Abstract
Different immunohistochemical algorithms for the classification of the activated B-cell (ABC) and germinal center B-cell (GCB) subtypes of diffuse large B-cell lymphoma (DLBCL) are applied in different laboratories. In the present study, 127 patients with DLCBL were investigated, all treated with rituximab and cyclophosphamide, hydroxydaunorubicin, oncovin and prednisone (CHOP) or CHOP-like regimens between April 2004 and December 2010. Multi-tumor tissue microarrays were prepared and were tested according to 4 algorithms: Hans; modified Hans; Choi; and modified Choi. For 39 patients, the flow cytometric quantification of CD19 and CD20 antigen expression was performed and the level of expression presented as molecules of equivalent soluble fluorochrome units. The Choi algorithm was demonstrated to be prognostic for OS and classified patients into the GCB subgroup with an HR of 0.91. No difference in the expression of the CD19 antigen between the ABC and GCB groups was observed, but the ABC subtype exhibited a decreased expression of the CD20 antigen compared with the GCB subtype.
Keywords: diffuse large B-cell lymphoma, Choi algorithm, cluster of differentiation 19 expression
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common mature B-cell lymphoma and accounts for 38–50% of incident lymphomas each year (1). The prognosis of patients with DLBCL was predominately estimated according to the International Prognostic Index (IPI) (2), which was proposed prior to the rituximab era of treatment. The IPI was also confirmed as a prognostic tool for predicting overall survival (OS), progression-free survival (PFS) and event-free survival (EFS) (3) following the introduction of rituximab, even though certain studies have claimed that it has lost its value in the rituximab era (4) and requires modifications (5). At present, the IPI divides patients into four groups (low-risk group, low-intermediate risk group, high-intermediate risk group and high-risk group), with different survival rates (1).
Subsequent to investigating the cell-of-origin, Hans et al (6) published an algorithm based on immunohistochemistry, classifying DLBCL by the cell-of-origin into germinal center B-cell (GCB) and non-GCB activated B-cell (ABC and type III) subtypes (6). When comparing the immunohistochemically-determined non-GCB and GCB subtypes with the results of gene expression profiling (6,7), the positive predictive value of this algorithm was 87% for the GCB group and 73% for the non-GCB group, and the concordance with the gene expression profile (GEP) was 86% (6). Following this, Choi et al (8) developed an additional algorithm with a higher accuracy, as 93% of algorithm predictions matched their GEP. Meyer et al (9) published a study in 2011 comparing several established algorithms, and also constructed 2 novel ones; the modified Hans and the modified Choi algorithms. The Choi algorithm exhibited an 87% concordance with GEP, the Hans algorithm exhibited an 86% concordance and the modified Hans and modified Choi algorithm each exhibited an 87% concordance with GEP. The 2 modified algorithms omitted the B-cell lymphoma 6 protein (Bcl-6) antigen determination, yet retained their concordance with GEP (9). The 2016 revision of the WHO classification of lymphoid neoplasms recommends the Hans algorithm for the classification of GCB and ABC subtypes, but also allows the application of other algorithms (10).
The ABC subtype is definitely associated with inferior survival, as demonstrated by Hans, Choi, Meyer and Alizadeh (6–9). Previously, numerous studies have addressed this subject, aiming to identify a more aggressive front-line treatment for the ABC subtype (11). At present, the majority of patients are treated with the standard rituximab and cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP) therapies (1). However, novel agents including bortezomib (12), lenalidomide (13) and ibrutinib (14) are being widely investigated in the treatment of the ABC subtype of DLBCL.
The aim of the present study was to assess the accuracy of each of the 4 most commonly-used and relatively easy-to-perform algorithms in classifying patients into the ABC and GCB subgroups, and to also evaluate which of the algorithms more proficiently stratified patients into the GCB subgroup according to the OS. The expression of cluster of differentiation (CD)19 and CD20 antigens on the ABC and GCB subtypes was also evaluated.
Patients and methods
Patients
A total of 127 patients with de novo DLBCL were included in the present study from April 2004 to December 2010. All were >18 years of age, HIV-negative and had a histological tissue sample removed for accurate histological diagnosis prior to any treatment either with surgical resection of involved tissue (16 patients) or lymph node biopsy (111 patients). They were treated with R-CHOP (intravenous cyclophosphamide 750 mg/m2, doxorubicin 50 mg/m2, vincristine 1.4 mg/m2; maximum dose 2 mg; rituximab 375 mg/m2 on day 1 and oral prednisolone 40 mg/m2 on days 1–5, administered every 21 days) or CHOP-like regimens (etoposide 100 mg/m2 instead of doxorubicin) for 2–10 treatment cycles between April 2004 and December 2010 at the Institute of Oncology (Ljubljana, Slovenia). Detailed data of their age at diagnosis, stage of the disease, number of treatment cycles and consolidation with radiotherapy was obtained from the records of the patients. The IPI was also determined. Ki-67 was determined on the available samples (96 patients, as other samples were too damaged for accurate analyses), and subgroups were created for each 10% measurement. For each patient, the response to first-line treatment was defined as complete remission (CR), partial remission (PR), stable disease (SD) or progressive disease (PD) based on the revised criteria of Cheson et al (15), and the PFS and OS were calculated. The PFS was defined as the period from the end of first-line treatment (either chemotherapy or consolidation radiotherapy) until verified progression or mortality by any cause for patients achieving CR or PR. The OS was defined as the period from the first day of treatment until mortality by any cause for all patients. Data on the cause and dates of mortality was obtained from the Cancer Registry of the Republic of Slovenia on 15 July, 2016, so that each patient had a minimum of 5.5 years observation time.
All patients signed an informed consent form to participate in the study, and the study was conducted in accordance with the Declaration of Helsinki. The present study was approved by the Republic of Slovenia National Medical Ethics Committee.
Immunohistochemistry
For the tissue microarray (TMA), hematoxylin and eosin-stained [H&E; stained in Leica automatic Multistainer ST5020 (Leica Microsystems, Inc., Buffalo Grove, IL, USA) by use of Mayer's hematoxylin (Merck & Co, Inc., Whitehouse Station, NJ, USA) for 10 min, Scott's solution (Merck & Co, Inc.) for 1 min and eosin-floxine (Merck & Co, Inc.) for 3 min at room temperature] sections from each paraffin-embedded, formalin-fixed block were used to define the diagnostic areas. A total of two representative 2 mm cores were obtained from each sample and inserted into a recipient block using a manual tissue arrayer (Beecher Instruments Inc., Silver Springs MD, USA. A 3–4 µm thick section was cut from each TMA and stained by H&E, as aforementioned. They were also subject to antigen retrieval and antibody staining. The immunoperoxidase stains were performed on either a Benchmark XT (Ventana Medical Systems, Inc., Tucson, AZ, USA) using a Cell conditioning solution for antigen unmasking (CC1; Ventana Medical Systems, Inc.) and Ultraview universal diaminobenzidine detection kits (Ventana Medical Systems, Inc.) or an Labvision 720 autostainer (Thermo Fisher Scientific, Inc., Waltham, MA, USA) using the Envision Flex High pH visualisation system (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Antibodies used in the present study were as follows: B-cell lymphoma 2 (Bcl2; clone 124; host, mouse; Dako, Agilent Technologies, Inc.; cat no. M0887; antigen retrieval using Flex; 1:40; incubated for 30 min at room temperature), Bcl6 (clone, GI 191E/A8; host, mouse; Cell Marque, Sigma-Aldrich, Merck KGaA, Darmstadt, Germany; cat no. 227M; antigen retrieval using CC1; 1:200; incubation for 60 min at 37°C), CD5 (clone, 4C7; host, mouse; Novocastra, Leica Microsystems, Inc.; cat no. CD5-4C7-L-CE; antigen retrieval using CC1; 1:400; incubation for 60 min at 37°C), CD10 (clone, 56C6; host, mouse; Novocastra, Leica Microsystems, Inc.; cat no. CD10-270-L-CE; antigen retrieval using CC1; 1:20; incubation for 60 min at 37°C), CD20 (clone, L26; host, mouse; Dako, Agilent Technologies, Inc.; cat no. M0755; antigen retrieval using Flex; 1:50; incubation for 30 min at room temperature), proliferation marker protein Ki-67 (Ki67; clone, MIB1; host, mouse; Dako, Agilent Technologies, Inc.; cat no. M7240; antigen retrieval with CC1; 1:200; incubation for 60 min at 37°C), multiple myeloma oncogene 1 (MUM1; clone, MUM1p; host, mouse; Dako, Agilent Technologies, Inc.; cat no. M7259; antigen retrieval using Flex; 1:100; incubation for 30 min at room temperature), Forkhead box protein P1 (FOXp1; clone, SP133; host, rabbit; Cell Marque, Sigma-Aldrich, Merck KGaA; cat no. 350R; antigen retrieval with CC1; 1:200; incubation for 60 min at 37°C) and serpin A9 (GCET1; clone, RAM 341; host, mouse; Abcam, Cambridge, UK; cat no. ab68889; antigen retrieval with CC1; incubation for 60 min at 37°C).
Antigens retrieved using CC1 (Bcl6, CD5, CD10, Ki-67 and GCET1) were treated for 88 min at 98°C and FOXp1 was treated for 56 min at 98°C, then blocked using an OptiView Peroxidase Inhibitor (Ventana Medical System, Inc.; 3%) at 37°C for 4 min. Antigens retrieved using the Envision Flex high pH Retrieval solution were treated for 10 min at 100°C in a microwave and blocked using Peroxidase-Blocking Solution (Dako; Agilent Technologies, Inc.) at room temperature for 8 min. All of the assays were validated using proper negative and positive controls by use of mutitumor tissue blocks consisting of tonsil, appendix, liver, melanoma, mantle cell lymphoma and classical Hodgkin's lymphoma tissue. Results were evaluated using a light microscope (Olympus BX51) at a ×200 magnification on at least randomly selected five fields. For each case, the core with a higher percentage of stained tumor cells was used for the analysis. Scoring of the antibodies was estimated visually in 10% increments. The intensity of staining was also evaluated but was not used to determine positivity as the variability in tissue fixation and processing appeared to affect the intensity of staining. GCET1 and FOXp1 were considered positive if ≥80% of the tumor cells were stained. The MUM1 was considered positive if ≥30% of the tumor cells were positive for the Hans and modified Hans algorithms and ≥80% for the Choi and modified Choi algorithms. The positive cut-off for all other antibodies was considered to be 30% (8). The Ki-67 proliferative index was evaluated as a percentage value calculated by scoring 500 tumor cell nuclei. The TMA evaluations and the classification of patients according to the algorithms were all performed by a hematopathologist (Gorana Gašljević, MD, PhD, Institute of Oncology, Ljubljana, Slovenia) who was blinded to all clinical data.
Cytological samples and flow cytometric quantification of CD19 and CD20 expression
For 39 patients, the cytological sample obtained via fine needle aspiration prior to the first therapy was available. A total of 1 skin lesion sample, 1 liver lesion sample, 1 nasal tumor sample and 36 lymph node samples were obtained for the flow cytometric immunophenotyping (FCI) analyses. The preparation of samples for FCI was performed according to the in-house protocol, previously described by Prevodnik et al (16). The FACSCalibur flow-cytometer and FACS Canto II flow-cytometer were used (BD Biosciences) and the CellQuest program version 5.1 (BD Biosciences) was applied for the analyses. SPHERO Rainbow calibration beads (Spherotech, Inc., Illinois, USA) were used for the quantification of the expression of CD19 and CD20. The beads have been routinely used in our laboratory for monitoring the stability and linear performance of the cytometer since 2001 (16). As these beads and the cytological samples included in the present study were tested with equal flow-cytometric settings, the beads were available for use for the quantification of CD19 and CD20 expression. The level of CD19 and C20 expression was determined with molecules of equivalent soluble fluorochrome using PMT Linearity QC Record software version RCP-30-5a (Spherotech, Inc.) (17).
Statistical analysis
The median age, number of treatment cycles, stage at the time of diagnosis and proliferation marker protein Ki-67 (Ki-67) index were determined. The PFS and OS were estimated using the Kaplan-Meier method. To compare the response to first-line treatment, a χ2 test was performed comparing two groups; CR and PR vs. SD and PD, with the exception of Ki-67, which was analyzed using logistic regression. Regression models were used to compare PFS and OS by testing one algorithm individually. The present study focused on the subgroup of patients who were differently classified according to the 4 algorithms; this subgroup was referred to as ‘heterogeneous’ (those who were classified as GCB by one and as ABC by other algorithms, and vice versa) and it was observed which of the 4 algorithms was best in identifying the patients in this subgroup that actually have the same OS as the subgroup unanimously classified as the GCB by all the four algorithms. For example, the algorithm that will have the HR of this subgroup as close to 1 as possible. The Mann-Whitney test was used for comparing the numeric measurements of CD19 and CD20 expression. P<0.05 was considered to indicate a statistically significant difference. R Statistical Software (version 3.2.2., R Foundation for Statistical computing, Vienna, Austria) and the GraphPad Prism program (version 3.02, GraphPad Software, Inc., La Jolla, CA, USA) were used for the analyses.
Results
Whole cohort analysis
The patient characteristics are summarized in Table I. For 5 patients, the response to first-line treatment was not able to be unequivocally determined. Relapse was documented in 41 patients (32%) during the course of follow-up. A total of 52 patients (41%) succumbed: 11 from a non-lymphoma-associated condition (heart failure, myocardial infarction, road accident, different infections and sepsis) and 3 of unknown causes.
Table I.
Patient characteristics.
| Patient characteristics | N |
|---|---|
| Age, median (range) | 62 (24–84) |
| Sex, n (%) | |
| Male | 65 (51) |
| Female | 62 (49) |
| Stage, n (%) | |
| I | 20 (16) |
| II | 29 (23) |
| III | 34 (27) |
| IV | 44 (35) |
| Disease presentation, n (%) | |
| B | 39 (31) |
| X | 29 (23) |
| S | 26 (20) |
| Treatment cycles, median (range) | 8 (2–10)a |
| Consolidation radiotherapy | 57 (45) |
| Treatment response, n (%) | |
| CR | 103 (81) |
| PR | 8 (6) |
| SD | 1 (1) |
| PD | 10 (8) |
| IPI Group, n (%) | |
| Low | 37 (29) |
| Low intermediate | 36 (28) |
| High intermediate | 30 (24) |
| Highb | 24 (19) |
| Ki-67 expression, median % (range) | 80 (50–95) |
one patient received 10 cycles: 2 cycles of cyclophosphamide, vincristine, prednisone, and 8 cycles of rituximab and cyclophosphamide, hydroxydaunorubicin, vincristine and prednisone following final staging
based on IPI score. B, constitutional symptoms; X, bulky disease; S, spleen involvement; CR, complete remission; PR, partial remission; SD, stable disease; PD, progressive disease; Ki-67, proliferation marker protein Ki-67; IPI, International Prognostic Index.
The median PFS was not reached during the course of the present study [95% confidence interval (CI), 109-not yet reached) and the median OS was 133 months, 95% CI, 111-not yet reached (Figs. 1 and 2). The median observation period was 6 years and 11 months.
Figure 1.
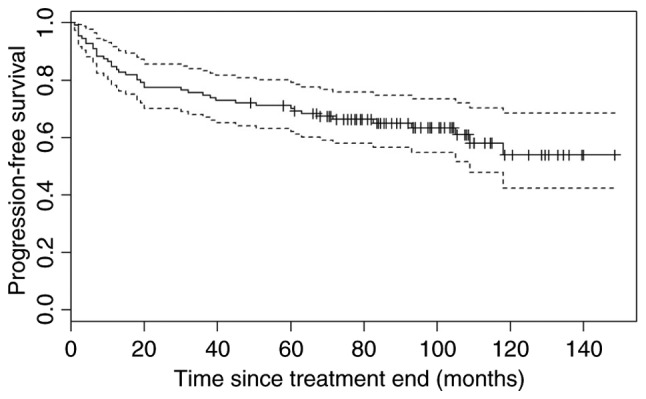
Progression-free survival for all patients achieving partial response and complete response (n=111). Dotted lines indicate the upper and lower confidence intervals.
Figure 2.
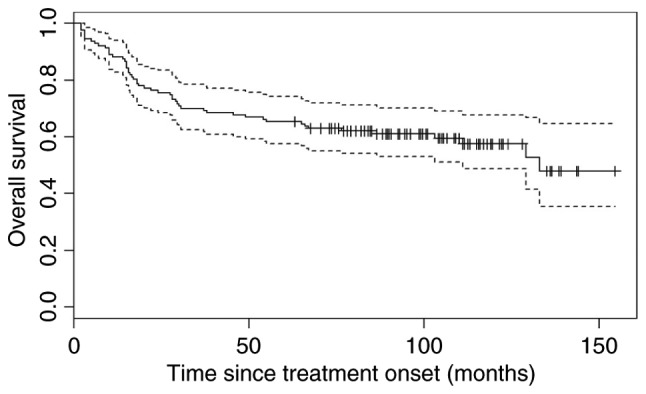
Overall survival for all patients (n=127). Dotted lines indicate the upper and lower confidence interval.
Due to relatively small number of cases exhibiting a bad treatment response (SD or PD), the power of any algorithm to distinguish between cases, was very low. Original immunohistochemistry data and flow cytometric data are presented in Table II.
Table II.
Original immunohistochemical and flow cytometric data.
| Sample no. | Hans algorithm | Modified Hans algorithm | Choi algorithm | Modified Choi algorithm | CD20 | CD19 |
|---|---|---|---|---|---|---|
| 1 | ABC | GCB | ABC | ABC | ||
| 2 | GCB | GCB | ABC | ABC | ||
| 3 | ABC | ABC | ABC | ABC | ||
| 4 | ABC | ABC | ABC | ABC | ||
| 5 | GCB | GCB | GCB | GCB | ||
| 6 | GCB | GCB | GCB | GCB | 183927 | 19560 |
| 7 | ABC | GCB | ABC | ABC | ||
| 8 | ABC | ABC | ABC | ABC | 49558 | 35198 |
| 9 | GCB | GCB | GCB | GCB | ||
| 10 | ABC | ABC | ABC | ABC | ||
| 11 | GCB | GCB | GCB | ABC | 68587 | 9570 |
| 12 | GCB | GCB | GCB | GCB | 129675 | 6525 |
| 13 | ABC | ABC | ABC | ABC | 75860 | 5571 |
| 14 | GCB | GCB | GCB | GCB | ||
| 15 | GCB | GCB | GCB | GCB | ||
| 16 | GCB | GCB | GCB | ABC | 169656 | 6498 |
| 17 | GCB | GCB | GCB | ABC | 30038 | 7991 |
| 18 | GCB | GCB | GCB | ABC | 35145 | 12937 |
| 19 | ABC | GCB | ABC | ABC | 74789 | 9174 |
| 20 | ABC | ABC | GCB | ABC | ||
| 21 | ABC | GCB | ABC | ABC | ||
| 22 | ABC | ABC | ABC | ABC | ||
| 23 | ABC | GCB | ABC | ABC | ||
| 24 | GCB | GCB | GCB | ABC | ||
| 25 | GCB | GCB | GCB | GCB | ||
| 26 | GCB | GCB | GCB | GCB | ||
| 27 | ABC | ABC | ABC | GCB | ||
| 28 | GCB | GCB | GCB | GCB | 150369 | 7713 |
| 29 | GCB | GCB | GCB | ABC | 11070 | 8838 |
| 30 | GCB | GCB | ABC | GCB | 173921 | 34158 |
| 31 | ABC | ABC | ABC | GCB | ||
| 32 | GCB | GCB | GCB | GCB | 63821 | 42365 |
| 33 | GCB | GCB | GCB | ABC | 149328 | 50691 |
| 34 | ABC | GCB | ABC | ABC | ||
| 35 | ABC | ABC | ABC | ABC | 4460 | 979 |
| 36 | GCB | GCB | GCB | ABC | ||
| 37 | GCB | GCB | GCB | GCB | 69712 | 6555 |
| 38 | ABC | ABC | ABC | ABC | ||
| 39 | ABC | GCB | ABC | ABC | ||
| 40 | ABC | ABC | ABC | GCB | ||
| 41 | GCB | GCB | GCB | GCB | 177781 | 14775 |
| 42 | ABC | ABC | ABC | ABC | ||
| 43 | ABC | GCB | ABC | ABC | ||
| 44 | ABC | ABC | ABC | ABC | ||
| 45 | GCB | GCB | GCB | GCB | ||
| 46 | ABC | ABC | ABC | ABC | 44264 | 3753 |
| 47 | ABC | ABC | ABC | ABC | 30091 | 2520 |
| 48 | GCB | GCB | GCB | GCB | 55441 | 6266 |
| 49 | ABC | ABC | ABC | ABC | ||
| 50 | GCB | GCB | GCB | GCB | 23355 | 2495 |
| 51 | GCB | GCB | GCB | GCB | ||
| 52 | ABC | ABC | ABC | ABC | ||
| 53 | GCB | GCB | GCB | GCB | ||
| 54 | ABC | ABC | ABC | GCB | ||
| 55 | GCB | GCB | GCB | GCB | ||
| 56 | ABC | ABC | ABC | ABC | ||
| 57 | GCB | GCB | GCB | GCB | ||
| 58 | GCB | GCB | GCB | GCB | 221029 | 20212 |
| 59 | GCB | GCB | GCB | ABC | ||
| 60 | GCB | GCB | ABC | ABC | ||
| 61 | GCB | GCB | ABC | ABC | ||
| 62 | GCB | GCB | ABC | ABC | ||
| 63 | ABC | ABC | GCB | GCB | ||
| 64 | ABC | ABC | ABC | GCB | ||
| 65 | GCB | GCB | ABC | ABC | ||
| 66 | ABC | ABC | ABC | ABC | ||
| 67 | GCB | GCB | GCB | GCB | ||
| 68 | ABC | GCB | ABC | ABC | ||
| 69 | GCB | GCB | GCB | GCB | ||
| 70 | GCB | GCB | GCB | GCB | ||
| 71 | GCB | GCB | GCB | GCB | 141135 | 22678 |
| 72 | GCB | GCB | ABC | ABC | 42681 | 13042 |
| 73 | GCB | GCB | GCB | GCB | 87892 | 12640 |
| 74 | GCB | GCB | GCB | ABC | 123789 | 5534 |
| 75 | ABC | ABC | GCB | ABC | 24403 | 2203 |
| 76 | GCB | GCB | GCB | ABC | 120392 | 3452 |
| 77 | GCB | GCB | GCB | GCB | 31825 | 10442 |
| 78 | ABC | ABC | ABC | ABC | 56768 | 12963 |
| 79 | GCB | GCB | GCB | GCB | 28606 | 41482 |
| 80 | GCB | GCB | GCB | ABC | ||
| 81 | ABC | ABC | ABC | ABC | 106388 | 34158 |
| 82 | GCB | GCB | GCB | GCB | ||
| 83 | GCB | GCB | ABC | ABC | ||
| 84 | GCB | GCB | ABC | ABC | ||
| 85 | ABC | ABC | GCB | GCB | ||
| 86 | GCB | GCB | GCB | GCB | ||
| 87 | ABC | ABC | ABC | ABC | ||
| 88 | ABC | ABC | ABC | ABC | ||
| 89 | GCB | GCB | GCB | GCB | ||
| 90 | ABC | ABC | ABC | ABC | ||
| 91 | ABC | ABC | ABC | ABC | 4474 | 3320 |
| 92 | ABC | ABC | ABC | ABC | ||
| 93 | ABC | ABC | ABC | ABC | ||
| 94 | GCB | GCB | ABC | ABC | ||
| 95 | GCB | GCB | GCB | GCB | 160240 | 37501 |
| 96 | ABC | ABC | ABC | ABC | ||
| 97 | GCB | GCB | GCB | GCB | ||
| 98 | ABC | ABC | ABC | ABC | 1444 | 6066 |
| 99 | ABC | ABC | ABC | GCB | ||
| 100 | ABC | ABC | ABC | ABC | 21125 | 18333 |
| 101 | GCB | GCB | GCB | ABC | ||
| 102 | GCB | GCB | GCB | GCB | ||
| 103 | ABC | ABC | GCB | ABC | ||
| 104 | GCB | GCB | GCB | GCB | ||
| 105 | ABC | ABC | ABC | ABC | ||
| 106 | GCB | GCB | GCB | GCB | ||
| 107 | ABC | ABC | GCB | ABC | 147365 | 21812 |
| 108 | GCB | GCB | GCB | GCB | ||
| 109 | GCB | GCB | ABC | ABC | ||
| 110 | GCB | GCB | GCB | ABC | ||
| 111 | ABC | ABC | ABC | ABC | ||
| 112 | ABC | ABC | ABC | ABC | ||
| 113 | GCB | GCB | GCB | GCB | ||
| 114 | GCB | GCB | GCB | GCB | ||
| 115 | GCB | GCB | GCB | ABC | ||
| 116 | GCB | GCB | GCB | ABC | ||
| 117 | GCB | GCB | GCB | ABC | ||
| 118 | GCB | GCB | GCB | ABC | ||
| 119 | GCB | GCB | GCB | GCB | ||
| 120 | ABC | ABC | ABC | GCB | ||
| 121 | ABC | GCB | GCB | / | ||
| 122 | / | GCB | GCB | GCB | ||
| 123 | GCB | GCB | ABC | ABC | 2960 | 664 |
| 124 | GCB | GCB | GCB | GCB | ||
| 125 | ABC | ABC | ABC | ABC | ||
| 126 | ABC | ABC | ABC | ABC | ||
| 127 | ABC | ABC | ABC | GCB | 299806 | 24765 |
/, the algorithm was not performed. CD20, expression in MESF; CD19, expression in MESF. MESF, molecules of equivalent soluble fluorochrome; ABC, activated B-cell; GCB, germinal center B-cell.
Hans algorithm
Patients with the ABC subtype (56 patients) did not exhibit an inferior response to first-line treatment (P=0.345) when compared with the GCB type (70 patients). The difference in OS was not significant [P=0.083; hazard ratio (HR)=1.61 (95% CI, 0.36–1.07)], and PFS was not significantly different among patients with the ABC and GCB subtypes of DLBCL [P=0.339; HR=1.3 (95% CI, 0.42–1.41)].
Modified Hans algorithm
Patients with the ABC subtype (46 patients) did not exhibit an inferior response to first-line treatment (P=0.543) when compared with the GCB type (81 patients). There was no difference observed in the OS [P=0.282; HR=1.35 (95% CI, 0.43–1.28)] or PFS [P=0.632; HR=1.17 (95% CI, 0.46–1.60)] between the two subtypes.
Choi algorithm
There was no difference in response to front-line treatment between the ABC (61 patients) and the GCB subtype (66 patients) (P=0.116). In the ABC subtype, the OS was significantly shorter [5-year OS, 73% in the GCB subtype and 5-year OS, 57% in the ABC subtype; P=0.019, HR=1.91 (95% CI, 0.30–0.91)]; the PFS was also shorter (5-year PFS in the GCB subtype, 77% and the 5-year PFS in the ABC subtype, 63%; HR=1.56, 95% CI, 0.35–1.18), but the difference was not statistically significant (P=0.151).
Modified Choi algorithm
In the ABC subtype (74 patients), the response to front-line treatment did not differ when compared to the GCB subtype (52 patients; P=0.364). The difference in the OS was not significant [P=0.047; HR=1.8 (95% CI, 0.31–1.00)], but no statistical difference was identified between the PFS in the two groups [P=0.903; HR=1.75 (95% CI, 0.29–1.10)].
According to the results of all 4 algorithms, patients were subdivided into three categories, as previously described above. These are indicated in Fig. 3, and have been divided as such: When all of the algorithms classified patients as having the GCB type; when all of the algorithms classified the patients as having the ABC type; and ‘heterogeneous’ types. The OS of the three groups is presented in Fig. 4.
Figure 3.
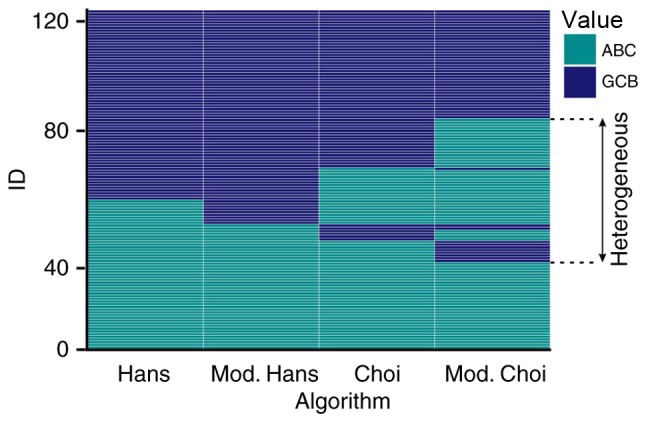
Classification of patients with diffuse large B-cell lymphoma in ABC (turquoise) and GCB groups (blue) according to the Hans, modified Hans, Choi and modified Choi algorithms. ID=identifier which represents a single patient-meaning that each row represents one patient, who is assigned to either the ABC or GCB subtype, by one of the four algorithms applied. ABC, activated B-cell; GCB, germinal center B-cell.
Figure 4.
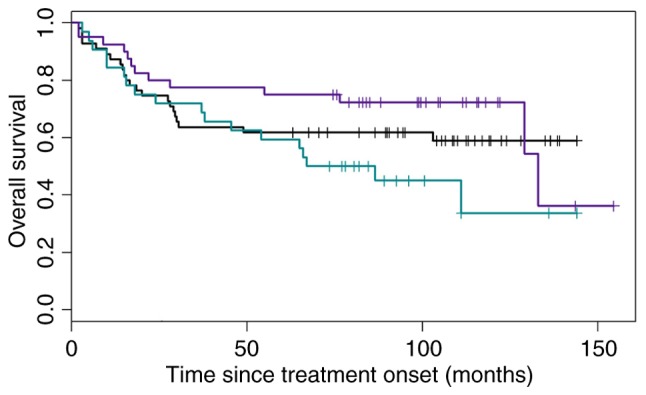
Overall survival of patients categorized according to Hans, modified Hans, Choi and modified Choi algorithms into the ABC (32 patients, turquoise), GCB (40 patients, blue) and ‘heterogeneous’ (55 patients, black) groups. ABC, activated B-cell; GCB, germinal center B-cell.
Furthermore, the accuracy of the Choi and modified Choi algorithms (as they presented a difference in OS between ABC and GCB subgroups) in classifying the ‘heterogeneous’ subgroup into the GCB subtype according to the OS, presuming that these patients have the same risk as the patients in the GCB subgroup and an HR that was as close to 1 as possible, was examined. The HR for the Choi algorithm in the whole cohort was 0.91, but the confidence interval was large (0.38–2.21; Fig. 5).
Figure 5.
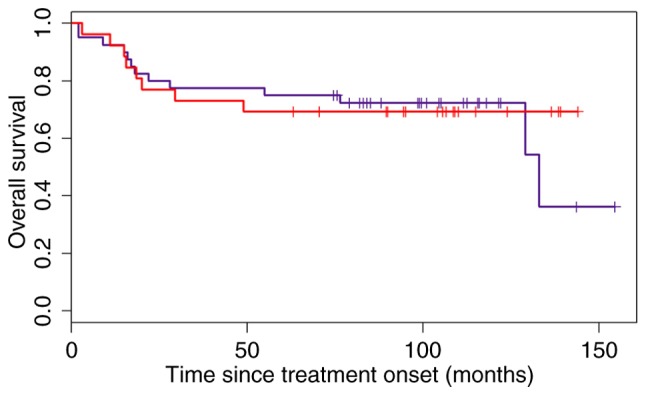
Overall survival of patients of the GCB and ‘heterogeneous’ groups categorized according to the Choi algorithm. The blue line denotes patients with the GCB subtype, and the red line represents patients in the ‘heterogeneous’ subgroup who were allocated to the GCB subgroup according to the Choi algorithm. GCB, germinal center B-cell.
The modified Choi algorithm was not as successful in classifying the ‘heterogeneous’ subgroup into the GCB subtype as the Choi algorithm with an HR=0.82 (95% CI 0.23–2.88; Fig. 6).
Figure 6.
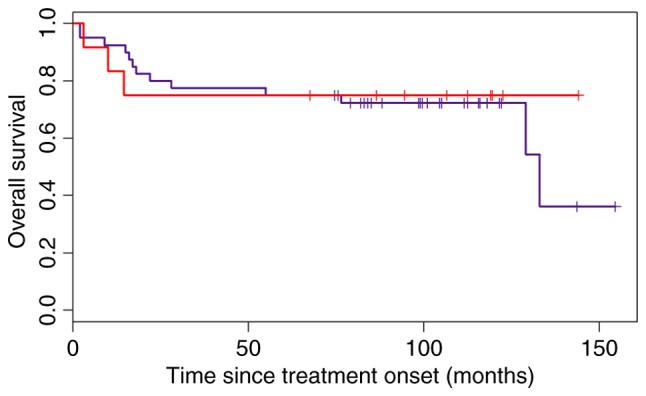
Overall survival of patients in the GCB and ‘heterogeneous’ groups categorized according to the Choi algorithm. The blue line represents patients in the GCB subgroup, and the red line denotes the patients in the ‘heterogeneous’ subgroup who were allocated to the GCB subgroup according to the modified Choi algorithm. GCB, germinal center B-cell.
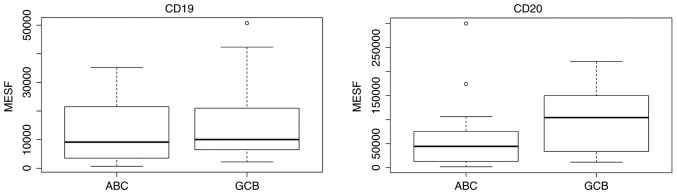
For the CD19 and CD20 expression studies, patients were divided into the GCB and ABC groups according to the Choi algorithm, as it was identified to be the most accurate in our analyses. The results are demonstrated in Fig. 7. There was no difference in the CD19 expression (P=0.427) observed, but the CD20 expression was lower in the ABC subtype (P=0.058) compared with the GCB subtype.
Figure 7.
CD19 and CD20 expression in the two subtypes of DLBCL, based on the Choi algorithm. MESF, molecules of equivalent soluble fluorochrome; CD, cluster of differentiation. ABC, activated B-cell; GCB, germinal center B-cell.
IPI
A group of patients with an IPI <2 (low risk patients) were compared with those with an IPI ≥2 (all other risk groups). The response (CR+PR) to first-line treatment was observed in 97.3% of patients with an IPI<2, and in 86.2% of those with an IPI ≥2, which was not significant (P=0.065). Patients with an IPI ≥2 had a higher risk of progression, but it was not statistically significant [P=0.161; HR=1.83 (95% CI, 0.7–4.25)]. Patients with an IPI ≥2 exhibited a higher risk of lymphoma-associated mortality [P=0.032; HR=2.59 (95% CI, 1.09–6.19)]. The prognosis for OS according to the IPI was also significant when assessing the risk of mortality by any cause [P=0.018; HR=2.36 (95% CI, 1.15–4.85)].
Ki-67
A higher expression of Ki-67 had no effect on the response to first-line treatment (P=0.130). There was also no difference in the PFS or OS regarding the Ki-67 expression [P=0.815; HR (for a 10% difference)=1.04 (95% CI, 0.73–1.5)] and [P=0.164; HR (for a 10% difference)=1.28 (95% CI 0.91–1.8)], respectively.
Discussion
When comparing the 4 selected immunohistochemical algorithms (Hans, Choi, modified Hans and modified Choi) in the present study, only the Choi algorithm was identified to be significantly prognostic of OS. The modified Choi algorithm was close to significance in terms of OS, while the modified Hans and Hans algorithm were the least reliable in discriminating between the two groups in the cohort of the present study. The Choi algorithm was also quite accurate in predicting the GCB subtype, but with a large confidence interval, therefore, it is not accurate to state that it is precise enough to classify the GCB subtype definitely. The modified Choi algorithm was less accurate in our series in terms of predicting the GCB subtype compared with the Choi algorithm. However, the overall treatment response was very high, so statistical power of this subdivision is low.
The present study focused on the determination of the GCB subtype due to the existence of novel studies with novel drugs included in the front-line treatment of the ABC subtype (11–14). Therefore, a patient classified as having the ABC subtype of DLBCL is supposed to be treated more aggressively and with novel drugs, while one classified as exhibiting the GCB type will receive the standard R-CHOP regimen. Therefore, the primary concern of the present study was the potential insufficient treatment of future patients who may be classified as GCB, but were actually the ABC subtype.
As the revised WHO lymphoma classification (10) advises the use of the Hans algorithm, the results of the present study may provide an interesting contrast. At present, there have been a number of studies suggesting that the Hans algorithm does not have prognostic value in terms of OS (18–21). A study by Meyer et al (9) also concluded that the Choi algorithm had the highest predictive power when compared with gene-expression profiling and in terms of OS, but that it is the least user-friendly as it includes 2 antibodies (GCET1 and FOXP1) not routinely applied in a number of laboratories. This study also suggested the use of Tally's algorithm for prognostic purposes, as it was identified to have the ability to predict OS slightly less accurately than Choi (9). The present study did not assess the Tally algorithm, as it includes LMO2 antibody staining, which was not performed as it is not routinely performed at the Institute of Oncology Ljubljana, Slovenia for diffuse large B-cell lymphoma.
To the best of our knowledge, there has been no FCI quantification of the CD19 and CD20 expression using the quantitative measurements of fluorescence intensity on the ABC or GCB subtype of DLBCL. At present, the majority of studies applied the immunohistochemical CD20 staining, while Johnson et al (22) used the semi-quantitative evaluation of CD19 and CD20 expression in mean fluorescence intensity units. They subdivided their study population to the ‘bright’ and ‘dim’ subgroups of CD19 and CD20 expression. They categorized patients with dim CD20 expression and bright CD19 expression into a ‘discordant CD20 group’. The authors also noticed that a high proportion of these ‘discordant’ patients exhibited the ABC subtype, and stated that the cell-of-origin may be a confounding factor in the prognostic effect of the discordant CD20 expression. However, they did not evaluate the CD19 and CD20 expression quantitatively. The ABC subtype exhibited a lower expression of CD20 when compared with the GCB subtype in the sample cohort of the present study, and additional studies are required to confirm this result.
The present study confirmed the prognostic value of IPI in the group of patients regarding the OS, but not the PFS. Ziepert et al (3) clearly demonstrated that IPI is a prognostic factor of PFS, EFS and OS. Certain other studies also confirmed these data (20,23).
The Ki-67 index was identified not to be prognostic in any aspect, which is inconsistent with the results of several larger studies (20,24) and meta-analyses (25). Yoon et al (24) set a cut-off level of 85%, and Salles et al (20) at 75%, to differentiate between the two subgroups (lower vs. higher expression) with a different OS. In the largest meta-analysis in this field of study by He et al (25), the cut-off level was not set, but simply confirmed that a higher Ki-67 expression was associated with inferior survival. The present study observed a trend of shorter survival associated with higher Ki-67 values, but the association was not significant.
Based on the results of the present study, it may be concluded that only the Choi algorithm significantly predicts OS in the ABC and GCB subgroups. The Choi algorithm also appeared to be quite accurate in defining the GCB subtype according to the OS, but additional studies with larger cohorts of patients are required. A lower expression of CD20 was observed in the ABC subtype. The IPI was confirmed to be prognostic for OS, but the Ki-67 index was not identified to be prognostic for the OS, PFS or response rate.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Ministry of Science and Technology, Slovenia (grant no. P3-0321).
Availability of data and materials
The datasets generated and analyzed in the present study are stored at the authors' institution (Institute of Oncology Ljubljana, Slovenia) and may be obtained upon reasonable request from the corresponding author.
Authors' contributions
LB wrote the manuscript and gathered the clinical data, VKP gathered the cytological data and revised the manuscript, MPP performed the statistical analyses and revised the manuscript, GG performed the pathology work and revised the manuscript, BJN provided the design of the study, clinical data and revised the manuscript.
Ethics and consent to participate
All participants provided written informed consent for participation in the present study. The present study was approved by the Republic of Slovenia National Medical Ethics Committee.
Consent for publication
All participants provided informed consent for the publication of this data.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tilly H, da Silva Gomes M, Vitolo U, Jack A, Meignan M, Lopez-Guillermo A, Walewski J, André M, Johnson PW, Pfreundschuh M, et al. Diffuse large B-cell lymphoma: ESMO clinical practice guidelines, treatment and follow-up. Ann Oncol. 2015;26(Suppl 5):v116–v125. doi: 10.1093/annonc/mdv304. [DOI] [PubMed] [Google Scholar]
- 2.International non-Hodgkin's lymphoma prognostic factors project: A predictive model for aggressive non-Hodgkin's lymphoma. N Engl J Med. 1993;329:987–994. doi: 10.1056/NEJM199309303291402. [DOI] [PubMed] [Google Scholar]
- 3.Ziepert M, Hasenclever D, Kuhnt E, Glass B, Schmitz N, Pfreundschuh M, Loeffler M. Standard International prognostic index remains a valid predictor of outcome for patients with aggressive CD20+ B-cell lymphoma in the rituximab era. J Clin Oncol. 2010;28:2373–2380. doi: 10.1200/JCO.2009.26.2493. [DOI] [PubMed] [Google Scholar]
- 4.Ngo L, Hee SW, Lim LC, Tao M, Quek R, Yap SP, Loong EL, Sng I, Hwan-Cheong TL, Ang MK, et al. Prognostic factors in patients with diffuse large B cell lymphoma: Before and after the introduction of rituximab. Leuk Lymphoma. 2008;49:462–469. doi: 10.1080/10428190701809156. [DOI] [PubMed] [Google Scholar]
- 5.Sehn LH, Berry B, Chhanabhai M, Fitzgerald C, Gill K, Hoskins P, Klasa R, Savage KJ, Shenkier T, Sutherland J, et al. The revised international prognostic index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood. 2007;109:1857–1861. doi: 10.1182/blood-2006-08-038257. [DOI] [PubMed] [Google Scholar]
- 6.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 7.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 8.Choi WW, Weisenburger DD, Greiner TC, Piris MA, Banham AH, Delabie J, Braziel RM, Geng H, Iqbal J, Lenz G, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15:5494–5502. doi: 10.1158/1078-0432.CCR-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer PN, Fu K, Greiner TC, Smith LM, Delabie J, Gascoyne RD, Ott G, Rosenwald A, Braziel RM, Campo E, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-Cell lymphoma treated with rituximab. J Clin Oncol. 2011;29:200–207. doi: 10.1200/JCO.2010.30.0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swerdlow SH, Campo E, Pileri SA, Harris NL, Stein H, Siebert R, Advani R, Ghielmini M, Salles GA, Zelenetz AD, Jaffe ES. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina TJ, Canioni D, Copie-Bergman C, Recher C, Brière J, Haioun C, Berger F, Fermé C, Copin MC, Casasnovas O, et al. Young patients with non-germinal center B-cell-like diffuse large B-cell lymphoma benefit from intensified chemotherapy with ACVBP plus rituximab compared with CHOP plus rituximab: Analysis of data from the groupe d'Etudes des lymphomes de l'Adulte/lymphoma study association phase III trial LNH 03-2B. J Clin Oncol. 2014;32:3996–4003. doi: 10.1200/JCO.2013.54.9493. [DOI] [PubMed] [Google Scholar]
- 12.Ruan J, Martin P, Furman RR, Lee SM, Cheung K, Vose JM, Lacasce A, Morrison J, Elstrom R, Ely S, et al. Bortezomib plus CHOP-rituximab for previously untreated diffuse large B-cell lymphoma and mantle cell lymphoma. J Clin Oncol. 2011;29:690–697. doi: 10.1200/JCO.2010.31.1142. [DOI] [PubMed] [Google Scholar]
- 13.Nowakowski GS, LaPlant B, Macon WR, Reeder CB, Foran JM, Nelson GD, Thompson CA, Rivera CE, Inwards DJ, Micallef IN, et al. Lenalidomide combined with R-CHOP overcomes negative prognostic impact of non-germinal center B-cell phenotype in newly diagnosed diffuse large B-Cell lymphoma: A phase II study. J Clin Oncol. 2015;33:251–257. doi: 10.1200/JCO.2014.55.5714. [DOI] [PubMed] [Google Scholar]
- 14.Younes A, Thieblemont C, Morschhauser F, Flinn I, Friedberg JW, Amorim S, Hivert B, Westin J, Vermeulen J, Bandyopadhyay N, et al. Combination of ibrutinib with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) for treatment-naive patients with CD20-positive B-cell non-Hodgkin lymphoma: A non-randomised, phase 1b study. Lancet Oncol. 2014;15:1019–1026. doi: 10.1016/S1470-2045(14)70311-0. [DOI] [PubMed] [Google Scholar]
- 15.Cheson BD, Fisher RI, Barrington SF, Cavalli F, Schwartz LH, Zucca E, Lister TA. Alliance, Australasian Leukaemia and Lymphoma Group; Eastern Cooperative Oncology Group: Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: The Lugano classification. J Clin Oncol. 2014;32:3059–3068. doi: 10.1200/JCO.2013.54.8800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prevodnik VK, Lavrenčak J, Horvat M, Novakovič BJ. The predictive significance of CD20 expression in B-cell lymphomas. Diagn Pathol. 2011;6:33. doi: 10.1186/1746-1596-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz A, Gaigalas AK, Wang L, Marti GE, Vogt RF, Fernandez-Repollet E. Formalization of the MESF unit of fluorescence intensity. Cytometry B Clin Cytom. 2004;57:1–6. doi: 10.1002/cyto.b.10066. [DOI] [PubMed] [Google Scholar]
- 18.Benesova K, Forsterova K, Votavova H, Campr V, Stritesky J, Velenska Z, Prochazka B, Pytlik R, Trneny M. The Hans algorithm failed to predict outcome in patients with diffuse large B-cell lymphoma treated with rituximab. Neoplasma. 2013;60:68–73. doi: 10.4149/neo_2013_010. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Lunning MA, Zhang Z, Migliacci JC, Moskowitz CH, Zelenetz AD. Excellent outcomes and lack of prognostic impact of cell of origin for localized diffuse large B-cell lymphoma in the rituximab era. Br J Haematol. 2015;171:776–783. doi: 10.1111/bjh.13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salles G, de Jong D, Xie W, Rosenwald A, Chhanabhai M, Gaulard P, Klapper W, Calaminici M, Sander B, Thorns C, et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: A study from the lunenburg lymphoma biomarker consortium. Blood. 2011;117:7070–7078. doi: 10.1182/blood-2011-04-345256. [DOI] [PubMed] [Google Scholar]
- 21.Castillo JJ, Beltran BE, Song MK, Ilic I, Leppa S, Nurmi H, Seki R, Uccella S, Li JM, Treaba DO, et al. The Hans algorithm is not prognostic in patients with diffuse large B-cell lymphoma treated with R-CHOP. Leuk Res. 2012;36:413–417. doi: 10.1016/j.leukres.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Johnson NA, Boyle M, Bashashati A, Leach S, Brooks-Wilson A, Sehn LH, Chhanabhai M, Brinkman RR, Connors JM, Weng AP, Gascoyne RD. Diffuse large B-cell lymphoma: Reduced CD20 expression is associated with inferior survival. Blood. 2009;113:3773–3780. doi: 10.1182/blood-2008-09-177469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu K, Weisenburger DD, Choi WW, Perry KD, Smith LM, Shi X, Hans CP, Greiner TC, Bierman PJ, Bociek RG, et al. Addition of rituximab to standard chemotherapy improves the survival of both the germinal center B-cell-like and non-germinal center B-cell-like subtypes of diffuse large B-cell lymphoma. J Clin Oncol. 2008;26:4587–4594. doi: 10.1200/JCO.2007.15.9277. [DOI] [PubMed] [Google Scholar]
- 24.Yoon DH, Choi DR, Ahn HJ, Kim S, Lee DH, Kim SW, Park BH, Yoon SO, Huh J, Lee SW, Suh C. Ki-67 expression as a prognostic factor in diffuse large B-cell lymphoma patients treated with rituximab plus CHOP. Eur J Haematol. 2010;85:149–157. doi: 10.1111/j.1600-0609.2010.01467.x. [DOI] [PubMed] [Google Scholar]
- 25.He X, Chen Z, Fu T, Jin X, Yu T, Liang Y, Zhao X, Huang L. Ki-67 is a valuable prognostic predictor of lymphoma but its utility varies in lymphoma subtypes: Evidence from a systematic meta-analysis. BMC Cancer. 2014;14:153. doi: 10.1186/1471-2407-14-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed in the present study are stored at the authors' institution (Institute of Oncology Ljubljana, Slovenia) and may be obtained upon reasonable request from the corresponding author.