Abstract
Osteosarcoma is the most frequent primary malignant bone tumor. An increasing body of evidence has suggested that microRNAs (miRNA/miRs) have emerged as critical regulators in the initiation and progression of osteosarcoma. The present study explored the biological function of miR-192-5p and ubiquitin-specific protease 1 (USP1), and investigated whether miR-192-5p could directly interact with USP1 in osteosarcoma. The results revealed that miR-192-5p was significantly downregulated in osteosarcoma tissues and cell lines, while a reverse expression profile of USP1 was observed. Ectopic expression of miR-192-5p restrained cell proliferation, apoptosis, migration and invasion. In addition, it increased the sensitivity of osteosarcoma cells to cisplatin. USP1 was also observed to be a direct target gene of miR-192-5p in osteosarcoma. Overexpression of USP1 promoted cell proliferation, apoptosis, migration and invasion, and decreased cell chemo-sensitivity; however, it could be partially reversed via the overexpression of miR-192-5p in osteosarcoma cell lines. Taken together, the present study demonstrated that miR-192-5p suppressed the initiation and progression of osteosarcoma by targeting USP1. Therefore, miR-192-5p may serve as a valuable biomarker and the miR-192-5p/USP1 axis may function as a novel therapeutic target for osteosarcoma.
Keywords: osteosarcoma, microRNA, microRNA-192-5p, ubiquitin-specific protease 1, targeted therapy
Introduction
Osteosarcoma is the most frequent primary malignant bone tumor, which mainly occurs in children and adolescents (1–3). Previous study has shown that osteosarcoma accounts for approximately 19 and 5% of all malignant bone tumors and childhood neoplasm, respectively (4). Despite the recently advances in multi-modal therapeutics, the prognosis and 5 years survival rate of osteosarcoma remains unsatisfactory (5). Therefore, it is urgent for us to explore the molecular mechanisms and find new therapeutic strategies to target this disease.
MicroRNAs (miRNAs/miRs) are short, endogenous noncoding and highly conserved RNAs, which can restrain the expression of target genes through binding to the 3′-untranslated region (3′-UTR) mRNA (6–8). According to previous studies, miRNAs have taken part in a great deal of cellular processes, including cell proliferation, invasion, apoptosis, chemo-resistance (9–11). Furthermore, many studies have already proved that abnormal expression of miRNAs play significantly parts in the occurrence and progression of cancers, including hepatocellular carcinoma, breast cancer, gastric carcinoma, prostate cancer, and also osteosarcoma (12–14). Numerous researches have suggested that miR-192-5p was dramatically downregulated in malignant tumors, while overexpression of miR-192-5p could inhibit tumorigenesis through different mechanisms (15,16). In human osteosarcoma, miR-192-5p functioned as a major role in inhibiting the tumorigenesis in osteosarcoma (17). Nevertheless, the potential mechanisms of miR-192-5p in regulating the development and progression of osteosarcoma remains largely unknown.
Ubiquitination is a critical posttranslational modification, which modulates cellular processes, including cell cycle regulation, chromatin remodeling, DNA damage response and so forth (18). Ubiquitination modification is a dynamic reversible process, which is catalyzed by deubiquitinases (DUBs) (19). Ubiquitin-specific protease 1 (USP1), one of the best characterized member of the DUBs family, is famous for its regulation of cellular response to DNA damage (20). Recently, more and more studies reported that USP1 played an important role in oncogenesis and tumorigenesis in human malignant cancers, including osteosarcoma (21–24).
Given the crucial parts of miR-192-5p and USP1 in regulating the initiation and progression of osteosarcoma, we performed this study to identify whether miR-192-5p could negatively regulates osteosarcoma by directly targeting USP1.
Materials and methods
Tissue collection
A total of 25 samples of osteosarcoma and matched the adjacent non-tumor tissues were collected from surgical resection between June 2009 and June 2012 in Renmin Hospital of Wuhan University. The parameters of patients were showed in Table I. Tissues were obtained and then frozen in liquid nitrogen immediately and stored at −80°C until being used. All patients were further followed up every 3–5 months until 5 years. The patients participated in this study did not undergo any chemotherapy, radiotherapy or immunotherapy before surgery. This study was approved by the Medical Ethics Committee of Renmin Hospital of Wuhan University. All patients and their families provided written informed consent to take part in this research. The study did not contain any identifying information about any participants. All the data was kept by the administrator of the study team in a confidential manner and was not used by any other purposes. We confirm that all experiments were performed in accordance with relevant guidelines and regulations.
Table I.
Associations between microRNA-192-5p levels and clinicopathological variables of osteosarcoma patients.
| Relative miR-192-5p expression | ||||
|---|---|---|---|---|
| Variable | Total no. (n=25) | Low (n=13) | High (n=12) | P-value |
| Sex | 0.870 | |||
| Male | 15 | 8 | 7 | |
| Female | 10 | 5 | 5 | |
| Age (years) | 0.568 | |||
| <20 | 18 | 10 | 8 | |
| ≥20 | 7 | 3 | 4 | |
| Histologic subtype | 0.238 | |||
| Osteoblastic | 13 | 6 | 7 | |
| Chondroblastic | 9 | 5 | 4 | |
| Fibroblastic | 3 | 2 | 1 | |
| Anatomical site | 0.920 | |||
| Femure | 11 | 6 | 5 | |
| Tibia | 9 | 5 | 4 | |
| Humerus | 3 | 1 | 2 | |
| Other | 2 | 1 | 1 | |
| Tumor grade | 0.007b | |||
| Low | 8 | 1 | 7 | |
| High | 17 | 12 | 5 | |
| Enneking stage | 0.009b | |||
| I | 6 | 1 | 5 | |
| II | 12 | 7 | 5 | |
| III | 7 | 5 | 2 | |
| Tumor size (cm) | 0.027a | |||
| <8 | 13 | 4 | 9 | |
| ≥8 | 12 | 9 | 3 | |
P<0.05
P<0.01. miR, microRNA.
Cell culture and transfection
143B and U2OS (human osteosarcoma cell lines) and hFOB (normal human osteoblast cell line) were used in the present study, which were gained from Cell Bank of Type Culture Collection (Shanghai, China). We used DMEM (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 U/ml penicillin and 100 µg/ml streptomycin to culture the above cells in an atmosphere of 5% CO2 at 37°C. miR-192-5p mimic was purchased from GeneCreate Biological Engineering Co., Ltd. (Wuhan, China). Then we transfected miR-192-5p mimic and miR-NC (negative control) into cells at 60% confluence by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Overexpression USP1 plasmid (pCDNA3.1-USP1) or empty plasmid (GeneCreate Biological Engineering Co., Ltd.) were transfected into cells at 80% confluence using Lipofectamine 2000 in accordance with the manufacturer's instructions.
Cell proliferation assay
CCK-8 (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay was performed to evaluate cell proliferation capacity. Briefly, transfected cells were seeded into individual well plates at 1×105 cells per/well, then incubated for 0, 24, 48 and 72 h. Subsequently, microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to detect the OD value at 450 nm.
Cell cycle assay
For cell cycle assay, transfected cells were inoculated in 6-well plates for 24 h. Then 143B and U2OS cells were fixed with 75% cold ethanol at 4°C for 24 h. Subsequently, both cell lines were stained with a propidium iodide (PI; BD Biosciences, San Jose, CA, USA) for 30 min in the dark. Then, stained cells were analyzed by using flow cytometer (FACS Calibur; BD Biosciences). At last, ModFitLT V2.0 software (BD Biosciences) was applied to analyze the above data.
Cell apoptosis assay
Apoptosis was assessed by flow cytometric. Cells were harvested and washed with ice-cold PBS twice. Then we resuspended cells with 300 µl of binding buffer. After being stained with Annexin V-FITC and PI (BD Biosciences), FACS Calibur was performed to analyze cell apoptosis.
Cell invasion assay
In order to investigate the cell invasion ability, Matrigel (BD Biosciences) were precoated into 8 micron Transwells (Corning Incorporated, Corning, NY, USA), and then 143B and U2OS cells were added into the upper chambers with serum free medium. DMEM (500 µl) with 10% FBS was placed into the lower chamber for chemical induction. After incubating for 24 h, we carefully wiped out remaining cells which did not invade. Matrigel membranes were fixed with paraformaldehyde and then stained with crystal violet solution. The invaded cells were counted under a phrase contrast microscope (Olympus, Tokyo, Japan).
Cell migration assay
Cell migration was evaluated by wound healing assay. The transfected cells were cultured in 6-well plates for 24 h. Then, artificial wound was scratched in confluent cell monolayers by sterile 10 ul pipette tip. Photographs were taken at 0 and 24 h by using inverse microscope.
Luciferase reporter assay
The mutant (mut) or wild-type (wt) 3′UTR of USP1 was inserted into pGL4 luciferase promoter vector (Promega Corporation, Madison, WI, USA). Subsequently, miR-192-5p mimics or miR-NC and the vectors carrying USP1 mut or wt 3′UTR were co-transfected into 143B and U2OS cells by lipofectamine 2000 (Invitrogen). After 24 h, the dual luciferase reporter assay system (Promega Corporation) was performed to determine luciferase values.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were extract from tissues and cell lines using TRIzol reagent (Invitrogen). Reverse transcription reactions were conducted via Takara RNA PCR kit (Takara, Kyoto, Japan). qPCR was performed using a SYBR-Green detection system (Takara). Relative gene expression was calculated using 2−ΔΔCq method. The expression of U6 and β-actin were acted as the internal control for the expression of miR-192-5p and USP1, respectively. The primers for miR-192-5p were forward, 5′-GCGGCGGCTGACCTATGAATTG-3′ and reverse, 5′-ATCCAGTGCAGGGTCCGAGG-3′; U6 forward, 5′-TCCGATCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; USP1 mRNA forward, 5′-AGGTTGCTAGTACAGCGTTTGC-3′ and reverse, 5′-CACTGGATTCCTTGTTTCTATCAGA-3′; and β-actin forward, 5′-GGCACTCTTCCAGCCTTCC-3′ and reverse, 5′-GAGCCGCCGATCCACAC-3′.
Western blot analysis
We extracted total proteins from transfected cells by using RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China). All proteins were separated by 10% SDS-PAGE gel and transferred them onto the PVDF membrane (EMD Millipore Corp., Bedford, MA, USA). The membrane were blocked in 10% non-fat dried milk for 2 h and then incubated with primary anti-USP1 and GAPDH (both from Abcam, Cambridge, UK) at 4°C overnight. After being washed with TBST five times, the membrane was incubated with secondary antibody for 1 h at room temperature. Then the membrane was washed three times with TBST again. Finally, the proteins were detected by using enhanced chemiluminescent. GAPDH was performed as an endogenous control.
Statistical analysis
All statistical analyses were conducted by using SPSS 20.0 (IBM Corp., Armonk, NY, USA) and Graph Pad Prism 5 software (GraphPad Software, Inc., San Diego, CA, USA). Data which were followed the Gaussian distribution, calculated as mean ± SD. Differences between two groups were measured by Student's t-test (for the migration, invasion, apoptosis and cell cycle assays), while one-way ANOVA (for CCK-8 assay) followed by Tukey's post hoc test was performed for comparisons between more than two groups. Correlation between miR-192-5p levels and clinicopathological variables of osteosarcoma patients were assessed by Chi-square test. The Pearson' procedure method was used in the patients. And the Kaplan-Meier survival analysis was applied in the survival experiments. P<0.05 was considered as statistically significant. Each experiment was performed three times.
Results
Reduced expression of miR-192-5p and elevated expression of USP1 in osteosarcoma tissues and cell lines
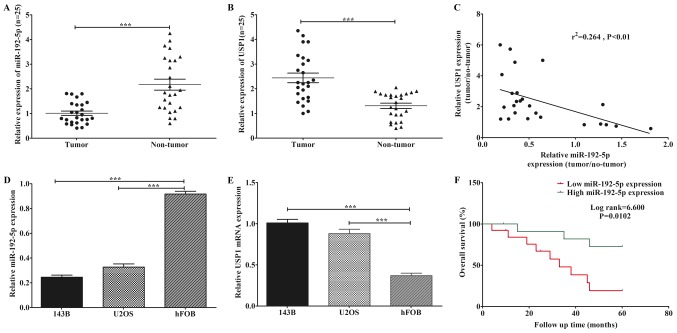
The expression levels of miR-192-5p and USP1 mRNA were detected in tissues and cell lines of osteosarcoma. MiR-192-5p was distinctly downregulated in osteosarcoma tissues (Fig. 1A). Whereas, USP1 mRNA showed an opposite trend (Fig. 1B). Moreover, Pearson's correlation assay indicated that the expression levels of USP1 and miR-192-5p were inversely correlated in osteosarcoma tissues (Fig. 1C). Next, two classic osteosarcoma cell lines: 143B and U2OS were performed to further confirm the findings above. Here we found that lower expressions of miR-192-5p were observed in both cell lines compared to hFOB (Fig. 1D). In contrast, the expression levels of USP1 mRNA was markedly higher in both osteosarcoma cell lines compared to hFOB (Fig. 1E). Furthermore, to evaluate the association between the expression level of miR-192-5p and clinicopathological variables, we divided all patients into two groups (low expression group and high expression group) based on the median expression level of tumor tissues. As showed in Table I, the low expression of miR-192-5p was statistically correlated with tumor grade, Enneking stage (25) and tumor size, while not associated with sex, age, histologic subtype and anatomical site in patients with osteosarcoma. Besides, the Kaplan-Meier survival analysis suggested that osteosarcoma patients with low expression of miR-192-5p presented to have a shorter overall survival (Fig. 1F). These finding indicated that the low expression of miR-192-5p was closely associated with the high expression of USP1.
Figure 1.
Reduced expression of miR-192-5p and elevated expression of USP1 in osteosarcoma tissues and cell lines. (A) miR-192-5p expression was statistically lower in osteosarcoma tissues than in adjacent non-tumor tissues. (B) USP1 expression was significantly higher in osteosarcoma tissues when compared with adjacent non-tumor tissues. (C) There was a negative correlation between miR-192-5p and USP1 in osteosarcoma tissues. (D) The expression of miR-192-5p was downregulated in the osteosarcoma cell lines (143B and U2OS) when compared with the normal human osteoblast cell line (hFOB). (E) The expression of USP1 was increased in the 143B and U2OS cell lines compared with that of osteosarcoma. (F) Kaplan-Meier survival analysis suggested that osteosarcoma patients with low expression of miR-192-5p presented a shorter overall survival. The results are presented as the mean ± standard deviation of 3 independent assays. ***P<0.001, as indicated. miR, microRNA; USP1, ubiquitin-specific protease 1.
Upregulation of miR-192-5p inhibited osteosarcoma cell proliferation
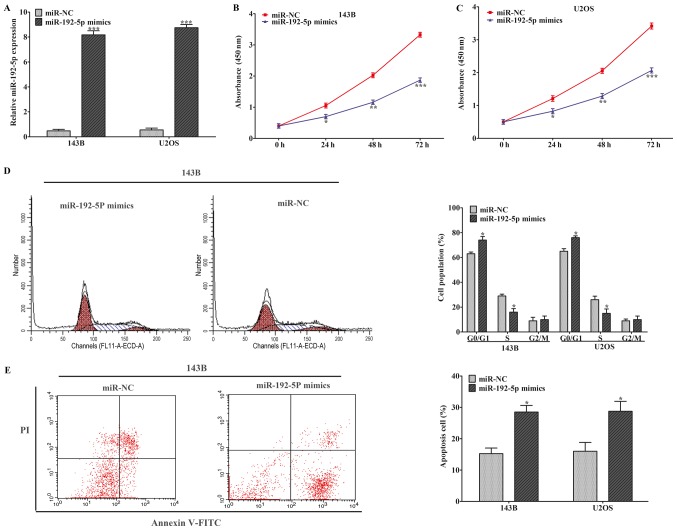
We transfected 143B and U2OS cells with miR-192-5p mimics or miR-NC. After being cultured, we found that the expression profile of miR-192-5p was dramatically increased compared to miR-NC group (Fig. 2A). Then, CCK-8 assay was conducted to evaluate the ability of cell proliferation in 143B and U2OS cells. The results indicated that cell proliferation was statistically suppressed when miR-192-5p was upregulated (Fig. 2B and C). Flow cytometric assay was used to further identify whether cell proliferation was inhibited by miR-192-5p through altering cell-cycle progression or inducing cell apoptosis. Cell cycle analysis showed that miR-192-5p mimics brought about a higher G0⁄G1-phase and a lower S-phase arrest in 143B and U2OS cells compared to miR-NC group (Fig. 2D). Subsequently, cell apoptosis analysis was performed to explore the influence of miR-192-5p mimics on cell apoptosis, which revealed that cell apoptosis was remarkably induced in 143B and U2OS cells compared to miR-NC group (Fig. 2E). These findings demonstrated that overexpression of miR-192-5p repressed osteosarcoma cell proliferation through regulating cell cycle during G1 to S phase and inducing cell apoptosis.
Figure 2.
Upregulation of miR-192-5p inhibited osteosarcoma cell proliferation and induced cell cycle at the G0/G1 stage and cell apoptosis. (A) Reverse transcription-quantitative polymerase chain reaction was performed to measure the expression of miR-192-5p in 143B and U2OS cells following transfection with miR-192-5p-mimics. Overexpression of miR-192-5p significantly restrained the proliferation of (B) 143B and (C) U2OS cells. (D) The cell cycle assay demonstrated that miR-192-5p mimics could prevent the progression of the cell cycle from the G1 to S phase in 143B and U2OS cells. (E) Elevated expression of miR-192-5p markedly induced 143B and U2OS cells apoptosis. Data are presented as the mean ± standard deviation of 3 independent assays. *P<0.05, **P<0.01 and ***P<0.001 vs. miR-NC. miR, microRNA; NC, negative control; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Overexpression of miR-192-5p suppressed osteosarcoma cell migration and invasion and enhanced the sensitivity of osteosarcoma cells to cisplatin
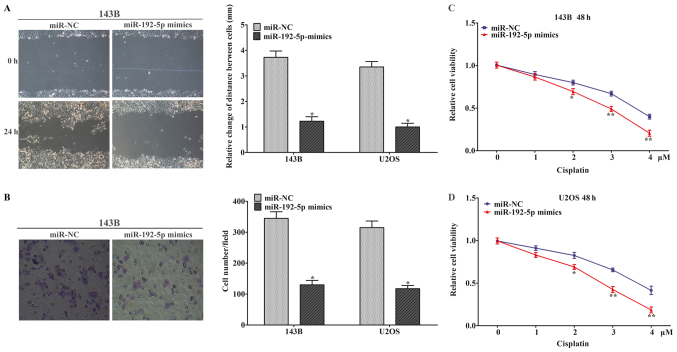
Wound-healing and Transwell chamber assays were performed to elucidated cell migration and invasion in 143B and U2OS cells. Wound-healing assay showed that the migration capacity of cells in the miR-192-5p mimics group was obviously inhibited after wounding (Fig. 3A). Transwell chamber assay showed that ectopic expression of miR-192-5p inhibited cell invasion (Fig. 3B). These findings suggested that upregulation of miR-192-5p weaken the migration and invasion capability of 143B and U2OS cells. CCK-8 assay was performed to evaluate the effect of miR-192-5p mimics on the chemo-sensitivity of osteosarcoma cells. The response of 143B and U2OS cells to cisplatin enhanced after treated with the miR-192-5p mimic compared to miR-NC group (Fig. 3C and D). The data demonstrated that miR-192-5p mimics reduced chemo-resistance of osteosarcoma cells to cisplatin. And it had been reported that the cisplatin drug was presented the chemio-sensitivity in osteosarcoma cells (26).
Figure 3.
Overexpression of miR-192-5p suppressed osteosarcoma cell migration and invasion, and enhanced the sensitivity of osteosarcoma cells to cisplatin. (A) Wound-healing assay revealed that the migratory ability of 143B and U2OS cells was suppressed by miR-192-5p mimics (magnification, ×100). (B) Transwell assay was conducted to evaluate the invasion of 143B and U2OS cells following treatment with mR-192-5p, which suggested that the capacity of invasion was significantly decreased in the treatment group (magnification, ×100). (C) The response of 143B cells to cisplatin enhanced following transfection with the miR-192 mimic compared to the negative control. (D) The response of U2OS cells to cisplatin enhanced following transfection with the miR-192 mimic compared with the negative control. Data are presented as the mean ± standard deviation of 3 independent assays. *P<0.05 and **P<0.01 vs. miR-NC. miR, microRNA; NC, negative control.
USP1 was a direct target of miR-192-5p in osteosarcoma cell
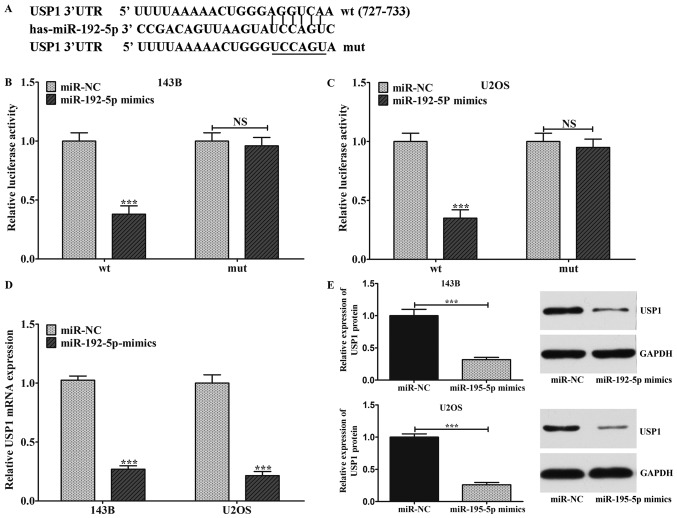
In order to verify the relationships between miR-192-5p and USP1, TargetScan tools was performed to predict the target sites for miR-192-5p. It was demonstrated that there was a combination of target sequences between the USP1 3′-UTR and miR-192-5p (Fig. 4A). Then, dual-luciferase reporter system assay was carried out to affirm the above prediction. In detail, the 3′-UTR of USP1 containing wild-type (wt) or mutant-type (mut) miR-192-5p target sequences was inserted into the plasmid. Then, we co-transfected 143B and U2OS cells with these reporters plasmid and miR-192-5p. Luciferase activity was detected 48 h after transfection. As showed in Fig. 4B and C, miR-192-5p mimics significantly restrained wild-type 3′UTR-USP1 reporter activity while there was no repression on the mutant 3′UTR-USP1 reporter activity, which revealed that miR-192-5p most likely suppressed gene expression via miR-192-5p binding sequences at the 3′-UTR of USP1. Besides, we observed that Ectopic expression of miR-192-5p statistically inhibited the expression of USP1 on mRNA and protein level (Fig. 4D and E).
Figure 4.
USP1 was a direct target of miR-192-5p in osteosarcoma cells. (A) Bioinformatics study suggested that there was a putative binding site between miR-192-5p and USP1. Dual-luciferase reporter system assay indicated that miR-192-5p mimics significantly restrained wild-type 3′UTR-USP1 reporter activity, while there was no repression effect on the mutant 3′UTR-USP1 reporter activity in (B) 143B and (C) U2OS cells. (D) Ectopic expression of miR-192-5p decreased the USP1 mRNA expression in 143B and U2OS cells. (E) Overexpression of miR-192-5p also reduced the expression of USP1 at the protein level. Data are presented as the mean ± standard deviation of 3 independent assays. ***P<0.001 vs. miR-NC. miR, microRNA; NC, negative control; USP1, ubiquitin-specific protease 1; UTR, untranslated region; wt, wild type; mut, mutant.
miR-192-5P regulated osteosarcoma cell proliferation, apoptosis, migration, invasion and chemo-sensitivity through USP1
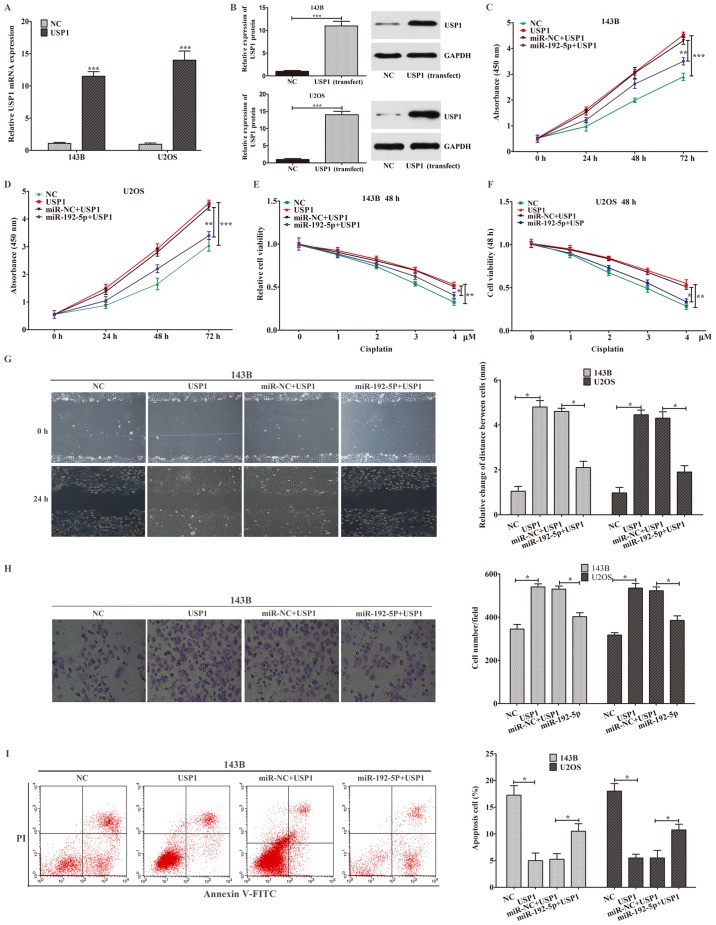
To further ascertain whether upregulation of miR-192-5p inhibited OS cells biological functions through USP1. We transfected USP1 vector into 143B and U2OS cells. After cultivated in vitro, the USP1 mRNA and protein expressions were restored in 143B and U2OS cells (Fig. 5A and B). Ectopic expression of USP1 statistically enhanced cell proliferation, migration and invasion abilities and decreased cell apoptosis and chemo-sensitivity in 143B and U2OS cells. However, when co-transfected with miR-192-5p mimics and USP1, overexpression of miR-192-5p partially abolished the effects of USP1 on cells proliferation, chemo-sensitivity, migration, invasion and apoptosis (Fig. 5C-I). Taken together, miR-192-5p inhibited osteosarcoma cell biological functions primarily through downregulating USP1.
Figure 5.
miR-192-5p regulated osteosarcoma cell proliferation, apoptosis, migration, invasion and chemo-sensitivity through USP1. (A) The USP1 mRNA expression was detected by reverse transcription-quantitative polymerase chain reaction. (B) The USP1 protein expression was detected by western blot analysis. (C-I) Ectopic expression of USP1 statistically enhanced cell (C and D) proliferation, decreased cell (E and F) chemo-sensitivity, promoted cell (G) migration and (H) invasion (magnification, ×100), and reduced cell (I) apoptosis in 143B and U2OS cells, which could be partially abolished by miR-192-5p following co-transfection with miR-192-5p mimics and USP1. Data are presented as the mean ± standard deviation of 3 independent assays. *P<0.05, **P<0.01 and ***P<0.001 vs. NC, or as indicated. miR-NC. miR, microRNA; NC, negative control; USP1, ubiquitin-specific protease 1; PI, propidium iodide.
Discussion
Increasing evidence have suggested that miRNAs could regulate cell proliferation, apoptosis, migration, invasion and chemo-sensitivity in malignant tumors, including osteosarcoma (27–30). For instance, Chen et al (31) revealed that miR-211-5p was downregulated in triple-negative breast cancer (TNBC), which inhibit TNBC cell biological functions via targeting SETBP1. Our previous study also showed that miR-335 was statistically downregulated in osteosarcoma stem cells. Moreover, overexpression of miR-335 suppressed stem cell-like characteristics by targeting POU5F1 (32). Our study indicated that miR-192-5p was significantly downregulated and USP1 was remarkably upregulated in 25 osteosarcoma samples and tow cell lines. Pearson's correlation assay indicated that the expression levels of miR-192-5p was inversely associated with USP1. Moreover, low expression of miR-192-5p in patients was statistically correlated with tumor grade, Enneking stage and tumor size. And osteosarcoma patients with low expression of miR-192-5p presented to have a shorter overall survival. Then, biological functions of miR-192-5p were explored in 143B and U2OS. The results shown that upregulation of miR-192-5p inhibited cell proliferation through cell cycle arrest and inducing cell apoptosis, suppressed cell migration and invasion and enhanced cell chemo-sensitivity in osteosarcoma cells. To gain insight into the detail relationship between miR-192-5p and USP1, we used TargetScan software (33) to predict that there was a highly conservative binding site between USP1 and miR-192-5p. Then the prediction was further proved by luciferase activity assay. Furthermore, we observed that USP1 acted as an opposite role in regulating cell biological function compared that with miR-192-5p. Moreover, when co-transfected with miR-192-5p mimics and USP1 simultaneously, overexpression of miR-192-5p partially abolished the effects of USP1 on cells proliferation, apoptosis, migration, invasion and chemo-sensitivity in osteosarcoma cells.
Aberrant expression of miR-192 played a crucial role in the development and progression of multiple malignant tumors (34–36). Previous studies have proved that miR-192 was significantly downregulated in many cancers, and miR-192 also has been reported to regulate cell biological functions in tumors, including proliferation, migration, invasion and apoptosis (37,38). For instance, Feng et al (15) suggested that miR-192-5p was significantly low in lung cancer. They further demonstrated that miR-192-5p suppressed cell proliferation and induced cell apoptosis through RB1. Lian et al (16) also proved that miR-192-5p reduced tumor metastasis by targeting the SLC39A6/SNAIL pathway in HCC cells. Although previous study had researched the effect of miR-192 on human osteosarcoma, which indicated that miR-192 was downregulated in osteosarcoma and miR-192 could suppressed the progression of osteosarcoma (17,39), the exact molecular mechanism remained largely unclear. Consistent with the above studies, we identified that upregulation of miR-192-5p inhibited cell proliferation by preventing cell cycle from G1 to S phase and inducing cell apoptosis in osteosarcoma cells. We also found that miR-192-5p repressed cell migration and invasion, and increased cells more sensitivity to cisplatin in osteosarcoma cells. Taken together, we came to the conclusion that miR-192-5p played an important role in suppressing osteosarcoma.
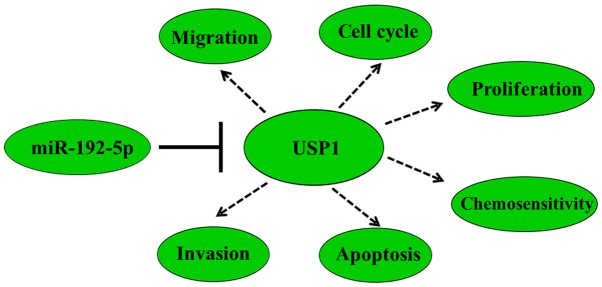
To clarify the potential molecular mechanism about miR-192-5p regulates cell biological function in osteosarcoma. Based on open-target prediction programs (TargetScan software), we found that USP1 may be a target gene of miR-192-5p. Recently, several studies indicated that USP1 had been found to be upregulated in many kinds of tumors, and they also found that deregulated USP1 could suppress cell proliferation, migration, invasion, chemo-resistance (40,41). Increasing studies suggested that USP1 played a role part in osteosarcoma. For example, Williams et al (24) indicated that USP1 was upregulated in osteosarcoma cells, which promoted cell proliferation, suppressed osteoblastic differentiation and stabilized ID proteins. Liu et al (23) confirmed that USP1 was upregulated in osteosarcoma. Silencing of USP1 inhibited cell proliferation and invasion through reducing expression of some downstream proteins, including Notch signaling pathway. Previous studies of our team suggested that Notch signaling pathway played a key role in the development and progression in osteosarcoma. Moreover Notch pathway could negatively regulated osteosarcoma stem cell-like properties, like cell proliferation, apoptosis, chemo-resistance (42,43). In the present research, we initially used the TargetScan software to suggest that USP1 was a putative binding site of miR-192-5p. Subsequently, overexpression of miR-192-5p restrained wild-type 3′UTR-USP1 reporter activity, while not in mutant 3′UTR-USP1 reporter activity in U2OS and 143B cells. Overexpression of miR-192-5p significantly repressed the USP1 expression on mRNA and protein level. Moreover, we also found that ectopic expression of USP1 promoted cell proliferation and migration, decreased cell chemo-sensitivity, which could be partially reversed by overexpression of miR-192-5p. All above data support that ectopic expression of miR-192-5p repressed OS cell proliferation, migration and invasion and increased the sensitivity of osteosarcoma cells to cisplatin via targeting USP1. The mechanism of how the miR-192-5p/USP1 axis regulates the initiation and progression of osteosarcoma was presented in Fig. 6. Considering that Notch signal pathway act as important downstream pathway of USP1, further research should be performed to explore whether miR-192-5p can suppress osteosarcoma oncogenicity by targeting USP1 through inactivation of Notch signal pathway.
Figure 6.
Schematic of the mechanism underlying the miR-192-5p/USP1 axis in 143B and U2OS cells. miR, microRNA; USP1, ubiquitin-specific protease 1.
However, there are still some limitations in our study. First of all, our sample size is too small, and we need a larger sample size experiment to verify the conclusions in our study. Secondly, transient transfection of miR-192-5p instead of stable expression is used in our research. Thirdly, in order to make the composing of figure more concise, we only put the pictures of 143B cell line instead of two cell lines in figures. Forthly, in our study, we only explored the expression of USP1 at the level of mRNA. And we will further investigate its expression at the level of protein, which will further demonstrate our point of view. Finally, the downstream signal pathway of USP1 in our study need to be further illustrated.
In conclusion, in addition to the above limitations, our study elaborates the relationship between miR-192-5p and USP1 in osteosarcoma for the first time. Moreover, we provide evidence to prove that miR-192-5p inhibited the progression of osteosarcoma by targeting USP1. Therefore, miR-192-5p may serve as a valuable biomarker and miR-192-5p/USP1 axis may function as a novel therapeutic target for osteosarcoma.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by a grant from the Natural Science Foundation of China (grant no. 81502575).
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Authors' contributions
SZ, WG, MX, JC and GD performed the experiments. SZ, LY, ZZ and GD analyzed the data. SZ, ZZ and LY contributed reagents, materials and analytical tools. SZ, JC and WG reviewed all data and wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Medical Ethics Committee of Renmin Hospital of Wuhan University (Hubei, China). All patients and their families provided written informed consent to take part in this research. All of the data was kept by the administrator of the study team in a confidential manner and was not used for any other purposes. We confirm that all experiments were performed in accordance with relevant guidelines and regulations.
Consent for publication
All participants provided written informed consent for the publication of their data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Geng S, Gu L, Ju F, Zhang H, Wang Y, Tang H, Bi Z, Yang C. MicroRNA-224 promotes the sensitivity of osteosarcoma cells to cisplatin by targeting Rac1. J Cell Mol Med. 2016;20:1611–1619. doi: 10.1111/jcmm.12852. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Zhang M, Wang D, Zhu T, Yin R. miR-214-5p targets ROCK1 and suppresses proliferation and invasion of human osteosarcoma cells. Oncol Res. 2017;25:75–81. doi: 10.3727/096504016X14719078133401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z, Zhang M, Chen Q, Zhang Q. Downregulation of microRNA-145 promotes epithelial-mesenchymal transition via regulating Snail in osteosarcoma. Cancer Gene Ther. 2017;24:83–88. doi: 10.1038/cgt.2017.1. [DOI] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B, Huang Z, Zhang Y, Chen Y, Li Z. MicroRNA-145 suppresses osteosarcoma metastasis via targeting MMP16. Cell Physiol Biochem. 2015;37:2183–2193. doi: 10.1159/000438575. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Wu W, Dang S, Feng Q, Liang J, Wang Y, Fan N. MicroRNA-542-3p inhibits the growth of hepatocellular carcinoma cells by targeting FZD7/Wnt signaling pathway. Biochem Biophys Res Commun. 2017;482:100–105. doi: 10.1016/j.bbrc.2016.10.136. [DOI] [PubMed] [Google Scholar]
- 8.Tormo E, Adam-Artigues A, Ballester S, Pineda B, Zazo S, González-Alonso P, Albanell J, Rovira A, Rojo F, Lluch A, Eroles P. The role of miR-26a and miR-30b in HER2+ breast cancer trastuzumab resistance and regulation of the CCNE2 gene. Sci Rep. 2017;7:41309. doi: 10.1038/srep41309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang T, Zhao P, Rong Z, Li B, Xue H, You J, He C, Li W, He X, Lee RJ, et al. Anti-tumor efficiency of lipid-coated cisplatin nanoparticles co-loaded with MicroRNA-375. Theranostics. 2016;6:142–154. doi: 10.7150/thno.13130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng JF, Ma XQ, Wang LP, Wang W. MicroRNA-145 exerts tumor-suppressive and chemo-resistance lowering effects by targeting CD44 in gastric cancer. World J Gastroenterol. 2017;23:2337–2345. doi: 10.3748/wjg.v23.i13.2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei R, Yang Q, Han B, Li Y, Yao K, Yang X, Chen Z, Yang S, Zhou J, Li M, et al. microRNA-375 inhibits colorectal cancer cells proliferation by downregulating JAK2/STAT3 and MAP3K8/ERK signaling pathways. Oncotarget. 2017;8:16633–16641. doi: 10.18632/oncotarget.15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou H, Jia L. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci Rep. 2017;7:41942. doi: 10.1038/srep41942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan X, Zhu Z, Xu S, Yang LN, Liao XH, Zheng M, Yang D, Wang J, Chen D, Wang L, et al. MicroRNA-140-5p inhibits hepatocellular carcinoma by directly targeting the unique isomerase Pin1 to block multiple cancer-driving pathways. Sci Rep. 2017;7:45915. doi: 10.1038/srep45915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Zhang S, Shen H, Li C. MicroRNA-144-3p suppresses gastric cancer progression by inhibiting epithelial-to-mesenchymal transition through targeting PBX3. Biochem Biophys Res Commun. 2017;484:241–247. doi: 10.1016/j.bbrc.2017.01.084. [DOI] [PubMed] [Google Scholar]
- 15.Feng S, Cong S, Zhang X, Bao X, Wang W, Li H, Wang Z, Wang G, Xu J, Du B, et al. MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res. 2011;39:6669–6678. doi: 10.1093/nar/gkr232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lian J, Jing Y, Dong Q, Huan L, Chen D, Bao C, Wang Q, Zhao F, Li J, Yao M, et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7:2672–2683. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhang S, Xu Y, Zhang Y, Guan H, Li X, Li Y, Wang Y. Upregulation of miR-192 inhibits cell growth and invasion and induces cell apoptosis by targeting TCF7 in human osteosarcoma. Tumour Biol. 2016;37:15211–15220. doi: 10.1007/s13277-016-5417-z. [DOI] [PubMed] [Google Scholar]
- 18.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang Q, Dexheimer TS, Zhang P, Rosenthal AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, et al. A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat Chem Biol. 2014;10:298–304. doi: 10.1038/nchembio.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sourisseau T, Helissey C, Lefebvre C, Ponsonnailles F, Malka-Mahieu H, Olaussen KA, André F, Vagner S, Soria JC. Translational regulation of the mRNA encoding the ubiquitin peptidase USP1 involved in the DNA damage response as a determinant of Cisplatin resistance. Cell Cycle. 2016;15:295–302. doi: 10.1080/15384101.2015.1120918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Santisteban I, Peters GJ, Giovannetti E, Rodríguez JA. USP1 deubiquitinase: Cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol Cancer. 2013;12:91. doi: 10.1186/1476-4598-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JK, Chang N, Yoon Y, Yang H, Cho H, Kim E, Shin Y, Kang W, Oh YT, Mun GI, et al. USP1 targeting impedes GBM growth by inhibiting stem cell maintenance and radioresistance. Neuro-Oncol. 2016;18:37–47. doi: 10.1093/neuonc/nov091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Zhu H, Zhong N, Jiang Z, Xu L, Deng Y, Jiang Z, Wang H, Wang J. Gene silencing of USP1 by lentivirus effectively inhibits proliferation and invasion of human osteosarcoma cells. Int J Oncol. 2016;49:2549–2557. doi: 10.3892/ijo.2016.3752. [DOI] [PubMed] [Google Scholar]
- 24.Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 25.Enneking WF, Spanier SS, Goodman MA. A system for the surgical staging of musculoskeletal sarcoma. Clin Orthop Relat Res. 1980:106–120. [PubMed] [Google Scholar]
- 26.Zhang Z, Yu L, Dai G, Xia K, Liu G, Song Q, Tao C, Gao T, Guo W. Telomerase reverse transcriptase promotes chemoresistance by suppressing cisplatin-dependent apoptosis in osteosarcoma cells. Sci Rep. 2017;7:7070. doi: 10.1038/s41598-017-07204-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei CH, Wu G, Cai Q, Gao XC, Tong F, Zhou R, Zhang RG, Dong JH, Hu Y, Dong XR. MicroRNA-330-3p promotes cell invasion and metastasis in non-small cell lung cancer through GRIA3 by activating MAPK/ERK signaling pathway. J Hematol Oncol. 2017;10:125. doi: 10.1186/s13045-017-0493-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Mahati S, Xiao L, Yang Y, Mao R, Bao Y. miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem Biophys Res Commun. 2017;486:732–737. doi: 10.1016/j.bbrc.2017.03.110. [DOI] [PubMed] [Google Scholar]
- 29.Xiao Q, Yang Y, An Q, Qi Y. MicroRNA-100 suppresses human osteosarcoma cell proliferation and chemo-resistance via ZNRF2. Oncotarget. 2017;8:34678–34686. doi: 10.18632/oncotarget.16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang P, Tang WM, Zhang H, Li YQ, Peng Y, Wang J, Liu GN, Huang XT, Zhao JJ, Li G, et al. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer. 2017;117:525–534. doi: 10.1038/bjc.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen LL, Zhang ZJ, Yi ZB, Li JJ. MicroRNA-211-5p suppresses tumour cell proliferation, invasion, migration and metastasis in triple-negative breast cancer by directly targeting SETBP1. Br J Cancer. 2017;117:78–88. doi: 10.1038/bjc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X, Yu L, Zhang Z, Dai G, Gao T, Guo W. miR-335 negatively regulates osteosarcoma stem cell-like properties by targeting POU5F1. Cancer Cell Int. 2017;17:29. doi: 10.1186/s12935-017-0398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E, Peterson KJ. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet. 2015;49:213–242. doi: 10.1146/annurev-genet-120213-092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Botla SK, Savant S, Jandaghi P, Bauer AS, Mücke O, Moskalev EA, Neoptolemos JP, Costello E, Greenhalf W, Scarpa A, et al. Early epigenetic downregulation of microRNA-192 expression promotes pancreatic cancer progression. Cancer Res. 2016;76:4149–4159. doi: 10.1158/0008-5472.CAN-15-0390. [DOI] [PubMed] [Google Scholar]
- 35.Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Are C, Brattain M, Wang J. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene. 2014;33:5332–5340. doi: 10.1038/onc.2013.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr, Hofmainster C, Alder H, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2016;30:349–351. doi: 10.1016/j.ccell.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Yang SY, Choi SA, Lee JY, Park AK, Wang KC, Phi JH, Koh EJ, Park WY, Park SH, Hwang DW, et al. miR-192 suppresses leptomeningeal dissemination of medulloblastoma by modulating cell proliferation and anchoring through the regulation of DHFR, integrins, and CD47. Oncotarget. 2015;6:43712–43730. doi: 10.18632/oncotarget.6227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J, Fan Z, Lu S, Yang J, Hao T, Huo Q. miR-192 suppresses the tumorigenicity of prostate cancer cells by targeting and inhibiting nin one binding protein. Int J Mol Med. 2016;37:485–492. doi: 10.3892/ijmm.2016.2449. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Jia LS, Yuan W, Wu Z, Wang HB, Xu T, Sun JC, Cheng KF, Shi JG. Low miR-34a and miR-192 are associated with unfavorable prognosis in patients suffering from osteosarcoma. Am J Transl Res. 2015;7:111–119. [PMC free article] [PubMed] [Google Scholar]
- 40.Das DS, Das A, Ray A, Song Y, Samur MK, Munshi NC, Chauhan D, Anderson KC. Blockade of deubiquitylating enzyme USP1 inhibits DNA repair and triggers apoptosis in multiple myeloma cells. Clin Cancer Res. 2017;23:4280–4289. doi: 10.1158/1078-0432.CCR-16-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhiqiang Z, Qinghui Y, Yongqiang Z, Jian Z, Xin Z, Haiying M, Yuepeng G. USP1 regulates AKT phosphorylation by modulating the stability of PHLPP1 in lung cancer cells. J Cancer Res Clin Oncol. 2012;138:1231–1238. doi: 10.1007/s00432-012-1193-3. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y, Yu L, Dai G, Zhang S, Zhang Z, Gao T, Guo W. Cinobufagin induces apoptosis of osteosarcoma cells through inactivation of Notch signaling. Eur J Pharmacol. 2017;794:77–84. doi: 10.1016/j.ejphar.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 43.Yu L, Fan Z, Fang S, Yang J, Gao T, Simões BM, Eyre R, Guo W, Clarke RB. Cisplatin selects for stem-like cells in osteosarcoma by activating Notch signaling. Oncotarget. 2016;7:33055–33068. doi: 10.18632/oncotarget.8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.