Abstract
Background
Alterations in the intestinal microbiome are prospectively associated with the development of asthma; less is known regarding the role of microbiome alterations in food allergy development.
Methods
Intestinal microbiome samples were collected at age 3–6 months in children participating in the follow-up phase of an interventional trial of high-dose vitamin D given during pregnancy. At age 3, sensitization to foods (milk, egg, peanut, soy, wheat, walnut) was assessed. Food allergy was defined as caretaker report of healthcare provider-diagnosed allergy to the above foods prior to age 3 with evidence of IgE sensitization. Analysis was performed using Phyloseq and DESeq2; P-values were adjusted for multiple comparisons.
Results
Complete data were available for 225 children; there were 87 cases of food sensitization and 14 cases of food allergy. Microbial diversity measures did not differ between food sensitization and food allergy cases and controls. The genera Haemophilus (log2 fold change −2.15, P=.003), Dialister (log2 fold change −2.22, P=.009), Dorea (log2 fold change −1.65, P=.02), and Clostridium (log2 fold change −1.47, P=.002) were underrepresented among subjects with food sensitization. The genera Citrobacter (log2 fold change −3.41, P=.03), Oscillospira (log2 fold change −2.80, P=.03), Lactococcus (log2 fold change −3.19, P=.05), and Dorea (log2 fold change −3.00, P=.05) were underrepresented among subjects with food allergy.
Conclusions
The temporal association between bacterial colonization and food sensitization and allergy suggests that the microbiome may have a causal role in the development of food allergy. Our findings have therapeutic implications for the prevention and treatment of food allergy.
Keywords: Dorea, food allergy, food sensitization, microbiome
1 | INTRODUCTION
Food allergies cause life-threatening anaphylactic reactions in affected children and adults and lead to significant morbidity, quality of life impairment, and healthcare costs.1,2 The prevalence of food allergy has markedly increased over a recent time period.3 Although the cause of food allergy is not currently known, it is generally accepted that changes in microbial exposures in line with the hygiene hypothesis of allergic disease, or the “gut microbial deprivation hypothesis,” may in part explain the recent rise in prevalence of food allergy.4–7
To date, several studies have investigated the relationship between the microbiome and development of allergic disease. Both diversity of microbial exposures and specific taxa are thought to be protective against development of allergy and asthma.8 The study of the microbiome and food allergy specifically has been less fruitful, in part because of the lower prevalence of food allergy, and challenges with accurate phenotyping of this disease. In a small prospective study, richness (a measure of microbial diversity) of the intestinal microbiota was inversely associated with the development of food sensitization, while the Enterobacteriaceae/Bacteroidaceae ratio was positively associated with the development of food sensitization at age 1 year.9 Animal models, which can elucidate disease mechanisms, highlight a protective role for members of the class Clostridia in food sensitization via induction of IL-22 production by innate lymphoid cells with subsequent reduction of allergen exposure to the circulation.10 Clostridia were also enriched in the stool of infants with milk allergy who outgrew their allergy by age 8 years, compared to those whose allergy did not resolve by that time.11
We performed a microbiome-wide association study of the infant intestinal flora and the development of food sensitization and food allergy in a prospective birth cohort of 216 children. Our objective was to determine whether gut microbial composition was associated with food sensitization and allergy and whether these associations might be along the causal pathway for food allergy or explained by other environmental exposures. To our knowledge, this is the largest prospective study of the microbiome and development of food sensitization and allergy performed to date.
2 | METHODS
2.1 | Source of subjects
The Vitamin D Antenatal Asthma Reduction Trial (VDAART) is a randomized, controlled trial of high (4000IU)- vs standard-dose (400IU) daily vitamin D supplementation during pregnancy. The study design and primary outcome have been published previously.12,13 Briefly, VDAART enrolled 881 pregnant women at weeks 10–18 of pregnancy at three clinical sites in the United States (Boston, MA; St. Louis, MO; and San Diego, CA). Inclusion criteria included a personal history of asthma or allergy in the pregnant woman or in the father of the fetus. Smoking, other chronic disease, multiple gestation pregnancy, and assisted reproduction were exclusion criteria. The intervention phase occurred during pregnancy, and after birth, infants were followed every 3 months for the development of respiratory disease and other outcomes, including diagnosis of food allergy by a healthcare provider.
An ancillary study of the child intestinal microbiome was initiated during follow-up, after roughly half of the children reached 3 months of age. Stool samples were collected from infants between ages 3 and 6 months. Caretakers were instructed to collect ½ teaspoon of child stool from a diaper using a tongue depressor 1–2 days prior to a study visit and to store the sample in the freezer at home until bringing it to the research clinic in a freezer pack. Antibiotic use by the infant in the past 7 days was an exclusion criterion for stool collection.
2.2 | Variable definitions
At age 3 years, laboratory testing for serum-specific IgE to milk, egg, peanut, soy, wheat, and walnut was performed (Thermo Scientific/Phadia Immunology Reference Laboratory, Uppsala, Sweden). A child was considered to have food sensitization if any food-specific IgE was greater than 0.35 kU/L. Two children had missing data for walnut IgE and had sensitization results for the other foods. Neither child reported a history of walnut allergy and were considered nonsensitized to walnut.
Telephone or in-person questionnaires were performed every 3 months following birth. Caretakers were asked “since [birth/the last time we talked], has a health care provider said that [CHILD] has food allergies?” If the answer was affirmative, the caretaker was asked “which foods did the health care provider say [CHILD] is allergic to?” Possible responses included milk, egg, peanut, wheat, soy, other nuts, shellfish, fish, with the option to report other food allergies as free text. A child was considered to have food allergy if there was caretaker report of healthcare provider-diagnosed allergy to milk, egg, peanut, wheat, soy, or other nut allergy prior to age 3 years with evidence of IgE sensitization to that food (>0.1 kU/L) at age 3 years.
Other study variables included self-reported race and ethnicity of the child (categorized as African American, Hispanic, White, and Other), maternal vitamin D supplementation treatment assignment, child gender, mode of delivery (vaginal vs C-section), age of child at stool collection, and whether the child was ingesting formula and/or breast milk and solid food at the time of the stool collection. Per study protocol, questions regarding the child diet were asked beginning at age 6 months. Because the stool collection occurred between the ages of 3 and 6 months, we used questionnaire responses obtained at age 6 months (or later if 6-month data were incomplete) to determine whether the child was on breast milk, infant formula, or solid food at the time of stool collection. For additional details on dietary variables, please see the Appendix S1. Subjects were included in this study if infant gut microbial characterization and sensitization data were available. Missing data were considered missing at random.
2.3 | Stool sample sequencing and taxonomic identification
Sequencing of the bacterial 16S V3 to V5 hypervariable regions was performed using the Roche 454 Titanium platform. Filtering, trimming, and chimera checking were performed as previously described.14,15 We used closed-reference operational taxonomic unit (OTU) classification in Qiime to group sequences according to sequence similarity and match to taxonomy.16 We excluded samples with total read counts <1000 and OTUs identified less than 10 times or in fewer than 10 subjects.
2.4 | Statistical analysis
Distributions of potential confounders were compared by food sensitization or food allergy status using the chi-square test. We calculated richness (Chao1) and diversity (Shannon index) at the genus level and compared these between sensitization and allergy groups using the t test.
We performed a microbiome-wide analysis to identify bacterial genera that were differentially abundant between cases and controls. We used a negative binomial model without rarefying to account for variability in read depth between samples following the recommendations of McMurdie and Holmes.17 Reported P-values were corrected for multiple comparisons using the method of Benjamini and Hochberg.18 Analyses were performed in Stata 12 (College Station, TX, USA) and the Phyloseq and DESeq2 packages of R.19,20 We stratified our analyses by potential confounders in multiple sensitivity analyses.
3 | RESULTS
A total of 810 children were born to VDAART participants who were randomized to treatment. The 216 subjects in this study included those with data on food sensitization and infant gut microbial characteristics, and were similar demographically to subjects who did not have these data available (data not shown). Complete phenotypic data were available on all study subjects with the exception of missing data for breastfeeding (n=3) and solid food introduction (n=2) at the time of stool collection (Table 1).
TABLE 1.
Demographics of 216 children providing evaluable stool samples for food sensitization and food allergy analysis
| Food sensitization N=85 |
No food sensitization N=131 |
P | Food allergy N=14 |
No food allergy N=202 |
P | |
|---|---|---|---|---|---|---|
| Milk | 67 (79) | 3 (21) | ||||
| Egg | 48 (56) | 9 (64) | ||||
| Peanut | 28 (33) | 8 (57) | ||||
| Soy | 15 (18) | 2 (14) | ||||
| Wheat | 33 (39) | 3 (21) | ||||
| Walnut | 11 (13) | 2 (14) | ||||
| Race | ||||||
| African American | 47 (55) | 46 (35) | .02 | 7 (50) | 86 (43) | .67 |
| Hispanic | 16 (19) | 45 (34) | 3 (21) | 58 (29) | ||
| White | 16 (19) | 33 (25) | 4 (29) | 45 (22) | ||
| Other | 6 (7) | 7 (5) | 0 | 13 (6) | ||
| Vitamin D Treatmenta | 48 (57) | 77 (59) | .74 | 8 (57) | 117 (58) | .96 |
| Female | 38 (45) | 65 (50) | .48 | 8 (57) | 98 (47) | .46 |
| C-section | 25 (29) | 44 (34) | .52 | 8 (57) | 61 (30) | .04 |
| Formula Fed | 63 (74) | 84 (64) | .12 | 9 (64) | 138 (68) | .75 |
| Breast fed | 32 (38) | 56 (44) | .38 | 7 (50) | 81 (41) | .50 |
| Solid food | 45 (54) | 53 (41) | .05 | 8 (57) | 90 (45) | .38 |
| Age at infant stool collection (mean, range) | 148 (92–208) | 140 (78–206) | .95 | 153 (115–208) | 142 (78–206) | .87 |
| Read counts of microbial sample (mean, range) | 5282 (1726–12 655) | 5310 (1236–19 917) | .48 | 5351 (2281–9387) | 5295 (1236–19 917) | .53 |
High-dose vitamin D supplementation during pregnancy, as part of the Vitamin D Antenatal Asthma Reduction Trial study.
There were 85 subjects with food sensitization and 131 subjects without food sensitization. The majority (79%) of food-sensitized subjects were sensitized to milk, with egg being the next most common sensitization with 56% sensitized. Food-sensitized subjects tended to be African American (55%) while nonsensitized subjects were more evenly distributed across race and ethnicity (35% African American, 34% Hispanic, 25% White; P=.02). A greater percentage of food-sensitized subjects had been introduced to solid food by the time of stool collection compared to nonsensitized subjects (54% vs 41%, P=.05).
There were 14 subjects with food allergy. These subjects reported a history of healthcare provider-diagnosed milk, egg, peanut, wheat, soy, or other nut allergy with onset between the ages of 9 months and 3 years and had evidence of sensitization to those foods at age 3 years. Egg and peanut allergy were the most common food allergies, reported in nine and eight subjects, respectively, while allergy to the other foods occurred in two to three subjects each. Subjects with food allergy were in general similar to subjects without food allergy, although more subjects with food allergy had a history of C-section delivery (57% vs 30%, P=.04).
3.1 | Microbiome diversity, food sensitization, and allergy
Measures of intestinal diversity were similar between subjects with and without food sensitization (mean Chao1 diversity index 31 vs 31, P=.26; mean Shannon diversity index 2.13 vs 2.21; P=.11) and with and without food allergy (mean Chao1 diversity index 32 vs 31, P=.67; mean Shannon diversity index 2.19 vs 2.17; P=.54; Table 2).
TABLE 2.
Intestinal microbial diversity and food sensitization and allergy
| Outcome | Diversity measure | Cases | Controls | P-value |
|---|---|---|---|---|
| Food sensitization | Chao1 | 31 (13–55) | 31 (9–52) | .26 |
| Shannon | 2.13 (0.70–3.15) | 2.21 (0.50–2.99) | .11 | |
|
| ||||
| Food allergy | Chao1 | 32 (23–44) | 31 (9–55) | .67 |
| Shannon | 2.19 (1.51–2.65) | 2.17 (0.50–3.15) | .54 | |
Values presented are mean (range).
3.2 | Microbiome-wide analysis of food sensitization and allergy
Prominent bacterial families across all groups included Bacteroidaceae, Enterobacteriaceae, and Lachnospiraceae (Figure 1). Microbiome-wide analysis was performed at the genus taxonomic rank. Four genera were statistically significantly associated with food sensitization; all had lower read counts in cases compared to controls: Haemophilus (log2 fold change −2.15, PBH=.003), Dialister (log2 fold change −2.22, PBH=.009), Dorea (log2 fold change −1.65, PBH=.02), and Clostridium (log2 fold change −1.47, PBH=.02; Figure 2, Table 3). No genera had statistically significantly increased relative abundance in cases compared to controls; there was a trend toward higher read counts for Dysgonomonas in those with food sensitization compared to controls (log2 fold change 2.26, PBH=.29).
FIGURE 1.
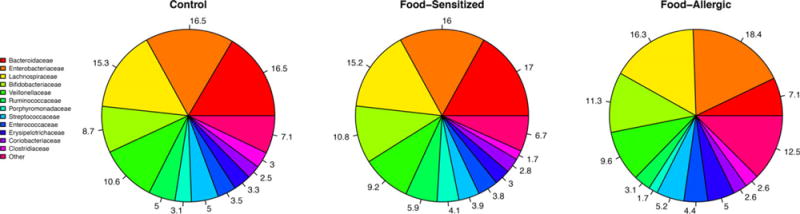
Bacterial relative abundances at the family taxonomic rank by food allergy or sensitization status
FIGURE 2.
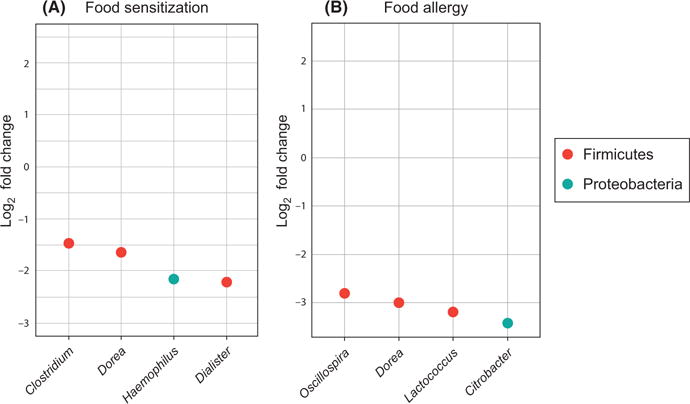
Significant associations between food sensitization A, and food allergy B, and bacterial genera. The log2 fold change is plotted for each bacterial genus significantly associated with food sensitization or food allergy
TABLE 3.
Significant associations between microbial genera and food sensitization
| Rank | Phylum | Family | Genus | Base mean | log2 Fold Change | Padj |
|---|---|---|---|---|---|---|
| 1 | Proteobacteria | Pasteurellaceae | Haemophilus | 12.52 | −2.15 | .003 |
| 2 | Firmicutes | Veillonellaceae | Dialister | 2.04 | −2.22 | .009 |
| 3 | Firmicutes | Lachnospiraceae | Dorea | 22.9 | −1.65 | .02 |
| 4 | Firmicutes | Clostridiaceae | Clostridium | 32.6 | −1.47 | .02 |
Four genera were statistically significantly associated with food allergy; again, all had lower read counts in cases compared to controls: Citrobacter (log2 fold change −3.41, PBH=.03), Oscillospira (log2 fold change −2.80, PBH=.03), Lactococcus (log2 fold change −3.19, PBH=.05), and Dorea (log2 fold change −3.00, PBH=.05; Figure 2, Table 4). No genera had statistically significantly increased relative abundance in cases compared to controls; there was a trend toward higher read counts for SMB53 (candidate genus of the family Clostridiaceae) and Stenotrophomonas in those with food allergy compared to controls (log2 fold change 2.29 and 2.20, respectively, and PBH=.05 and .37, respectively).
TABLE 4.
Significant associations between microbial genera and food allergy
| Rank | Phylum | Family | Genus | Base mean | log2 Fold Change | Padj |
|---|---|---|---|---|---|---|
| 1 | Proteobacteria | Enterobacteriaceae | Citrobacter | 6.14 | −3.41 | .03 |
| 2 | Firmicutes | Ruminococcaceae | Oscillospira | 23.2 | −2.80 | .03 |
| 3 | Firmicutes | Streptococcaceae | Lactococcus | 8.3 | −3.19 | .05 |
| 4 | Firmicutes | Lachnospiraceae | Dorea | 22.8 | −3.00 | .05 |
3.3 | Sensitivity analyses
Because methods to perform an adjusted microbiome-wide association study in the prediction of food allergy are to our knowledge currently limited, we repeated our initial analyses stratified by potential confounders of the relationship between the microbiome and food sensitization and allergy that were identified in Table 1: race/ethnicity and solid food feeding for food sensitization and C-section history for food allergy. We found in general that among subgroups of subjects, the direction and magnitude of point estimates for the association between Haemophilus, Dialister, Dorea, and Clostridium and food sensitization, and between Citrobacter, Oscillospira, Lactococcus, and Dorea and food allergy were similar to those identified in the larger cohort (Tables 5–6). However, the association between food sensitization and Clostridium was reduced in magnitude and changed direction in the African American subgroup as well as the subgroup that had been introduced to solid food. The associations between food allergy and Citrobacter, Oscillospira, Lactococcus, and Dorea persisted after stratification by mode of delivery.
TABLE 5.
Sensitivity analyses for food sensitization
| Subset | n | Rank | Phylum | Family | Genus | Base mean | log2 Fold Change | Padj |
|---|---|---|---|---|---|---|---|---|
| African American | 93 | 65 | Proteobacteria | Pasteurellaceae | Haemophilus | 4.29 | −0.34 | .99 |
| 5 | Firmicutes | Veillonellaceae | Dialister | 2.71 | −1.81 | .33 | ||
| 6 | Firmicutes | Lachnospiraceae | Dorea | 23.60 | −1.22 | .62 | ||
| 56 | Firmicutes | Clostridiaceae | Clostridium | 8.52 | 0.42 | .99 | ||
|
| ||||||||
| Non-African American | 123 | 2 | Proteobacteria | Pasteurellaceae | Haemophilus | 16.10 | −2.37 | .02 |
| 4 | Firmicutes | Veillonellaceae | Dialister | 1.03 | −2.39 | .12 | ||
| 6 | Firmicutes | Lachnospiraceae | Dorea | 22.00 | −1.95 | .16 | ||
| 7 | Firmicutes | Clostridiaceae | Clostridium | 45.88 | −1.49 | .19 | ||
|
| ||||||||
| Solid Food | 98 | 11 | Proteobacteria | Pasteurellaceae | Haemophilus | 6.97 | −1.40 | .41 |
| 26 | Firmicutes | Veillonellaceae | Dialister | 0.94 | −0.94 | .94 | ||
| 12 | Firmicutes | Lachnospiraceae | Dorea | 21.04 | −1.17 | .42 | ||
| 27 | Firmicutes | Clostridiaceae | Clostridium | 8.41 | 0.58 | .94 | ||
|
| ||||||||
| No solid food | 118 | 1 | Proteobacteria | Pasteurellaceae | Haemophilus | 14.68 | −2.33 | .15 |
| 3 | Firmicutes | Veillonellaceae | Dialister | 3.00 | −2.35 | .17 | ||
| 4 | Firmicutes | Lachnospiraceae | Dorea | 24.91 | −2.06 | .17 | ||
| 5 | Firmicutes | Clostridiaceae | Clostridium | 39.45 | −1.54 | .27 | ||
TABLE 6.
Sensitivity analyses for food allergy
| Subset | n | Rank | Phylum | Family | Genus | Base mean | log2 Fold Change | Padj |
|---|---|---|---|---|---|---|---|---|
| C-section | 69 | 1 | Proteobacteria | Enterobacteriaceae | Citrobacter | 6.23 | −4.15 | .09 |
| 4 | Firmicutes | Ruminococcaceae | Oscillospira | 27.15 | −2.25 | .39 | ||
| 6 | Firmicutes | Streptococcaceae | Lactococcus | 7.47 | −2.81 | .43 | ||
| 13 | Firmicutes | Lachnospiraceae | Dorea | 35.88 | −1.82 | .75 | ||
|
| ||||||||
| No C-section | 147 | 44 | Proteobacteria | Enterobacteriaceae | Citrobacter | 4.92 | −2.00 | .99 |
| 3 | Firmicutes | Ruminococcaceae | Oscillospira | 18.33 | −3.95 | .03 | ||
| 8 | Firmicutes | Streptococcaceae | Lactococcus | 8.36 | −2.53 | .99 | ||
| 2 | Firmicutes | Lachnospiraceae | Dorea | 7.93 | −4.68 | .03 | ||
4 | DISCUSSION
We conducted a microbiome-wide association study of the infant gut microbiome and food sensitization and food allergy in early childhood. After correction for multiple comparisons, we identified four bacterial genera present in the infant intestinal microbiome that were significantly inversely associated with the development of food sensitization at the age of 3 years and four genera that were significantly inversely associated with the development of food allergy by age 3 years. Notably, the genus Dorea was associated with both food sensitization and food allergy. We completed a sensitivity analysis indicating that our results are generally robust to potential confounders, although associations between Clostridia and food sensitization may be confounded by race and diet.
We identified seven individual genera with significant inverse associations with food sensitization and/or allergy. Two members of the phylum Proteobacteria were inversely associated with food sensitization (Haemophilus) or allergy (Citrobacter). These gram-negative bacteria are better known for their role in disease than in health, although commensal members are known to exist. Additionally, prior studies investigating the association of the intestinal microbiome during infancy and allergic disease in humans have demonstrated similar inverse associations between Haemophilus and eczema,21 and Citrobacter and allergic sensitization and asthma.22
Five members of the phylum Firmicutes, predominantly gram-positive bacteria which make up a majority of the intestinal microbiome, were inversely associated with food sensitization or allergy.23 The Firmicutes include the class Clostridia, which has recently been identified as protective against peanut sensitization in a mouse model of food allergy.10 Clostridia may also speed the resolution of milk allergy.11 The class Clostridia includes the genera Dorea, which was significantly reduced among children with food sensitization and allergy in the current study; Clostridium reduced in children with food sensitization, and Oscillospira reduced among children with food allergy. The Clostridia are widely appreciated for their ability to cause disease in Clostridium difficile infection, and indeed the presence of C. difficile in the infant microbiome has been positively associated with atopic outcomes in later childhood.24–26 However, Clostridia class members are also increasingly recognized as important in health promotion.27 Many have anti-inflammatory properties through their ability to produce short-chain fatty acids28,29 or via other mechanisms leading to reduced IL-12 and IFN-γ levels and secretion of IL-10.30 Dialister was formerly recognized as a member of the class Clostridia, as it is closely related to this class. Lactococcus, of the class Bacilli, is the only non-Clostridial Firmicute associated with food allergy in the current study and has anti-inflammatory properties such as reduction of TNF-α.31 Similar relationships between the Firmicutes identified in our analysis and other allergic disorders have been seen in prior studies of the human intestinal microbiome during infancy: Dorea has been inversely associated with both eczema and allergic sensitization and asthma,21,22 Dialister has been inversely associated with eczema,21 and Lactococcus, Clostridium, and Oscillospira have been inversely associated with atopy and asthma.22,32 While the Firmicutes mentioned above do have biological activity suggesting that a potential role in susceptibility to food sensitization or allergy is plausible, further work is necessary in humans and animal models to understand the strength and mechanism of these associations.
Although we did identify individual genera that were significantly associated with food sensitization and allergy, global microbiome measures of diversity were similar between cases and controls. This was surprising in light of several reports of inverse associations between intestinal microbial diversity and risk of allergic diseases including eczema33 and asthma34–36 using methods similar to ours. The one study that reported an inverse association between infant intestinal microbial diversity at age 3 months and food sensitization at age 1 year used family taxonomic rank measurements and included only 12 cases of food sensitization, factors that may in part underlie the discrepancy in findings.9
The strengths of our study include our large and diverse study population, prospective study design, and analysis of the entire metagenome using sequencing-based methods. Although the number of food allergy cases (defined by history and detectible IgE at age 3 years) was low, this is similar to what has been published in case-control studies without the benefit of our prospective design.37
Our study has several limitations. Unlike traditional genomewide association studies, the methodology for analyzing the microbiome as a predictor of disease while accounting for potential confounding by other exposures has not, to our knowledge, been standardized. We therefore evaluated the effect of each known potential confounder independently of other confounders using stratified analyses, as we were methodologically unable to complete a true multivariate association study. We found that our overall findings were relatively robust to confounding, except perhaps to race; however, we may be limited by unmeasured confounders that we could not address with our dataset. Further, we chose to analyze the microbiome at the genus level as we did not have the power to identify meaningful associations when over 1000 individual taxa (species or OTUs) were included in the analysis. Therefore, we are limited in the ability to fully resolve the identity of taxa that may be important to the development of food allergy. Finally, although we are reasonably confident in our case definitions of food sensitization and food allergy, some children who did develop and then resolve food allergy may have been misclassified as controls, which may have biased our results toward the null.
In summary, we identified a bacterial genus, Dorea, which is reduced in the intestinal microbiomes of infants who later develop food sensitization and food allergy. Our results suggest that Dorea may promote or protect against food sensitization and food allergy. Further work to identify the specific relevant species and strains may lead to prevention or treatment of food allergy through altering the developing infant microbiome.
Supplementary Material
Acknowledgments
Funding information
R01HL091528, R01HL108818, and K23AI110522.
Footnotes
Edited by: Antonella Muraro
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
SUPPORTING INFORMATION
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- 1.Sicherer SH, Sampson HA. Food allergy: epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol. 2014;133:291–307. doi: 10.1016/j.jaci.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 2.Gupta R, Holdford D, Bilaver L, Dyer A, Holl JL, Meltzer D. The economic impact of childhood food allergy in the United States. JAMA Pediatr. 2013;167:1026–1031. doi: 10.1001/jamapediatrics.2013.2376. [DOI] [PubMed] [Google Scholar]
- 3.Keet CA, Savage JH, Seopaul S, Peng RD, Wood RA, Matsui EC. Temporal trends and racial/ethnic disparity in self-reported pediatric food allergy in the United States. Ann Allergy Asthma Immunol. 2014;112:222–229. doi: 10.1016/j.anai.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strachan DP. Hay fever, hygiene and household size. Br Med J. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marrs T, Bruce KD, Logan K, et al. Is there an association between microbial exposure and food allergy? A systematic review. Pediatr Allergy Immunol. 2013;24:311–320. doi: 10.1111/pai.12064. [DOI] [PubMed] [Google Scholar]
- 6.Wold AE. The hygiene hypothesis revised: is the rising frequency of allergy due to changes in the intestinal flora? Allergy. 1998;53(46 Suppl):20–25. doi: 10.1111/j.1398-9995.1998.tb04953.x. [DOI] [PubMed] [Google Scholar]
- 7.West CE, Renz H, Jenmalm MC, et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. J Allergy Clin Immunol. 2015;135:3–13. doi: 10.1016/j.jaci.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Legatzki A, Rösler B, von Mutius E. Microbiome diversity and asthma and allergy risk. Curr Allergy Asthma Rep. 2014;14:1–9. doi: 10.1007/s11882-014-0466-0. [DOI] [PubMed] [Google Scholar]
- 9.Azad MB, Konya T, Guttman DS, et al. Infant gut microbiota and food sensitization: associations in the first year of life. Clin Exp Allergy. 2015;45:632–643. doi: 10.1111/cea.12487. [DOI] [PubMed] [Google Scholar]
- 10.Stefka AT, Feehley T, Tripathi P, et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci U S A. 2014;111:13145–13150. doi: 10.1073/pnas.1412008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunyavanich S, Shen N, Grishin A, et al. Early-life gut microbiome composition and milk allergy resolution. J Allergy Clin Immunol. 2016;138:1122–1130. doi: 10.1016/j.jaci.2016.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litonjua A, Lange N, Carey V, Al E. The Vitamin D Antenatal Asthma Reduction Trial (VDAART): rationale, design, and methods of a randomized, controlled trial of vitamin D supplementation in pregnancy for the primary prevention of asthma and allergies in children. Contemp Clin trials. 2014;38:37–50. doi: 10.1016/j.cct.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Litonjua AA, Carey VJ, Laranjo N, et al. Effect of prenatal supplementation with vitamin D on asthma or recurrent wheezing in offspring by age 3 years: the VDAART randomized clinical trial. JAMA. 2016;315:362–370. doi: 10.1001/jama.2015.18589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.HMP Consortium. Notes S. A framework for human microbiome research. Nature. 2012;486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas BJ, Gevers D, Earl AM, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamini Y, Hochberg Y, Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 19.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng H, Liang H, Wang Y, et al. Altered gut microbiota composition associated with eczema in infants. PLoS One. 2016;11:e0166026. doi: 10.1371/journal.pone.0166026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 24.van Nimwegen F. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128:948–955. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 25.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 26.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M, Björksten B. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol. 2001;108:516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 27.Velasquez-Manoff M. Gut microbiome: the peacekeepers. Nature. 2015;518:S3–S11. doi: 10.1038/518S3a. [DOI] [PubMed] [Google Scholar]
- 28.Klampfer L, Huang J, Sasazuki T, Shirasawa S, Augenlicht L. Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol Cancer Res. 2003;1:855–862. [PubMed] [Google Scholar]
- 29.Lee W-J, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 30.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han K. Anticancer and anti-inflammatory activity of probiotic Lactococcus lactis NK34. J Microbiol Biotechnol. 2015;25:1697–1701. doi: 10.4014/jmb.1503.03033. [DOI] [PubMed] [Google Scholar]
- 32.Arrieta M-C, Stiemsma LT, Dimitriu PA, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 33.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012:434–434. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 34.Bisgaard H, Li N, Bonnelykke K, et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol. 2011;128:646–652. doi: 10.1016/j.jaci.2011.04.060. [DOI] [PubMed] [Google Scholar]
- 35.Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy. 2014;44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 36.Ege MJ, Mayer M, Normand A-C, et al. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 37.Ling Z, Li Z, Liu X, et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


