Abstract
Background
Lack of vitamin D (VD) has been associated with colorectal cancer (CRC). VD has anti-inflammatory effects and regulates several cellular pathways by means of its receptor, including epigenetic modifications. Adipose tissue dysfunction has been related to low-grade inflammation, which is related to diseases like cancer. The aim of this study was to explore the relationship between serum 25-hydroxyvitamin D (25(OH)D), adipose tissue gene expression of VD receptor (VDR), pro-inflammatory markers, and the epigenetic factor DNA methyltransferase 3a (DNMT3A) as well as VDR promoter methylation in CRC.
Methods
Blood and visceral adipose tissue from 57 CRC and 50 healthy control subjects were collected. CRC subjects had lower serum 25(OH)D levels and higher VDR gene expression, and these were negatively correlated in the CRC group.
Results
Adipose tissue NFκB1, IL6, and IL1B gene expression were higher in the CRC subjects than in the control subjects. 25(OH)D correlated negatively with NFκB1 and CRP. In turn, CRP correlated positively with NFκB1, IL6, IL1B, and VDR gene expression as well as NFκB1 that correlated positively with IL6 and IL1B. DNMT3A mRNA was negatively correlated with serum 25(OH)D and positively correlated with VDR DNA methylation. VDR DNA methylation at position 4 had lower levels in the CRC group. Global NFκB1 methylation at dinucleotide 3 was lower in the CRC group.
Conclusion
Our results suggest that adipose tissue may be a key factor in CRC development. The low 25(OH)D levels and high adipose tissue VDR expression in CRC may, at least in part, mediate this relationship by modifying adipose tissue DNA methylation and promoting inflammation.
Electronic supplementary material
The online version of this article (10.1186/s13148-018-0493-0) contains supplementary material, which is available to authorized users.
Keywords: Vitamin D, VDR, DNA methylation, Low-grade inflammation, Colorectal cancer, Adipose tissue
Background
Colorectal cancer (CRC) has become one of the most important health issues of our time due to its elevated prevalence, increasing incidence, morbidity, associated costs, and mortality rates. Several risk factors have been linked to CRC development: population aging, lack of physical activity, obesity, low fruit and vegetable intake, tobacco use, alcohol consumption, and other unknown factors [1].
Vitamin D (VD) can be incorporated from diet (vitamin D2 or D3) or synthetized by photoactivation of 7-dehydrocholesterol to previtamin D3 in the skin, a process that is mediated by sunlight [2]. Then, vitamin D2/D3 is converted to 25-hydroxyvitamin D (25(OH)D) by CYP2R1 in the liver, which is the main form of plasma vitamin D. The conversion of 25(OH)D to the active form, the 1,25-hydroxyvitamin D (1,25(OH)D), is carried out by the enzyme CYP24A1 in the kidneys [2]. Vitamin D functions primarily through the VD receptor (VDR), which is regulated by environment, genetics, and epigenetics [2].
Several studies have related plasma 25-hydroxyvitamin D (25(OH)D) levels with CRC [3–5], finding that low circulating 25(OH)D levels have been associated with CRC [4], and high 25(OH)D levels correlate with low risk for the onset of CRC [5]. A number of studies have reported the benefits of VD in processes such as metabolic modulation, autoimmunity, cardiovascular function, and cancer [6]. In fact, it has been proposed that calcitriol has anti-tumor effects [7, 8] as well as a direct effect on tumor development by acting as a tumor repressor in many solid tumors including CRC [9]. In a clinical trial carried out with patients who suffered from colorectal adenoma, the administration of VD together with calcium was able to reduce the expression of genes implicated in CRC development [10].
A number of studies have noted the relationship between CRC and low-grade inflammation [11]. Specifically, CRC patients have dysfunctional adipose tissue that might be a key contributor to the inflammatory state, by the secretion of several detrimental molecules such as tumor necrosis factor alpha (TNFA), interleukin-6 (IL6), and nuclear factor κ-light-chain-enhancer of activated B cells 1 (NFκB1) [12]. Adipose tissue has traditionally only been considered as an energy storage organ. Nevertheless, the importance of this tissue in systemic physiology and especially in systemic inflammation has been pointed out in recent years [13]. Adipose tissue expresses proteins related to VD metabolism [14], and it has been proposed that it can act as VD storage tissue [15]. The active form of VD, 1,25-dihydroxyvitamin D3 (1,25(OH)2D3), is able to modify adipocyte and adipose tissue physiology via the VDR [16, 17], decreasing the expression of pro-inflammatory cytokines in adipose tissue [18]. Therefore, VD might be a key factor for the higher risk of CRC in obese subjects, since low serum 25(OH)D levels or impaired adipose tissue responsiveness to VD might lead to a higher inflammatory state, which is directly implied in cancer development. However, the precise mechanism by which VD leads to a decrease in inflammation is not completely clear.
DNA methylation is an epigenetic regulatory process in which cytosine residues are methylated normally within CpG dinucleotides, referred to as CpG, and it is usually associated with gene repression, although it has also been related to gene activation in some cases [2, 19, 20]. Epigenetic mechanisms are susceptible to environmental factors such as diet, exercise, smoking, and hormones, and could be the basis for factors associated with an increased risk of cancer development such as obesity, inflammation, diabetes, or metabolic syndrome, as well as tumor onset or development [21–26]. DNA methylation is carried out by DNA methyltransferases DNMT1, DNMT3A, and DNMT3B, being DNMT1 implied in the maintenance of DNA methylation and DNMT3A and DNMT3B in de novo DNA methylation processes. Furthermore, the relationship between vitamin D and DNA methylation has been analyzed in several studies, and 25(OH)D levels appear to control DNA methylation or demethylation [27], a mechanism in which DNA methyltransferases could be involved [28]. Furthermore, adipose tissue DNMT3A overexpression provokes a rise in adipose tissue inflammation in mice, which could be involved in adipose tissue-related diseases and CRC [29].
Therefore, we hypothesized that dysfunctional adipose tissue may play a major role in CRC development. 25(OH)D could be involved in epigenetic changes in adipose tissue triggering a change in the inflammatory profile, which could promote CRC onset and/or development. Thus, the aim of this study was to test serum levels of 25(OH)D, as well as the gene expression of the epigenetic factor DNMT3A and inflammatory markers in adipose tissue in CRC. We also studied VDR and NFκB1 DNA methylation in adipose tissue to determine the possible role of the VD system in the epigenetic regulation of VDR and NFκB1 gene.
Methods
Subjects
A total of 57 participants with CRC who underwent colorectal surgery and 50 control subjects who underwent hiatal hernia surgery or cholecystectomy were recruited from the Virgen de la Victoria University Hospital (Málaga, Spain) during 2012–2013.
Patients were excluded if they had cardiovascular disease, arthritis, acute inflammatory disease, infectious disease, and renal disease, and were receiving drugs that could alter the lipid or glucose profile, were undergoing treatment with calcium or vitamin D supplements, or if they consumed > 20 g ethanol per day at the time of inclusion in the study. The study was conducted in accordance with the guidelines laid down in the Declaration of Helsinki. All participants gave their written informed consent (0311/PI7), and the study was reviewed and approved by the Ethics and Research Committee of Virgen de la Victoria Hospital.
Epiploic visceral adipose (VAT) tissue was obtained during surgery, washed in physiological saline solution, and immediately frozen in liquid nitrogen. Biopsy samples were maintained at − 80 °C until analysis.
Laboratory measurements
Before surgery and after an overnight fast, blood samples were obtained from the antecubital vein and placed in vacutainer tubes (BD vacutainer™). The serum was separated by centrifugation for 15 min at 4000 rpm and immediately frozen at − 80 °C until analysis. Serum glucose, cholesterol, triglycerides, HDL cholesterol (HDL-C), and C-reactive protein (CRP) were measured in a Dimension autoanalyzer (Dade Behring Inc.) by enzymatic methods (Randox Laboratories Ltd.). LDL cholesterol (LDL-C) was calculated using the Friedewald equation. Insulin was quantified by radioimmunoassay supplied by BioSource International Inc., Camarillo, CA, USA. The homeostasis model assessment of insulin resistance (HOMA-IR) was calculated with the following equation: HOMA-IR = fasting insulin (μIU/mL) × fasting glucose (mmol/L)/22.5. Serum 25(OH)D and parathyroid hormone levels were determined by enzyme immunoassay (ELISA) kits (Immundiagnostik and DRG Diagnostics, respectively). Corrected calcium was calculated using the following equation: fasting calcium (mg/dl) + 0.8 × (4-fasting albumin (g/dl)).
Visceral adipose tissue RNA isolation and real-time quantitative PCR
Total RNA isolation from VAT was obtained using RNeasy Lipid Tissue Mini Kit (Qiagen GmbH, Hilden, Germany). The purity of the RNA was determined by the 260/280 absorbance ratio on the NanoDrop. The integrity of the total purified RNA was checked by denaturing agarose gel electrophoresis and ethidium bromide staining. For first strand cDNA synthesis, a constant amount of 1 μg of total RNA was reverse transcribed using random hexamers as primers and Transcriptor Reverse Transcriptase (Roche, Mannheim, Germany). Gene expression was assessed by real-time PCR using an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Darmstadt, Germany) with TaqMan technology as previously described [30]. The commercially available and prevalidated TaqMan primer/probe sets used were as follows: VDR (Hs01045840_m1, RefSeq. NM_000376.2, NM_001017535.1 and NM_001017536.1), NFκB1 (Hs00765730_m1, RefSeq. NM_001165412.1 and NM_003998.3), DNMT3A (NM_001320893.1, NM_022552.4, NM_153759.3, NM_175629.2), IL6 (Hs00174131_m1; RefSeq. NM_000600.4, NM_001318095.1), IL1B (Hs00174097_m1; NM_000576.2), and PPIA (4326316E, RefSeq. NM_021130.3), used as endogenous control for the target gene in each reaction.
Protein extraction and western blot
For total protein extract preparation, adipose tissue samples were washed once in erythrocyte lysis buffer (sacarose 320 mM, Tris-HCl pH 7.5 10 mM, MgCl2 5 mM, Tritonx-100 1%) and PBS for 20 min at 4° with agitation. Samples were homogenized using T-PER tissue protein extraction reagent (Thermofisher, USA) and Ultra Turrax Homogenizator and then centrifuged to discard the pellet and the upper fatty layer. Samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane to perform the Western blotting. A mouse monoclonal anti-VDR (sc 13133, Santa Cruz Biotechnology) was used as primary antibody and a goat anti-mouse IgG-HRP (sc-2005, Santa Cruz Biotechnology) as secondary antibody. Clarity Western ECL substrate (Bio-Rad, USA) was used for detection, and the target protein was determined by using the total protein determined by ponceau staining to normalize the quantification. All experiments were performed in duplicate.
Pyrosequencing
The DNA methylation status was determined by pyrosequencing using the PyroMarkTMQ96 ID Pyrosequencing System (Qiagen). We used a premade Pyromark CpG assay for VDR (PM00051443) and NFκB1 (PM00110908). An overview of the analyzed regions for VDR and NFκB1 is depicted in Supplementary Additional files 1 and 2: Figures S1 and S2, respectively. Briefly, NFκB1 is located at chromosome 4. The region 103,423,139-103,423,177 (39 bp length) was analyzed, which contains more than 30 transcription factor binding sites (TFBS) determined by ChiP experiments by the ENCODE project [31]. Besides, the assay was located inside additional TFBS according to the Open Regulatory Annotation database (ORegAnno) [32], as SMARCA4, SPL1, STAT1, RBL2, RB1, and ETS1. VDR is located at chromosome 12. The region 48,299,419-48,299,455 (37 bp length) was analyzed, which contains several TFBS determined by ChiP experiments by the ENCODE Project [31], as POLR2A, ATF2, CTCF, EZH2, E2F6, GATA2, GATA3, CEBPB, or POL2. Additional TFBS are present according to ORegAnno [32], as SMARCA4, SPL1, STAT1, RBL2, RB1, and ETS1. DNA methylation analyses were performed using bisulfite-treated DNA followed by a highly quantitative analysis based on PCR-based pyrosequencing. The bisulfite conversion was conducted with 2-μg genomic DNA isolated from VAT using Qiazol (Qiagen) and 0.1 μM citrate ethanol solution. Then, the PCR was performed in a total volume of 25 μL, with a final primer concentration of 0.2 μM. One of the primers was biotinylated in order to purify the final PCR product using Sepharose beads. The biotinylated PCR amplification was purified using the Pyrosequencing Vacuum Prep Tool (Qiagen). Finally, 15 μL of the PCR products was pyrosequenced using the PyroMarkTMQ96 ID Pyrosequencing System, using a 0.4-μM sequencing primer.
The methylation level was expressed as the percentage of methylated cytosine over the sum of methylated and unmethylated cytosines. Non-CpG cytosine residues were used as built-in controls to verify bisulfite conversion. The values are expressed as the mean for all the sites and individually for six CpGs at the VDR gene promoter and seven CpGs at the NFκB1 promoter. We also included unmethylated and methylated DNA as controls in each run (New England Biolabs). Inter-assay precision (% CV) was < 2.5%; intra-assay (% CV) was < 1.0%.
Statistical analysis
The results are given as the mean ± SD (Table 1) and as a box plot with the minimum and maximum value (control case comparisons shown in Figs. 1, 2, and 3). Student’s t test was used for comparisons of the anthropometric and biochemical characteristics as well as serum 25(OH)D levels between the CRC and control groups. Mann-Whitney U test was performed for comparisons of serum CRP and PTH levels as well as for VDR methylation, mRNA, and protein expression levels and for NFκB1 methylation and mRNA levels between the CRC and the control groups. Spearman’s correlation analyses were performed to study the correlations between 25(OH)D and VDR mRNA and to study the correlations for NFκB1 DNA methylation. Partial correlation analyses corrected by gender were used to study the correlation among the study gene expressions and plasma levels of 25(OH)D and CRP. For the analysis, non-normal distribution variables were log-transformed. The analyses were performed with SPSS (Version 15.0 for Windows; SPSS Iberica, Spain). Values were considered to be statistically significant when p < 0.05.
Table 1.
Anthropometric and biochemical variables of the study groups
| Control (n = 57) |
CRC (n = 50) |
|
|---|---|---|
| Age (years) | 64.94 ± 8.84 | 68.035 ± 8.43 |
| Male/Female (%)* | 68/32 | 45/55 |
| BMI (kg/m2) | 28.51 ± 4.21 | 27.61 ± 3.91 |
| Waist (cm) | 96.55 ± 11.64 | 97 ± 12.74 |
| Glucose (mg/dl) | 111.72 ± 28.77 | 125.035 ± 46.87 |
| Insulin (μUI/ml)** | 11.638 ± 6.54 | 6.23 ± 5.19 |
| Triglycerides (mg/dl)* | 142.3 ± 70.69 | 172.821 ± 87.54 |
| Cho (mg/dl)** | 220.68 ± 39.84 | 169.625 ± 43.57 |
| HDL-C (mg/dl)** | 53.28 ± 14.4 | 40.053 ± 15.12 |
| LDL-C (mg/dl)** | 136.61 ± 29.80 | 101.58 ± 35.67 |
| Corrected calcium (mg/dl)** | 8.99 ± 0.44 | 9.67 ± 0.65 |
| Alkaline phosphatase (U/L) | 72.67 ± 21.63 | 64.66 ± 22.81 |
CRC Colorectal Cancer Group, BMI Body Mass Index, DM Diabetes Mellitus, Cho Total Cholesterol, HDL-C High Density Lipoprotein Cholesterol, LDL-C Low density Lipoprotein Cholesterol
Results are presented as means ± S.D. *p < 0.05 CRC vs. Control; **p < 0.01 CRC vs. control according to t student’s test and Chi squared test for variables expressed as percentage
Fig. 1.
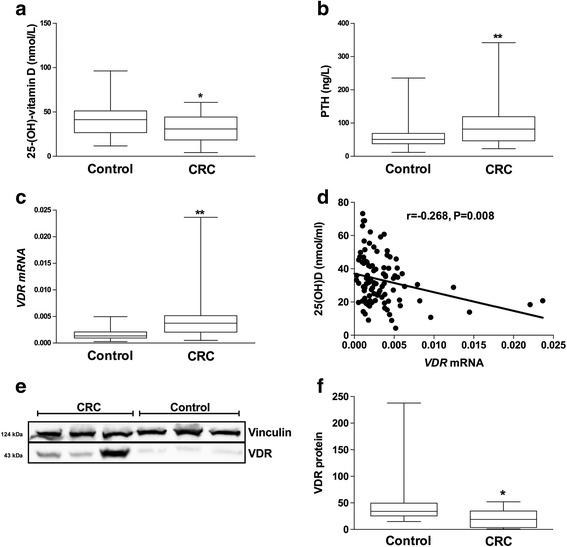
Comparison of serum 25(OH)D and PTH levels and adipose tissue VDR mRNA and protein in CRC patients and controls. Comparisons were performed using Student t test (for 25(OH)D) and Mann-Whitney U test (for serum PTH and adipose tissue VDR mRNA and VRD protein). Serum levels of a 25(OH)D, and b PTH was measured by ELISA in both the control and CRC group. c Adipose tissue VDR mRNA expression was measured by qPCR (n = 107), and Spearman’s correlation (d) between serum 25(OH)D and adipose tissue VDR mRNA in the whole study population was performed. Comparison of adipose tissue VDR protein (e, f) analyzed by Western blot (n = 18). * and ** mean p < 0.05 and p < 0.01, respectively. Parathyroid hormone (PTH), vitamin D receptor (VDR), colorectal cancer (CRC)
Fig. 2.
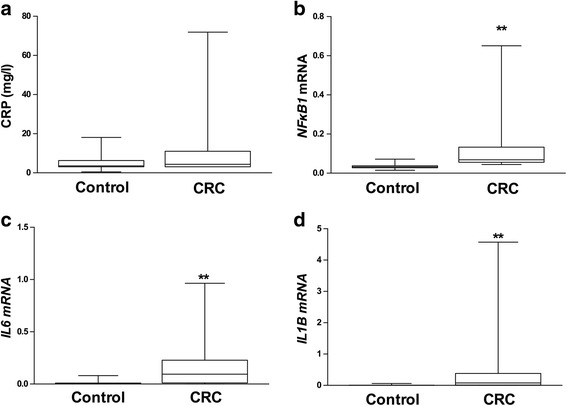
Serum and adipose tissue inflammatory markers. Comparisons were performed using non-parametric test (Mann-Whitney U test). Serum CRP levels (a) and adipose tissue NFκB1 (b), IL6 (c), and IL1B (d) gene expression in the control and CRC groups. ** means p < 0.01. C-reactive protein (CRP), nuclear factor kappa B subunit 1 (NFκB1), interleukin 6 (IL6), interleukin 1 beta (IL1B); colorectal cancer (CRC)
Fig. 3.
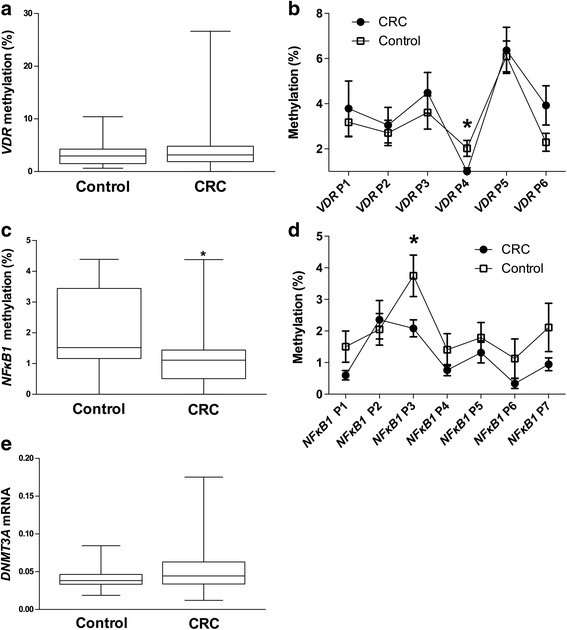
Methylation analyses at specific CpG dinucleotides for VDR and NFκB1 promoters and gene expression of the epigenetic factor DNMT3A were performed to compare both the control and CRC groups. Comparisons of the global VDR methylation (a) and among the CpG dinucleotides analyzed (b) by Mann-Whitney U test. Non-parametric (Mann-Whitney U test) comparison for the global NFκB1 methylation (c) and at specific CpG dinucleotides (d), as well as for the methyltransferase DNMT3A gene expression (e). * means p < 0.05. Vitamin D receptor (VDR), DNA methyltransferase 3a (DNMT3A), colorectal cancer (CRC)
Results
Anthropometric and biochemical variables
Table 1 shows the biochemical and anthropometric characteristics of the study groups. There were no differences in age, BMI, or gender between the control and CRC groups. The CRC group had lower levels of insulin, total cholesterol, HDL-C, and LDL-C than the control group. In contrast, the CRC group presented higher levels of plasma triglycerides when compared with the control group.
Serum 25(OH)D levels and adipose tissue VDR gene and protein expression
Our results showed that 12% in the control group and 26% in the CRC group were vitamin D deficient (25(OH)D lower than 20 nmol/L) according to the Endocrine Society Clinical Practice Guideline [33], although no significant differences were found according to Fisher’s test. Serum 25(OH)D levels were significantly lower in the CRC group than in the control group (Fig. 1a), while parathyroid hormone levels showed an inverse result (Fig. 1b). Contrary to serum 25(OH)D levels, adipose tissue VDR mRNA levels were higher in the CRC group than in the control group (Fig. 1c) which in turn correlated negatively with 25(OH)D (r = − 0.268; p = 0.008) (Fig. 1d). This correlation was maintained using a partial correlation analysis corrected by gender (r = − 0.273, p = 0.01). Accordingly, mRNA levels were translated to higher VDR protein levels in the CRC group with regard to the control group (Fig. 1e, f).
Inflammatory status and relationship with circulating vitamin D
We checked the systemic inflammatory status by measuring serum CRP, as well as the inflammatory status of adipose tissue by determining mRNA levels of NFκB1, IL6, and IL1B gene expression. We found higher levels of adipose tissue NFκB1, IL6, and IL1B mRNA levels (Fig. 2b–d) in the CRC group with regard to the control group.
25(OH)D correlated negatively with adipose tissue NFκB1 mRNA (Table 2). Concordantly, we observed that serum CRP levels were negatively correlated with serum 25(OH)D levels and positively correlated with both VDR and NFκB1 gene expression in adipose tissue (Table 3A). In turn, there was a positive correlation between NFκB1 mRNA and both IL6 and IL1B mRNA levels (Table 3B). Furthermore, a positive correlation was found between NFκB1 mRNA and VDR (Table 3B).
Table 2.
Partial correlation between serum 25(OH)D and adipose tissue NFκB1 mRNA, IL6 mRNA and IL1B mRNA corrected by gender in the whole population
| 25(OH)D | ||
|---|---|---|
| r | p | |
| Log(NFκB1 mRNA) | −0.232 | 0.041 |
| Log(IL6 mRNA) | −0.125 | 0.251 |
| Log(IL1B mRNA) | −0.106 | 0.339 |
25(OH)D 25-hydroxy-vitamin D, NFκB1 Nuclear Factor Kappa B subunit 1, IL6 Interleukin 6, IL1B Interleukin 1 beta
Table 3.
Partial correlations of C-reactive protein (A) with serum 25(OH)D, adipose tissue VDR mRNA, NFκB1 mRNA, IL6 mRNA, and IL1B mRNA corrected by gender. Partial correlations corrected by gender of adipose tissue NFκB1 mRNA (B) with IL 6 mRNA, IL1B mRNA and VDR mRNA
| A | Log(C-reactive protein) | |
| r | p | |
| Log(25(OH)D) | −0.270 | 0.011 |
| Log(VDR mRNA) | 0.219 | 0.049 |
| Log(NFκB1 mRNA) | 0.284 | 0.016 |
| Log(IL6 mRNA) | 0.245 | 0.029 |
| Log(IL1B mRNA) | 0.272 | 0.016 |
| B | NFκB1 mRNA | |
| r | p | |
| Log(IL6 mRNA) | 0.688 | 0.000 |
| Log(IL1B mRNA) | 0.778 | 0.000 |
| Log(VDR mRNA) | 0.761 | 0.000 |
25(OH)D 25-hydroxy-vitamin D, VDR vitamin D receptor, NFκB1 Nuclear Factor Kappa B subunit 1, IL6 Interleukin 6, IL1B Interleukin 1 beta
VDR and NFκB1 methylation and association between the epigenetic factor DNMT3A and 25(OH)D
The DNA methylation status of the VDR promoter was determined by pyrosequencing, but no differences between the control and CRC groups were found (Fig. 3a). When individual VDR CpG positions were compared, significant lower VDR methylation at position 4 (VDR P4) was found in the CRC group when compared with the control group (Fig. 3b). NFκB1 global methylation was lower in the CRC group than in the control group. A comparative analysis at each NFκB1 CpG analyzed showed that NFκB1 at position 3 (NFκB1 P3) presented a lower methylation level in CRC with regard to the control group. Moreover, a negative trend (r = − 0.252, p = 0.061) was observed between NFκB1 mRNA levels and NFκB1 P3 and between VDR mRNA and the global NFκB1 methylation (r = − 0.228; p = 0.064). Additionally, a negative and significant correlation was found between VDR mRNA and NFκB1 at position 3 (NFκB1 P3) (r = − 0.296; p = 0.015) and at position 4 (NFκB1 P4) (r = − 0.327; p = 0.007). Furthermore, we analyzed DNMT3A gene expression in adipose tissue, which was higher in the CRC group than in the control group but without getting statistic significance (Fig. 3e). We also found a negative correlation (corrected by gender) between the gene expression of the epigenetic factor DNMT3A and serum 25(OH)D levels (r = − 0.264, p = 0.013). There was a positive correlation between adipose tissue DNMT3A gene expression and adipose tissue VDR DNA methylation in a partial correlation corrected by gender (r = 0.256, p = 0.034). We also found a positive correlation (Spearman’s correlation) between DNMT3A mRNA and NFκB1 mRNA (r = 0.279, p = 0.009).
Discussion
In this study, we found lower levels of serum 25(OH)D and higher levels of CRP, which is in accordance with previous studies [34, 35]. In addition, to our knowledge, this is the first study which has aimed at analyzing the putative relationship between the VDR in adipose tissue and CRC, taking into consideration the anti-inflammatory role that has been attributed to VD [18, 36]. This is based on the hypothesis that the pro-inflammatory profile of adipose tissue could contribute to the systemic inflammation which has been described to be related to CRC [24]. According to this hypothesis, we found that, apart from lower plasma 25(OH)D levels, CRC patients had higher adipose tissue mRNA levels of pro-inflammatory mediators. Interestingly, we also found significant differences in VDR gene expression between the CRC and the control group, which suggest that in fact, VD may be mediating an anti-inflammatory role in the adipose tissue of CRC patients [37]. Our study shows that adipose tissue DNMT3A mRNA correlates negatively with 25(OH)D and positively with adipose tissue VDR and NFκB1 methylation, suggesting that VD could be involved in epigenetic modifications in both genes in adipose tissue by mechanisms involving the DNA-methyltransferase DNMT3A. Interestingly, the VDR CpGs analyzed were located in the promoter region of the VDR gene (Additional file 1: Figure S1) and inside several TFBS as POLR2A, ATF2, CTCF, EZH2, E2F6, GATA2, GATA3, CEBPB, or POL2, which some of them has been related to DNA methyltransferases recruitment [38, 39]. Changes in adipose tissue methylation status could be related to its pro-inflammatory profile, as previous studies have described a positive relationship between DNMT3A and inflammation in a murine model [29] as we have described in our study population.
It has been found that circulating CRP is associated with a higher risk of CRC [40], and high levels of serum CRP have been related to low levels of 25(OH)D [10], which is in agreement with our results showing a negative correlation between CRP and 25(OH)D levels. Adipose tissue dysfunction might play a crucial role in the promotion of different diseases including insulin resistance, diabetes, and cancer [41, 42]. Several mechanisms have been proposed to explain this relationship, including an altered adipokine secretion profile [24]. Some of these alterations may provoke the development of a chronic low-grade inflammatory state due to the production and secretion of pro-inflammatory cytokines by adipose tissue [41, 43, 44], which has been regarded as a favorable environment for tumor development [24]. Recently, it has been proven that cytokines secreted by adipose tissue have a direct effect on tumor aggressiveness and cancer cell migration in prostate cancer [45], and on colon cancer in mice [46]. Interestingly, it has been described that calcitriol decreases the adipose tissue chronic pro-inflammatory status by downregulating pro-inflammatory cytokine production in a process in which NFκB1 and VDR are involved [47]. Indeed, NFκB has been implied in cancer development [12]. Our results concur with this idea since CRC patients showed higher adipose tissue NFκB1 gene expression in comparison with control subjects, and NFκB1 transcription levels correlated positively to VDR, IL6, and IL1B mRNA levels, confirming the relationship between VDR and inflammation [47]. These facts support the idea that low circulating 25(OH)D promotes a chronic low-grade inflammatory state in adipose tissue, the effects of which could be the release of pro-inflammatory cytokines inducing or stimulating colon tumors.
Calcitriol action is mediated by its receptor, VDR. VDR has also been associated with CRC. Specifically, a higher tumor expression of VDR in CRC correlates with a better prognosis, and there is a direct relation between the tumor differentiation level and VDR gene expression level [48, 49]. However, to our knowledge, there are no previous studies analyzing the relationship between adipose tissue VDR gene expression and CRC. Here, we found that there were higher adipose tissue VDR mRNA and protein levels in the CRC group when compared with the control group. These high VDR levels in adipose tissue might be due to a compensatory mechanism in response to the low 25(OH)D levels in these subjects, and it could be a sign of VD insufficiency. Moreover, it has been reported that the inflammatory factor TNF can activate VDR gene expression [50]. So, a lack of 25(OH)D might lead to an inflammatory process which could subsequently promote VDR expression as we observed in our study. High VDR expression without its ligand, calcitriol, has been shown to have an opposite effect in gene expression regulation and could alter epigenetic marks [51, 52].
In addition, although previous studies described a relationship between methylation status and 25(OH)D [27], which agrees with our observation between DNMT3A and 25(OH)D levels, this phenomenon has not previously been analyzed in adipose tissue. Therefore, to our knowledge, the present study is the first one to report a negative association between 25(OH)D levels and adipose tissue DNMT3A gene expression. Concordantly, DNMT3A mRNA levels were positively correlated with global VDR promoter methylation. This could suggest an epigenetic effect in adipose tissue via VD action. Interestingly, when the methylation of key individual CpGs was analyzed, we found that methylation levels at VDR P4 were significantly lower in CRC than in the control subjects. However, it should also be taken into consideration that other mechanisms might be involved in the regulation of gene expressions such as histone modifications or microRNAs. Therefore, further studies will be necessary to clarify the relevance of VDR methylation in the regulation of its expression in adipose tissue as well as the consequences that these epigenetic changes could have in CRC. We also found a possible regulation of NFκB1 gene expression through DNA methylation. Furthermore, we saw an association between VDR mRNA levels and NFκB1 DNA methylation, which could agree with previous studies although a deeper approach would be necessary to clarify the possible relationship between VDR and the VD signaling with the epigenetic control in NFκB1 gene expression.
Conclusions
Our results suggest that adipose tissue may be a key factor in CRC development. The low 25(OH)D levels in CRC and high adipose tissue VDR expression may, at least in part, mediate this relationship by modifying adipose tissue DNA methylation and promoting inflammation. Although more studies are needed to discover the precise mediators and mechanisms that determine this relationship, the possible mediation of adipose tissue in CRC should be borne in mind to create new treatments and preventive strategies for CRC.
Additional files
Figure S1. VDR promoter overview generated by UCSC genome Browser (https://genome.ucsc.edu). The sequence analyzed is highlighted in light blue, showing that is in the promoter region of VDR and inside a CpG island and several transcription factor binding sites (POLR2A, ATF2, CTCF, EZH2, E2F6, GATA2, GATA3, CEBPB and POL2) all of them determined by experimental procedures (ENCODE project). (DOCX 188 kb)
Figure S2. NFκB1 promoter overview generated by UCSC genome Browser (https://genome.ucsc.edu). The sequence analyzed is highlighted in light blue, showing that is in the promoter region of NFκB1 and inside a CpG island and several transcription factor binding sites (SMARCA4, SPL1, STAT1, RBL2, RB1 and ETS1) all of them determined by experimental procedures (ENCODE project). (DOCX 341 kb)
Acknowledgements
The authors thank all the subjects for their important contribution and to Richard Carlsson for his help in the English grammar correction.
Funding
This study was supported by “Centros de Investigación En Red” (CIBER, CB06/03/0018) of the “Instituto de Salud Carlos III” (ISCIII) and grants from ISCIII (PI11/01661, PI15/0114) and co-financed by the European Regional Development Fund (FEDER). DCC was the recipient of a FPU grant from Education Ministry, Madrid, Spain (13/04211). MMG was the recipient of the Nicolas Monardes Programme from the “Servicio Andaluz de Salud, Junta de Andalucia”, Spain (C-0029-2014).
Abbreviations
- 25(OH)D
25-Hydroxyvitamin D
- CRC
Colorectal cancer
- CRP
C-reactive protein
- DNMT3A
DNA methyltransferase 3a
- HOMA-IR
Homeostasis model assessment of insulin resistance
- IL1
Interleukin-1
- IL6
Interleukin-6
- NFκB1
Nuclear factor κ-light-chain-enhancer of activated B cells
- TNFA
Tumor necrosis factor alpha
- VAT
Visceral adipose tissue
- VD
Vitamin D
- VDR
Vitamin D receptor
Authors’ contributions
The authors’ responsibilities were as follows. DCC, SM, MCP, and MMG contributed to the project conception and leadership of overall research plan, writing of the first draft of the manuscript, and primary responsibility for the final content of the manuscript. DCC, SM, and MCP performed the statistical analysis. DCC, SM and ABC contributed to the bisulfite genomic sequencing and pyrosequencing assessment. DCC and MCP performed the functional and gene expression analysis. FJT, JCF, and EO contributed to the sampling and clinical data collection. MCP, JCF, EO, and FJT contributed to the interpretation of data and critical revision of the manuscript. MMG and FJT acquired funding for this study. All authors were involved in the writing of the manuscript and approving the final version of this article.
Ethics approval and consent to participate
All participants gave their written informed consent, and the study was reviewed and approved by the Local Ethics and Research Committee of the Virgen de la Victoria University Hospital.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13148-018-0493-0) contains supplementary material, which is available to authorized users.
Contributor Information
Mercedes Clemente-Postigo, Phone: +34 951032648, Email: mer.cp@hotmail.com.
Manuel Macias-Gonzalez, Phone: +34 951032648, Email: mmacias.manuel@gmail.com.
References
- 1.Lippi G, Mattiuzzi C, Cervellin G. Meat consumption and cancer risk: a critical review of published meta-analyses. Crit Rev Oncol Hematol. 2015; 10.1016/j.critrevonc.2015.11.008. [DOI] [PubMed]
- 2.Saccone D, Asani F, Bornman L. Regulation of the vitamin D receptor gene by environment, genetics and epigenetics. Gene. 2015;561:171–180. doi: 10.1016/j.gene.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 3.Platz EA, Hankinson SE, Hollis BW, Colditz GA, Hunter DJ, Speizer FE, et al. Plasma 1, 25-Dihydroxy- and 25-hydroxyvitamin D and adenomatous polyps of the distal colorectum 1. Cancer Epidemiol Biomarkers Prev. 2000;9:1059–65. [PubMed]
- 4.Song M, Konijeti GG, Yuan C, Ananthakrishnan AN, Ogino S, Fuchs CS, et al. Plasma 25-hydroxyvitamin D, vitamin D binding protein, and risk of colorectal cancer in the nurses’ health study. Cancer Prev Res. 2016; 10.1158/1940-6207.CAPR-16-0053. [DOI] [PMC free article] [PubMed]
- 5.Giovannucci E, Liu Y, Rimm EB, Hollis BW, Fuchs CS, Stampfer MJ, et al. Prospective study of predictors of vitamin D status and cancer incidence and mortality in men. J Natl Cancer Inst. 2006;98:451–459. doi: 10.1093/jnci/djj101. [DOI] [PubMed] [Google Scholar]
- 6.Mason RS, Sequeira VB, Gordon-Thomson C. Vitamin D: the light side of sunshine. Eur J Clin Nutr. 2011;65:986–993. doi: 10.1038/ejcn.2011.105. [DOI] [PubMed] [Google Scholar]
- 7.Peehl DM, Skowronski RJ, Leung GK, Wong ST, Stamey TA, Feldman D. Antiproliferative effects of 1,25-dihydroxyvitamin D3 on primary cultures of human prostatic cells. Cancer Res. 1994;54:805–810. [PubMed] [Google Scholar]
- 8.Flynn G, Chung I, Yu W-D, Romano M, Modzelewski RA, Johnson CS, et al. Calcitriol (1,25-dihydroxycholecalciferol) selectively inhibits proliferation of freshly isolated tumor-derived endothelial cells and induces apoptosis. Oncology. 2006;70:447–457. doi: 10.1159/000098872. [DOI] [PubMed] [Google Scholar]
- 9.Neska J, Swoboda P, Przybyszewska M, Kotlarz A, Bolla N, Miłoszewska J, et al. The effect of analogues of 1α,25-dihydroxyvitamin D2 on the regrowth and gene expression of human colon cancer cells refractory to 5-fluorouracil. Int J Mol Sci. 2016;17:903. doi: 10.3390/ijms17060903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hopkins MH, Owen J, Ahearn T, Fedirko V, Flanders WD. Effects of supplemental vitamin D and calcium on biomarkers of inflammation in colorectal adenoma patients: a randomized, controlled clinical trial. Cancer Prev Res (Phila). 2011;4(10):1645–655. [DOI] [PMC free article] [PubMed]
- 11.Liu Z, Brooks RS, Ciappio ED, Kim SJ, Crott JW, Bennett G, et al. Diet-induced obesity elevates colonic TNF- α in mice and is accompanied by an activation of Wnt signaling: a mechanism for obesity-associated colorectal cancer. J Nutr Biochem. 2012;23:1207–1213. doi: 10.1016/j.jnutbio.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6:203–208. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol. 2005;115:911–920. doi: 10.1016/j.jaci.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 14.Wamberg L, Christiansen T, Paulsen SK, Fisker S, Rask P, Rejnmark L, et al. Expression of vitamin D-metabolizing enzymes in human adipose tissue—the effect of obesity and diet-induced weight loss. Int J Obes. 2012;37:651–657. doi: 10.1038/ijo.2012.112. [DOI] [PubMed] [Google Scholar]
- 15.Davis CD, Dwyer JT. The “sunshine vitamin”: benefits beyond bone? J Natl Cancer Inst. 2007;99:1563–1565. doi: 10.1093/jnci/djm211. [DOI] [PubMed] [Google Scholar]
- 16.Wood RJ. Vitamin D and adipogenesis: new molecular insights. Nutr Rev. 2008;66:40–46. doi: 10.1111/j.1753-4887.2007.00004.x. [DOI] [PubMed] [Google Scholar]
- 17.Ding C, Gao D, Wilding J, Trayhurn P, Bing C. Vitamin D signalling in adipose tissue. Br J Nutr. 2012;2:1–9. doi: 10.1017/S0007114512003285. [DOI] [PubMed] [Google Scholar]
- 18.Lira FS, Rosa JC, Cunha CA, Ribeiro EB, do Nascimento CO, Oyama LM, et al. Supplementing alpha-tocopherol (vitamin E) and vitamin D3 in high fat diet decrease IL-6 production in murine epididymal adipose tissue and 3T3-L1 adipocytes following LPS stimulation. Lipids Health Dis. 2011;10:37. doi: 10.1186/1476-511X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki MM, Bird A. DNA methylation landscapes provocative inside from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 20.Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, et al. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc Natl Acad Sci U S A. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campión J, Milagro F, Martínez JA. Epigenetics and obesity. Prog Mol Biol Transl Sci. 2010;94:291–347. doi: 10.1016/B978-0-12-375003-7.00011-X. [DOI] [PubMed] [Google Scholar]
- 22.Sabatino L, Fucci A, Pancione M, Colantuoni V. PPARG epigenetic deregulation and its role in colorectal tumorigenesis. PPAR Res. 2012;2012:687492. doi: 10.1155/2012/687492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sinha G. Homing in on the fat and cancer connection. J Natl Cancer Inst. 2012;104:966–967. doi: 10.1093/jnci/djs306. [DOI] [PubMed] [Google Scholar]
- 24.Riondino S, Roselli M, Palmirotta R, Della-Morte D, Ferroni P, Guadagni F. Obesity and colorectal cancer: role of adipokines in tumor initiation and progression. World J Gastroenterol. 2014;20:5177–5190. doi: 10.3748/wjg.v20.i18.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali O, Cerjak D, Kent JW, James R, Blangero J, Carless MA, et al. An epigenetic map of age-associated autosomal loci in northern European families at high risk for the metabolic syndrome. Clin Epigenetics. 2015;7:12. doi: 10.1186/s13148-015-0048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Uzunlulu M, Telci Caklili O, Oguz A. Association between metabolic syndrome and cancer. Ann Nutr Metab. 2016;178:173–179. doi: 10.1159/000443743. [DOI] [PubMed] [Google Scholar]
- 27.Zhu H, Bhagatwala J, Huang Y, Pollock NK, Parikh S, Raed A, et al. Race/ethnicity-specific association of vitamin D and global DNA methylation: cross-sectional and interventional findings. PLoS One. 2016;11:2–10. doi: 10.1371/journal.pone.0152849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue J, Schoenrock SA, Valdar W, Tarantino LM, Ideraabdullah FY. Maternal vitamin D depletion alters DNA methylation at imprinted loci in multiple generations. Clin Epigenetics. 2016;8:107. doi: 10.1186/s13148-016-0276-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamei Y, Suganami T, Ehara T, Kanai S, Hayashi K, Yamamoto Y, et al. Increased expression of DNA methyltransferase 3a in obese adipose tissue: studies with transgenic mice. Obesity (Silver Spring) 2010;18:314–321. doi: 10.1038/oby.2009.246. [DOI] [PubMed] [Google Scholar]
- 30.Clemente-Postigo M, Muñoz-Garach A, Serrano M, Garrido-Sánchez L, Bernal-López MR, Fernández-García D, et al. Serum 25-Hydroxyvitamin D and adipose tissue vitamin D receptor gene expression: relationship with obesity and type 2 diabetes. J Clin Endocrinol Metab. 2015; 10.1210/jc.2014-3016. [DOI] [PubMed]
- 31.Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan K, Cheng C, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;488:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesurf R, Cotto KC, Wang G, Griffith M, Kasaian K, Jones SJM, et al. ORegAnno 3.0: a community-driven resource for curated regulatory annotation. Nucleic Acids Res. 2016;44:D126–D132. doi: 10.1093/nar/gkv1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holick MF, Binkley NC, Bischoff-Ferrari HA, Gordon CM, Hanley DA, Heaney RP, et al. Evaluation, treatment, and prevention of vitamin D deficiency: an endocrine society clinical practice guideline. J Clin Endocrinol Metab. 2011;96:1911–1930. doi: 10.1210/jc.2011-0385. [DOI] [PubMed] [Google Scholar]
- 34.Klampfer L. Vitamin D and colon cancer. World J Gastrointest Oncol. 2014;6(11): 430–37. [DOI] [PMC free article] [PubMed]
- 35.Allin KH, Bojesen SE, Nordestgaard BG. Inflammatory biomarkers and risk of cancer in 84,000 individuals from the general population. Int J Cancer. 2016;139:1493–1500. doi: 10.1002/ijc.30194. [DOI] [PubMed] [Google Scholar]
- 36.Krishnan AV, Feldman D. Mechanisms of the anti-cancer and anti-inflammatory actions of vitamin D. Annu Rev Pharmacol Toxicol. 2011;51:311–336. doi: 10.1146/annurev-pharmtox-010510-100611. [DOI] [PubMed] [Google Scholar]
- 37.Abbas MA. Physiological functions of vitamin D in adipose tissue. J Steroid Biochem Mol Biol. 2017;165:369–381. doi: 10.1016/j.jsbmb.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 38.Velasco G, Hubé F, Rollin J, Neuillet D, Philippe C, Bouzinba-segard H. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. 2010;107:9281–6. [DOI] [PMC free article] [PubMed]
- 39.Chou R, Yu Y, Hung M. The roles of EZH2 in cell lineage commitment. 2011;3:243–50. [PMC free article] [PubMed]
- 40.Zhou B, Shu B, Yang J, Liu J. C-reactive protein, interleukin-6 and the risk of colorectal cancer: a meta-analysis. 2014;1397–1405. [DOI] [PubMed]
- 41.Van Kruijsdijk RCM, Van Der Wall E, Visseren FLJ. Obesity and cancer: the role of dysfunctional adipose tissue. Cancer Epidemiol Biomark Prev. 2009;18:2569–2578. doi: 10.1158/1055-9965.EPI-09-0372. [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto S, Nakagawa T, Matsushita Y, Kusano S, Hayashi T, Irokawa M, et al. Visceral fat area and markers of insulin resistance in relation to colorectal neoplasia. Diabetes Care. 2010;33:184–189. doi: 10.2337/dc09-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sikalidis AK, Varamini B. Roles of hormones and signaling molecules in describing the relationship between obesity and colon cancer. Pathol Oncol Res. 2011;17:785–790. doi: 10.1007/s12253-010-9352-9. [DOI] [PubMed] [Google Scholar]
- 44.Aleksandrova K, Nimptsch K, Pischon T. Influence of obesity and related metabolic alterations on colorectal cancer risk. Curr Nutr Rep. 2013;2:1–9. doi: 10.1007/s13668-012-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurent V, Guérard A, Mazerolles C, Le Gonidec S, Toulet A, Nieto L, et al. Periprostatic adipocytes act as a driving force for prostate cancer progression in obesity. Nat Commun. 2016;7:10230. doi: 10.1038/ncomms10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao L, Liu X, Lin ED, Wang C, Choi EY, Riban V, et al. Environmental and genetic activation of a brain-adipocyte BDNF/leptin axis causes cancer remission and inhibition. Cell. 2010;142:52–64. doi: 10.1016/j.cell.2010.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marcotorchino J, Gouranton E, Romier B, Tourniaire F, Astier J, Malezet C, et al. Vitamin D reduces the inflammatory response and restores glucose uptake in adipocytes. Mol Nutr Food Res. 2012;56:1771–1782. doi: 10.1002/mnfr.201200383. [DOI] [PubMed] [Google Scholar]
- 48.Shabahang M, Buras RR, Davoodi F, Schumaker LM, Nauta RJ, Evans SR. 1,25-Dihydroxyvitamin D3 receptor as a marker of human colon carcinoma cell line differentiation and growth inhibition. Cancer Res. 1993;53:3712–3718. [PubMed] [Google Scholar]
- 49.Evans SR, Nolla J, Hanfelt J, Shabahang M, Nauta RJ, Shchepotin IB. Vitamin D receptor expression as a predictive marker of biological behavior in human colorectal cancer. Clin Cancer Res. 1998;4:1591–1595. [PubMed] [Google Scholar]
- 50.Ziv E, Koren R, Zahalka MA, Ravid A. TNF-α increases the expression and activity of vitamin D receptor in keratinocytes: role of c-Jun N-terminal kinase. Dermatoendocrinol. 2016;8:e1137399. doi: 10.1080/19381980.2015.1137399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fetahu IS, Höbaus J, Kállay E. Vitamin D and the epigenome. Front Physiol. 2014;5:164. 10.3389/fphys.2014.00164. [DOI] [PMC free article] [PubMed]
- 52.Lee SM, Pike JW. The vitamin D receptor functions as a transcription regulator in the absence of 1,25-dihydroxyvitamin D3. J Steroid Biochem Mol Biol. 2015:4–9. 10.1016/j.jsbmb.2015.08.018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. VDR promoter overview generated by UCSC genome Browser (https://genome.ucsc.edu). The sequence analyzed is highlighted in light blue, showing that is in the promoter region of VDR and inside a CpG island and several transcription factor binding sites (POLR2A, ATF2, CTCF, EZH2, E2F6, GATA2, GATA3, CEBPB and POL2) all of them determined by experimental procedures (ENCODE project). (DOCX 188 kb)
Figure S2. NFκB1 promoter overview generated by UCSC genome Browser (https://genome.ucsc.edu). The sequence analyzed is highlighted in light blue, showing that is in the promoter region of NFκB1 and inside a CpG island and several transcription factor binding sites (SMARCA4, SPL1, STAT1, RBL2, RB1 and ETS1) all of them determined by experimental procedures (ENCODE project). (DOCX 341 kb)


