Abstract
Osteosarcoma (OS) is an aggressive malignant neoplasm that arises from primitive transformed cells of mesenchymal origin, and that exhibits osteoblastic differentiation and produces malignant osteoid. MicroRNA (miR)-542-3p has been reported to serve a crucial role in the initiation and development of several types of human cancer. However, whether miR-542-3p is involved in tumorigenesis and tumor progression of OS remains unclear. In the present study, the expression of miR-542-3p in OS cells and patients with OS, and its functional mechanism in OS were investigated. The data demonstrated that expression of miR-542-3p was significantly decreased in OS tissues and cell lines, and that restoration of miR-542-3p expression in OS cells inhibited cell proliferation and induced cell apoptosis. Through bioinformatics analysis and luciferase reporter assays, the present study identified that miR-542-3p directly targeted Smad2 mRNA and negatively regulated the expression of Smad2 at the protein level in OS cells. Furthermore, it was confirmed that OS tumor tissues with a low expression of miR-542-3p exhibited markedly higher Smad2 expression. Finally, through the use of gain of function and rescue experiments, the present study demonstrated that restoration of miR-542-3p was able to suppress the growth and proliferation of OS cells through directly targeting Smad2. To the best of our knowledge, this is the first study to demonstrate that decreased expression of miR-542-3p serves a role in tumor suppression in OS pathogenesis through targeting Smad2. These results will aid in elucidating the functions of miR-542-3p, and suggest that miR-542-3p may serve as a tumor suppressor gene and a promising therapeutic target of OS.
Keywords: osteosarcoma, miR-542-3p, cell proliferation, cell apoptosis, Smad2
Introduction
Osteosarcoma (OS), which is the most common histological form of primary bone cancer that arises from primitive transformed cells of mesenchymal origin, exhibits osteoblastic differentiation and produces malignant osteoid (1). OS is an aggressive malignant neoplasm primarily identified in teenagers and young adults, with an incidence of 4–5 cases/million people (2). OS is the eighth-most common form of childhood cancer, comprising 2.4% of all malignancies in pediatric patients and ~20% of all primary bone cancer cases in the United States in 2008 (3). Despite the success of chemotherapy and other therapeutic options that have been reported for the treatment of OS, studies have demonstrated that the 5-year overall survival rate remains ~70% (4–6). Therefore, elucidation of the mechanism underlying the initiation and progression of OS is urgent, and of great interest.
MicroRNAs (miRNAs/miRs) are a class of short (~22 nucleotides) and highly conserved non-coding RNA sequences, which are able to regulate gene expression via targeting promoters of mRNAs for transcriptional inhibition or translational repression (7). miRNAs are important modulators of key regulatory networks and signaling pathways through their influence on target genes (8). The accumulating scientific and clinical evidence has suggested that miRNAs are involved in tumorigenesis and tumor progression, serving the roles of tumor suppressors or oncogenes, and have become potential biomarkers for tumor diagnosis, therapy and prognosis (9).
miR-542-3p, located in Xq26.3, has been reported to serve a crucial role in the initiation and development of several types of cancer, including astrocytoma (10), neuroblastoma (11), hepatocellular carcinoma (12), breast cancer (13), colorectal cancer (14) and melanoma (15). miR-542-3p has also been reported to be downregulated in non-small cell lung cancer and breast cancer, and directly targets survivin or angiopoietin-2, respectively, leading to cell growth, arrest and the inhibition of angiogenesis (16,17).
However, whether miR-542-3p is involved in the tumorigenesis and tumor progression of OS remains unclear. In the present study, the biological function and underlying mechanism of miR-542-3p in OS were investigated, and the expression of miR-542-3p was decreased in OS tissues and cell lines. Through bioinformatics analysis and luciferase reporter assays, it was identified that miR-542-3p directly targeted Smad2 mRNA and negatively regulated the expression of Smad2 at the protein level. The present study demonstrated that restoration of miR-542-3p was able to suppress the growth and proliferation of OS cells through directly targeting Smad2, and revealed the mechanism of its tumor-suppressive role in OS pathogenesis. Therefore miR-542-3p may serve as a promising therapeutic target of OS.
Materials and methods
Cell lines and tissue samples
The human osteoblastic cell line hFOB, OS cell lines (U2OS, MG-63, and SAOS-2) and 293T cells were purchased from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C in a humidified 5% CO2 atmosphere.
A total of 48 OS tissues and self-matched non-tumor tissues were collected from 48 patients with OS between February 2014 and September 2015. In total, 40 cases were <18 years old and 8 cases were >18 years old, and 22 cases were women and 26 cases were men. All tissue samples were obtained via surgical resection and stored in liquid nitrogen until use in further analyses. All OS tissues were pathologically confirmed, and OS specimens that received chemotherapy, radiotherapy, or other types of therapy were excluded from the present study. The collection and the use of all tissue samples in the present study were approved by the Research Ethics Committee of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine (Wenzhou, China). Written informed consent was obtained from all participating patients.
RNA quantification and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from OS cell lines, OS tissues and self-matched adjacent normal tissues using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Purified mRNA and miRNAs were quantified by a RT-qPCR assay using an All-in-One™ miRNA RT-qPCR Detection kit (GeneCopoeia, Inc., Rockville, MD, USA) according to the manufacturer's protocol. All primers are listed in Table I. The reverse primer sequence of miR-542-3p was provided by All-in-one miRNA RT-qPCR Detection kit (GeneCopoeia, Inc.). U6 small RNA was used as an internal control for normalization, miR-542-3p expression were calculated as 2−[(Ct miR-542-3p)-(Ct U6)], where Ct represents the threshold cycle for each transcript.
Table I.
Sequence of primers and RNA fragments used in the present study.
| Name | Sequence (5′-3′) | |
|---|---|---|
| U6 | F | 5′-GTGCTCGCTTCGGCAGCACAT-3′ |
| R | 5′-AATATGGAACGCTTCACGAAT −3′ | |
| β-actin | F | 5′-AGAGCTACGAGCTGCCTGAC-3′ |
| R | 5′-AGCACTGTGTTGGCGTACAG-3′ | |
| Smad2 (reporter) | F | 5′-AAACTAGTTCTTGTAGAGGTTGTGT-3′ |
| R | 5′-GGAAGCTTGCAGATTTCCTTCTGCC-3′ | |
| Smad2 (pGL3) | F | 5′-AAAGGTACCACATGTCGTCCATCTTGCCATTCACG-3′ |
| R | 5′-AAACTCGAGTGGGACTTGATTGGTGAAGCTTTATGAC-3′ | |
| miR-542-3p | F | 5′-TGTGACAGATTGATAACTGAAA-3′ |
The reverse primer sequence of miR-542-3p was provided by the All-in-one miRNA qRT-PCR Detection kit (GeneCopoeia). F, forward; R, reverse; miR, microRNA; Smad2 (reporter), pMIR-Smad2 luciferase reporter; Smad2 (pGL3), pGL3-Smad2 vector.
Oligonucleotide transfection
RNA oligos were chemically synthesized and purified by Shanghai Genepharma Co., Ltd., (Shanghai, China). The sense sequence of the human miR-542-3p mimics was 5′-UGUGACAGAUUGAUAACUGAAA-3′, and the antisense sequence was 5′-UUUCAGUUAUCAAUCUGUCACA-3′. The sequences of the negative control oligonucleotides were 5′-AAUUCUCCGAACGUGUCACTT-3′ and 5′-GUGACACGUUCGGAGAAUUTT-3′. In total, 1×105 cells were seeded in a 6-well plate, and the transfections were performed with INTERFER in reagent (Polyplus-transfection SA, Illkirch, France) according to the manufacturer's protocol. The final concentration of miRNA was 50 nM.
To construct a pGL3-Smad2 vector, the sequence of Smad2 mRNA was amplified (primers are listed in Table I). The Smad2 fragments were inserted into pGL3 by the designed cutting sites: KpnI and XhoI. In total, 1×105 cells were seeded in a 6-well plate, the transfections were performed with Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The final concentration of plasmids was 100 ng.
Luciferase reporter assays
The pMIR-Smad2 luciferase reporter vector was constructed by cloning a human Smad2 mRNA sequence into a pMIR-Report construct (Ambion; Thermo Fisher Scientific, Inc.). A 60-bp Smad2 mRNA fragment (5,084–5,143 bp of Smad2 mRNA), which is the predicted target of miR-542-3p, was amplified and cloned into the luciferase reporter via SpeI and HindIII sites. All sequences are listed in Table I. Luciferase reporter assays were performed as follows: 5×103 293T cells were seeded in a 96-well plate, then co-transfected with 50 nM single-stranded miR-542-3p mimics or negative control oligonucleotides, 10 ng of pMIR-Smad2 luciferase reporter, pMIR luciferase reporter and 3 ng of pRL-TK (Promega Corporation, Madison, WI, USA) using the JetPRIME® reagent (Polyplus-transfection SA). Cells were collected 36 h after transfection and analyzed using a Dual-Luciferase Reporter assay system (Promega Corporation). pRL-TK was cotransfected as an internal control to correct the differences in both transfection and harvest efficiencies. The firefly luciferase activity of each sample was normalized to the Renilla luciferase activity.
Cell proliferation assay
The proliferation of OS cells was measured by MTT assay. A total of 5,000 cells were seeded into each well of 96-well plates and transfected with miR-542-3p mimics or negative control oligonucleotides at a final concentration of 50 nM. At 24, 48 and 72 h after transfection, the medium was replaced with 100 µl fresh medium containing MTT (0.5 mg/ml), and the plates were incubated for 4 h at 37°C. The medium was replaced by 100 µl DMSO (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and plates were agitated at room temperature for 10 min. The absorbance was measured at 570 nm.
Cell apoptosis analyses
Cell apoptosis analyses were performed using a Phycoerythrin-Annexin V Apoptosis Detection kit I (BD Pharmingen; BD Biosciences, San Jose, CA, USA). For cell apoptosis analysis, 8×105 cells were seeded in each well of 6-well plates. At 78 h after transfection, cells were harvested and labeled with Annexin V for 15 min. Subsequently, 50 µg/ml propidium iodide was added for 1 h at 37°C to each sample prior to flow cytometry using the BD LSR II (BD Biosciences).
Prediction of candidate miRNA targets
The possible targets of miR-542-3p were screened by the DIANA-TarBase web platform (version 7; http://diana.imis.athena-innovation.gr/), which included >500,000 experimentally confirmed miRNA-mRNA interactions utilizing cell types from 24 species (18).
Western blot analysis
MG-63 and U2OS cells were washed with pre-chilled PBS three times. The cells were then solubilized in 1% Nonidet P-40 lysis buffer [20 mM Tris, pH 8.0, 137 mM NaCl, 1% Nonidet P-40, 10% glycerol, 1 mM CaCl2, 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium fluoride, 1 mM sodium orthovanadate, and phosphatase inhibitor cocktail II (5 mg/ml, Sigma-Aldrich; Merck KGaA)] for 0.5 h at 4°C, and protein concentration was determined using a bicinchoninic acid assay (BCA Protein Assay Kit, Pierce; Thermo Fisher Scientific, Inc.). Proteins (40 µg) were separated on a 12% SDS-PAGE gel and then transferred to a nitrocellulose membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membrane was blocked with 5% non-fat milk in PBS with 0.1% Tween-20 for 2 h and washed three times with PBS with 0.1% Tween-20 at 4°C, then incubated with anti-Smad2 antibody (1:1,000 dilution) or anti-β-actin antibody (1:5,000 dilution) (Sigma-Aldrich; Merck KGaA). Following extensive washing, a goat anti-mouse secondary antibody (1:1,000 dilution) (Pierce; Thermo Fisher Scientific, Inc.) was added to the system. The proteins were detected using enhanced chemiluminescence reagents (Pierce; Thermo Fisher Scientific, Inc.). Protein bands were quantified using Pearson's correlation coefficient analysis (LabWorks software version 4.0; Image Acquisition; UVP, Inc., Upland, CA, USA).
Statistical analysis
All statistical analyses were performed using the SPSS 16.0 statistical software (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± standard deviation. Data of miR-542-3p expression levels in OS specimens compared to matched adjacent normal tissues were subjected to analysis by paired Student's t-test. Data of miR-542-3p expression levels in several cell lines and MTT analysis were analyzed by one-way analysis of variance followed by the Student-Newman-Keuls post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of miR-542-3p is decreased in OS
In order to identify the role of miR-542-3p in OS carcinogenesis, the expression levels of miR-542-3p in 48 OS samples and three OS cell lines were analyzed. Total RNAs were extracted from OS tissues and adjacent normal tissues. The expression levels of miR-542-3p were analyzed using RT-qPCR and normalized to an endogenous control (U6 RNA). As shown in Fig. 1A, the expression of miR-542-3p was significantly decreased in OS tissues vs. adjacent normal tissues (0.0066±0.0033 vs. 0.0141±0.0040). The results also demonstrated that the expression of miR-542-3p was significantly downregulated in OS cell lines, U2OS, MG-6 and SAOS-2, compared with which in human osteoblastic cell line hFOB (Fig. 1B). This indicates that miR-542-3p may function as an oncosuppressor gene in OS carcinogenesis.
Figure 1.
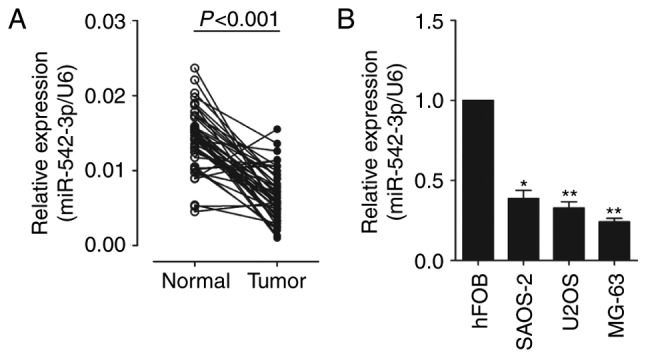
Expression of miR-542-3p decreased in OS samples and cell lines. (A) Expression of miR-542-3p was measured in 48OS samples and matched adjacent normal tissues by RT-qPCR, and the expression levels of miR-542-3p were normalized to U6 RNA expression. The data were subjected to paired Student's t-test (P<0.001). (B) The expression levels of miR-542-3p were measured in human hFOB cells and three OS cell lines (SAOS-2, U2OS and MG-63) by RT-qPCR, and the expression levels of miR-542-3p were normalized to U6 RNA expression. *P<0.05, **P<0.01 vs. hFOB cells by one-way analysis of variance followed by the Student Newman-Keuls test. OS, osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA.
Overexpression of miR-542-3p inhibits cell growth and induces apoptosis in OS cells
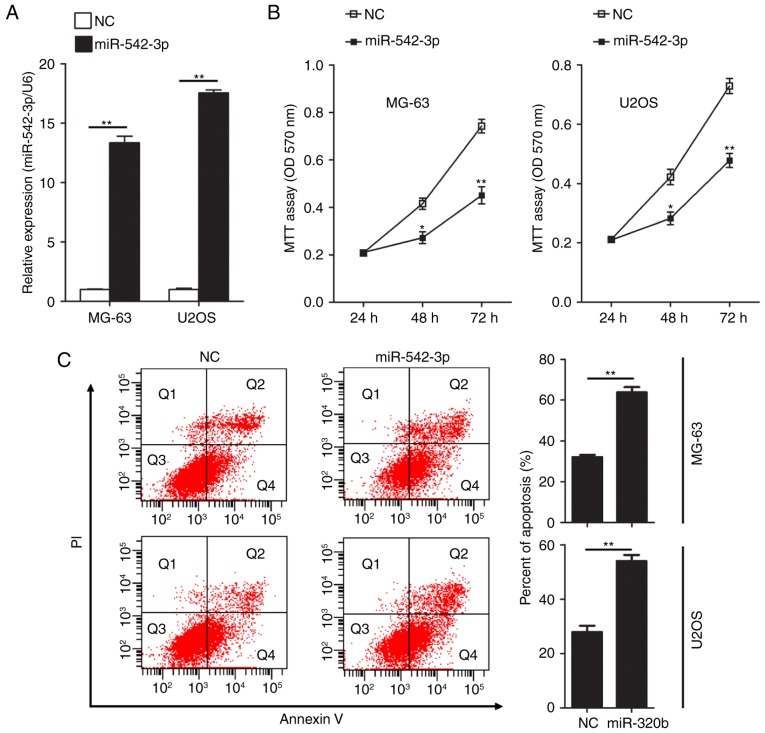
To further investigate the effect of miR-542-3p on the proliferation ability of OS cells, gain of function studies were performed in two human OS cell lines, MG-63 and U2OS. miR-542-3p mimics or negative control oligonucleotides were transiently transfected into MG-63 and U2OS cells. Expression of miR-542-3p was detected by RT-qPCR. The results confirmed that expression levels of miR-542-3p in MG-63 and U2OS cells were significantly increased following transfection of miR-542-3p mimics (Fig. 2A). Next, the functional role of miR-542-3p in cell proliferation in OS cells was investigated by performing MTT assays. Restoration of miR-542-3p expression in MG-63 and U2OS cells resulted in significant suppression of cell proliferation (Fig. 2B). Using flow cytometry, the influence of overexpression of miR-542-3p on apoptosis in U2OS and MG63 cells was evaluated following transfection of miR-542-3p mimics. Overexpression of miR-542-3p significantly increased the rate of apoptosis in MG-63 (63.77 vs. 32.07% in the control group; P<0.01) and U2OS cells (54.03 vs. 27.9% in the control group; P<0.01; Fig. 2C).
Figure 2.
Overexpression of miR-542-3p suppresses cell proliferation and promotes apoptosis in OS cells. (A) Expression of miR-542-3p was quantified by RT-qPCR analysis. Expression of miR-542-3p increased significantly in MG-63 and U2OS cells following transfection with miR-542-3p mimics compared with the NC group. The data were subjected to Student's t-test. **P<0.01. (B) Proliferation ability of OS cells was observed following restoration of miR-542-3p expression. MG-63 or U2OS cells were seeded into 96-well plates and incubated in the presence of miR-542-3p mimics or NC oligonucleotides. Cell proliferation was measured by MTT analysis. *P<0.05, **P<0.01 by one-way analysis of variance followed by the Student Newman-Keuls post-hoc test. (C) Apoptotic cells were analyzed by fluorescence-activated cell sorting analysis. MG-63 or U2OS cells were transfected with miR-542-3p mimics or NC oligonucleotides. The Annexin V-positive cells were regarded as apoptotic cells. The data were subjected to Student's t-test. **P<0.01 vs. NC group. OS osteosarcoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control; miR, microRNA.
Smad2 as miR-542-3p direct target in OS cells
After confirmation of function action of miR-542-3p in OS cells, the underlying mechanism of the effects of miR-542-3p in OS was investigated. Recently published DIANA-TarBase v7.0 data included over half a million experimentally confirmed miRNA-mRNA interactions utilizing cell types from 24 species (18). Following this analysis, many potential miR-542-3p targeting genes were identified (data not shown). Among these targeting genes, the present study focused on Smad2, which is a member of the Smad family. Smad2 has been previously identified in the regulation of cell proliferation and in apoptosis as a key element in TGF-β signaling (19,20). In addition, mirPath (http://www.microrna.gr/miRPathv2) was used to perform the pathway analysis of the target genes of miR-542-3p. Enrichment analysis identified the 12 most significant pathways, which included the ‘TGF-β signaling pathway’ (P=0.003; Table II).
Table II.
Pathway analysis of potential miR-542-3p targeting genes.
| KEGG pathway | Count (target genes) | P-value |
|---|---|---|
| Prion diseases | 1 | 1.46×10−36 |
| Lysine degradation | 4 | 3.56×10-3 |
| Biosynthesis of unsaturated fatty acids | 1 | 3.56×10−3 |
| TGF-β signaling pathway | 7 | 3.56×10-3 |
| Cell cycle | 10 | 4.64×10−3 |
| Ubiquitin mediated proteolysis | 13 | 1.17×10-2 |
| Adherens junction | 7 | 1.43×10−2 |
| Viral carcinogenesis | 13 | 1.43×10-2 |
| Endocytosis | 13 | 2.28×10−2 |
| Central carbon metabolism in cancer | 5 | 2.28×10-2 |
| ECM-receptor interaction | 4 | 4.70×10−2 |
| Proteoglycans in cancer | 13 | 4.70×10-2 |
KEGG, Kyoto encyclopedia of genes and genomes; TGF, transforming growth factor; ECM, extracellular matrix; miR, microRNA.
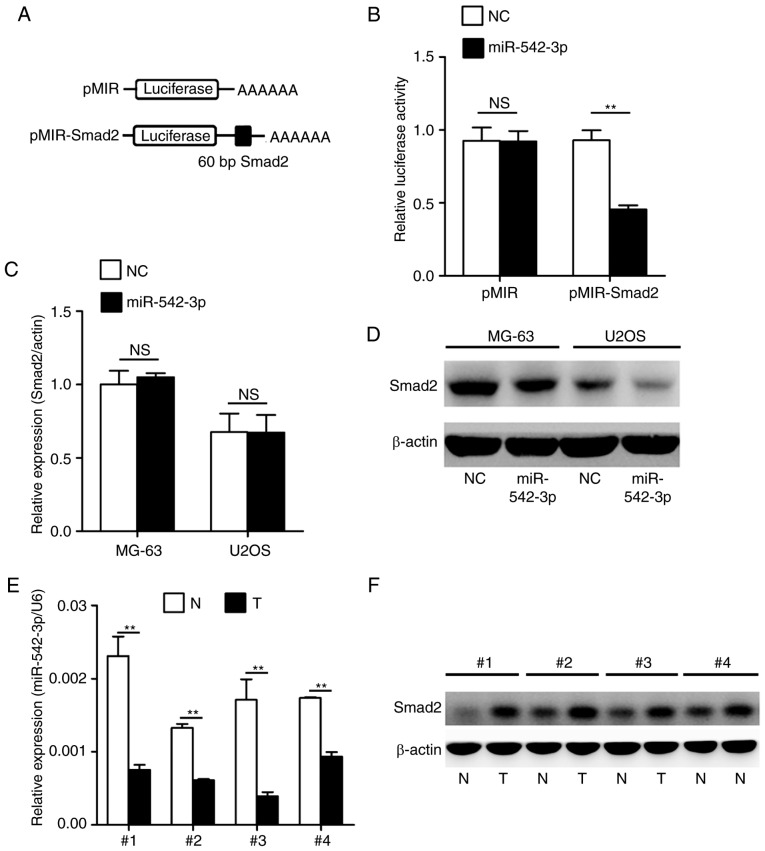
Consequently, the present study established whether Smad2 was a genuine target of miR-542-3p by performing a set of experiments. To confirm whether miR-542-3p regulated the expression of Smad2 gene, a luciferase reporter assay was performed in 293T cells. As indicated in Fig. 3A, a targeting sequence of Smad2 mRNA was cloned into a luciferase reporter plasmid. These luciferase reporter plasmids were co-transfected into 293T cells with miR-542-3p mimics or negative control oligonucleotides. The luciferase activities in transfected cells were measured after 36 h. As shown in Fig. 3B, overexpression of miR-542-3p caused a significant decrease in luciferase activity in cells transfected with the reporter plasmid containing the miR-542-3p-targeted sequence of Smad2 mRNA, whereas overexpression of miR-542-3p produced no significant change in luciferase activity in cells transfected with the reporter plasmid without the targeted sequence of Smad2. Subsequently, the present study investigated whether endogenous Smad2 in OS cells was similarly regulated. MG-63 and U2OS cells were transfected with miR-542-3p mimics or negative control oligonucleotides. The mRNA and protein levels of Smad2 were examined by RT-qPCR and western blotting, respectively. The level of Smad2 protein was consistently and substantially downregulated by miR-542-3p; however, the level of Smad2 mRNA was not affected by miR-542-3p (Fig. 3C and D). Finally, the expression levels of miR-542-3p and Smad2 were analyzed in four representative OS tumor tissues and adjacent non-tumor tissues by RT-qPCR and western blotting. The results confirmed that OS tumor tissues with low expression of miR-542-3p exhibited markedly higher Smad2 expression compared with adjacent non-tumor tissues (Fig. 3E and F).
Figure 3.
miR-542-3p directly targets and decreases Smad2 expression. (A) Schematic representation of construction of pMIR-Smad2 firefly luciferase reporter vector. 60 bp of the miR-542-3p target sequence in the Smad2/3′-UTR was cloned into a pMIR firefly luciferase reporter. (B) Analysis of luciferase activity. HEK 293T cells were co-transfected with pMIR-Smad2 firefly luciferase reporter plasmids, pTK-Renilla luciferase plasmids, together with miR-542-3p mimics and NC oligonucleotides. Firefly luciferase activity was measured and normalized by Renilla luciferase activity after 36 h. (C and D) Effects of miR-542-3p mimics on the endogenous expression of Smad2. MG-63 or U2OS cells were co-transfected with miR-542-3p mimics or NC oligonucleotides. Cells were isolated and the expression of Smad2 was analyzed by (C) RT-qPCR and (D) western blotting after 48 h. (E) miR-542-3p expression in four OS tissues was examined by RT-qPCR analysis. (F) Smad2 expression in four OS tissues was analyzed by western blotting. The data were subjected to Student's t-test. **P<0.01. NC, negative control oligonucleotides; N, normal tissues; T, tumor tissues; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; OS, osteosarcoma; miR, microRNA.
Taken together, these results demonstrated that Smad2 is a direct target of miR-542-3p in OS, and that miR-542-3p directly regulates Smad2 expression at the protein level.
Restoration of miR-542-3p inhibits Smad2-induced OS cell proliferation
In order to further confirm that miR-542-3p exerts tumor-suppressor activity in OS pathogenesis through targeting Smad2, gain of function and rescue experiments were performed in OS cells. It was investigated whether the proliferation of OS cells was further promoted following transfection with Smad2 cDNA, and whether this effect could be attenuated by miR-542-3p mimics. Smad2 cDNA was transiently transfected into MG-63or U2OS cells with and without miR-542-3p mimics. Subsequently cell proliferation assays were performed.
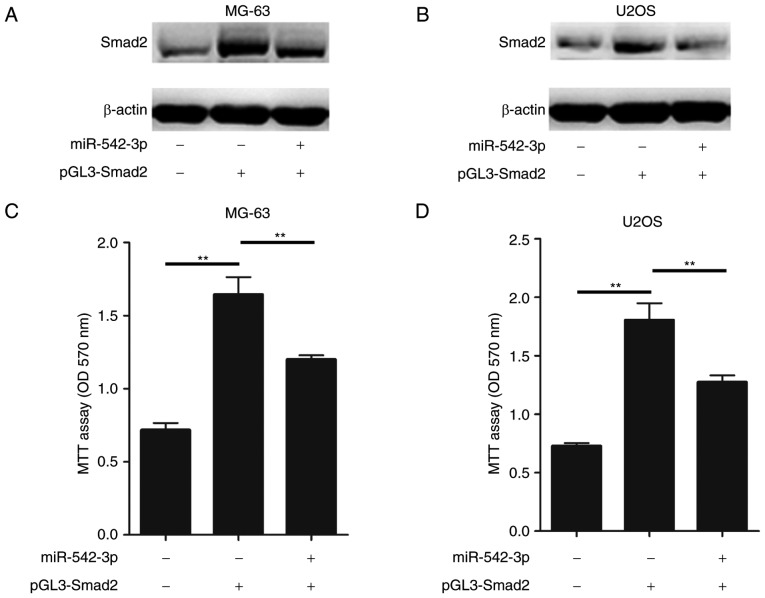
As hypothesized, ectopic expression of Smad2 in MG-63 cells enhanced the accumulation of Smad2, while restoration of the miR-542-3p expression in MG-63 cells markedly inhibited the expression of Smad2 (Fig. 4A). Furthermore, restoration of miR-542-3p inhibited the Smad2 expression in U2OS cells (Fig. 4B). In conjunction with these results, the overexpression of miR-542-3p significantly attenuated the Smad2-induced increase in cell proliferation in MG-63 and U2OS cells (Fig. 4C and D). Taken together, these results demonstrated that restoration of miR-542-3p was able suppress the growth and proliferation of OS cells through directly targeting Smad2.
Figure 4.
Restoring miR-542-3p expression in osteosarcoma cells inhibits Smad2-dependent cell proliferation. (A and B) miR-542-3p inhibits Smad2 expression in OS cells. (A) MG-63 and (B) U2OS cells were transfected with pGL3-Smad2 with and without miR-542-3p, and the expression levels of Smad2 were analyzed by western blotting. (C and D) miR-542-3p inhibits Smad2-dependent cell proliferation. (C) MG-63 and (D) U2OS cells were transfected with pGL3-Smad2 with and without miR-542-3p, and the cell proliferation was measured by MTT analysis. **P<0.01 by one-way analysis of variance followed by the Student Newman-Keuls post-hoc test. miR, microRNA; OS, osterosarcoma.
Discussion
OS is the most frequently occurring type of primary bone cancer. Although multiple treatments options, including chemotherapy, are available, the 5-year survival rate of OS remains poor due to the occurrence of drug resistance among patients (21–23). The OS-associated high mortality rate is attributed to the difficulty of early diagnosis and the lack of efficient therapeutic approaches for OS. Therefore, it is essential to elucidate the underlying mechanism that mediates the initiation and progression of OS, and to identify diagnostic biomarker or potential therapeutic targets for the treatment of this disease (24).
miR-542-3p has been reported to serve crucial roles in the initiation and development of multiple types of cancer. These studies reported that miR-542-3p suppressed tumor cell growth, invasion and metastasis via targeting the AKT serine/threonine kinase signaling pathway (10), the frizzled class receptor 7/Wnt pathway (11), serine/threonine protein kinases (15), survivin (16) or angiopoietin-2 (17). Previous studies reported that miR-542-3p expression was decreased in colorectal cancer cells, and that the restoration of miR-542-3p was able to inhibit the growth and invasion of colorectal cancer cells through decreasing the expression of cortactin as a target (14). Another study indicated that miR-542-3p suppressed cellular proliferation of bladder cancer cells through post-transcriptionally regulating survivin (25). Kureel et al (26) also reported that miR-542-3p suppressed osteoblast cell proliferation and differentiation, and inhibited bone formation through targeting of bone morphogenetic protein 7 signaling (26). However, the involvement of miR-542-3p is involved in the tumorigenesis and progression of OS remains unclear.
In the present study, the biological function and underlying mechanism of miR-542-3p in OS was investigated. This demonstrated that the expression of miR-542-3p was significantly decreased in OS tissues and cell lines, and that overexpression of miR-542-3p in OS cells significantly inhibited cell proliferation, and induced cell apoptosis. Through bioinformatics analysis, 190 potential miR-542-3p targeting genes were identified, with a member of the Smad family, Smad2, serving as the primary focus. Furthermore, mirPath was used to perform pathway analysis of the predicted target genes of miR-542-3p. Enrichment analysis identified the 12 most significant pathways, which included the ‘TGF-β signaling pathway’. The TGF-β signaling pathway is involved in a number of cellular processes, including cell growth, differentiation, apoptosis and homeostasis, amongst other cellular functions. TGF-β signaling is mediated by a complex of membrane-bound type I and type II receptors, and Smad proteins function as intracellular mediators (27,28). Binding of the TGF-β ligand with its receptors leads to the phosphorylation of Smad2 and Smad3. This activity enables the association of Smad2 and Smad3 with Smad4. Subsequently, the complex of Smad2/3 and Smad4 is able to translocate to the nucleus, and bind directly to Smad-binding elements, in addition to a number of co-activators, to directly modulate TGF-β-regulated gene expression. Tumor cells may exhibit resistance to TGF-β-induced growth inhibition and apoptosis if the functional inactivation of TGF-β receptors and Smads occurs (29,30). In order to verify this, the present study used luciferase reporter assays to establish that Smad2 is a genuine target of miR-542-3p. Furthermore, it was confirmed that OS tumor tissues with low expression of miR-542-3p exhibited markedly higher Smad2 expression. In order to demonstrate that miR-542-3p serves the role of tumor suppression in OS pathogenesis through the targeting of Smad2, gain of function and rescue experiments in OS cells were performed. The present study demonstrated that restoration of miR-542-3p was able to suppress the growth and proliferation of OS cells through directly targeting Smad2.
In conclusion, the findings of the present study suggest that miR-542-3p may serve as a tumor suppressor gene in OS pathogenesis, and that miR-542-3p may be a promising therapeutic target in the treatment of OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by Rising Star in Medicine of Zhejiang (grant no. 201505403).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YWu and JY contributed equally to the study. YWu and YWa conceived and designed the study. YWu, JY, FL and FW performed the experiments. YWu and JY wrote the paper. YWu, JY, FL, FW and YWa reviewed and edited the manuscript. All authors read and approved the manuscript.
Ethics approval and consent to participate
The collection and the use of all tissue samples in the present study were approved by the Research Ethics Committee of Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine (Wenzhou, China). Written informed consent was obtained from all participating patients.
Consent for publication
Written informed consent was obtained from all participating patients.
Competing interests
The authors declare that they have no competing interests.
Authors' information
YWu, JY and YWa, Department of Orthopedic Surgery, Wenzhou Hospital of Integrated Traditional Chinese and Western Medicine, Wenzhou, Zhejiang 325000, P.R. China
References
- 1.Luetke A, Meyers PA, Lewis A, Juergens H. Osteosarcoma treatment-where do we stand? A state of the art review. Cancer Treat Rev. 2014;40:523–532. doi: 10.1016/j.ctrv.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin. 2014;64:83–103. doi: 10.3322/caac.21219. [DOI] [PubMed] [Google Scholar]
- 3.Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3–13. doi: 10.1007/978-1-4419-0284-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Kleinerman E. Maximum benefit of chemotherapy for osteosarcoma achieved-what are the next steps? Lancet Oncol. 2016;17:1340–1342. doi: 10.1016/S1470-2045(16)30270-4. [DOI] [PubMed] [Google Scholar]
- 5.Cates JM. Utility of examination of biopsy tracts in osteosarcoma resection specimens. Am J Clin Pathol. 2016;146:324–327. doi: 10.1093/ajcp/aqw119. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins RM, Cullen JW, Odom L, Jamroz BA, Cullen PM, Fink K, Peck SD, Stevens SL, Kelly CM, Camozzi AB. Superior survival in treatment of primary non-metastatic pediatric osteosarcoma of the extremity. Ann Surg Oncol. 2003;10:498–507. doi: 10.1245/ASO.2003.03.061. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Lionetti M, Agnelli L, Mosca L, Fabris S, Andronache A, Todoerti K, Ronchetti D, Deliliers GL, Neri A. Integrative high-resolution microarray analysis of human myeloma cell lines reveals deregulated miRNA expression associated with allelic imbalances and gene expression profiles. Genes Chromosomes Cancer. 2009;48:521–531. doi: 10.1002/gcc.20660. [DOI] [PubMed] [Google Scholar]
- 9.Visone R, Croce CM. MiRNAs and cancer. Am J Pathol. 2009;174:1131–1138. doi: 10.2353/ajpath.2009.080794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai J, Zhao J, Zhang N, Xu X, Li R, Yi Y, Fang L, Zhang L, Li M, Wu J, Zhang H. MicroRNA-542-3p suppresses tumor cell invasion via targeting AKT pathway in human astrocytoma. J Biol Chem. 2015;290:24678–24688. doi: 10.1074/jbc.M115.649004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Althoff K, Lindner S, Odersky A, Mestdagh P, Beckers A, Karczewski S, Molenaar JJ, Bohrer A, Knauer S, Speleman F, et al. miR-542-3p exerts tumor suppressive functions in neuroblastoma by downregulating Survivin. Int J Cancer. 2015;136:1308–1320. doi: 10.1002/ijc.29091. [DOI] [PubMed] [Google Scholar]
- 12.Wu W, Dang S, Feng Q, Liang J, Wang Y, Fan N. MicroRNA-542-3p inhibits the growth of hepatocellular carcinoma cells by targeting FZD7/Wnt signaling pathway. Biochem Biophys Res Commun. 2017;482:100–105. doi: 10.1016/j.bbrc.2016.10.136. [DOI] [PubMed] [Google Scholar]
- 13.Venkatadri R, Muni T, Iyer AK, Yakisich JS, Azad N. Role of apoptosis-related miRNAs in resveratrol-induced breast cancer cell death. Cell Death Dis. 2016;7:e2104. doi: 10.1038/cddis.2016.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long HC, Gao X, Lei CJ, Zhu B, Li L, Zeng C, Huang JB, Feng JR. miR-542-3p inhibits the growth and invasion of colorectal cancer cells through targeted regulation of cortactin. Int J Mol Med. 2016;37:1112–1118. doi: 10.3892/ijmm.2016.2505. [DOI] [PubMed] [Google Scholar]
- 15.Rang Z, Yang G, Wang YW, Cui F. miR-542-3p suppresses invasion and metastasis by targeting the proto-oncogene serine/threonine protein kinase, PIM1, in melanoma. Biochem Biophys Res Commun. 2016;474:315–320. doi: 10.1016/j.bbrc.2016.04.093. [DOI] [PubMed] [Google Scholar]
- 16.Yoon S, Choi YC, Lee S, Jeong Y, Yoon J, Baek K. Induction of growth arrest by miR-542-3p that targets survivin. FEBS Lett. 2010;584:4048–4052. doi: 10.1016/j.febslet.2010.08.025. [DOI] [PubMed] [Google Scholar]
- 17.He T, Qi F, Jia L, Wang S, Song N, Guo L, Fu Y, Luo Y. MicroRNA-542-3p inhibits tumour angiogenesis by targeting angiopoietin-2. J Pathol. 2014;232:499–508. doi: 10.1002/path.4324. [DOI] [PubMed] [Google Scholar]
- 18.Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA: mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: Differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101:9–33. doi: 10.1002/jcb.21255. [DOI] [PubMed] [Google Scholar]
- 20.Bao Y, Chen Z, Guo Y, Feng Y, Li Z, Han W, Wang J, Zhao W, Jiao Y, Li K, et al. Tumor suppressor microRNA-27a in colorectal carcinogenesis and progression by targeting SGPP1 and smad2. PLoS One. 2014;9:e105991. doi: 10.1371/journal.pone.0105991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagmay JP, Krailo MD, Dang H, Kim A, Hawkins DS, Beaty O, III, Widemann BC, Zwerdling T, Bomgaars L, Langevin AM, et al. Outcome of patients with recurrent osteosarcoma enrolled in seven phase II trials through children's cancer group, pediatric oncology group, and children's oncology group: Learning from the past to move forward. J Clin Oncol. 2016;34:3031–3038. doi: 10.1200/JCO.2015.65.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann S, Hennet T. Collagen accumulation in osteosarcoma cells lacking GLT25D1 collagen galactosyltransferase. J Biol Chem. 2016;291:18514–18524. doi: 10.1074/jbc.M116.723379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelini A, Mavrogenis AF, Trovarelli G, Ferrari S, Picci P, Ruggieri P. Telangiectatic osteosarcoma: A review of 87 cases. J Cancer Res Clin Oncol. 2016;142:2197–2207. doi: 10.1007/s00432-016-2210-8. [DOI] [PubMed] [Google Scholar]
- 24.Buondonno I, Gazzano E, Jean SR, Audrito V, Kopecka J, Fanelli M, Salaroglio IC, Costamagna C, Roato I, Mungo E, et al. Mitochondria-targeted doxorubicin: A new therapeutic strategy against doxorubicin-resistant osteosarcoma. Mol Cancer Ther. 2016;15:2640–2652. doi: 10.1158/1535-7163.MCT-16-0048. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Wang S, Han F, Li J, Yu L, Zhou P, Chen Z, Xue S, Dai C, Li Q. MicroRNA-542-3p suppresses cellular proliferation of bladder cancer cells through post-transcriptionally regulating survivin. Gene. 2016;579:146–152. doi: 10.1016/j.gene.2015.12.048. [DOI] [PubMed] [Google Scholar]
- 26.Kureel J, Dixit M, Tyagi AM, Mansoori MN, Srivastava K, Raghuvanshi A, Maurya R, Trivedi R, Goel A, Singh D. miR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014;5:e1050. doi: 10.1038/cddis.2014.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massagué J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 28.Mishra L, Derynck R, Mishra B. Transforming growth factor-beta signaling in stem cells and cancer. Science. 2005;310:68–71. doi: 10.1126/science.1118389. [DOI] [PubMed] [Google Scholar]
- 29.Bierie B, Moses HL. Tumour microenvironment: TGFbeta: The molecular Jekyll and Hyde of cancer. Nat Rev Cancer. 2006;6:506–520. doi: 10.1038/nrc1926. [DOI] [PubMed] [Google Scholar]
- 30.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.