Abstract
Previous studies have explored the functions of microRNA (miR)-146a in different types of cancer through mediating different targets. However, the roles of miR-146a in malignant melanoma (MM) cell migration and invasion remain largely elusive. In the present study, the potential molecular function of miR-146a in MM was investigated. Reverse transcription-quantitative polymerase chain reaction was utilized to detect miR-146a expression in MM tissues and cell lines. A Transwell assay was performed to confirm the ability of migration and invasion. A luciferase assay and biological analysis were used to predict and determine the targets of miR-146a. The expression of miR-146a was upregulated in melanoma tissues and cell lines. Clinicopathological analysis indicated that the miR-146a level was negatively correlated with the progression of melanoma. Abnormal expression of miR-146a promoted cell migration and invasion in MM cells. Additionally, it was also observed that Mothers against decapentaplegic homolog 4 (SMAD4) was a novel target of miR-146a in MM. SMAD4 was negatively associated with miR-146a in MM and abnormal expression of SMAD4 attenuated miR-146a-mediated promotion of cell migration and invasion. In conclusion, miR-146a functioned as an oncogene by directly targeting SMAD4 and it may be a novel diagnostic and therapeutic marker of MM.
Keywords: invasion, miR-146a, migration, melanoma, mothers against decapentaplegic homolog 4
Introduction
Malignant melanoma (MM) is an aggressive form of human skin cancer and arises from malignant transformation of melanocytes. It usually metastasizes to other organs, for example to brain and bone; metastasis to lung can occur, and the lungs can also be the primary site as in previous reports (1,2). Metastatic melanoma occurs when tumor cells dissociate from the primary lesion, migrate through the surrounding stroma, and invade blood vessels and lymphatics to form a tumor at a distant site (3). The incidence of MM has risen in the recent past, and the survival rate has been observed to be very low (4,5). Therefore, it becomes very important to find out the exact molecular mechanisms behind MM and the right diagnostic strategies for the detection of MM.
In recent years, microRNAs (miRNAs/miRs) have been reported in various cancers. miRNAs are endogenous, small non-coding RNAs containing 18–25 nucleotides (6). Usually, miRNAs mediate gene expression by targeting to the 3′UTR of mRNA to repress target gene expression (7). Increasing numbers of reports have indicated that miRNA is involved in various types of developments and progressions in carcinomas, including metastasis, differentiation and cell proliferation (8–10). The miRNAs function as oncogene or tumor suppressors in cancers through targeting different genes (11–13). Among these miRNAs, miR-146a has been reported to be involved in different kinds of tumors, including MM. However, the effect of miR-146a in mediating cell migration and invasion ability of MM cells remains largely elusive.
Mothers against decapentaplegic homolog 4 (SMAD4) was identified on human chromosome 18q21.1 and is a member of the Smad family. SMAD4 is a tumor suppressor and a key mediator of TGF-β signaling pathway (14,15). Previous studies indicated that loss of SMAD4 frequently occurred in multiple cancers, such as lung, colon, breast and gastric cancers (16–19). Moreover, SMAD4 plays critical roles in cell differentiation, migration, invasion and apoptosis in various cancers (14,19,20). Although the roles of SMAD4 in various cancers are important, the regulation effect of SMAD4 mediated by miRNA in MM is still unclear.
In this study, we observed that miR-146a was frequently upregulated in MM tissues compared with the adjacent normal tissues and the same result was found in MM cell lines. The abnormal expression of miR-146a could promote cell migration and invasion by targeting the 3′UTR of SMAD4. Additionally, the protein expression level of SMAD4 was decreased in MM tissues, and SMAD4 expression was negatively associated with miR-146a in MM. Moreover, SMAD4 expression could attenuate miR-146a-mediated promotion on MM cell migration and invasion. Therefore, our results illustrated that miR-146a functioned as an oncogene in MM and miR-146a-SMAD4 may be the novel therapeutic strategies in MM.
Materials and methods
Patients and tissues
We collected 55 pairs of human MM tissues and the matched normal tissues from patients with MM who underwent surgery at the Central Hospital of Zibo (Zibo, China) from 2014 to 2016. All the tissues were stored at −80°C. This study was approved by the Ethics Committee of the Central Hospital of Zibo, and all patients provided written informed consent.
Cell culture and transfection
Human melanoma cells A375, WM115, M14 and G361 were purchased from American Type Culture Collection (ATCC; Rockville, MD, USA). Human normal skin cell HACAT was obtained from Zibo Traditional Chinese Medicine Hospital. All cells were cultured in RPMI-1640 (Gibco-BRL, Karlsruhe, Germany) supplemented with 15% fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cultures were maintained at 37°C with 5% CO2.
miR-146a inhibitor or miR-146a mimic and the control were purchased from RiboBio (Guangzhou, China). Human melanoma cell lines A375 and G361 were transfected with miRNA inhibitor or mimics using Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.. 72 h after transfections, the cells were collected for further assays. The sequences for miR-146a mimics and inhibitor are as follows: miR-146a mimics 5′-UGAGAACUGAAUUCCAUGGGUU-3′; 5′-CCCAUGGAAUUCAGUUCUCAUU-3′; miR-146a inhibitor 5′-AACCCAUGGAAUUCAGUUCUCA-3′; the control is a kind of scrambled miR-control.
RNA extraction and RT-qPCR
The total RNA in MM cell lines or human samples was extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. cDNA was synthesized from 2 µg total RNA with random primers or miRNA-specific primers and MMLV reverse transcription (RT) (Fermentas; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). The expression of genes was detected by RT-quantitative polymerase chain reaction (RT-qPCR) with SYBR-Green kit (Takara Biotechnology Co., Ltd., Dalian, China) using Bio-Rad CFX96TM Real-Time PCR System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). U6 was used as internal controls for miRNA-146a quantification and β-actin was used as internal controls for mRNA quantification. All of the reactions were performed three times and the relative expression of mRNA was analysed using 2−ΔΔCq. Primers were designed for miR-146a (forward 5′-CATGGGTTGTGTCAGTGTCAGAGCT-3′3 and reverse 5′-TGCCTTCTGTCTCCAGTCTTCCAA-3′), SMAD4 (forward 5′-CGGACATTACTGGCCTGTTC-3′ and reverse 5′-TAGGGCAGCTTGAAGGAAACC-3′), U6 (forward 5′-CTCGCTTCGGCAGCACA-3′ and reverse 5′-AACGCTTCACGAATTTGCGT-3′) and GAPDH (forward 5′-CAGCCTCAAGATCATCAGCA-3′ and reverse 5′-TGTGGTCATGAGTCCTTCCA-3′). The conditions for PCR were as follows: 95°C for 5 min, 40 cycles of denaturation at 95°C (15 sec), 50°C (30 sec) and 72°C (30 sec).
Western blot analysis
Cells were lysed with RIPA lysis buffer supplemented with protease and phosphatase inhibitors (Roche Diagnostics, Basel, Switzerland). The concentration of protein was determined by Pierce® BCA Protein Assay kit (Thermo Fisher Scientific, Inc., Shanghai, China). The proteins were separated with 10% sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto the polyvinylidene difluoride (PVDF) membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked for 1 h with 5% bovine serum albumin in TBS at room temperature. Then we incubated the PVDF membranes with rabbit anti-SMAD4 polyclonal antibody (1:500; cat. no. ab110175; Abcam, Cambridge, UK,) and anti-GAPDH polyclonal antibody (1:1,200; cat. no. ab9485; Abcam) in TBST (0.1% Tween-20 in TBS) at 4°C overnight, followed by incubation with rabbit antibody for 2 h at room temperature. GAPDH was used as an endogenous reference for normalization. The proteins were detected using chemiluminescence method (ECL; EMD Millipore).
Transwell assay
Transwell assay was performed to detect the migration and invasion abilities of MM cells. Firstly, Transwell Assay was performed to detect the migration and invasion abilities of A375 and G361 transfected with miR-146a mimics or miR-146a inhibitor. At the same time, the migration and invasion abilities of A375 cells transfected with SMAD4 and miR-146a mimics were also detected by Transwell Assays. Melanoma cells A375 and G361 were seeded into 6-well plates with the different transfection treatments. 48 h after the transfection, the cells were harvested and re-suspended with DMEM culture medium with FBS. A total of 1×105 cells/ml was prepared in DMEM, 150 µl cell suspensions was added into the upper 24-well transwell chamber (EMD Millipore) with or without Matrigel (Clontech Laboratories, Inc., Mountainview, CA, USA). Then, we added 600 µl DMEM with 10% FBS to the lower chambers. After a 24 h incubation period at 37°C, the cotton swabs were used to wipe out the non-migratory or non-invasive cells on the upper chambers. The cells on the lower chambers were fixed with 100% methanol for 20 min, after that, the cells were stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min. The cells were counted under an inverted microscope (Olympus Corporation, Tokyo, Japan).
Luciferase assay
The 3′-UTR sequence of SMAD4 gene containing miR-146a binding sites was amplified by PCR. The 3′-UTR of SMAD4 was cloned into the pMIR-REPORT luciferase vector named pMIR-SMAD4-wild type. The mutation of the binding site (CAAGA) in 3′UTR of SMAD4 was amplified by PCR and cloned into the plasmid named pMIR-SMAD4-mutant. miR-146a mimics and luciferase plasmid pMIR-SMAD4 (wild-type/mutant, wt/mut) were transfected into A375 and G361 cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) on the basis of the manufacture's instruction. Being cultivated for 48 h, the cells were harvested and lysed. Luciferase activity in the cell lysates was measured by the Dual Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA). Firefly luciferase activity was normalized to renilla luciferase activity. The experiments were performed in triplicate.
Statistical analysis
Statistical analysis was performed using SPSS statistical software, version 17.0 (SPSS, Inc., Chicago, IL, USA) with Student's t-test or ANOVA and Scheffe post-hoc test. P<0.05 was considered to indicate a statistically significant difference. Correlation between mRNA and miRNA were estimated using the Spearman's correlation method.
Results
miR-146a expression was upregulated in melanoma and involved in the tumor progression
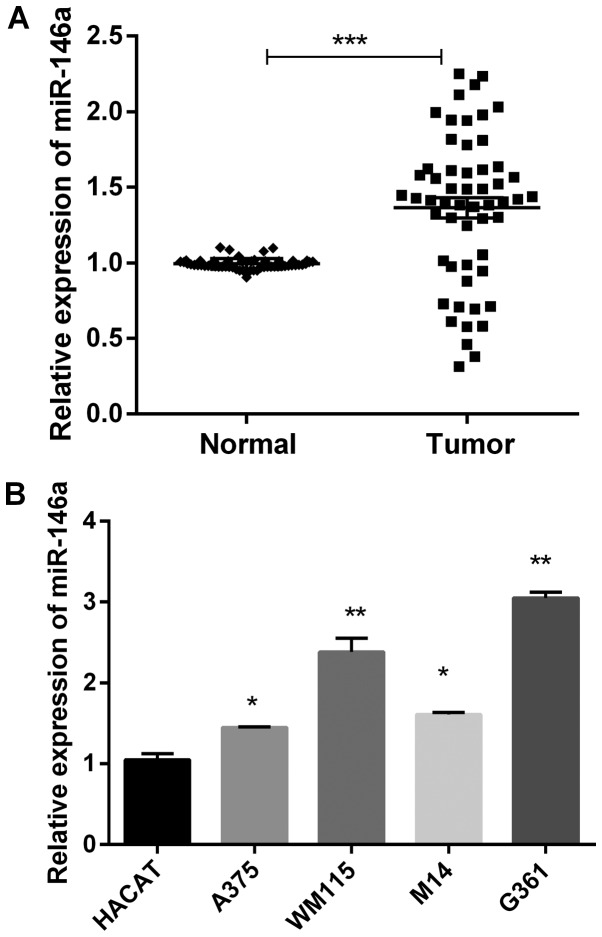
To explore the function of miR-146a in MM in vitro, we detected miR-146a expression in 55 paired of MM tissues and the adjacent normal tissues using RT-qPCR. The results demonstrated that miR-146a was significantly highly expressed in MM tissues compared to adjacent normal tissues (Fig. 1A). And then, we detected expression level of miR-146a in MM cell lines. As shown in Fig. 1B, miR-146a expression was remarkably high in four MM cells (A375, WM115, M14 and G361) compared with the human normal skin cells HACAT.
Figure 1.
miR-146a expression was upregulated in melanoma. (A) The mRNA expression of miR-146a in 55 paired of MM tissues and corresponding non-cancerous tissues. ***P<0.001. (B) The mRNA expression of miR-146a in MM cells (HACAT cell line was human normal skin cells). *P<0.05 and **P<0.01 vs. HACAT group. MM, malignant melanoma; miR, microRNA.
Additionally, the relationship between miR-146a expression and clinicopathological characteristics in those patients was analyzed. The increased levels of miR-146a were notably related to TNM stage and lymph node metastasis of MM (Table I). To investigate the potential associations between miR-146a expression and patients' clinicopathological variables, we divided the patients with melanoma into two groups based on mean value (1.323) of miR-146a expression: High expression group (>1.323, n=40), and low expression group (<1.323, n=15). Those results indicated that miR-146a was related to the progression of MM.
Table I.
Clinicopathological features and miR-146a expression in patients with malignant melanoma.
| miR-146a expression | ||||
|---|---|---|---|---|
| Clinicopathological features | Cases (n=55) | High (n=40) | Low (n=15) | P-value |
| Gender | 0.2048 | |||
| Male | 26 | 21 | 5 | |
| Female | 29 | 19 | 10 | |
| Age (years) | 0.8257 | |||
| >60 | 27 | 20 | 7 | |
| ≤60 | 28 | 20 | 8 | |
| Tumor thickness (mm) | 0.2164 | |||
| ≥1.0 | 33 | 22 | 11 | |
| <1.0 | 22 | 18 | 4 | |
| TNM stage | 0.0039 | |||
| I + II | 19 | 10 | 9 | |
| III | 36 | 30 | 6 | |
| Lymph node metastasis | 0.0020 | |||
| Yes | 33 | 29 | 4 | |
| No | 22 | 11 | 11 | |
TNM, tumor-node-metastasis.
Ectopic expression of miR-146a could promote MM cell migration and invasion
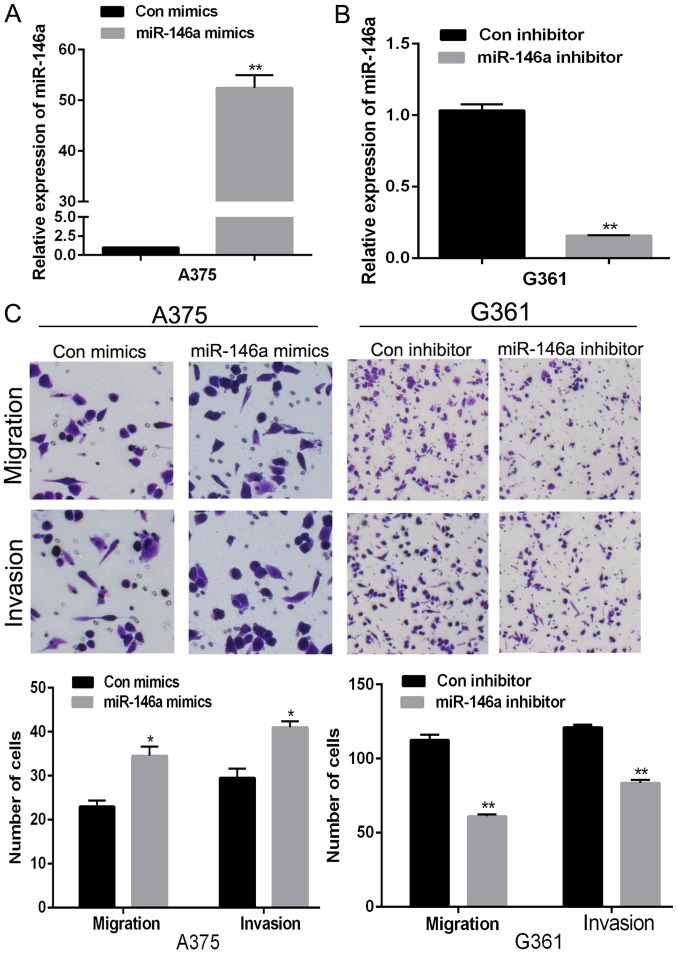
Clinicopathological analysis indicated that miR-146a was related to the progression of MM. Among all the selected MM cell lines, the expression of miR-146a in A375 was the lowest while G361 has the highest miR-146a expression. To better and full understand the roles of miR-146a and SMAD4, we selected A375 and G361 for further assays. In addition, as the expression of miR-146a in A375 cells was low and in G361 was high, we transfected miR-146a mimics into A375 and miR-146a inhibitor into G361 to further understand the biological roles of miR-146a in MM cells. The transfection efficiency was detected using RT-qPCR (Fig. 2A and B). And then, we detected the migration and invasion abilities by transwell assays, the results showed that overexpression of miR-146a dramatically raised A375 cells migration and invasion (Fig. 2C). Knockdown of miR-146a by miR-146a inhibitor decreased cell migration and invasion in G361 (Fig. 2C). Those data suggested that miR-146a could promote cell migration and invasion of MM cells.
Figure 2.
miR-146a promoted malignant melanoma cell migration and invasion. (A) The relative expression of miR-146a in A375 cell transfected miR-146a mimics or control. (B) The relative expression of miR-146a in G361 cell transfected with miR-146a inhibitor or control. (C) Transwell assay was performed to measure the migration and invasion ability in A375 and G361 cells transfected with miR-146a mimics or inhibitor. *P<0.05 and **P<0.01 vs. the control. miR, microRNA; con, control.
miR-146a downregulated the expression of SMAD4 through directly targeting the 3′UTR of SMAD4
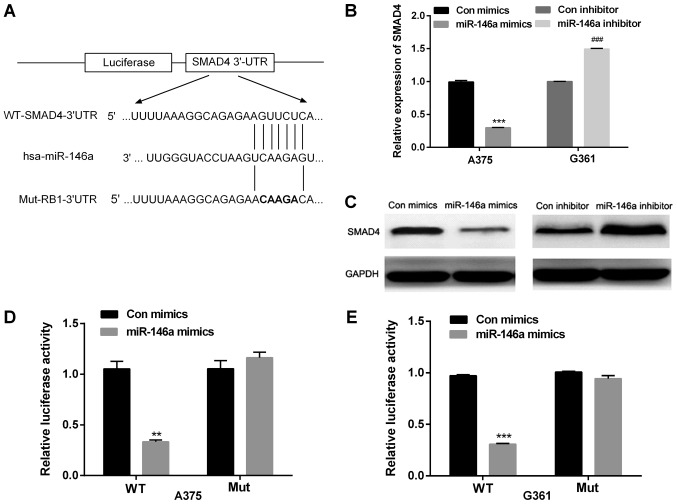
To explore the underlying effects of miR-146a on cell migration and invasion in MM, Target Scan and miRanda were used to analyze the predicted targets of miR-146a. We found that one target gene for miR-146a was SMAD4, one putative binding site was found at 390–397 and this is highly conserved across species. As shown in Fig. 3A, the wild type or mutant 3′UTR of SMAD4 was cloned into a luciferase vector. To further confirm that whether SMAD4 was a target of miR-146a, mimics or inhibitor of miR-146a were transfected in A375 cells, and the effect of miR-146a on SMAD4 was observed through detecting the mRNA and protein levels of SMAD4. The findings indicated that miR-146a mimics downregulated SMAD4 mRNA and protein levels in A375 cells', whereas miR-146a inhibitor increased the mRNA and protein levels of SMAD4 in G361 cells (Fig. 3B and C).
Figure 3.
miR-146a decreased SMAD4 expression by directly targeting the 3′UTR of SMAD4. (A) The putative binding site of miR-146a in the SMAD4 3′UTR region. The WT or Mut sequence was cloned into luciferase reporter vector. (B and C) Reverse transcription-quantitative polymerase chain reaction and western blot analysis were used to measure mRNA and protein level of SMAD4 in A375 cells transfected with miR-146a mimics and G361 cells transfected with miR-146a inhibitor. (D and E) MM cells were co-transfected with miR-146a mimics and luciferase reporter plasmid containing WT or Mut SMAD4 3′UTR. **P<0.01 and ***P<0.001 vs. Con mimics; ###P<0.001 vs. Con inhibitor. WT, wild type; Mut, mutant; con, control; miR, microRNA; SMAD4, Mothers against decapentaplegic homolog 4; UTR, untranslated region.
Additionally, the luciferase reporter assay was applied to confirm the predications. We constructed the wild type (WT) 3′UTR and the mutated type (Mut) 3′UTR of SMAD4 luciferase report vectors. A375 and G361 cells were co-transfected with miR-146a mimic and WT or Mut luciferase reporter vector. The relative luciferase activity was significantly reduced in MM cells co-transfected with miR-146a mimics and WT luciferase vector (Fig. 3D). The activity of Mut luciferase reporter activity was not affected by miR-146a mimics in A375 and G361 cell lines. Therefore, miR-146a directly binds to 3′UTR of SMAD4 of MM cells (Fig. 3D).
The expression of SMAD4 was decreased and negatively associated with miR-146a in MM tissues
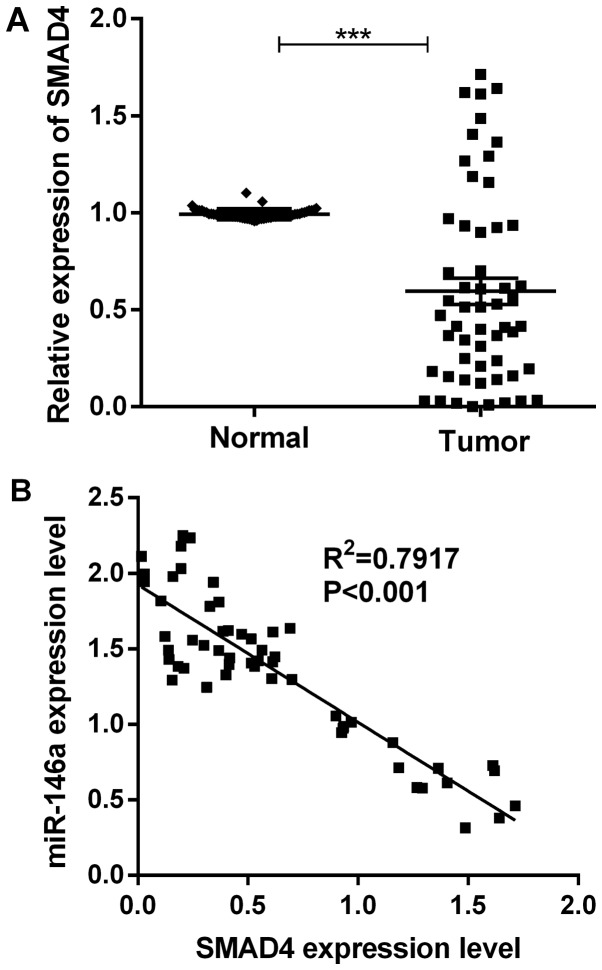
To further investigate the relationship between SMAD4 and miR-146a in MM tissues, we looked into the SMAD4 expression level of 55 paired of MM tissues and adjacent normal tissues using RT-qPCR. The results showed that SMAD4 was remarkably downregulated in MM tissues (Fig. 4A). Interestingly, statistical analysis showed that miR-146a expression was negatively associated with SMAD4 mRNA expression in MM tissues (Fig. 4B).
Figure 4.
SMAD4 was upregulated in MM tissues. (A) The mRNA expression of SMAD4 in 55 paired of MM tissues and corresponding non-cancerous tissues. (B) Statistical analysis of the correlation between miR-146a and SMAD4 in MM tissues. ***P<0.001. SMAD4, Mothers against decapentaplegic homolog 4; MM, malignant melanoma; miR, microRNA.
Restoration of SMAD4 expression abrogated miR-146a-mediated cell migration and invasion
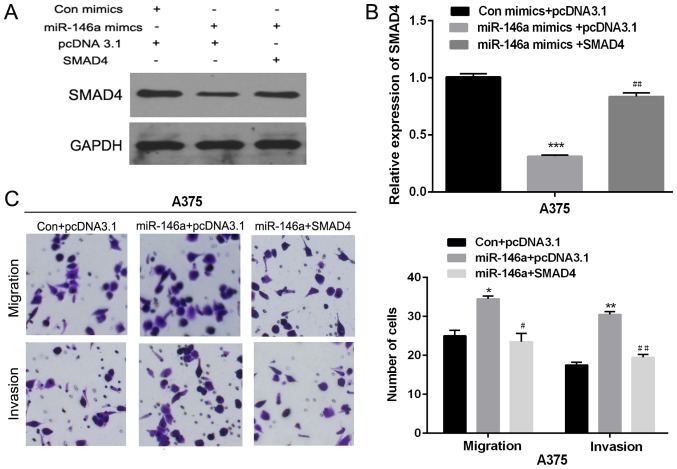
Previous studies indicated that SMAD4 was associated with cell migration and invasion. Next, we investigated the significance of SMAD4 in miR-146a mediated cell migration and invasion. Overexpression vector of SMAD4 and miR-146a mimic were co-transfected into A375 cells. RT-qPCR and western blot were used to detect the SMAD4 expression (Fig. 5A and B). And then, transwell assay was used to detect the migration and invasion abilities of MM cells, after the co-transfection with SMAD4 and miR-146a mimics. As shown in Fig. 5C, the migration and invasion abilities of A375 cells co-transfected with miR-146a mimics and SMAD4 were remarkably lower than that only transfected with miR-146a mimics. Therefore, those results suggested that ectopic expression of SMAD4 partial attenuated miR-146a mediated promotion on cell migration and invasion.
Figure 5.
Restoration of SMAD4 expression abrogated miR-146a-mediated cell migration and invasion. (A and B) The SMAD4 mRNA or protein expression level was detected using western blotting or reverse transcription-quantitative polymerase chain reaction in A375 cells co-transfected with SMAD4 overexpression vector and miR-146a mimics. (C) Transwell assay was performed to detect migration and invasion ability in A375 cells co-transfected with SMAD4 overexpression vector and miR-146a mimics. *P<0.05, **P<0.01 and ***P<0.001 vs. Con+pcDNA3.1 group; #P<0.05 and ##P<0.01 vs. miR-146a mimics+pcDNA3.1 group. SMAD4, Mothers against decapentaplegic homolog 4; miR, microRNA; Con, control.
Discussion
Recently, large numbers of studies indicated that miRNAs play critical roles in various cancers and participated in many biological processes (21,22). Due to the important roles of miRNAs in the progression and development of cancers, more and more researchers are starting to pay attention to miRNAs (23). Accumulating evidence suggested that miR-146a acted as a tumor suppressor or an oncogene in multiple cancers. For example, miR-146a had a low expression in HCC tissues (24), breast cancer (25). On the contrary, miR-146a was found to be elevated in colorectal cancer (26), oral carcinoma (27). However, the biological function of miR-146a in MM remains unknown. In present report, we firstly measured miR-146a expression in 55 paired MM tissues and the corresponding non-cancerous tissues. We found that miR-146a was significantly upregulated in MM tissues. Moreover, we also detected the expression level of miR-146a in MM cells. The results showed that miR-146a was increased in MM cells compared to the normal cell lines.
Previous studies demonstrated that miR-146a was involved in the development and metastasis in cancers (26,28). Here, clinicopathological analysis suggested that the increased levels of miR-146a were notably related with TNM stage and lymph node metastasis in patients with MM. Subsequently, we studied the roles of miR-146a on tumor migration and invasion. The result showed that miR-146a abnormal expression increased cell migration and invasion in MM cell lines, and the deletion of miR-146a by transfecting cells with a specific miR-146a inhibitor, decreased cell migration and invasion. Moreover, we identified SMAD4 as a novel target of miR-146a in MM cell lines. SMAD4 belongs to the SMAD family, functioning as a tumor suppressor and playing important roles in the progress (14,15). Zhang et al (28) indicated that SMAD4 inhibited cell migration through suppressing the activity of JNK in human pancreatic carcinoma cells. Yan et al (17) reported that SMAD4 loss was associated with poor survival in colon cancer. In the current study, we also measured the protein levels of SMAD4 in MM tissues. The results showed that SMAD4 was downregulated in MM tissues compared with non-cancerous tissues. Interestingly, statistical analysis showed that miR-146a was correlated with the SMAD4 expression in MM tissues. To further understand the functions of miR-146a and SMAD4 in MM, we performed rescue assay. The results suggested that abnormal expression of SMAD4 could attenuate the miR-146a-mediated promotion on cell migration and invasion in MM. Although there were several studies about the correlation of miR-146a and SMAD4 (29), there was no relevant study about the correlation of them in MM. The focus of our work was on the correlation of miR-146a and SMAD4 in MM, which was very different from previous studies, and this was also the major contribution of our study.
In conclusion, our study demonstrated that miR-146a was frequently over expressed and SMAD4 was downregulated in MM tissues and miR-146a has a negatively association with SMAD4 in MM tissues. Ectopic expression of miR-146a could promote MM cell migration and invasion. miR-146a negatively regulates SMAD4 expression by targeting the 3′UTR of SMAD4. Moreover, the over-expression of SMAD4 could abrogate miR-146a-mediated cell migration and invasion in MM cells. Therefore, the results suggested that miR-146a functioned as an oncogene by directly targeting SMAD4 and it may be a novel diagnostic and therapeutic marker in MM.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- UTR
untranslated region
- MM
malignant melanoma
- FBS
fetal bovine serum
- TNM
tumor-node-metastasis
- WT
wild type
- M
mutated type
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
WP conceived and designed the study. YS and QS collected, interpreted and analyzed the data. YS contributed significantly to the preparation of the manuscript. QS wrote the manuscript. WP and XY performed the analysis with constructive discussions. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Central Hospital of Zibo (Zibo, China), and all patients provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 2.Brose MS, Volpe P, Feldman M, Kumar M, Rishi I, Gerrero R, Einhorn E, Herlyn M, Minna J, Nicholson A, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62:6997–7000. [PubMed] [Google Scholar]
- 3.Haass NK, Smalley KS, Li L, Herlyn M. Adhesion, migration and communication in melanocytes and melanoma. Pigment Cell Res. 2005;18:150–159. doi: 10.1111/j.1600-0749.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 4.Baroudjian B, Pagès C, Lebbè C. Melanoma, from diagnosis to treatment. Rev Infirm. 2016:16–18. doi: 10.1016/j.revinf.2015.12.022. (In French) [DOI] [PubMed] [Google Scholar]
- 5.Barth A, Wanek LA, Morton DL. Prognostic factors in 1,521 melanoma patients with distant metastases. J Am Coll Surg. 1995;181:193–201. [PubMed] [Google Scholar]
- 6.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Ebert MS, Sharp PA. MicroRNA sponges: Progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Fang W, Fan Y, Fa Z, Xu J, Yu H, Li P, Gu J. microRNA-625 inhibits tumorigenicity by suppressing proliferation, migration and invasion in malignant melanoma. Oncotarget. 2017;8:13253–13263. doi: 10.18632/oncotarget.14710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, Sultmann H, Lyko F. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67:1419–1423. doi: 10.1158/0008-5472.CAN-06-4074. [DOI] [PubMed] [Google Scholar]
- 13.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 14.Miyaki M, Kuroki T. Role of Smad4 (DPC4) inactivation in human cancer. Biochem Biophys Res Commun. 2003;306:799–804. doi: 10.1016/S0006-291X(03)01066-0. [DOI] [PubMed] [Google Scholar]
- 15.Yang G, Yang X. Smad4-mediated TGF-beta signaling in tumorigenesis. Int J Biol Sci. 2010;6:1–8. doi: 10.7150/ijbs.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W, Liu Z, Huang JA. MicroRNA-205 targets SMAD4 in non-small cell lung cancer and promotes lung cancer cell growth in vitro and in vivo. Oncotarget. 2016;8:30817–30829. doi: 10.18632/oncotarget.10339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yan P, Klingbiel D, Saridaki Z, Ceppa P, Curto M, McKee TA, Roth A, Tejpar S, Delorenzi M, Bosman FT, Fiocca R. Reduced expression of SMAD4 is associated with poor survival in colon cancer. Clin Cancer Res. 2016;22:3037–3047. doi: 10.1158/1078-0432.CCR-15-0939. [DOI] [PubMed] [Google Scholar]
- 18.Wang LH, Kim SH, Lee JH, Choi YL, Kim YC, Park TS, Hong YC, Wu CF, Shin YK. Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res. 2007;13:102–110. doi: 10.1158/1078-0432.CCR-06-1467. [DOI] [PubMed] [Google Scholar]
- 19.Stuelten CH, Buck MB, Dippon J, Roberts AB, Fritz P, Knabbe C. Smad4-expression is decreased in breast cancer tissues: A retrospective study. BMC Cancer. 2006;6:25. doi: 10.1186/1471-2407-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, Perry SR, Labrot ES, Wu X, Lis R, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–273. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng X, Zhao Y, Wang J, Gao Z, Geng Q, Liu X. Regulatory roles of miRNA-758 and matrix extracellular phosphoglycoprotein in cervical cancer. Exp Ther Med. 2017;14:2789–2794. doi: 10.3892/etm.2017.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei YT, Guo DW, Hou XZ, Jiang DQ. miRNA-223 suppresses FOXO1 and functions as a potential tumor marker in breast cancer. Cell Mol Biol (Noisy-le-grand) 2017;63:113–118. doi: 10.14715/cmb/2017.63.5.21. [DOI] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Zu Y, Yang Y, Zhu J, Bo X, Hou S, Zhang B, Qiu J, Zheng J. miR-146a suppresses hepatocellular carcinoma by downregulating TRAF6. Am J Cancer Res. 2016;6:2502–2513. [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Wang W, Yang X, Zhao D, Li F, Wang H. MicroRNA-146a inhibits cell migration and invasion by targeting RhoA in breast cancer. Oncol Rep. 2016;36:189–196. doi: 10.3892/or.2016.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu D, Yao Q, Zhan C, Le-Meng Z, Liu H, Cai Y, Tu C, Li X, Zou Y, Zhang S. MicroRNA-146a promote cell migration and invasion in human colorectal cancer via carboxypeptidase M/src-FAK pathway. Oncotarget. 2017;8:22674–22684. doi: 10.18632/oncotarget.15158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hung PS, Liu CJ, Chou CS, Kao SY, Yang CC, Chang KW, Chiu TH, Lin SC. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS One. 2013;8:e79926. doi: 10.1371/journal.pone.0079926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Cao J, Pei Y, Zhang J, Wang Q. Smad4 inhibits cell migration via suppression of JNK activity in human pancreatic carcinoma PANC-1 cells. Oncol Lett. 2016;11:3465–3470. doi: 10.3892/ol.2016.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao B, Zhu ED, Li N, Lu DS, Li W, Li BS, Zhao YL, Mao XH, Guo G, Yu PW, Zou QM. Increased miR-146a in gastric cancer directly targets SMAD4 and is involved in modulating cell proliferation and apoptosis. Oncol Rep. 2012;27:559–566. doi: 10.3892/or.2011.1514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.