Abstract
The quinazolin-4(3H)-one structural motif possesses a wide spectrum of biological activities. DNA gyrase play an important role in induction of bacterial death. It has been shown that many quinazolin-4(3H)-one derivatives have antibacterial effects through inhibition of DNA gyrase. Based on this information we decided to synthesize novel quinazolinone Schiff base derivatives in order to evaluate their antibacterial effects. A series of novel quinazolinone Schiff base derivatives were designed and synthesized from benzoic acid. The potential DNA gyrase inhibitory activity of these compounds was investigated using in silico molecular docking simulation. All new synthesized derivatives were screened for their antimicrobial activities against three species of Gram-negative bacteria including Escherichia coli, Pseudomonas aeruginosa, Salmonella entritidis and three species of Gram-positive bacteria comprising of Staphylococcus aurous, Bacillus subtilis, Listeria monocitogenes as well as for antifungal activities against Candida albicans using the conventional micro dilution method. Most of the compounds have shown good antibacterial activities, especially against E. coli at 128 µg/mL concentration while no remarkable antifungal activities were observed for these compounds. All the synthesized compounds exhibit dock score values between -5.96 and -8.58 kcal/mol. The highest dock score among them was -8.58 kcal/mol for compound 4c.
Keywords: Schiff base, Synthesis, Quinazoline-4(3H)-ones, Antibacterial activity, Docking study
INTRODUCTION
Due to multiple resistances to antibiotics, many researchers around the world have focused their efforts on the development of new antimicrobial agents (1,2,3). DNA gyrase is an enzyme within the class of topoisomerase that catalyze changes in the topology of DNA. The presence of DNA gyrase in all bacteria except higher eukaryotes, makes it a drug target for antibacterial studies (4,5). Over the past decade, the quinazolin-4(3H)-one structural motifs containing azomethine (-C=N) functional group has become one of the main areas of interest due to possessing a wide spectrum of biological activities including antibacterial, antifungal, anticonvulsant, anti-inflammatory, antiviral, antihyperlipidemic, anti-tubercular, CNS depressant, anti-tumor, analgesic, antimalarial, and anti-cancer effects (6,7). Azomethine functional groups are formed when a primary amine reacts with a carbonyl compound (8,9). Quinazolinone as an important pharmacophore for a range of inhibitors such as tyrosine kinase, thymidylate synthase and 5HT7 receptor could be grafted by azomethine functional group to exhibit better pharmacological properties. Gefitinib (ZD-1839, Iressa_) and erlotinib (OSI-774, Tarceva), two quinazoline based structures, are already in clinic as important epidermal growth factor receptor inhibitors (Scheme 1) (10,11,12).
Scheme 1.
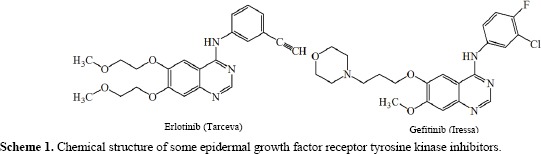
Chemical structure of some epidermal growth factor receptor tyrosine kinase inhibitors.
The molecular docking technique is one of the most interesting tools applied in pharmaceutical research. In this method one can dock small molecules into receptor targets which helps to predict ligand conformation and orientation within a targeted binding site (2,3). Moreover, to the best of our knowledge, no docking study of 3-(arylidene-amino)-2-methyl quinazolin-4(3H)-one with DNA-gyrase has been reported so far.
In the present work we have synthesized some new derivatives of 3-(arylidene-amino)-2-methyl quinazolin-4(3H)-one containing chlorine atoms at positions 6 and 8, methyl group at position 2 and various aromatic aldehydes at position 3 (Scheme 2).
Scheme 2.
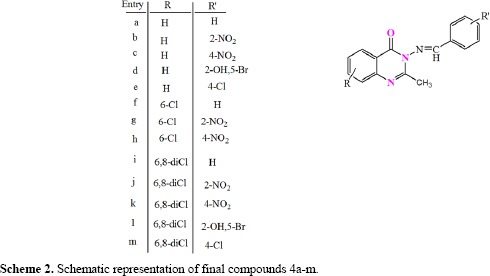
Schematic representation of final compounds 4a-m.
In vitro antimicrobial activities of these compounds were evaluated against both Gram-positive and Gram-negative bacteria as well as fungal strains. The synthesized compounds were docked into the binding sites of DNA gyrase and their binding energies were calculated.
MATERIALS AND METHODS
All the chemicals were of synthetic grade and solvents used in this study were purchased from Merck Co. (Merck, Germany). The reactions were monitored by thin-layer chromatography on silica gel (F245 Merck plates, Merk, Germany). Melting points were recorded in the open capillaries using electrothermal 9200 melting point apparatus and uncorrected. 1H NMR spectra were obtained on a Bruker 400 MHz spectrometer (United States) in deuterated dimethyl sulfoxide (DMSO-d6) using tetramethylsilane as an internal reference. Mass spectra were measured on a Shimadzu mass spectrometer (Japan). The infrared radiation spectra were determined on a WQF-510 Fourier-transform infrared (FT-IR) spectrophotometer (BRAIC Co., China) using the KBr disk method. For in silico protein-ligand docking simulation, AutoDock 4.2, Discovery studio 2.5, Hyper Chem 7.0, and Lig Plot software packages were used. Microorganism in antibacterial test was purchased from the Persian Type Culture Collection (Iran). Muller Hinton agar and Sabouraud dextrose agar were purchased from Merck (Germany). Standard antifungal drug (ketoconazole) and antibacterial drug (ciprofloxacin) (Farabi, Iran) were used for comparison.
Molecular docking studies
The novel quinazolinone Schiff base derivatives were subjected to dock in the active site of DNA gyrase enzyme using Autodock 4 software. We investigated the theoretical binding mode of 13 ligands at the chlorobiocin binding site using molecular docking modeling. Molecular docking studies were performed for these ligands to understand the ligand-receptor possible intermolecular interactions in detail. Chlorobiocin is an amino coumarin antibiotics that act by an inhibition of the DNA gyrase enzyme involved in the cell division of bacteria (13,14,15,16).
The crystal structure of the DNA gyrase (PDB code 1KZN) with resolution 2.3 Å was chosen as the protein model for the present study (17). Water molecules and ligand were removed from the protein file. The resulting crystallography structure was imported in AutoDock. The binding features of 13 synthesized compounds with DNA gyrase were evaluated in the same manner of binding of chlorobiocin as a well-known enzyme inhibitor (18).
The structures of the ligands were optimized using HyperChem 7.0 software (version 7.0; Hypercube, Inc., Gainesville, FL, USA; http://www.hyper.com). Using the MM+ molecular mechanical force field, 3D geometry optimization calculations for each ligand were performed. The ultimate conformations were calculated with the semi empirical parameterized model number 3 (PM3) method. The molecular structures were optimized using the Polak-Ribiere algorithm until the root mean square gradient was 0.01 kcal/mol/Å. Geometry optimization was run many times with different starting points of each 13 ligand (19).
Docking was performed using the routine procedure and default parameters of molecular docking AutoDock 4.2 software and implemented empirical free energy function (19). Only polar hydrogens were added to the protein and all water molecules were removed from the protein file in AutoDock Tools. In the docking protocol, ligands were assumed to be flexible molecules and the docking software was allowed to rotate all rotatable bonds of the ligands to obtain the best and optimized conformer of the ligands within the active site of the enzyme. The native ligand, chlorobiocin, was redocked to the binding site. The grid box was centered with the coordinates x = 19.259, y = 29.159, z = 42.461 for DNA gyrase (PDB code 1KZN). Grid box dimensions were 46 × 46 × 46 with a 0.375 Å grid points spacing. Grid maps were calculated by Autogrid4. A lamarckian genetic algorithm program with an adaptive whole method search in the Autodock was used to calculate the different ligand conformers (19). At the end of docking experiment with 200 runs, a cluster analysis was performed. Conformations were clustered according to the root mean square deviation tolerance of 2.0 Å and were ranked according to the binding free energy (20). Among the various conformations of these ligands obtained from the docking procedure, the conformation with the best scored pose and with the lowest binding energy was selected for these ligands (19,20). LigPlot software was used to investigate the hydrophobic and hydrogen bonding interactions between the ligand and the enzyme.
Chemistry
Quinazolinone Schiff base derivatives were prepared according to the main procedure introduced by Rezvan Rezaee Nasab, et al. (21).
Antimicrobial activity
The antimicrobial activities of the synthesized compounds were studied using broth micro dilution method against three Gram-positive bacteria of Staphylococcus aureus PTCC 1337, Bacillus subtilis PTCC 1023, and Listeria monocitogenes PTCC 1165 and three Gram-negative bacteria including Escherichia coli PTCC 1338, Pseudomonas aeruginosa PTCC 1074, and Salmonella entritidis PTCC 1091 and antifungal activity was screened against Candida albicans PTCC 5027 obtained from Persian Type Culture Collection. Using the micro plate alamar blue assay, minimum inhibitory concentration (MIC) was determined (22,23).
In this test dimethyl sulfoxide (DMSO) was used as a solvent for stock solution and diluted with water. Therefore, the final concentration of DMSO was no greater than 1%. A single 96-well micro dilution plate was used. Muller-Hinton broth and RPMI 1640 were used as media for bacterial and fungal growth, respectively (24,25).
RESULTS
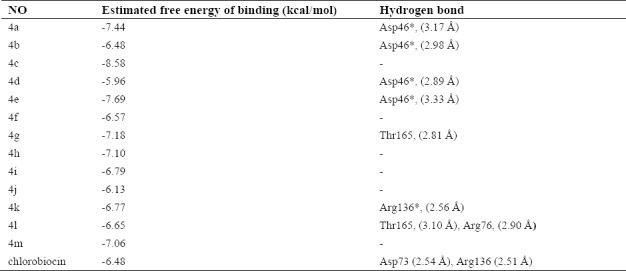
Table 1. summarizes the binding characterizations between synthesized compounds and DNA gyrase. According to the data presented in this table ligands interact with the DNA-gyrase binding site through hydrogen bonding. Amongst various conformations of the ligands obtained from the docking procedure the conformation with the best scored pose, with the lowest binding energy (~ -6 – -9 kcal/mol), and as the most populated cluster was selected. However, the best scored pose for 4d and 4l (-6.84 and -6.67 kcal/mol, respectively) was not the most populated cluster. The next cluster (-5.96 and -6.65 kcal/mol) had a similar position and orientation with the reference structure and at the same time was the most populated cluster (number of neighbors = 143 and 118, respectively).
Table 1.
Energy-based interactions and hydrogen bonds for 13 novel quinazolinone Schiff base derivatives and chlorobiocin docked into DNA gyrase
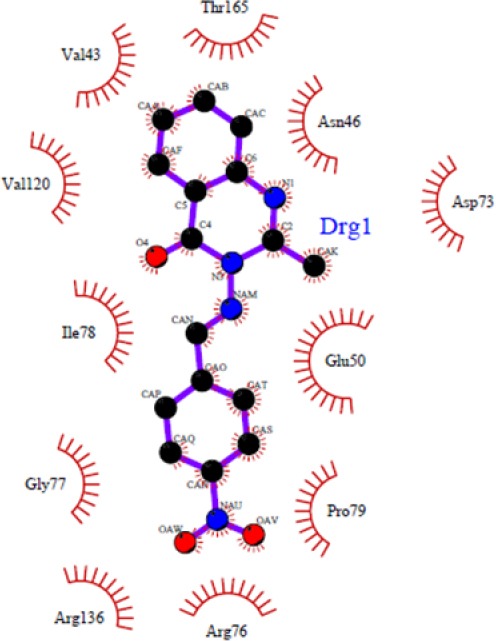
Figures 1–4 show 3D schematic presentations of compounds 4a-4l as well as chlorobiocin while docked into the chlorobiocin binding site and supports the idea that the compounds are well incorporated into the binding pocket. Dock pose of each ligand was analyzed for interactions with the receptor. These hydrophobic sites of the ligands are conserved in the majority of the structures (Figs. 1–4). The residue that interacts through a hydrogen bonding with the acceptor/donor site of the ligand differs depending on the ligand. The binding patterns of different ligands are also slightly different.
Figure 1.
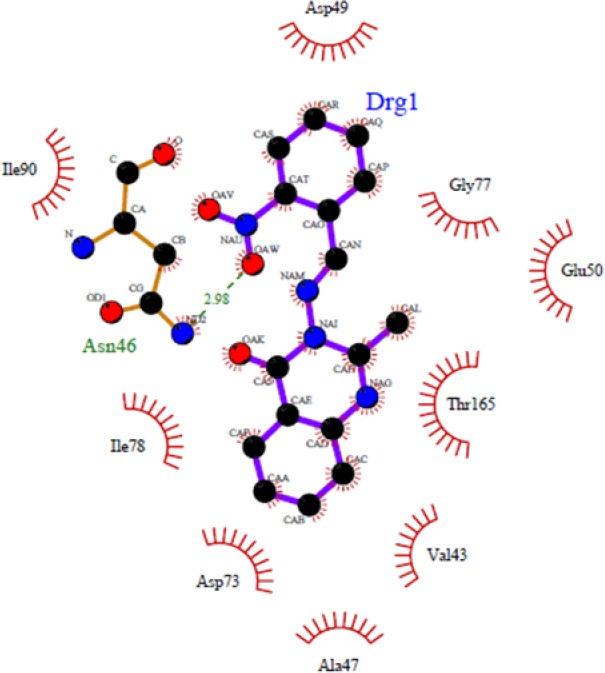
Docked conformation of compound 4b in the binding site of DNA-gyrase. Hydrogen bonds are shown by green dashed line.
Figure 4.
Binding model of compound 4c for the best docked pose in the DNA-gyrase active site
In this study, the results have shown that synthesized novel quinazolinone Schiff base derivatives followed Lipinski's RO5 (data not shown) (26).
The designed compounds were prepared through conventional synthetic pathways and characterized by spectroscopic methods. Structural properties of the newly synthesized compounds are shown below.
3-(2-nitrobenzylideneamino)-2-methyl-quinazolin-4(3H)-one (4b)
Light yellow crystals, yield: 76%. m.p: 245-246 °C, (MS: m/z (%): 308 (M+, 100), C16H12N4O3M.W. 308), FT-IR (KBr cm-1): 3064.33 (C-H arom, str.), 2962.13, 2925.48 (C-H aliph, str.), 1667.16 (N-C=O, str.), 1610.27 (C=N, str.).1H NMR (400 MHz, DMSO-d6) δ in ppm: 9.4 (1H, s, -N=CH-), 8.2 (1H, t, j = 4 Hz, Ar-H), 8.19 (1H, d, j = 3.2 Hz, Ar-H), 8.16 (1H, s, Qu-H), 7.9 (1H, t, j = 7.6 Hz, Ar-H), 7.86 (1H, t, j = 7.6 Hz, Ar-H), 7.84 (1H, t, j = 8 Hz, Qu-H), 7.6 (1H, d, j = 7.2 Hz, Qu-H), 7.5 (1H, t, j.= 8 Hz, Qu-H), 2.5 (3H, s, CH3).
3-(5-bromo-2-hydroxybenzylideneamino)- 2 -methylquinazolin-4(3H)-one (4d)
White crystals yield: 70%. m.p: 235-236 °C, (MS: m/z (%): 357 (M+, 100), C16H12BrN3O2M.W. 357), FT-IR (KBr cm-1): 3398.92 (OH, str.), 3066.26 (C-H arom, str.), 2924.52 (C-H aliph, str.), 1683.55 (N-C=O, str.), 1605.45 (C=N, str.). 1H NMR (400 MHz, DMSO-d6) δ in ppm:10.911 (1H, broad, OH,), 9.1 (1H, s, -N=CH-), 8.2 (1H, d, j = 8 Hz, Qu-H), 8.1 (1H, s, Ar-H), 7.9 (1H, t, j = 8 Hz, Qu-H), 7.7 (1H, d, j = 8 Hz, Ar-H), 7.5(1H, t, j = 8 Hz, Qu-H), 7.0 (1H, d, j = 8.8 Hz, Qu-H), 2.6 (3H, s, CH3).
3-(4-chlorobenzylideneamino)-2-methyl-quinazolin-4(3H)-one (4e)
Pale yellow crystals, yield: 90%. m.p: 203-231 °C, (MS: m/z (%): 297 (M+, 100), 299 (M+2) C16H12ClN3OM.W. 297.07), FT-IR (KBr cm-1): 3087.48 (C-H arom, str.), 2928.38 (C-H aliph, str.), 1679.69 (N-C=O, str.), 1598.7 (C=N, str.). 1H NMR (400 MHz, DMSO-d6) δ in ppm: 9.0 (1H, s, -N=CH-), 8.1 (1H, d, j = 8 Hz, Qu-H), 8.0 (2H, d, j = 7.2 Hz, Ar-H), 7.8 (1H, t, j = 8 Hz, Qu-H), 7.6 (2H, d, j = 8 Hz, Ar-H), 7.5(1H, t, j = 8 Hz, Qu-H), 2.5(3H, s, CH3).
3-(benzylideneamino)-6, 8-dichloro-2-methyl-quinazolin-4(3H)-one (4i)
Pale yellow crystals, yield: 89%. M.p: 211-212 °C, (MS: m/z(%): 331 (M+, 100), 333 (M+2), 335 (M+4) C16H11Cl2N3O M.W. 331), FT-IR (KBr cm-1): 2379.73 (C-H arom, str.), 2358.52, 2348.87 (C-H aliph, str.), 1672.95 (N-C=O, str.), 1595.81 (C=N, str.). 1H NMR (400 MHz, DMSO-d6) δ in ppm: 8.9 (1H, s, -N=CH-), 8.1 (1H, d, j = 2 Hz, Qu-H), 8.0 (1H, d, j = 2 Hz, Qu-H), 7.9 (1H, d, j = 6.8 Hz, Ar-H), 7.6 (1H, t, j = 6.8 Hz, Qu-H), 7.5 (1H, t, j = 6.8 Hz, Ar-H), 2.5 (3H, s, CH3).
3-(5-bromo-2-hydroxybenzylideneamino)-6, 8-dichloro-2-methylquinazolin-4(3H)-one (4l)
Yellow solid, yield: 80%. M.p: 220-222 °C, (MS: m/z (%): 424 (M+, 100), 426 (M+2), 428 (M+4) C16H10BrCl2N3O2M.W. 424), FT-IR (KBr cm-1): 3477.03 (C-H arom, str.), 3416.28 (OH, str.), 3071.08, 2924.52 (C-H aliph, str.), 1683.55 (N-C=O, str.), 1607.38 (C=N, str.). 1H-NMR (400 MHz, DMSO-d6) δ in ppm: 10.9 (1H, broad, OH), 9.1 (1H, s, -N=CH-), 8.19 (1H, d, j = 2.4Hz, Qu-H), 8.14 (1H, d, j = 2.4Hz, Qu-H), 8.09 (1H, d, j = 2.8Hz, Ar-H), 7.6 (1H, d, j = 8.8Hz, Ar-H), 7.0 (1H, d, j = 8.8 Hz, Ar-H), 2.6 (3H, s, CH3).
3-(4-chlorobenzylideneamino)-6, 8-dichloro-2-methylquinazolin-4(3H)-one (4m)
Light yellow crystals, yield: 90%. m.p: 210-211 °C, (MS: m/z (%): 364 (M+, 100), 366 (M+2), 368 (M+4) C16H10Cl3N3OM.W. 364), FT-IR (KBr cm-1): 3219.7 (C-H arom, str.), 3001.1, 2900.2 (C-H aliph, str.), 1679.69 (N-C=O, str.), 1598.7 (C=N, str.). 1H NMR (400 MHz, DMSO-d6) δ in ppm: 8.9 (1H, s, -N=CH-), 8.1 (1H, d, j = 2.4 Hz, Qu-H), 8.0 (1H, d, j = 2.4 Hz, Qu-H), 7.9 (2H, d, j = 8.4 Hz, Ar-H), 7.6 (2H, d, j = 8.4 Hz, Ar-H), 2.6 (3H, s, CH3).
All the synthesized compounds were tested for antibacterial activities against three species of Gram-negative bacteria and three species of Gram-positive and also for antifungal activities. The results of biological effects are shown in Table 2 and 3.
Table 2.
MIC (µg/mL) results of synthesized compounds against various bacteria
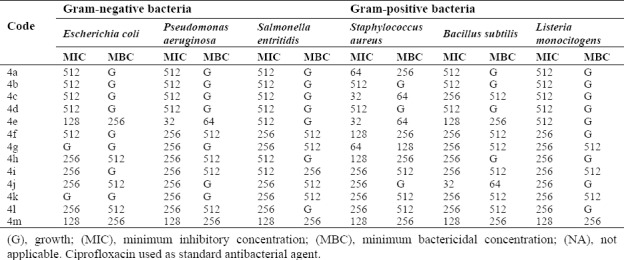
Table 3.
MIC (µg/mL) and results of synthesized compounds against Candida albicans

DISCUSSION
As shown in Fig. 1, some compounds (4b, 4d, 4e, and 4h) can create a strong hydrogen bond with Asn46 at distance 2.89 Å - 3.33 Å, which is consistent with the decomposition analysis of the electrostatic interaction. It is interesting that more complex stabilization might result from the hydrogen bonds between these ligands and Arg136 via nitro electron withdrawing substitution on the phenyl ring (for 4h and 4k) (Fig. 2). Although these interactions were also observed for these derivatives, these are different from those present in 4g (Thr165) and 4l (Thr165 and Arg76) (Fig. 3) which may be responsible for the activity variations. These results are consistent with the X-ray cocrystal structure and previous studies indicating the important roles of those residues (27,28,29,30). No hydrogen bonds interaction for 4a, 4c, 4f, 4i, 4j, 4l, and 4m was predicted (Fig. 4).
Figure 2.
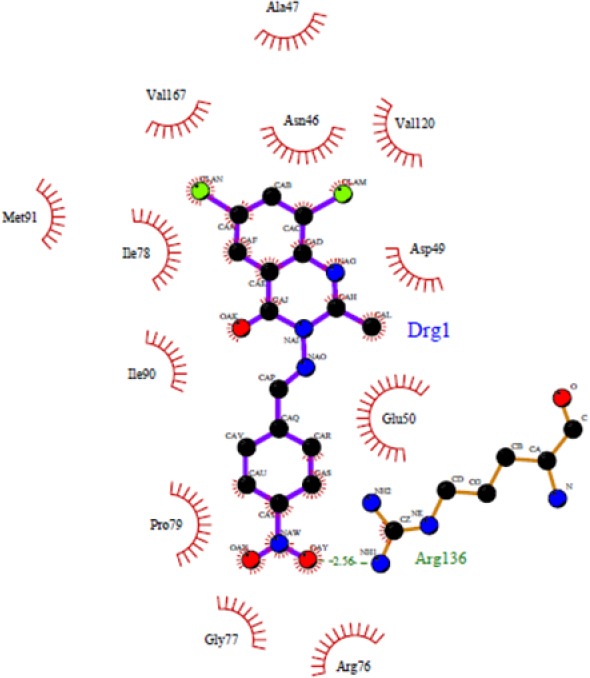
Docked conformation of ligand structure 4k in the binding site of DNA-gyrase. Hydrogen bonds are shown by green dashed line.
Figure 3.
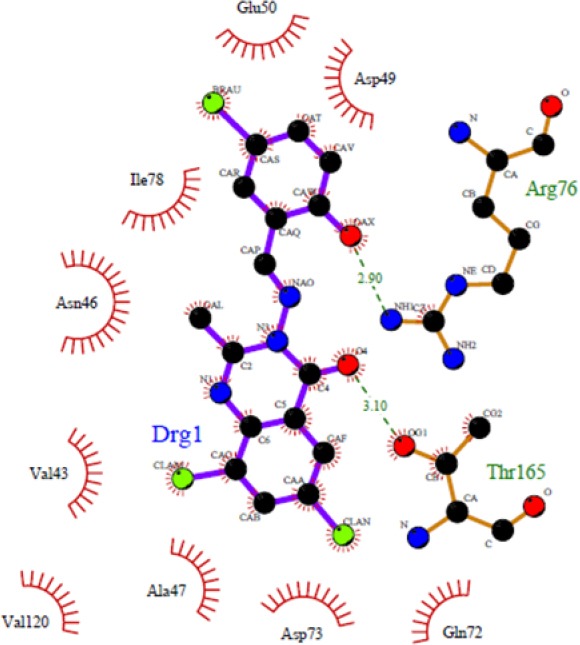
Docked conformation of ligand structure 4l in the binding site of DNA-gyrase. Hydrogen bonds are shown by green dashed line.
These compounds also significantly stabilize the DNA gyrase through hydrophobic contacts with Ala47, Glu50, Val71, Asp73, Arg76, Gly77, Ile78, Pro79, Met91, Val120, Thr163, Thr165, and Val167. Compounds were deeply embedded into the hydrophobic pocket. In all compounds it is clear that the hydrophobic pocket of the inhibitor binding site was occupied by quinazolinone or phenyl along with the groups substituted on these rings.
The docking protocol adopted in this investigation was validated by docking of chlorobiocin as a well-known inhibitor to the energy minimized DNA gyrase protein. The residues Asp73, Asn46, and Arg136 are important in making hydrogen bond (31,32,33). Most of the compounds in this study also showed a strong hydrogen bond with Asn46.
The highest dock score in docking protocol for these series was -8.58 kcal/mol for compound 4c. Rest of the molecules showed a proper dock scores ranging from -5.96 to -8.58 kcal/mol. Thus, the binding model reported here, suggests that these quinazolinone Schiff base derivatives behave as DNA gyrase inhibitors and show some key structural points to be considered in future optimization.
In the biological assay, most of the synthesized compounds showed potent activities against both the Gram-positive and Gram-negative bacterial species. Obtained results of screened compounds against Gram negative organisms demonstrated that compounds 4e and 4m had the highest activities against E.coli at 128 μg/mL. Compound 4e has shown the best activity against P. aeruginosa and S. aureus at 32 μg/mL. Compound 4c has shown the highest activity at 32 μg/mL against Staphylococcus aureus strain.
Structure-activity relationship studies based on the observed results showed that in 4a-4l, presence of electron-withdrawing groups (chlorine group) at the para positions of the phenyl ring could be responsible for good activities because of its inductive effect. Interestingly, compounds that are substituted with 6, 8-dichloro have shown better activity than 6-chloro substituted compounds. Furthermore, these compounds have better activity than unsubstituted quinazolinone derivatives (34,35). Results of antifungal study showed that almost all of the tested compounds have no antifungal activity against C. albicans, except for compound 4m which showed good activity at 128 μg/mL (Table 3).
CONCLUSION
All of the synthesized compounds showed good antibacterial activity, especially against E.coli at 128 μg/mL concentration while no remarkable antifungal activities were observed for these compounds. In the docking experiments, all compounds were able to interact in a manner similar to the known inhibitors with the residues located in the DNA-gyrase binding site. All derivatives exhibited the docking ΔG binding values lower than chlorobiocin except for 4d. The highest dock score in docking protocol for these series was -8.58 kcal/mol for compound 4c. This study indicates that quinazolinone Schiff base derivatives could be a suitable scaffold for DNA gyrase inhibitors and may be helpful in the search for novel classes of potent antibiotic agents.
ACKNOWLEDGEMENTS
The content of this paper is extracted from the Ph.D thesis No. 394158 submitted by Rezvan Rezaee Nasab which was financially supported by the Isfahan University of Medical Sciences, Isfahan, I.R. Iran.
REFERENCES
- 1.Asadi P, Khodarahmi GA, Jahanian-Najafabadi A, Saghaie L, Hassanzadeh F. Synthesis, characterization, molecular docking studies and biological evaluation of some novel hybrids based on quinazolinone, benzofuran and imidazolium moieties as potential cytotoxic and antimicrobial agents. Iran J Basic Med Sci. 2017;20(9):975–989. doi: 10.22038/IJBMS.2017.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collin F, Karkare S, Maxwell A. Exploiting bacterial DNA gyrase as a drug target: current state and perspectives. Appl Microbiol Biotechnol. 2011;92(3):479–497. doi: 10.1007/s00253-011-3557-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrov DA, Hernández Prada JA, Corsino PE, Finton KA, Le N, Rowe TC. Discovery of novel DNA gyrase inhibitors by high-throughput virtual screening. Antimicrob Agents Chemother. 2007;51(10):3688–3698. doi: 10.1128/AAC.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heddle J, Maxwell A. Quinolone-binding pocket of DNA gyrase: role of GyrB. Antimicrob Agents Chemother. 2002;46(6):1805–1815. doi: 10.1128/AAC.46.6.1805-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar R, Madhumathi BS, Nagaraja V. Molecular basis for the differential quinolone susceptibility of mycobacterial DNA gyrase. Antimicrob Agents Chemother. 2014;58(4):2013–2020. doi: 10.1128/AAC.01958-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rezaee Nasab R, Karami B, Khodabakhshi S. Selective solvent-free biginelli condensation using tungstate sulfuric acid as powerful and reusable catalyst. Bull Chem React EngCatalys. 2014;9(2):148–154. [Google Scholar]
- 7.Rakesh KP, Darshini N, Shubhavathi T, Mallesha N. Biological applications of quinazolinone analogues: A review. Org Med Chem. 2017;2(2):1–5. [Google Scholar]
- 8.Farag AA, Khalifa EM, Sadik NA, Abbas SY, Al-sehemi AG, Ammar YA. Synthesis, characterization, and evaluation of some novel 4(3H)-quinazolinone derivatives as anti-inflammatory and analgesic agents. Med Chem Res. 2013;22(1):440–452. [Google Scholar]
- 9.Agbo EN, Makhafola TJ, Choong YS, Maphahlele MJ, Ramasami P. Synthesis, biological evaluation and molecular docking studies of 6-aryl-2-styrylquinazolin-4(3H)-ones. Molecules. 2016;21(1):E28. doi: 10.3390/molecules21010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert IH. Inhibitors of dihydrofolate reductase in leishmania and trypanosomes. Biochim Biophys Acta. 2002;1587(2-3):249–257. doi: 10.1016/s0925-4439(02)00088-1. [DOI] [PubMed] [Google Scholar]
- 11.Jackman AL, Taylor GA, Gibson W, Kimbell R, Brown M, Calvert AH, et al. ICI D1694, a quinazoline antifolate thymidylate synthase inhibitor that is a potent inhibitor of L1210 tumor cell growth in vitro and in vivo: a new agent for clinical study. Cancer Res. 1991;51(20):5579–5586. [PubMed] [Google Scholar]
- 12.Sequist LV, Lynch TJ. EGFR tyrosine kinase inhibitors in lung cancer: an evolving story. Annu Rev Med. 2008;59:429–442. doi: 10.1146/annurev.med.59.090506.202405. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49(11):3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 14.Nanda AK, Ganguli S, Chakraborty R. Antibacterial activity of some 3-(arylideneamino)-2-phenylquinazoline-4(3H)-ones: synthesis and preliminary QSAR studies. Molecules. 2007;12(10):2413–2426. doi: 10.3390/12102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bansal S, Kumar S, Aggarwal V, Joseph A. Design, synthesis, docking study & antibacterial evaluation of 1, 3-diarylpyrazolyl substituted indolin-2-ones. Indo Glob J Pharm Sci. 2014;4(1):1–7. [Google Scholar]
- 16.Jayashree BS, Thomas S, Nayak Y. Design and synthesis of 2-quinolones as antioxidants and antimicrobials: a rational approach. Med Chem Res. 2010;19(2):193–209. [Google Scholar]
- 17.Lafitte D, Lamour V, Tsvetkov PO, Makarov AA, Klich M, Deprez P, et al. DNA gyrase interaction with coumarin-based inhibitors: the role of the hydroxybenzoate isopentenyl moiety and the 5’-methyl group of the noviose. Biochemistry. 2002;41(23):7217–7223. doi: 10.1021/bi0159837. [DOI] [PubMed] [Google Scholar]
- 18.Rezaee Nasab R, Hassanzadeh F, Khodarahmi GA, Rostami M, Mirzaei M, Jahanian-Najafabadi A, et al. Docking study, synthesis and antimicrobial evaluation of some novel 4-anilinoquinazoline derivatives. Res Pharm Sci. 2017;12(5):425–433. doi: 10.4103/1735-5362.213988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mansourian M, Fassihi A, Saghaie L, Madadkar-Sobhani A, Mahnam K, Abbasi M. QSAR and docking analysis of A2B adenosine receptor antagonists based on non-xanthine scaffold. Med Chem Res. 2015;24:394–407. [Google Scholar]
- 20.Mansourian M, Mahnam K, Madadkar-Sobhani A, Fassihi A, Saghaie L. Insights into the human A1 adenosine receptor from molecular dynamics simulation: Structural study in the presence of lipid membrane. Med Chem Res. 2015;24:3645–3659. [Google Scholar]
- 21.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639–1662. [Google Scholar]
- 22.Mansourian M, Madadkar-Sobhani A, Mahnam K, Fassihi A, Lotfollah Saghaie L. Characterization of adenosine receptor in its native environment: insights from molecular dynamics simulations of palmitoylated/glycosylated, membrane-integrated human A2B adenosine receptor. J Mol Model. 2012;18:4309–4324. doi: 10.1007/s00894-012-1427-y. [DOI] [PubMed] [Google Scholar]
- 23.Rezaee Nasab R, Hassanzadeh F, Khodarahmi GA, Mirzaei M, Rostami M, Jahanian-Najafabadi A. Synthesis, characterization, cytotoxic screeninig, and density functional theory studies of new derivatives of quinazolin-4(3H)-one Schiff bases. Res Pharm Sci. 2017;12(6):444–455. doi: 10.4103/1735-5362.217425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jayalakshmi B, Raveesha KA, Amruthesh KN. Evaluation of antibacterial and antioxidant potential of Euphorbia cotinifolia linn. leaf extracts. Chem Ind Chem Eng Q. 2014;20(1):19–28. [Google Scholar]
- 25.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 26.Baker CN, Banerjee SN, Tenover FC. Evaluation of Alamar colorimetric MIC method for antimicrobial susceptibility testing of gram-negative bacteria. J Clin Microbiol. 1994;32(5):1261–1267. doi: 10.1128/jcm.32.5.1261-1267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khodarahmi GA, Jafari E, Hakimelahi Gh, Abedi D, Rahmani Khajouei M, Hassanzadeh F. Synthesis of some new quinazolinone derivatives and evaluation of their antimicrobial activities. Iran J Pharm Res. 2012;11(3):789–797. [PMC free article] [PubMed] [Google Scholar]
- 28.Lipinski CA. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol. 2004;1(4):337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Kampranis SC, Gormley NA, Tranter R, Orphanides G, Maxwell A. Probing the binding of coumarins and cyclothialidines to DNA gyrase. Biochemistry. 1999;38(7):1967–1976. doi: 10.1021/bi982320p. [DOI] [PubMed] [Google Scholar]
- 30.Lewis RJ, Singh OM, Smith CV, Skarzynski T, Maxwell A, Wonacott AJ, et al. The nature of inhibition of DNA gyrase by the coumarins and the cyclothialidines revealed by X-ray crystallography. EMBO J. 1996;15(6):1412–1420. [PMC free article] [PubMed] [Google Scholar]
- 31.Blance SJ, Williams NL, Preston ZA, Bishara J, yth MS, Maxwell A. Temperature-sensitive suppressor mutations Smyth MS, Maxwell A. Temperature-sensitive suppressor mutations of the Escherichia coli DNA gyrase B protein. Protein Sci. 2009;9(5):1035–1037. doi: 10.1110/ps.9.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyapati S, Kulandaivelu U, Sangu S, Vanga MR. Synthesis, antimicrobial evaluation, and docking studies of novel 4‐substituted quinazoline derivatives as DNA‐gyrase inhibitors. Arch Pharm (Weinheim) 2010;343(10):570–576. doi: 10.1002/ardp.201000065. [DOI] [PubMed] [Google Scholar]
- 33.Mladenović M, Vuković N, Sukdolak S, Solujić S. Design of novel 4-hydroxy-chromene-2-one derivatives as antimicrobial agents. Molecules. 2010;15(6):4294–4308. doi: 10.3390/molecules15064294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahimi H, Najafi A, Eslami H, Negahdari B, Moosazadeh Moghaddam M. Identification of novel bacterial DNA gyrase inhibitors: An in silico study. Res Pharm Sci. 2016;11(3):250–258. [PMC free article] [PubMed] [Google Scholar]
- 35.Patil RB, Sawant SD. Synthesis, characterization, molecular docking and evaluation of antimicrobial activity of some 3-heteroaryl substituted chromen-2-one derivatives. Der Pharma Chemica. 2015;7(3):26–37. [Google Scholar]
- 36.Patel NB, Patel JC. Synthesis and antimicrobial activity of Schiff bases and 2-azetidinones derived from quinazolin-4(3H)-one. Arab J Chem. 2011;4(4):403–411. [Google Scholar]
- 37.Wang X, Yin J, Shi L, Zhang G, Song B. Design, synthesis, and antibacterial activity of novel Schiff base derivatives of quinazolin-4(3H)-one. Eur J Med Chem. 2014;77:65–74. doi: 10.1016/j.ejmech.2014.02.053. [DOI] [PubMed] [Google Scholar]


