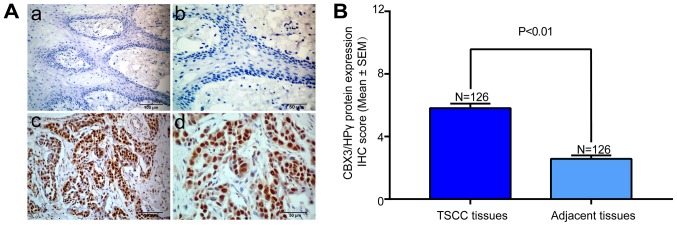
The CBX3/HP1γ protein expression of all TSCC tissues and adjacent non-tumor tissues were evaluated by IHC. The representative immunohistochemical staining of CBX3/HP1γ in TSCC and adjacent tongue mucosa was shown in Fig. 2A. High CBX3/HP1γ expression was mainly located in the nucleus areas rather than the cytoplasm in cancer cells, whereas weak or negative staining was observed in normal tongue epithelial cells. According to our IHC scoring regime, CBX3/HP1γ protein abundance in these primary TSCC and adjacent tissues was categorized, respectively. These data showed that CBX3/HP1γ was significantly overexpressed in TSCC than in the adjacent tissues (P<0.001; Fig. 2B).
Figure 2.
Immunohistochemical staining of CBX3/HP1γ protein in TSCC tissues and cancer-adjacent normal tissue. (Aa) Representative weak staining of CBX3/HP1γ (low expression) in cancer-adjacent normal tissue (magnification, ×200). Nuclei were counterstained with hematoxylin. (Ab) Magnified image from a (magnification, ×400). (Ac) Representative strong staining of CBX3/HP1γ (high expression) in primary TSCC sample (magnification, ×200). (Ad) Magnified image from c (magnification, ×400). CBX3/HP1γ expression was identified primarily in nuclei of cancer cells. (B) Statistical analysis of CBX3/HP1γ IHC scores in TSCC tissues and cancer-adjacent normal tissue. Data was presented as the mean with SEM.
Association between CBX3/HP1γ expression and clinicopathological characteristics
The association between CBX3/HP1γ expression and clinical pathological characteristics (age, gender, smoking, pathological grade, T stage, TNM stage, cervical node status) were summarized in Table I. Briefly, 82 male and 44 female patients were enrolled with mean age 54.1 years (25–83 years). The follow-up period ranged from 6.5 to 97.8 months with average 67.6 months. The results showed that cervical node metastasis (P=0.010), clinical stage (P=0.025) were related to high CBX3/HP1γ expression (Table I). There were no significant correlations between CBX3/HP1γ expression with patients' age, gender, smoking state, tumor size and pathological grade (Chi square test).
Table I.
Associations between CBX3/HP1γ expression and multiple clinicopathological parameters in primary TSCC.
| CBX3/HP1γ expression | ||||
|---|---|---|---|---|
| Variables | Cases | Lowa | High | χ2 P-value |
| Age | 0.182 | |||
| <60 | 81 | 28 | 53 | |
| ≥60 | 45 | 21 | 24 | |
| Gender | 0.273 | |||
| Male | 80 | 34 | 46 | |
| Female | 46 | 15 | 31 | |
| Smoking | 0.745 | |||
| Yes | 64 | 24 | 40 | |
| No | 62 | 25 | 37 | |
| Tumor size | 0.843 | |||
| T1–2 | 107 | 42 | 65 | |
| T3-4 | 19 | 7 | 12 | |
| Pathological grade | 0.824 | |||
| G1 | 99 | 38 | 61 | |
| G2-3 | 27 | 11 | 16 | |
| Cervical node metastasis | 0.010 | |||
| N- | 89 | 41 | 48 | |
| N+ | 37 | 8 | 29 | |
| Clinical stage | 0.025 | |||
| I–II | 80 | 37 | 43 | |
| III–IV | 46 | 12 | 34 | |
Both of patients with low and negative CBX3/HP1γ staining are stratified into low category for simplicity. TSCC, tongue squamous cell carcinoma.
Association between CBX3/HP1γ expression and overall survival of TSCC patients
The correlation between CBX3/HP1γ expression and patients' survival was analyzed to uncover the potential prognostic value of CBX3/HP1γ expression. Up to the last follow-up, 85 of 126 (67.5%) patients were alive, 41 (32.5%) patients died of cancer recurrence or metastases. The Kaplan-Meier survival analyses showed that high CBX3/HP1γ expression indicated unfavorable outcomes. As shown in Fig. 3, patients with low CBX3/HP1γ expression had significantly better overall survival than those with high expression (Log-rank test, P=0.004).
Figure 3.
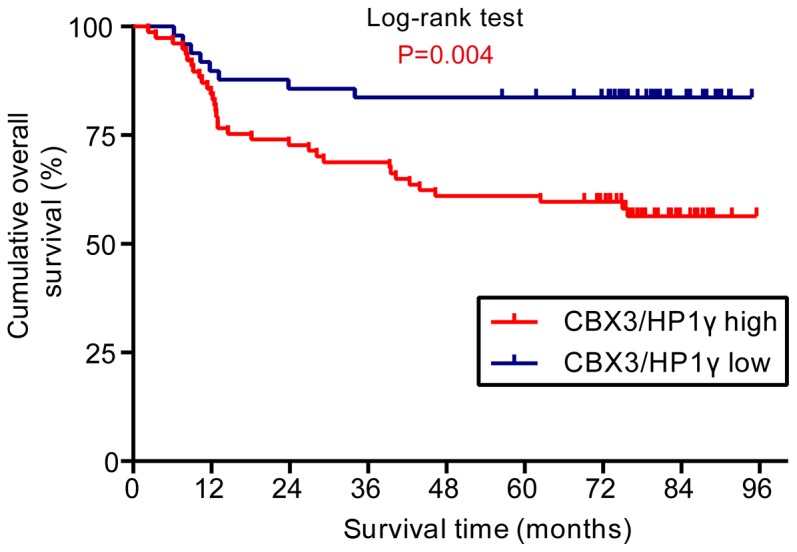
Overexpression of CBX3/HP1γ protein was associated with poor overall survival in patients with TSCC. Kaplan-Meier curves for overall survival rate in 126 patients with TSCC according to the expression status of CBX3/HP1γ. The staining score (6) served as a cut-off to divide patients into high and low expression. Blue line: Patients with low CBX3/HP1γ expression, red line: Patients with high CBX3/HP1γ expression. High CBX3/HP1γ protein expression was associated with significantly reduced overall survival (P=0.004, Log-rank test).
We conducted the univariate and multivariate survival analyses to further evaluate the clinical significance of CBX3/HP1γ expression as a prognostic predictor for TSCC patients. The univariate survival analysis revealed that CBX3/HP1γ expression status, cervical node metastasis and clinical stage were significantly associated with overall survival, while other clinicopathological parameters had not reached the statistical significance. Furthermore, multivariate survival analysis was also performed to exclude the confounding factors. Both the CBX3/HP1γ expression status (HR=2.461; 95% CI 1.128–5.370; P=0.024) and clinical stage (HR=3.663; 95% CI 1.322–10.146; P=0.013) were identified as an independent prognostic factor for the overall survival of patients with TSCC in this Cox regression model (Table II).
Table II.
Cox proportional hazard model analysis of variables affecting survival in patients with TSCC.
| Univariate survival analysis | Multivariate survival analysis | |||||
|---|---|---|---|---|---|---|
| Variables | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
| Age (<60 vs. ≥60) | 1.468 | 0.749–2.878 | 0.263 | |||
| Gender (male vs. female) | 1.947 | 0.954–3.973 | 0.067 | |||
| Smoking (yes vs. no) | 1.427 | 0.767–2.657 | 0.262 | |||
| Tumor size (T1-2 vs. T3-4) | 1.610 | 0.744–3.488 | 0.227 | |||
| Pathological grade (G1 vs. G2-3) | 0.802 | 0.393–1.636 | 0.544 | |||
| Cervical node metastasis (N- vs. N+) | 3.431 | 1.855–6.345 | 0.000a | 1.054 | 0.397–2.802 | 0.916 |
| Clinical stage (I–II vs. III–IV) | 4.257 | 2.249–8.057 | 0.000a | 3.663 | 1.322–10.146 | 0.013a |
| CBX3/HP1γ expression (lowa vs. high) | 2.967 | 1.370–6.426 | 0.006a | 2.461 | 1.128–5.370 | 0.024a |
Both of patients with low and negative CBX3/HP1γ staining are stratified into low category for simplicity. TSCC, tongue squamous cell carcinoma.
Discussion
Being a key hallmark of human cancer, aberrant epigenetic alternations including histone modification are significantly involved in multiple stages of oncogenesis (18). HP1, a well-known marker of histone modification for transcriptionally silenced heterochromatin (19), is reported to have various cellular functions including heterochromatin formation, chromatin remodeling, transcription regulation, DNA damage repairing and cancer progression (20–22). Previous studies have suggested that the CBX3/HP1γ, one paralog of HP1, might be an oncogene driving tumorigenesis and a novel diagnostic and prognostic biomarker in multiple cancers (11,23–25). Herein we investigated the expression pattern of CBX3/HP1γ in primary TSCC and its clinical pathological relevance and prognostic significance. We found that CBX3/HP1γ was aberrantly overexpressed in TSCC tissues than in normal tissues and its overexpression was significantly related to aggressive clinical pathological parameters and an unfavorable prognosis. This result was consistent with the trend in other cancers (11,25).
Tumorigenesis of TSCC is a multistep process driven by numerous abnormal gene alterations, such as oncogenes activation and tumor suppressor inactivation (26). Particularly, inactivation of tumor suppressor genes modulated by epigenetics is a major player in this multistep carcinogenesis and progression of tongue cancer (27,28). Among them, H3K9/HP1 is one of the key epigenetic silencing pathways for gene regulation (29). As an isoforms of HP1, CBX3/HP1γ has been proved to have various biological functions including RNA alternative splicing, DNA damage response, transcription elongation, cell growth and differentiation (7). Notably, highly elevated expression had been found in non-small cell lung cancer, prostate cancer and colorectal cancer (9,11,25) etc. In line with these previous finding, our study showed that CBX3/HP1γ was significantly overexpressed in human TSCC, thus indicating that the CBX3/HP1γ might be fostering tumorigenesis in multiple tissues including tongue. To the best of our knowledge, this might be the first study to uncover the abnormal expression pattern of CBX3/HP1γ in TSCC. However, the number of patients enrolled in this study was limited. Therefore, more cases from different institutions are needed to definitively confirm the expression pattern of CBX3/HP1γ as well as its diagnostic value in TSCC.
There is increasing evidence corroborating the notion that elevated expression of CBX3/HP1γ is significantly associated with aggressiveness in various types of cancers (13). For example, elevated CBX3/HP1γ is significantly associated with lymph node metastasis and clinical stages in non-small cell lung cancer or with high recurrence rate in prostate cancer (9,11). Similarly, our data showed that overexpression of CBX3/HP1γ was significantly associated with cervical lymph nodes metastasis and clinical stages in TSCC. Moreover, previous studies have demonstrated that CBX3/HP1γ can cooperate with MMP3 (matrix metalloproteinase 3), a well-known secretory endopeptidase that degrades extracellular matrices, in transcriptional regulation by serving as a nuclear MMP3-associated protein (NuMAP) (30). Taken together, we propose that CBX3/HP1γ might be an efficient biomarker for cervical node metastasis in TSCC. It will be interesting to explore the expression level of CBX3/HP1γ in the metastatic lesions and its clinical significance in further studies.
During the past decades the long term survival rate of TSCC remains disappointing despite the progress in diagnosis and treatment, suggesting that an accurate prognostic marker is highly desired and beneficial in the clinical practice (31). Previous researches showed that CBX3/HP1γ expression was associated with patients' survival and was identified as an independent prognostic predictor for patients with colorectal cancer, non-small cell lung cancer (11,25). For example, Slezak J et al (9) found that high expression of CBX3/HP1γ positively correlated with overexpression of Ki67 and predicted unfavorable prognosis in patients with prostate cancer. In agreement with these findings, our results indicated that patients with low CBX3/HP1γ expression had significantly longer survival comparing to those with high CBX3/HP1γ. Furthermore, univariate analysis identified CBX3/HP1γ expression as an independent prognostic factor affecting survival of patients with TSCC. Thus, the expression status of CBX3/HP1γ might offer valuable information for predicting patients' prognosis and effective follow-up management.
Increasing evidence had uncovered that CBX3/HP1γ was critically involved in tumorigenesis by promoting cell proliferation via regulating the expression of P21 or cyclin dependent kinases 6 (CDK6) during cell cycle (7,25). In addition, CBX3/HP1γ was demonstrated to have novel function in the epigenetic regulation of both cell differentiation and cancer development in various types of cancers (10). In particular, Michael Su et al found that reducing the levels of Cbx3/HP1γ could enhance tumor-killing capacity on CD8+ T cells (32). Taken these findings together, we suppose that Cbx3/HP1γ serve as not only a novel cancer biomarker of high value in diagnosis and prognosis, but also an effective target for gene therapy against various human cancers. Therefore, further researches are solely needed to uncover the roles and mechanisms of Cbx3/HP1γ during tongue tumorigenesis.
In conclusion, Cbx3/HP1γ is aberrantly overexpressed in TSCC tissues. CBX3/HP1γ expression is an independent prognostic factor of patients with TSCC, suggesting that Cbx3/HP1γ may be a candidate prognostic marker for TSCC patients. More studies are warranted to gain the insight into the mechanism of CBX3/HP1γ playing during the tumorigenesis and progression in TSCC.
Acknowledgements
The authors would like to thank Professor WuGuo Deng and Mr. DingBo Shi of The State Key Laboratory of Oncology in South China (Guangzhou, China) for their help during the experiment.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
AY designed the study. XS collected the patients' samples and performed the statistical analysis. HZ and XF performed the experiments and were major contributors in writing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The experimental protocol was established, according to the ethical guidelines of the Helsinki Declaration and was approved by the Human Ethics Committee of Sun Yan-sen University Cancer Centre. Written informed consent was obtained from individual or guardian participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma-an update. CA Cancer J Clin. 2015;65:401–421. doi: 10.3322/caac.21293. [DOI] [PubMed] [Google Scholar]
- 3.Duz MB, Karatas OF, Guzel E, Turgut NF, Yilmaz M, Creighton CJ, Ozen M. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: A pilot study. Cell Oncol (Dordr) 2016;39:187–193. doi: 10.1007/s13402-015-0259-z. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Wang Y, Qiu J, Li Q, Yuan C, Zhang W, Wang D, Ye J, Jiang H, Yang J, Cheng J. The polycomb group protein EZH2 is a novel therapeutic target in tongue cancer. Oncotarget. 2013;4:2532–2549. doi: 10.18632/oncotarget.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canzio D, Larson A, Narlikar GJ. Mechanisms of functional promiscuity by HP1 proteins. Trends Cell Biol. 2014;24:377–386. doi: 10.1016/j.tcb.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan C, Li Z, Qi B, Zhang W, Cheng J, Wang Y. High expression of the histone demethylase LSD1 associates with cancer cell proliferation and unfavorable prognosis in tongue cancer. J Oral Pathol Med. 2015;44:159–165. doi: 10.1111/jop.12220. [DOI] [PubMed] [Google Scholar]
- 7.Fan Y, Li H, Liang X, Xiang Z. CBX3 promotes colon cancer cell proliferation by CDK6 kinase-independent function during cell cycle. Oncotarget. 2017;8:19934–19946. doi: 10.18632/oncotarget.15253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 9.Slezak J, Truong M, Huang W, Jarrard D. HP1γ expression is elevated in prostate cancer and is superior to Gleason score as a predictor of biochemical recurrence after radical prostatectomy. BMC Cancer. 2013;13:148. doi: 10.1186/1471-2407-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takanashi M, Oikawa K, Fujita K, Kudo M, Kinoshita M, Kuroda M. Heterochromatin protein 1gamma epigenetically regulates cell differentiation and exhibits potential as a therapeutic target for various types of cance. Am J Pathol. 2009;174:309–316. doi: 10.2353/ajpath.2009.080148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Bi H, Zhan P, Chang C, Xu C, Huang X, Yu L, Yao X, Yan J. Overexpression of HP1γ is associated with poor prognosis in non-small cell lung cancer cell through promoting cell survival. Tumour Biol. 2014;35:9777–9785. doi: 10.1007/s13277-014-2182-8. [DOI] [PubMed] [Google Scholar]
- 12.Edge SB, Compton CC. The American joint committee on cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17:1471–1474. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 13.Zheng X, Jia B, Lin X, Han J, Qiu X, Chu H, Sun X, Hu W, Pan J, Chen J, Zhao J. FRMD4A: A potential therapeutic target for the treatment of tongue squamous cell carcinoma. Int J Mol Med. 2016;38:1443–1449. doi: 10.3892/ijmm.2016.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Talbot SG, Estilo C, Maghami E, Sarkaria IS, Pham DK, O-charoenrat P, Socci ND, Ngai I, Carlson D, Ghossein R, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65:3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 15.Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuriakose MA, Chen WT, He ZM, Sikora AG, Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, et al. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61:1372–1383. doi: 10.1007/s00018-004-4069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suvà ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng W, Ball AR, Jr, Yokomori K. HP1: Heterochromatin binding proteins working the genome. Epigenetics. 2010;5:287–292. doi: 10.4161/epi.5.4.11683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 21.Bártová E, Malyšková B, Komůrková D, Legartová S, Suchánková J, Krejčí J, Kozubek S. Function of heterochromatin protein 1 during DNA repair. Protoplasma. 2017;254:1233–1240. doi: 10.1007/s00709-017-1090-3. [DOI] [PubMed] [Google Scholar]
- 22.Dialynas GK, Vitalini MW, Wallrath LL. Linking heterochromatin protein 1 (HP1) to cancer progression. Mutat Res. 2008;647:13–20. doi: 10.1016/j.mrfmmm.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saini V, Hose CD, Monks A, Nagashima K, Han B, Newton DL, Millione A, Shah J, Hollingshead MG, Hite KM, et al. Identification of CBX3 and ABCA5 as putative biomarkers for tumor stem cells in osteosarcoma. PLoS One. 2012;7:e41401. doi: 10.1371/annotation/8c74aaee-897d-4682-b62d-d95a3506c210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomberk G, Bensi D, Christensen T, Salisbury J, Urrutia R. HP1 γ has a significant impact on cell cycle progression through the regulation of G2/M, centrosome biology and, chromosomal stability in pancreatic cancer cells. Pancreas. 2007;35:412. doi: 10.1097/01.mpa.0000297734.76617.e9. [DOI] [Google Scholar]
- 25.Liu M, Huang FF, Zhang D, Ju J, Wu XB, Wang Y, Wang Y, Wu Y, Nie M, Li Z, et al. Heterochromatin protein HP1γ promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75:4593–4604. doi: 10.1158/0008-5472.CAN-14-3735. [DOI] [PubMed] [Google Scholar]
- 26.Scully C, Bagan J. Oral squamous cell carcinoma overview. Oral Oncol. 2009;45:301–308. doi: 10.1016/j.oraloncology.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 27.González-Ramírez I, Soto-Reyes E, Sánchez-Pérez Y, Herrera LA, García-Cuellar C. Histones and long non-coding RNAs: The new insights of epigenetic deregulation involved in oral cancer. Oral Oncol. 2014;50:691–695. doi: 10.1016/j.oraloncology.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 28.Mascolo M, Siano M, Ilardi G, Russo D, Merolla F, De Rosa G, Staibano S. Epigenetic disregulation in oral cancer. Int J Mol Sci. 2012;13:2331–2353. doi: 10.3390/ijms13022331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nestorov P, Tardat M, Peters AH. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr Top Dev Biol. 2013;104:243–291. doi: 10.1016/B978-0-12-416027-9.00008-5. [DOI] [PubMed] [Google Scholar]
- 30.Eguchi T, Calderwood SK, Takigawa M, Kubota S, Kozaki KI. Intracellular MMP3 promotes HSP gene expression in collaboration with chromobox proteins. J Cell Biochem. 2017;118:43–51. doi: 10.1002/jcb.25607. [DOI] [PubMed] [Google Scholar]
- 31.Hu H, Wang Y, Li Z, Zhu Y, Zhang W, Wang D, Lin T, Yang J, Wang Y, Cheng J. Overexpression of suppressor of zest 12 is associated with cervical node metastasis and unfavorable prognosis in tongue squamous cell carcinoma. Cancer Cell Int. 2017;17:26. doi: 10.1186/s12935-017-0395-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun M, Ha N, Pham DH, Frederick M, Sharma B, Naruse C, Asano M, Pipkin ME, George RE, Thai TH. Cbx3/HP1γ deficiency confers enhanced tumor-killing capacity on CD8+ T cells. Sci Rep. 2017;7:42888. doi: 10.1038/srep42888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.