Abstract
Vanillin is the most important flavor compound in the vanilla pod. Vanilla planifolia vanillin synthase (VpVAN) catalyzes the conversion of ferulic acid and ferulic acid glucoside into vanillin and vanillin glucoside, respectively. Desorption electrospray ionization mass spectrometry imaging (DESI-MSI) of vanilla pod sections demonstrates that vanillin glucoside is preferentially localized within the mesocarp and placental laminae whereas vanillin is preferentially localized within the mesocarp. VpVAN is present as the mature form (25 kDa) but, depending on the tissue and isolation procedure, small amounts of the immature unprocessed form (40 kDa) and putative oligomers (50, 75 and 100 kDa) may be observed by immunoblotting using an antibody specific to the C-terminal sequence of VpVAN. The VpVAN protein is localized within chloroplasts and re-differentiated chloroplasts termed phenyloplasts, as monitored during the process of pod development. Isolated chloroplasts were shown to convert [14C]phenylalanine and [14C]cinnamic acid into [14C]vanillin glucoside, indicating that the entire vanillin de novo biosynthetic machinery converting phenylalanine to vanillin glucoside is present in the chloroplast.
Keywords: Chloroplasts, Immunolocalization, Phenyloplasts, Vanillin biosynthesis, Vanillin synthase, VpVAN
Introduction
In the last several decades, extensive studies have been carried out to gain a better understanding of the cellular machineries involved in efficient and channeled synthesis of plant natural products. One approach towards elucidation of the biosynthetic pathways involved and the physiological roles of the compounds produced is to determine the tissue, cellular and subcellular localization of their site of biosynthesis and accumulation. In this study, we set out to determine the biosynthetic production site of vanillin (4-hydroxy-3-methoxybenzaldehyde), the world’s most popular flavor and aroma compound.
Vanilla and its key flavor component vanillin are universally appreciated flavors. Vanilla is the complete alcohol extract of the mature vanilla seed capsule, commonly called a pod (Sinha et al. 2008). Vanilla plants belong to the Orchidaceae family, the genus Vanilla, tribe Vanilleae, subfamily Vanilloideae (Cameron et al. 1999, Cameron 2004, Cameron and Molina 2006). Among the approximately 110 vanilla species in the genus Vanilla, three species are used for production of vanilla extracts by local farmers as well as at an industrial scale: Vanilla planifolia, V. tahitensis and V. pompona. Vanilla planifolia is the most valued among these three species because of its high vanilla flavor quality, providing 95% of the world vanilla pod production (Odoux and Grisoni 2010).
Vanilla planifolia is a climbing perennial vine with a large, green and succulent stem that is photosynthetic. The plant produces oblong, smooth, bright green leaves, and adventitious aerial roots that grow opposite each leaf, aiding lateral support. The roots are associated with endotrophic mycorrhiza (Anuradha et al. 2013). The vanilla flowers are yellow, bisexual and develop towards the top of the plant when the vine is approximately 4–5 m long. When successful pollination occurs, each flower yields a single pod. The pod reaches its full size about 3 months after pollination. The immature green V. planifolia pods are almost odorless as the key flavor components are stored as glucosides. Mature pods are approximately 15 cm long and are pale green to yellow in color (Odoux and Brillouet 2009). Vanilla planifolia pods are harvested when they are 8-9-months-old, before the pods begin to dehisce. Freshly harvested pods are processed by curing to stop the natural vegetative processes, to initiate the enzymes responsible for the formation of the aromatic flavor constituents and to prevent microbial growth, thereby enabling long-term preservation (Odoux and Grisoni 2010).
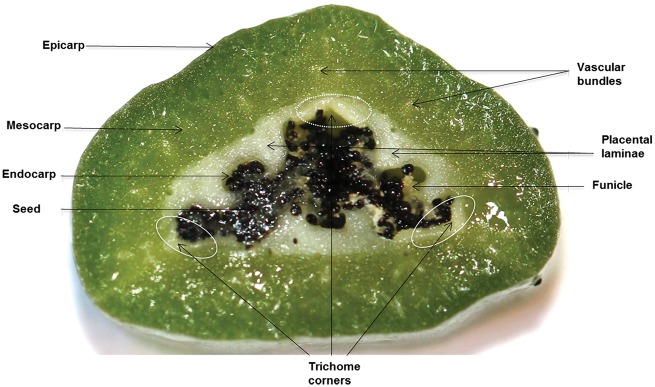
Fig. 1 shows a transverse section of a V. planifolia pod to demonstrate its anatomy and tissue terminology. The V. planifolia pod encompasses three areas, which are visually distinct; the outer part (greener area), inner part (white/yellowish green area) and seeds. The outer part includes the epicarp and outer mesocarp. The inner part includes the inner mesocarp, placental laminae, endocarp and seeds. In total, the mesocarp is formed by 15–20 layers of large cells. Seeds are localized towards the cavity of the pod (Fig. 1) (Odoux et al. 2003b, Odoux and Brillouet 2009).
Fig. 1.
Transverse section of a 6-month-old vanilla pod, with arrows pointing to the different tissues present.
The vanilla pod is known to produce >200 different flavor compounds (Sinha et al. 2008). Vanillin is the most abundant compound and provides the key flavor and aroma of the vanilla extract and of the cured V. planifolia pod (Sinha et al. 2008). The compound vanillin is suggested to have various physiological functions in the plant (Burri et al. 1989, Lopezmalo et al. 1995). As vanillin is toxic to living organisms in high concentrations (Boonchird and Flegel 1982), V. planifolia plants store vanillin as vanillin-β-d-glucoside, a conjugated form with glucose, commonly called vanillin glucoside or glucovanillin.
Vanilla planifolia pods are the prime plant organ source of vanillin and the site of vanillin glucoside biosynthesis and storage (Odoux et al. 2003b, Odoux and Brillouet 2009, Gallage et al. 2014). Vanillin glucoside starts to accumulate in the inner part of the pod when they are 3-month-old, and continues to do so until the pod is 7-8-months-old, reaching concentrations >300 mM in the water phase of the mesocarp cells (Odoux et al. 2003b, Odoux et al. 2006, Odoux and Brillouet 2009, Palama et al. 2009). Vanillin glucoside was shown by Odoux et al. (2003b) to accumulate in the inner part of the mesocarp and placental laminae, as the main sites. In contrast, Havkin-Frenkel and Dixon reported that vanillin glucoside was produced and accumulated in a unique hairy secretory tissue, the trichomes, and accumulated in the secretion around the seeds (Joel et al. 2003). In a subsequent and thorough study, the conclusions of the latter study were refuted (Odoux and Brillouet 2009). Vanillin is distributed in similar tissues to vanillin glucoside yet at an about 20- to 50-fold lower concentration (Odoux et al. 2003b, Odoux and Brillouet 2009). Vanillin and its glucoside are absent from seeds (Odoux and Brillouet 2009).
Vanillin production in V. planifolia may be divided into three modules: synthesis of vanillin via ferulic acid by a C−C chain shortening step, glucosylation of vanillin to vanillin glucoside (the non-toxic storage form) and hydrolysis of vanillin glucoside and liberation of the aromatic compound vanillin.
The biosynthetic pathway of vanillin in the pod of V. planifolia has recently been elucidated. Vanillin is synthesized via conversion of ferulic acid and ferulic acid glucoside to vanillin and vanillin glucoside, respectively (Negishi et al. 2009, Gallage et al. 2014). This reaction is catalyzed by a single enzyme, referred to as vanillin synthase (VpVAN, accession No. KP278240.1) as demonstrated by the ability of the enzyme to convert ferulic acid/ferulic acid glucoside into vanillin/vanillin glucoside following coupled transcription/translation of the VpVAN gene in in vitro assays, following transient expression of the gene in Nicotiana benthamiana and following stable expression in Hordeum vulgare and Saccharomyces cerevisiae (Gallage et al. 2014).
VpVAN has high amino acid sequence similarity to enzymes of the cysteine protease family (Gallage et al. 2014). Cysteine proteases are a large group of enzymes with versatile physiological functions and not very well defined substrate specificities (Storer and Menard 1996). Papain (EC 3.4.22.2) from the latex of Carica papaya is the best studied plant cysteine protease (Otto and Schirmeister 1997, Shindo and Van der Hoorn 2008). In general, cysteine proteases are expressed as immature proteins with an N-terminal endoplasmic reticulum (ER)-targeting signal peptide being part of a propeptide domain comprising 130–160 residues (Wiederanders et al. 2003, Cambra et al. 2012). In the mature protein, the propeptide sequence is removed either by a processing enzyme or by autocatalytic processing (Turk et al. 2012). Two putative protease cleavage sites in VpVAN were predicted at residue 61 (RFAR/RYGK) (Gallage et al. 2014) and residue 135 (VD/GVLPVT) (Yang et al. 2017). The in vitro transcription/translation experiments showed no evidence of autocatalytic processing of the VpVAN protein. This indicates that removal of the propeptide requires the action of a separate processing enzyme (Gallage et al. 2014). The physiological function of the VpVAN propeptide is not known. In general, it has been proposed that the propeptide sequence may control proper intracellular targeting, promote proper folding of the mature enzyme and/or serve to maintain the enzyme in an inactive form in the cell to balance its function according to physiological demands (Turk et al. 2012). In the cysteine protease papain, the presence of the N-terminal propeptide blocks the active site cleft directly by non-covalent interactions (Turk et al. 2012). Both the immature and mature forms of VpVAN were shown to be catalytically active, with the latter being more active (Gallage et al. 2014).
The glucosylation step resulting in the conversion of vanillin into its glucoside is much less defined. It is not known at which step glucose incorporation occurs in the biosynthetic pathway. If vanillin glucoside is preferentially made directly from ferulic acid glucoside, the glucosylation may proceed at the level of p-coumaric acid, caffeic acid or ferulic acid. In the green pod, vanillin is almost entirely stored as vanillin glucoside, demonstrating that the glucosylation of vanillin is highly efficient or, alternatively, that free vanillin is not involved as an intermediate in vanillin glucoside formation. Our previous proteomic and transcriptomic studies demonstrated that several UDP-glucosyltransferases (UGTs) are present in the V. planifolia pod. Of these, VpUGT72E1 has been shown to be a vanillin-specific glucosyltransferase (Gallage et al. 2014).
The third module in vanillin formation involves hydrolysis of vanillin glucoside and liberation of the aromatic compound. Hydrolysis of vanillin glucoside takes place when the cells are broken down as part of the pod maturation process or as a result of pathogen attack and under the curing process. The enzyme which is involved in hydrolyzing vanillin glucoside, vanillin β-d-glucosidase, has been well characterized (Odoux et al. 2003a, Odoux et al. 2003b). The β-d-glucosidase was purified to homogeneity and demonstrated to be a tetramer (201 kDa) composed of four identical subunits (50 kDa). The optimum pH was 6.5 and the Km value for vanillin glucoside was 20.0 mM; Vmax was 5.0 μkat mg−1 (Odoux et al. 2003a). The high Km value matches the high substrate concentration. At the tissue level, vanillin β-d-glucosidase activity was found to be distributed in the inner mesocarp and placental laminae in a similar manner to vanillin glucoside (Odoux et al. 2003b). The subcellular localization of the β-d-glucosidase throughout pod development was determined using a specific antibody and confocal microscopy (Brillouet et al. 2014), and it was demonstrated that the β-d-glucosidase was localized as in a corona around re-differentiating chloroplasts, probably being situated in the lumen between the inner and outer envelopes. In 4-month-old pods, few re-differentiating chloroplasts were visible among the photosynthetically active chloroplasts in the cytoplasm of cells of the inner mesocarp. In 8-month-old pods, almost all chloroplasts were re-differentiated, as demonstrated by the presence of a corona of β-d-glucosidase and complete loss of Chl red fluorescence. The re-differentiation of chloroplasts was followed by transmission electron microscopy during pod ontogeny and demonstrated a progressive dismantling of the grana thylakoids and thylakoid budding, resulting in the formation of circular membrane vesicles packed with ribosomes. At full pod maturity, internal membrane structures were no longer visible but the β-d-glucosidase remained localized as a corona around the re-differentiated chloroplasts (Brillouet et al. 2014).
Brillouet et al. also proceeded to investigate the subcellular localization of vanillin glucoside during pod ontogeny (Brillouet et al. 2014). Using multiple cell imaging approaches combining immunohistochemistry localization by confocal microscopy and fluorescence spectral analysis by multiphotonic microscopy, it was discovered that vanillin glucoside was progressively stockpiled in the inner volume of the re-differentiated chloroplasts as solid amorphous deposits. The vanillin glucoside concentration was estimated to exceed 4 M and the deposition as an amorphous solid was suggested to ensure homeostasis. At the end of the re-differentiation process, the vanillin glucoside deposits filled the entire plastidial volume (Brillouet et al. 2014). Based on the discovered function of the re-differentiated chloroplasts as a storage site for a phenolic plant-specialized metabolite, the re-differentiated chloroplasts were named phenyloplasts (Brillouet et al. 2014). In summary, the vanillin glucoside biosynthetic pathway, the storage site of vanillin glucoside in re-differentiated chloroplasts termed phenyloplasts as well as the β-d-glucosidase involved in aroma release have been elucidated.
In our current study, we set out to increase further our understanding of how V. planifolia orchestrates the biosynthesis of vanillin glucoside in such high concentrations by investigating the cellular and intracellular site of vanillin biosynthesis in the V. planifolia pod and the post-translational processing of VpVAN. Desorption electrospray ionization mass spectrometry imaging (DESI-MSI) was used to visualize the distribution of the main flavor components within the V. planifolia pod. A polyclonal antibody specific for the C-terminal sequence of VpVAN was used to demonstrate that the VpVAN protein and thus vanillin glucoside biosynthesis is localized in chloroplasts and in the re-differentiated chloroplasts termed phenyloplasts. Radiolabeling experiments support that chloroplasts de novo synthesize vanillin glucoside from phenylalanine and cinnamic acid.
Results
Vanillin glucoside is localized in the inner part of the vanilla pod where vanillin biosynthetic activity is detected
Administration of 14C-radiolabeled precursors to V. planifolia pods harvested at 3, 4, 5, 6, 7, 8 and 9-months following pollination demonstrated that the pods actively biosynthesized vanillin. The studies with radiolabeled precursors were carried out using V. planifolia pods shipped from La Reunion to Copenhagen by courier mail while still attached to their vine. In one set of experiments, [14C]phenylalanine, [14C]cinnamic acid, p-[14C]hydroxybenzaldehyde and [14C]vanillin were administered to the inner yellow fleshy region of the pod including the inner mesocarp, placental laminae and endocarp. In a second set of parallel experiments, the radiolabeled compounds were administered to the thick dark green outer part of the pod including mainly the epicarp and outer mesocarp. Radiolabeled metabolites formed were separated by thin-layer chromatography (TLC) (Supplementary Fig. S1). Upon administration of [14C]phenylalanine and [14C]cinnamic acid to the inner part of the pod, a radiolabeled product co-migrating with the vanillin glucoside authentic standard was observed, while administration of p-[14C]hydroxybenzaldehyde gave rise neither to formation of radiolabeled vanillin nor to vanillin glucoside. These results show that an active vanillin glucoside biosynthetic machinery is situated in the inner part of the V. planifolia pod throughout pod development. The time course study also demonstrated that administration of p-[14C]hydroxybenzaldehyde did not contribute to vanillin biosynthesis at any of the time points examined during V. planifolia pod development. It should be noted that the administration of radiolabeled [14C]vanillin resulted in formation of radiolabeled vanillin glucoside in the inner as well as the outer part of the vanilla pod, demonstrating that vanillin glucosyltransferase activity was also present in parts of the pod not catalyzing de novo synthesis of vanillin (Supplementary Fig. S1).
DESI-MSI was carried out to visualize the localization of the two main metabolites, vanillin and vanillin glucoside, in the V. planifolia pod using longitudinal and cross-sections of a 6-month-old V. planifolia pod (Fig. 2). In the recordings obtained, the color intensity of a pixel in each of the compound-specific images is scaled relative to the intensities for that compound in the rest of the image, and not relative to the signal of other detected compounds. MS images thus provide information about the distributions of individual compounds, but not about their mutual abundances. Comparison of absolute intensities obtained with different compounds in the imaging experiment cannot be translated into mutual abundances due to their differences in ionization efficiency.
Fig. 2.
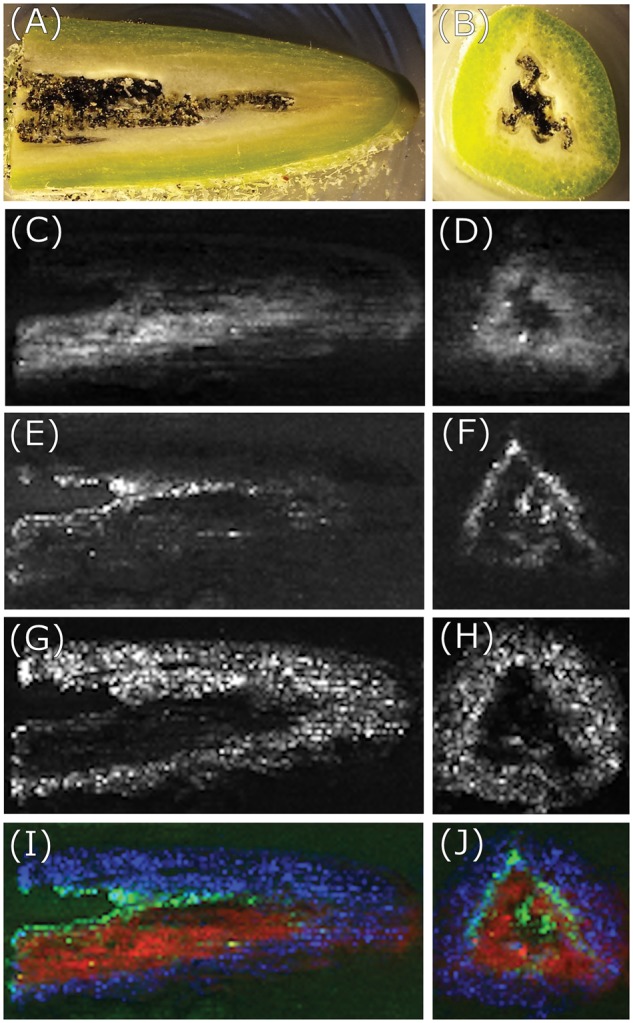
Images obtained by DESI-MSI of longitudinal (left column) and cross- (right column) sections of a 6-month-old vanilla pod. (A and B) Photo of vanilla pod tissue obtained immediately following longitudinal and cross cryosectioning, respectively. (C and D) Vanillin distribution (m/z 153). (E and F) Vanillin glucoside distribution (m/z 337). (G and H) Sucrose distribution (m/z 381). (I and J) Colored overlay of vanillin (red), vanillin glucoside (green) and sucrose (blue) distributions.
As shown in Supplementary Fig. S1, radiolabeling experiments based on administration of 14C-radiolabeled precursors demonstrated that vanillin biosynthesis takes place in the inner part of the pod. DESI-MS images (Fig. 2C, D and E, F) visualize the tissue distribution of vanillin and vanillin glucoside, respectively, in a 6-month-old pod. Both compounds are present in the tissues where biosynthetic activity was detected. Vanillin glucoside is specifically localized within the inner mesocarp, placental laminae and endocarp (Fig. 2E, F), while vanillin is distributed within the mesocarp (Fig. 2C, D). The partly non-superimposed distribution of vanillin and vanillin glucoside suggests that one or more vanillin β-glucosidases and/or vanillin glucosyltransferase activity are differentially distributed at this pod developmental stage (Fig. 2C–F).
Presence of VpVAN as demonstrated by Western blot analysis of V. planifolia protein extracts and following transient expression of VpVAN in tobacco leaves
The presence of the VpVAN protein was monitored during pod development using a specific VpVAN antibody prepared towards the C-terminal peptide (NH2-) CNWGDNGYFKMELGK (-CONH2) and used in a 1 : 5,000 dilution. No background reactions were observed using pre-immune serum from the same rabbit (Supplementary Fig. S2). In agreement with previous results (Gallage et al. 2014), the recognized protein bands at 25 and 40 kDa match the calculated molecular masses of 23.89 and 39.15 kDa of mature and immature VpVAN, respectively. These immunoblot patterns demonstrate that VpVAN undergoes post-translational maturation in V. planifolia as well as in tobacco where the propeptide is cleaved off, as generally observed for classical cysteine proteases (Wiederanders et al. 2003, Gallage et al. 2014). Comparing the strength of the band patterns, it is apparent that VpVAN is present almost entirely in its mature form in 7-month-old V. planifolia pods (Fig. 3). The highest concentration of vanillin and vanillin glucoside is found in 6- to 7-month-old pods. It is thus likely that the activity of the protein is stimulated upon maturation of VpVAN (Fig. 3).
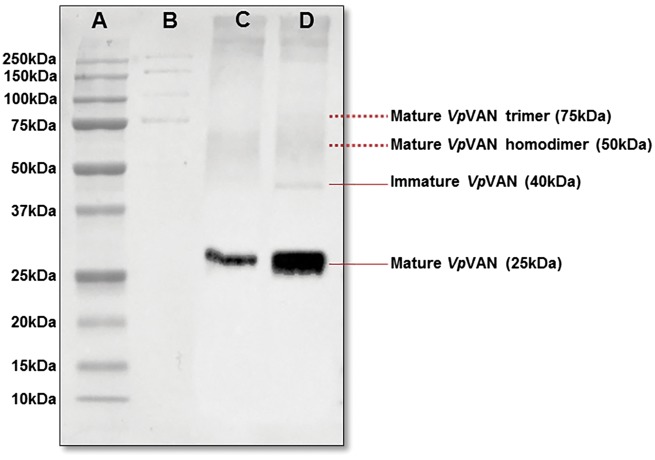
Fig. 3.
Western blot analysis of the presence of mature and immature forms of VpVAN in protein extracts. of vanilla pods and in N. benthamiana leaves following transient expression of VpVAN. Lane A: pre-stained Bio-Rad protein ladder. Lane B: unstained Bio-Rad protein ladder. Lane C: protein extract from N. benthamiana leaves following transient expression of VpVAN probed with an antibody specific to the C-terminal sequence of VpVAN. Lane D: protein extract of a 7-month-old vanilla pod probed with an antibody specific to the C-terminal sequence of VpVAN. A 10 μg aliquot of protein were applied to each lane.
The formation and post-translational modification of VpVAN was further investigated following Agrobacterium-mediated transient expression of the VpVAN-encoding gene in N. benthamiana (tobacco). Crude protein extracts of the VpVAN-expressing leaves were obtained 7 d after Agrobacterium infiltration and probed with the antibody specific to the C-terminal sequence of VpVAN at a 1 : 5,000 dilution (Fig. 3). Western blot analyses showed the presence of a dominant immunoreactive band with an apparent mass of 25 kDa representing mature VpVAN, indicating effective processing of VpVAN into the processed mature form after 7 d of transient expression in N. benthamiana (Fig. 3).
Mature VpVAN oligomers detected in the crude protein extracts from V. planifolia pods as demonstrated by Western blotting analysis
Cysteine proteinases are known to form dimers (Vincent and Brewin 2000), and dimerization has been associated with gain of optimal protease activity (Olsen et al. 2009). The possible occurrence of dimers of VpVAN was investigated by SDS–PAGE fractionation of a protein extract from a 7-month-old V. planifolia pod followed by Western blot analysis using a dilution series of the VpVAN C-terminal sequence-specific antibody. When the VpVAN C-terminal antibody was used at a 1 : 50 dilution, five protein bands migrating with apparent molecular masses of 25, 40, 50, 75 and 100 kDa were detected (Fig. 4). The VpVAN C-terminal antibody did not show cross-reactivity towards any of the major protein components in the crude protein extract from V. planifolia pods (Fig. 4). In agreement with previous results (Gallage et al. 2014), the 25 and 40 kDa bands match the calculated molecular masses of 23.89 and 39.15 kDa of mature and immature VpVAN, respectively, and the 50, 75 and 100 kDa bands possibly represent homodimers, trimers and tetramers of the mature VpVAN. When the Western blot analyses were carried out using antibodies applied at increasingly higher fold dilutions (Supplementary Fig. S3), the 50, 75 and 100 kDa bands vanished, possibly demonstrating that oligomeric forms of mature VpVAN had reduced binding affinity for the antibody compared with the monomer of mature VpVAN (Supplementary Fig. S3).
Fig. 4.
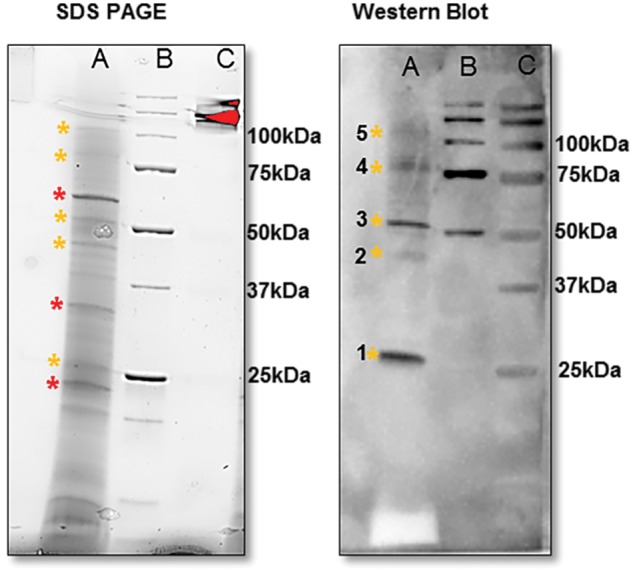
Mature VpVAN homodimer and putative oligomers detected in the crude V. planifolia extracts as demonstrated by SDS–PAGE followed by Western blot analysis. Lane A: crude protein extract from a 7-month-old vanilla pod from V. planifolia. Lane B: pre-stained Bio-Rad protein ladder. Lane C: unstained Bio-Rad protein ladder. 1, VpVAN mature protein (25 kDa); 2, VpVAN immature protein (39 kDa); 3, putative mature VpVAN homodimer (50 kDa); 4, putative mature VpVAN trimer (75 kDa); 5, putative mature VpVAN oligomers (100 kDa). The proteins present in a 7-month-old vanilla pod were separated by SDS–PAGE (12% Criterion™ TGX Stain-Free Precast Gels) and visualized using the ChemiDoc MP Imaging System (Bio-Rad). The major protein components present in the pod extracts are marked with * (red) while VpVAN and putative oligomers are marked with * (yellow). Western blot analysis was conducted using a C-terminal sequence-specific antibody against VpVAN in a 1 : 50 dilution. At this extremely high antibody concentration, the primary antibody demonstrated specificity towards VpVAN. No cross-reactions were observed with the major protein components present in the extract.
In attempts to convert the putative oligomers of mature VpVAN into the 25 kDa monomer, the protein extract from an 8-month-old vanilla pod was reduced with tris(2-carboxyethyl)phosphine (TCEP) before SDS–PAGE. The concentrations of TCEP tested varied from 5 to 50 mM. At none of these concentrations did the presence of TCEP result in reduced band intensities at 50, 75 and 100 kDa (Fig. 5; Supplementary Fig. S4). Oxidation of the protein extract with potassium ferricyanide also did not result in intensification of the 50, 75 and 100 kDa bands (Fig. 5). Based on the specificity of the antibody used, we propose that the immunoreactive bands at 50, 75 and 100 kDa represent oligomers of the 25 kDa monomer formed as artifacts during homogenization of the vanilla pod. The inner part of the vanilla pod contains high amounts of oleoresins and a highly viscous mucilaginous material impregnating the proteins upon homogenization of the vanilla pod, as previously reported (Odoux 2005). Random protein cross-linking reactions are thus likely also to result in the formation of oligomers of VpVAN not cleavable by TCEP. To reduce protein impregnation with the mucilaginous material and obtain sharper protein bands upon SDS–PAGE, the vanilla pod protein extracts used for Western blots in the current study were extracted in an optimized buffer composed of 10 mM MES, 2 mM dithiothreitol (DTT) at pH 5 as recommended for protein extraction of complex fruit tissues (Wang et al. 2008).
Fig. 5.
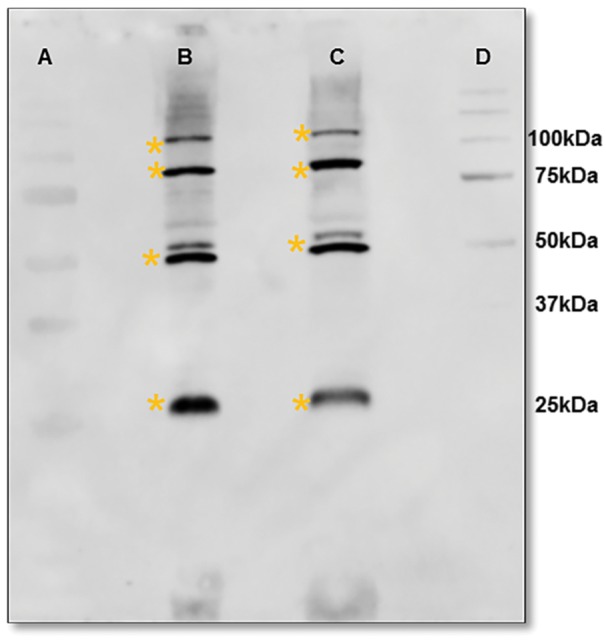
The ratio between monomeric and putative oligomeric forms of VpVAN as analyzed by treatments with reductant and oxidant as montiored by SDS–PAGE followed by Western blot analysis. Crude protein extracts from 8-month-old pods were analyzed. Protein bands recognized by the VpVAN antibody are marked with * (yellow). Lane A: pre-stained Bio-Rad protein ladder. Lane B: treatment with 50 mM TCEP for 15 min. Lane C: treatment with 50 mM TCEP for 15 min followed by 50 mM potasium fericyanide for 30 min. Lane D: unstained Bio-Rad protein ladder.
VpVAN is localized in the chloroplasts and phenyloplasts as demonstrated by immunohistochemical analyses
The tissue and cellular localization of VpVAN in fresh 7-month-old V. planifolia pods was examined by confocal microscopy. The immunohistochemical analyses were based on use of the specific antibody to the C-terminal sequence of VpVAN and thus targeted both the immature and the mature form of VpVAN. The images obtained by confocal microscopy clearly demonstrated that the localization of VpVAN was restricted to specific plastids present in multiple copies and situated within the cytoplasmic space of the cell (Fig. 6). Control images were obtained using pre-immune serum at the same dilution, and in these experiments no reactions mimicking those of the VpVAN antibody were observed (Supplementary Figs. S5, S6). The instrumental gain setting which gave a minimum fluorescence background when used with the pre-immune serum was also used in the experiments with the VpVAN antibody. The fluorescence signal observed using the VpVAN antibody thus represents specific immune labeling of VpVAN.
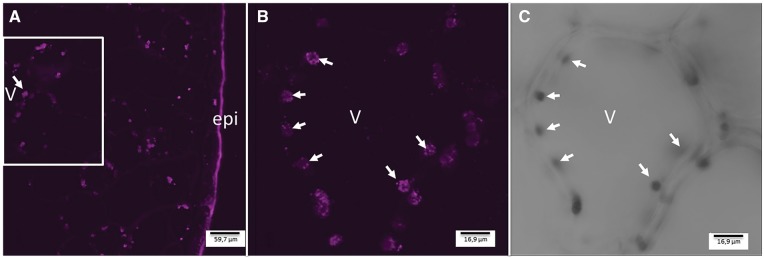
Fig. 6.
Immunohistochemical localization of VpVAN in cytoplasmic organelles in a 7-month-old V. planifolia pod as observed by confocal microscopy. (A) Transverse section of a 7-month-old V. planifolia pod providing an overview of VpVAN localization in the mesocarp. (B) Close-up of a single mesocarp cell showing the intracellular localization of VpVAN in plastids in the cytoplasm. (C) The same section as shown in (B) observed with translucent light. Images were obtained from transverse section of a 7-month-old V. planifolia pod using the VpVAN C-terminal antibody and a goat anti-rabbit antibody labeled with FITC. The images were recorded using a Leica SPII confocal scanning microscope. Arrows indicate the position of selected cytoplasmic plastids harboring VpVAN. Abbreviations: epi, epicarp; v, vacuole.
Light microscopy of transverse sections of 7-month-old V. planifolia pods demonstrated the presence of chloroplasts throughout the entire mesocarp layer (Fig. 7A, D, G). The chloroplasts were likewise detected by their red autofluorescence following excitation at 580 nm (Fig. 7C, F, I). The plastids present in the cytosolic matrix labeled by the specific VpVAN antibody were identified as chloroplasts in light microscopy- and fluorescence microscopy-based studies in which the chloroplasts were identified visually based on their Chl content and red autofluorescence in parallel with simultaneous superimposed immunohistochemical localization of VpVAN (Fig. 7A, D, and C, F).
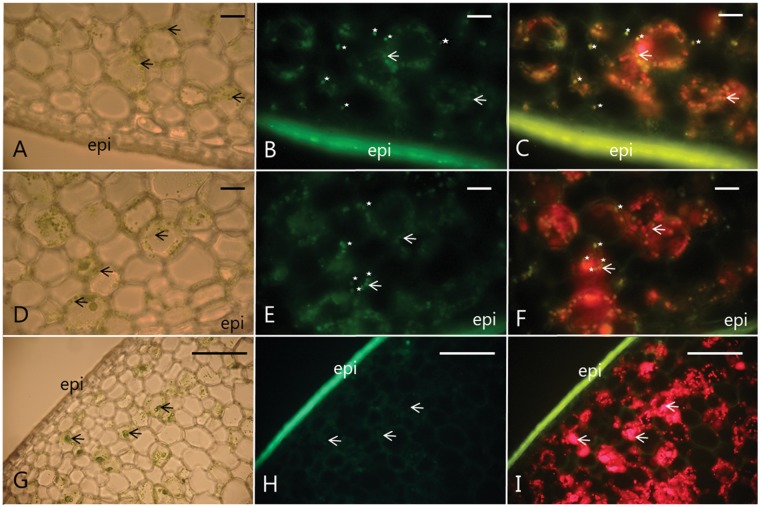
Fig. 7.
Immunolocalization of VpVAN to chloroplasts and phenyloplasts in 7-month-old V. planifolia pods using an antibody specific to the C-terminal sequence of VpVAN. (A–F) Data obtained from two transverse sections of two different pods are shown; V. planifolia pod 1 (A–C) and V. planifolia pod 2 (D–F). (A, D and G) Visualization of the examined transverse vanilla pod sections using light microscopy documenting the localization of chloroplasts. Chloroplasts are identified based on their Chl content. Selected chloroplasts are marked with black arrows. (B and E) Immunodetection of VpVAN in chloroplasts and phenyloplasts by FITC and fluorescence microscopy. The chloroplasts and phenyloplasts are stained green due to the antibody reaction. Selected phenyloplasts and chloroplasts are marked with white stars and white arrows, respectively. (C and F) Localization of VpVAN in chloroplasts (white arrows) and phenyloplasts (white stars) as monitored by fluorescence microscopy using a filter setting enabling simultaneous detection of FITC and Chl fluorescence. Chl fluorescence appears in red and demonstrates the localization of chloroplasts. Phenyloplasts that are totally free of Chl appear in green on (C) and (F) while those plastids retaining a small amount of Chl due to incomplete re-differentiation appear in yellow in (C) and (F). (G, H and I) Control panels; vanilla pod sections probed with pre-immune serum. No cross-reactions were observed. Chloroplasts: black arrows (A, D and G) and white arrows (B, C, E, F, H and I). Phenyloplasts: white stars. The same selected set of chloroplasts and phenyloplasts is labeled on the different panels to facilitate interpretation of the nature of the observed structures. Scale bars correspond to 100 μm. epi, epicarp.
It is evident, that some of the antibody-labeled plastids did not show the red autofluorescence characteristic of chloroplasts or only weak autofluorescence (Fig. 7B, E, and C, F). These plastids have the same overall dimensions as the chloroplasts. Independent of the Chl content and level of red autofluorescence, these plastids gave a positive immunochemical reaction with the specific antibody towards VpVAN. This indicated that VpVAN is localized in chloroplasts as well as in cytoplasmic-localized plastids that are devoid of Chl or with reduced Chl content. Re-differentiating chloroplasts have previously been reported to be present in vanilla pods and demonstrated to be the main storage sites of vanillin glucoside. The re-differentiated chloroplasts are referred to using the term phenyloplasts and are devoid of Chl (Brillouet et al. 2014). In the merged images (Fig. 7C, F), the chloroplasts harboring VpVAN are observed as orange (Fig. 7C, F, white arrows) whereas VpVAN-containing phenyloplasts completely devoid of Chl are observed as green (Fig. 7C, F, white stars). With the fluorescence filter settings used to record fluorescein isothiocyanate (FITC) labeling of the immunolocalized VpVAN as well as autofluorescence of chloroplasts, it is very likely that chloroplasts in the transition of being converted into phenyloplasts are observed as having a yellowish color due to reduced Chl content.
The localization of VpVAN was followed at 1 month intervals throughout pod development (3–9 months). Throughout the entire development phase, VpVAN was observed to locate strictly to chloroplasts and phenyloplasts (Supplementary Fig. S7). In fully mature 9-month-old pods, the signals became weak and diffuse (data not shown).
Agrobacterium-mediated transient expression of the gene encoding VpVAN in N. benthamiana leaves resulted in what would appear to represent a similar subcellular localization of VpVAN in chloroplasts (Supplementary Fig. S8). However, using this experimental system, strong background fluorescence was observed from the plant cell walls. This strong background fluorescence was also observed in control experiments in the absence of the antibody targeting the C-terminal sequence of VpVAN. The use of this analytical system for localization of VpVAN was therefore considered not to provide unambiguous conclusions and was not further pursued.
VpVAN detection and functional activity in isolated chloroplasts from V. planifolia
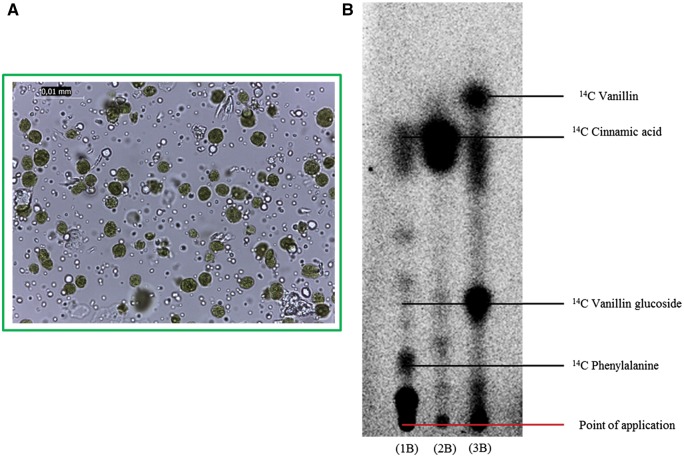
The demonstrated localization of VpVAN in chloroplasts prompted us to isolate intact chloroplasts from 8-month-old V. planifolia pods to test their biosynthetic activity. The isolated chloroplast preparations were examined by light microscopy to ascertain their purity and intactness (Fig. 8A). The chloroplasts were isolated following gentle homogenization of the pod tissue in 50 mM HEPES (pH 8.0)/0.33 M sorbitol and purified using Percoll gradients. Radiolabeled precursors were administrated to the isolated intact chloroplasts. Following incubation for 24 h, the radiolabeled products formed were separated by TLC. The isolated chloroplasts were functionally active, as demonstrated by the production of radiolabeled metabolites upon administration of [14C]phenylalanine, [14C]cinnamic acid and [14C]vanillin (Fig. 8). Administration of [14C]phenylalanine resulted in formation of [14C]cinnamic acid (Fig. 8, 1B). This conversion is catalyzed by phenylalanine ammonia lyase (PAL), and PAL is known to be localized in the chloroplasts (Sainders and McClure 1975). [14C]Vanillin was readily converted to [14C]vanillin glucoside, demonstrating the presence of vanillin glucosyltransferase activity (Fig. 8, 3B). Furthermore, a radiolabeled product co-migrating with [14C]vanillin glucoside was observed upon administration of [14C]phenylalanine and [14C]cinnamic acid (Fig. 8, 1B and 2B). As expected, the strongest labeling of [14C]vanillin glucoside was observed upon administration of [14C]cinnamic acid, a more specific vanillin glucoside precursor compared with phenylalanine.
Fig. 8.
Intact chloroplasts isolated from 8-month-old V. planifolia pods synthesize vanillin de novo. (A) The purity and integrity of the isolated chloroplasts as monitored by light microscopy. (B) Vanillin biosynthetic activity in isolated chloroplasts as monitored by administration of [14C]phenylalanine (lane 1B) and [14C-cinnamic acid (lane 2B) as substrates. An extract from the inner part of a V. planifolia pod disc incubated with [14C]vanillin was applied (lane 3B) to visualize the migration position of [14C]vanillin glucoside.
The presence of VpVAN in protein extracts obtained from the intact chloroplasts isolated from 8-month-old V. planifolia pods was analyzed by SDS–PAGE and Western blot experiments (Supplementary Figs. S9, S10). A specific antibody towards PSI subunit D (PSI-D) (Haldrup et al. 2003) was used as a positive control (Supplementary Fig. S9). PSI-D is an 18 kDa subunit of the PSI complex and localized on the stromal side of the PSI complex in the thylakoid membrane. In most experiments, immunoreactive bands at 25 and 40 kDa corresponding to the mature and immature forms of monomeric VpVAN were not observed using the antibody specific to the C-terminal sequence of VpVAN. Instead a strong immunoreaction was observed in the 50 kDa region as also observed on the Western blots with crude V. planifolia pod protein extracts when VpVAN C-terminal antibody was used in a dilution of 1 : 300 (Supplementary Fig. S10). Thus, in the isolated chloroplasts, the 25 kDa VpVAN monomer was converted into its oligomeric form. Oligomerization may have been favored by the extended procedure and the high pH of the isolation buffer required for isolation of intact chloroplasts. The isolation of intact chloroplasts from the vanilla pods was hampered by the high amounts of mucilaginous material and oleoresins present in the pods.
Discussion
When the current study was initiated, the enzyme VpVAN catalyzing vanillin glucoside synthesis from ferulic acid and the β-glucosidase catalyzing liberation of free vanillin had been identified. Likewise, re-differentiated chloroplasts named phenyloplasts had been shown to be the storage site of vanillin glucoside. In the present study, we demonstrate that the chloroplasts and phenyloplasts are also the site of accumulation of VpVAN. In agreement with those observations, tracer studies further support that isolated chloroplasts are able to synthesize vanillin glucoside de novo.
The VpVAN-catalyzed synthesis of vanillin glucoside from ferulic acid glucoside proceeds as a retro aldol elimination reaction resulting in C3 side chain shortening of ferulic acid glucoside (Gallage et al. 2014). Such a pathway was initially proposed by Zenk (1965) and subsequently by Negishi et al. (2009) based on radioactive precursor studies using [14C]ferulic acid. In the present study, radiolabeling studies confirmed that active vanillin glucoside biosynthesis takes place throughout pod development (3–9 months) and that p-hydroxybenzaldehyde did not serve as a vanillin precursor.
DESI-MSI requires minimal sample preparation and thereby minimizes contamination and formation of artifactual compounds. MSI is emerging as an excellent alternative to classical analytical methods where breakdown of plant constituents may occur in the course of preparation (Bjarnholt et al. 2014). In the present study, DESI-MSI showed localization of vanillin and vanillin glucoside in the inner part of the pod, where vanillin glucoside biosynthetic activity likewise was detected by radioactive precursor administration. The concentration of vanillin is about 20- to 50-fold lower than that of vanillin glucoside (Odoux 2005) and their distribution patterns are not superimposable. This indicates that β-glucosidase activities involved in vanillin glucoside hydrolysis or glucosyltransferase activities catalyzing its formation varies across the placental laminae and mesocarp. A radial distribution of vanillin glucoside and the β-glucosidase enzyme activity has previously been reported (Odoux et al. 2003b). The radiolabeling experiments based on administration of [14C]vanillin indeed showed the presence of glucosyltransferase activity in both the inner and outer parts of the pod. The localization in the V. planifolia pod of vanillin-specific glucosyltransferases has not been thoroughly studied.
To determine the subcellular localization of the vanillin biosynthetic machinery, we investigated the cellular and intracellular localization of the key enzyme of vanillin biosynthesis, VpVAN. Immunolocalization studies clearly demonstrated that VpVAN was localized in chloroplasts distributed within the inner mesocarp and placental laminae of the vanilla pod throughout V. planifolia pod development (3–9 months). Furthermore, studies of VpVAN demonstrated that the protein underwent a maturation step involving removal of the propeptide sequence corresponding to mobility shifts on SDS–PAGE from 40 to 25 kDa positions. We previously reported that removal of the propeptide sequence augmented the activity of VpVAN (Gallage et al. 2014). Putative dimers, trimers and tetramers of mature VpVAN were observed in crude protein extracts of the vanilla pods and especially in isolated intact chloroplasts. Self-association of proteins forming dimers or oligomers is a common phenomenon. Some cysteine proteases such as caspases are known to undergo dimerization as a requirement to gain catalytic activity and enzyme activation (Grzonka et al. 2001, Marianayagam et al. 2004, Mackenzie and Clark, 2012). In the present study, the oligomeric forms could not be converted into the monomeric form by treatment with a strong reductant such as TCEP. Thus, we conclude that these oligomers are artifacts formed as a result of a chemical reaction of the VpVAN monomer with the abundant mucilaginous material present in the pods.
Administration of [14C]phenylalanine to intact chloroplasts isolated from the vanilla pod resulted in formation of a radiolabeled product co-migrating with [14C]vanillin glucoside, suggesting that the entire vanillin biosynthetic machinery responsible for conversion of phenylalanine to vanillin glucoside operated in the chloroplasts. It is not known at which stage in the vanillin pathway the glucosylation takes place. If vanillin glucoside is preferentially made directly from ferulic acid glucoside, the glucosylation may proceed at the level of p-coumaric acid, caffeic acid or ferulic acid. The storage form of vanillin in the V. planifolia pod is the vanillin β-d-glucoside. High concentrations of vanillin are toxic to the cell (Boonchird and Flegel 1982). Accordingly, a vanillin-specific UGT would be expected to be co-localized with VpVAN to catalyze conversion of any vanillin formed into vanillin β-d-glucoside to avoid cell toxicity. A study has shown that UDP-glucose, which is the co-substrate for family 1 glucosyltransferases, is de novo synthesized in the chloroplasts of A. thaliana (Okazaki et al. 2009).
The classes of plant natural products that are synthesized in the chloroplasts are remarkably diverse. In addition to photosynthesis, chloroplasts are known to carry out many other essential functions such as synthesis of amino acids (Kirk and Leech 1972), fatty acids (Lippold et al. 2012), lipids (Wang and Benning 2012), plant hormones (Metraux 2002) and vitamins (Dellapenna and Pogson 2006). Most of these compounds are not only essential for chloroplasts to accomplish their metabolic role but at different stages of plant ontogeny are released from the chloroplast to serve directly as or be metabolized into signaling components for plant growth and development or as defense compounds against pathogens or herbivores (Joyard et al. 2009). Synthesis of aromatic amino acids in the chloroplasts would constantly provide phenylalanine as the initial substrate for vanillin biosynthesis. The phenylalanine precursors chorismic acid and shikimic acid are known to form esters with p-coumaric acid, which are intermediates in ferulic acid biosynthesis. In 1966, conversion of p-coumaric acid into caffeic acid was demonstrated in isolated chloroplasts from leaves of Saxifraga stolonifera, which shows 4-hydroxycinnamoyl transferase (4-HCL) activity in the chloroplasts (Sato 1966, Bassard et al. 2012).
While the chloroplast genome encodes about 80–100 proteins, between 2,500 and 3,500 nuclear-encoded proteins are predicted to be targeted to the chloroplast (Joyard et al. 2009). In general, chloroplast proteins encoded by the nuclear genome and imported into chloroplasts are synthesized as precursor proteins with cleavable N-terminal transit peptides that direct each protein to its final destination within the chloroplast subcompartments (Nielsen et al. 1994, Bruce 2000). Sequence analysis of the vanillin biosynthetic pathway genes (Gallage et al. 2014) encoding VpVAN and putative V. planifolia PAL, putative VpC4H (cinnamate-4-hydroxylase), putative Vp4-HCL, putative VpHCT (hydroxycinnamoyl transferase), putative VpC3H (cinnamoyl ester 3' hydroxylase), putative VpCOMT (caffeic acid O-methyl transferase) and putative VpUGT using the chloroplast transit peptide prediction program SignalP (http://www.cbs.dtu.dk/services/SignalP/) did not provide evidence for the presence of a cleavable transit peptides within the encoded proteins. However, the function of chloroplasts as an organelle harboring entire pathways for secondary metabolites is far from being understood, and further biochemical evidence may serve to upgrade the prediction software (Kiessling et al. 2000, McAndrew et al. 2001).
Phenyloplasts have been shown to be the storage site of vanillin glucoside (Brillouet et al. 2014). The phenyloplasts arise following re-differentiation of chloroplasts resulting in loss of their Chl content and photosynthetic abilities, and gain of the ability to store high concentrations of phenylpropanoid-derived glucosides (Brillouet et al. 2013, Brillouet et al. 2014). Re-differentiation of chloroplasts to different storage plastids is a well-known phenomenon (Weier 1936, Leyon 1953, Kutik 1998). Leucoplasts are colorless plastids that function as storage organelles. Leucoplasts comprise amyloplasts, oleoplasts and proteinoplasts, and these are known to store starch, lipids and proteins, respectively. In fleshy fruits such as tomatoes, ripening is associated with the re-differentiation of green fruit chloroplasts into ripe fruit chromoplasts (Klee and Giovannoni 2011, Llorente et al. 2016, Llorente et al. 2017). Chromoplasts contain carotenoid pigments that give the red, orange and yellow colors to the plant structure (Roberts 1946, Egea et al. 2010, Camara et al. 1982, Llorente et al. 2017). Young chromoplasts are metabolically active but contain fewer DNA copies than chloroplasts. Gerontoplasts are known to be the last ontogeny stage of chloroplasts, and these plastids no longer harbor functional DNA (Sitte 1977, Kutik 1998).
In analogy to the re-differentiation processes of chloroplasts described above resulting in formation of plastids accumulating primary metabolites (amyloplasts, oleoplasts and proteoplasts) as well as secondary metabolites in the form of pigments (chromoplasts), it is not surprising that re-differentiated chloroplasts may also store secondary metabolites as phenylpropanoids. Strong scientific documentation for this was first provided using the vanilla pod as the experimental system. Brillouet et al. (2014) coined the term phenyloplasts for this type of re-differentiated chloroplasts. The grana thylakoids of the vanilla chloroplasts were dismantled in the early phases of phenyloplast ontogeny, and no deposits of phenolics were observed within the thylakoidal lumen. Based on indirect histochemical data, phenyloplasts have earlier been suggested as storage sites for secondary metabolites (Saunders and McClure 1976, Zaprometov and Nikolaeva 2003, Liu et al. 2009). Thus, the storage of vanillin glucoside in phenyloplasts may not represent a unique case of subcellular sequestration of phenolics in the plant kingdom. Some secondary metabolites accumulate in massive amounts in certain plant tissues. This applies to the cyanogenic glucoside dhurrin which in the tip of etiolated seedlings of Sorghum bicolor (L.) constitutes up to 30% of the dry weight (Saunders and Conn 1977, Halkier and MØller 1989), and flavan-3-ols which in leaves of Camellia sinensis (L.) constitute up to 30% of the dry weight (Liu et al. 2009). Such high concentrations of secondary metabolites might be attractive to store in phenyloplasts (Gachon et al. 2005).
Biological research concerning the V. planifolia orchid, vanilla pods and the physiological role of vanillin formation has been paid relatively little attention by the research community in spite of the importance of the vanilla flavor (Gallage and MØller 2015). Production of vanilla from the vanilla orchid is highly labor intensive and a lengthy process not easily adapted to market demands. Global demand for vanilla was estimated at between 2,500 and 3,000 Mt annually in 1998, while the global demand for vanillin was estimated at around 16,000 Mt annually in 2010 and worth US$ 650 million in total on the world market (Smolarski 2012). However, only 0.25% of vanillin originates from cured pods of the vanilla orchid, V. planifolia (Gallage and MØller, 2015). Today 99% of all vanillin consumed worldwide is synthetically made, primarily using chemical synthesis based on petrochemicals, or chemically derived by acid hydrolysis of lignin. Market pull and consumer demand have promoted intensive research to develop sustainable biological production platforms for vanillin using microorganisms as a replacement for environmentally unsustainable chemical synthesis (Gallage and MØller 2015).
The demonstration that vanillin biosynthesis takes place in the chloroplasts would open the door for design of photosynthetic production platforms using algae, cyanobacteria or moss producing vanillin. When fully developed, such production platforms offer superior alternatives to classical biotechnological hosts such as bacteria and yeast for vanillin biosynthesis because they use carbon dioxide as the sole carbon source and sunlight as the energy source. Production in photosynthetic organisms such as cyanobacteria offers a sustainable alternative, because the carbon skeletons, energy and reducing power are derived from photosynthesis via CO2 fixation and light-driven electron transport. Using the cyanogenic glucoside dhurrin as a model system, such photosynthetic systems are now also being developed for synthesis of complex diterpenoids (MØller 2014, Lassen et al. 2014, Gnanasekaran et al. 2016, MØller 2017).
In a recent report including Hailian Yang, Daphna Havkin-Frenkel and Richard A. Dixon as authors (Yang et al. 2017), the catalytic activity of VpVAN was re-evaluated. It was concluded that the VpVAN protein catalyzes conversion of p-coumaric acid to p-hydroxybenzaldehyde or as an alternative that this conversion proceeds non-enzymatically (Yang et al. 2017). In a previous publication, we studied the catalytic activity of VpVAN in in vitro and in vivo experiments and documented that the VpVAN enzyme catalyzes the conversion of ferulic acid and ferulic acid glucoside to vanillin and vanillin glucoside, respectively (Gallage et al. 2014). This result has also been obtained in patent applications (Havkin-Frenkel et al. 2006, Havkin-Frenkel and Podstolski 2007, Gallage et al. 2014). The catalytic property of VpVAN as a vanillin synthase has been verified in independent studies demonstrating vanillin and vanillin glucoside formation in Capsicum frutescens (hot chili pepper) stably transformed with VpVAN (Chee et al. 2017), and further substantiated in the present study.
In the previous studies by Daphna Havkin-Frenkel and Richard A. Dixon, a 28 kDa protein was partially purified from an embryo culture extract of V. planifolia and used for preparation of an antibody. As already pointed out by Odoux et al. (2009), this antibody was not raised towards a single purified protein but based on a protein fraction with the ability to convert p-coumaric acid into p-hydroxybenzaldehyde. In the recent publication by Yang et al. (2017), the 28 kDa protein reported to be encoded by the same gene sequence as VpVAN was isolated from a crude V. planifolia embryo culture extract following immune purification using the antibody described above followed by elution of the protein at pH 2.6. The eluted and supposedly reconstituted protein was reported to convert p-coumaric acid to p-hydroxybenzaldehyde as monitored by HPLC analysis. No activity was obtained with ferulic acid as a substrate, in contrast to what was observed in previous studies (Havkin-Frenkel et al. 2006, Havkin-Frenkel and Podstolski 2007, Gallage et al. 2014, Chee et al. 2017). In our study, we obtained the native immature and mature VpVAN protein using a rabbit reticulocyte lysate-based transcription/translation system (Gallage et al. 2014). The mature and immature proteins migrated on SDS–PAGE with apparent molecular masses of 25 and 40 kDa, respectively. Using highly sensitive LC-MS (ion trap) analyses, VpVAN was demonstrated in vitro to catalyze the conversion of ferulic acid into vanillin. No production of p-hydroxybenzaldehyde could be detected using p-coumaric acid as substrate. These results have been confirmed and further substantiated in the current study.
In our previous publication on VpVAN (Gallage et al. 2014), in vivo studies were carried out by transient expression of the VpVAN-encoding gene in N. benthamiana and by stable expression of the gene in yeast and barley. In all these studies, a codon-optimized gene sequence was used to achieve efficient gene expression. Expression of the transgene was in all cases correlated with formation of vanillin or vanillin-derived metabolites, whereas formation of p-hydroxybenzaldehyde or metabolites thereof was not correlated with transgene expression. The studies reported by Yang and co-workers were carried out without codon optimizations. Heterologous expression of VpVAN in host systems such as yeast requires prior modification of endogenous background reactions in the host system to achieve production of vanillin glucoside. Guidance on how to achieve this in combination with sensitive analytics based on use of LC-MS ion trap instrumentation to monitor the activity of VpVAN was provided in our previous publication (Gallage et al. 2014).
In conclusion, in this study, we demonstrated the tissue and intracellular localization of the vanillin biosynthetic machinery in chloroplasts and re-differentiated chloroplasts termed phenyloplasts during pod development. Vanillin biosynthetic activity was demonstrated in isolated chloroplasts, and VpVAN was identified by immunolocalization in both chloroplasts and phenyloplasts, exactly the plastids that have previously been documented to store vanillin glucoside. Furthermore, VpVAN expression studies highlighted that VpVAN indeed undergoes a maturation step where the propeptide is cleaved off. With these results, we are one step closer to understanding how V. planifolia is able to produce vanillin in high concentrations during the 5–6 month period during which it develops from an immature to a mature pod.
Materials and Methods
Biological materials
Healthy vines of V. planifolia carrying foliage and green vanilla pods were harvested at different time points after pollination and were shipped from the Biological Resource Center (BRC), VATEL, CIRAD, Saint-Pierre, La Réunion, France, by courier carrier to Denmark while maintaining high humidity conditions.
Three-week-old N. benthamiana plants were used for the transient expression of the gene encoding VpVAN.
Agrobacterium strain
Agrobacterium tumefaciens strain AGL1 was used for transient and stable expression assays in planta and grown following standard procedures in LB medium with appropriate antibiotics (Bach et al. 2014).
Polyclonal antibody
A polyclonal antibody was obtained by immunizing rabbits with the VpVAN-specific peptide sequence (NH2-)CNWGDNGYFKMELGK(-CONH2) derived from the C-terminal amino acid sequence of VpVAN (amino acids 327–340) (Agrisera AB, http://agrisera.com). To test the specificity of the antibody, crude protein extracts from 7-month-old pods of V. planifolia were separated by SDS–PAGE and subjected to Western blotting.
The specificity of the C-terminal VpVAN-specific antibody was investigated using a series of antibody dilutions (1 : 50, 1 : 100, 1 : 500 and 1 : 1,000). When used in a 1 : 50 dilution, the antibody reacted with proteins migrating with apparent molecular masses of 25, 40, 50 and 100 kDa. In different series of experiments, the crude protein extract was either reduced by incubating with TCEP (5 to 50 mM, pH 3.5) for 10 min or oxidized by incubating with potassium ferricyanide (10–50 mM, pH 2.95) for 30 min before SDS–PAGE analysis to investigate the presence or formation of protein oligomers. Pre-immune serum from the rabbit used for the antibody production showed no cross-reaction to protein extracts from V. planifolia (Supplementary Fig. S2).
Vector constructs
Plant expression vectors for transient expression in tobacco were constructed using Gateway® cloning technology (Life Technologies) as previously described (Gallage et al. 2014).
Biosynthetic assays with green V. planifolia pods and isolated intact chloroplasts
The V. planifolia pods harvested at 3, 4, 5, 6, 7, 8 and 9 months following pollination were cut transversely into 1–2 mm thick discs (approximately 25 mg FW) using a scalpel and further dissected to separate the inner and outer part of the pod. Radiolabeled precursors (0.5 μCi) were administered to samples of the inner and outer part of the pod and incubated (30 °C) in 400 mM Tris–HCl pH 8, 20 mM MgCl2 for 6–48 h.
The 14C-labeled products formed in the experiments with fresh V. planifolia pods were extracted by 25% MeOH and applied to Silica Gel 60 F254 TLC plates (Merck, http//www.merck-chemicals.com). The plates were developed in ethyl acetate : acetone : dichloromethane : methanol : water (40 : 30 : 20 : 10 : 8, by vol.), dried, exposed (48 h) on phosphor-imaging screens (Molecular Dynamics, http://www.moleculardynamics.com) and the radiolabeled products visualized using a Storm 860 Molecular Imager (Molecular Dynamics). Identification of the radiolabeled compounds formed was guided by co-application of authentic standards.
The same experimental procedure was followed using isolated intact chloroplasts (10 μl, 0.5 μg Chl μl−1) obtained from 8-month-old V. planifolia pods.
Total protein extraction and Western blot analysis
Vanilla planifolia pods were ground in liquid nitrogen with a mortar and pestle. The resulting powder was homogenized in 100 μl of SDS running buffer (Laemmli buffer, Bio-Rad) and 300 μl of 10 mM MES buffer (pH 5–6) including 20 mM DTT. The homogenate was clarified by centrifugation (10,000Xg, 10 min, 4 °C). Crude protein extracts were incubated at 65°C for 30 min before being subjected to separation by SDS–PAGE (225 V, 30 min, 400 mA, 300 W) using 12% Criterion™ TGX Stain-Free Precast Gels (Bio-Rad) and Precision Plus Protein Stained and Unstained Standards (Bio-Rad) as molecular mass markers. Western blots of SDS–PAGE-separated proteins were performed using a Trans-Blot R Turbo Transfer Blotting instrument (Bio-Rad) and Trans-Blot R Turbo Midi Nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% (w/v) skimmed milk in PBS-T buffer (phosphate buffered saline with Tween-20) for 2 h. The VpVAN C-terminal sequence-specific antibody was applied in a dilution of 1 : 5,000. The presence of immunoreactive polypeptides was visualized using a horseradish peroxidase-conjugated goat anti-rabbit antibody (DakoCytomations, http://www.dako.com/dk) in a 1 : 5,000 dilution using the Super Signal West Dura extended duration substrate kit (Pierce). In the experiments with pre-sera, the same 1 : 5,000 dilution of pre-sera and horseradish peroxidase-conjugated goat anti-rabbit antibody was used as advised by the producer (Agrisera).
Blots were developed for 1–30 min with a ChemiDoc MP Imaging System (Bio-Rad) equipped with a cooled CCD camera (Bio-Rad) set to automatic exposure setting. Total protein was visualized on the membranes using the stain-free blot setting. The Precision Plus Protein unstained ladder (Bio-Rad) did not give visible bands below 50 kDa; therefore, the Precision Plus Protein Stained ladder was included in the gel. After blotting, the Precision Plus Protein Stained ladder can be seen with the naked eye. When imaging with the ChemiDoc MP Imaging System, a blot picture was taken before the settings were changed to visualize chemiluminescence. Both blot pictures were then merged to enable visualization of the entire Precision Plus Protein Stained ladder.
The same procedure was followed to extract total protein and to analyze the presence of VpVAN following transient expression of the VpVAN gene in tobacco.
Immunolocalization of immature and mature VpVAN in V. planifolia and N. benthamiana
To localize both the immature and mature forms of VpVAN, immunolocalization was performed as previously reported (Sanchez-Perez et al. 2012). Infiltrated tobacco leaves or V. planifolia pods at different ontogenies were cut into 1 cm pieces, embedded in 5% agarose and cut into 120 μm sections. The sections were placed in 5% skimmed milk in PBS at room temperature to block unspecific background. After 30 min incubation, the C-terminal sequence-specific VpVAN antibody was added at a 1 : 100 dilution (Kannangara et al. 2011). Following incubation for 2 h, the sections were washed three times with PBS. Secondary antibody (goat anti-rabbit with the FITC fluorophore) was added 1 : 160 in PBS and incubated for 2 h at room temperature. Thereafter, the sections were washed three times in PBS and left in PBS.
Sections were mounted with anti-fading agent and analyzed by confocal scanning microscopy using Leica SPII at 488 nm excitation for FITC detection or with a fluorescence microscope, Leica DMR using filters for specific observation of FITC and a combined filter for simultaneous detection of FITC and chloroplast autofluorescence.
Chloroplast isolation and fractionation
Intact chloroplasts were isolated from four 8-month-old V. planifolia pods essentially as previously reported (Robinson 2002). Pods were cut transversely into 2 cm long sections and homogenized in HS buffer [50 mM HEPES, KOH (pH 8) and 0.33 M sorbitol]. The homogenate was filtered through nylon mesh (44 μm) and centrifuged (3,330Xg, 2 min). The chloroplast pellet was gently re-suspended in HS buffer, layered onto pre-cooled Percoll pads and centrifuged (1,400Xg, 8 min). The intact chloroplast sediment was washed in HS buffer, re-sedimented (3,000Xg, 2 min) and re-suspended in 50 μl of HS buffer. The purity and integrity of the purified chloroplasts were checked under a Leica ICC50 HD microscope.
Immunoblot analysis of isolated chloroplasts
Samples (40 μl) were heated at 65°C for 30 min before separation by SDS–PAGE using 12% Criterion TGX Stain-Free pre-cast gels (Bio-Rad) and a Tris/glycine/SDS running buffer (Bio-Rad) at 240 V for 35 min. Proteins were transferred to a 0.2 μm PVDF (polyvinylidene fluoride) membrane using the Trans-Blot Turbo transfer system (Bio-Rad) according to the manufacturer’s protocol. The membrane was blocked for 20 min at room temperature with 5% (w/v) skimmed milk in PBS-T buffer and incubated for 1 h with the C-terminal sequence-specific VpVAN primary antibody at a 1 : 300 dilution in 2% (w/v) skimmed milk in PBS-T. The blot was washed with PBS-T buffer and incubated with a secondary swine anti-rabbit horseradish peroxidase-conjugated antibody (Dako) using a 1 : 5,000 dilution in PBS-T for 1 h at room temperature. The membrane was washed again with PBS-T buffer and the secondary antibody detected using SuperSignal West Dura Chemiluminescent Substrate (Pierce) and developed for 1–10 min with a ChemiDoc MP Imaging System (Bio-Rad). A specific antibody to the PSI-D (Haldrup et al. 2003) in a 1 : 10,000 dilution was used as a reference in quality tests of the isolated chloroplasts.
Desorption electrospray ionization mass spectrometry imaging (DESI-MSI)
Pieces of frozen V. planifolia pods were mounted on a cryo-microtome sample holder using water as the only adhesive. Using a Leica CM3050S cryo-microtome (Leica Microsystems), the tissue was cut into 40 μm thin sections, which were thaw-mounted on microscope glass slides and stored at −80°C until the time of analysis. Compared with most other plant tissues, the tissue was very fragile and difficult to section, and therefore a relatively high thickness was chosen. On the day of analysis, the sample slide was taken directly from the freezer to a vacuum desiccator for 10 min prior to DESI analysis.
Imaging was performed on a Thermo LTQ XL linear ion trap mass spectrometer (Thermo Scientific) equipped with a custom-built DESI ion source based on a motorized microscope stage by Märzhäuser Wetzla and controlled by in-house software. The imaging stage is described in detail elsewhere (Thunig et al. 2011). Imaging was performed in the positive ion mode using a 4 μl min−1 flow rate of methanol and water (80 : 20) containing 50 mM NaCl and 0.1% formic acid for enhanced generation of sodium adducts and protonated species. The nebulizer gas pressure was 6 bar. The pixel size was 250 μm. Vanillin was imaged at m/z 153 in its protonated state, vanillin glucoside was imaged at m/z 337 as its sodium adduct and sucrose (or an isomer) was imaged at m/z 381 as its potassium adduct. The raw data files were converted to imzML files (Schramm et al. 2012), and Data Cube Explorer (AMOLF, Amsterdam) was used to generate images. MATLAB was used to create colored overlaid images.
Supplementary Data
Supplementary data are available at PCP online.
Author Contributions
N.J.G. planned and designed the project and performed the 14C-radiolabeled precursor feeding experiments, TLC analysis, all the molecular biology analysis, tobacco expression studies, protein extractions and expression studies, and contributed to writing the manuscript. K.J. contributed in planning and carrying out all immunolocalization studies, designing the project and writing the manuscript. C.J. carried out the DESI analysis. A.J.Z.N. carried out isolation of intact chloroplasts. T.N. performed vanilla/tobacco expression studies. E.D. performed and optimized vanilla crude protein extraction and Western blotting analysis. L.D. carried out the vanilla pod cryo-sectioning. M.G. was in charge of vanilla pod sampling and courier shipments of fresh vanilla materials from La Reunion to Denmark. B.L.M. planned and designed the project, provided biochemical expertise and scientific mentoring, and contributed to writing the manuscript.
Funding
This work was supported by the VILLUM Foundation [a grant to the VILLUM Research Center ‘Plant Plasticity’] and the UCPH Excellence Programme for Interdisciplinary Research [Center for Synthetic Biology ‘bioSYNergy’].
Supplementary Material
Acknowledgments
We thank BRC, VATEL, CIRAD, La Reunion for providing Vanilla planifolia materials.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- Chl
chlorophyll
- d
days
- DESI-MESI
desorption electrospray ionization mass spectrometry imaging
- DTT
dithiothreitol
- FITC
fluorescein isothiocyanate
- PAL
phenylalanine ammonia lyase
- PBS
phosphate-buffered saline
- TCEP
tris(2-carboxyethyl)phosphine
- TLC
thin-layer chromatography
- UGT
UDP-glucosyltransferase
- VpVAN
Vanilla planifolia vanillin synthase
References
- Anuradha K., Shyamala B.N., Naidu M.M. (2013) Vanilla—its science of cultivation, curing, chemistry, and nutraceutical properties. Crit. Rev. Food. Sci. Nutr. 53: 1250–1276. [DOI] [PubMed] [Google Scholar]
- Bach S.S., Bassard J.E., Andersen-Ranberg J., Moldrup M.E., Simonsen H.T., Hamberger B. (2014) High-throughput testing of terpenoid biosynthesis candidate genes using transient expression in Nicotiana benthamiana. Methods Mol. Biol. 1153, 245–255. [DOI] [PubMed] [Google Scholar]
- Bassard J.E., Richert L., Geerinck J., Renault H., Duval F., Ullmann P., et al. (2012) Protein–protein and protein–membrane associations in the lignin pathway. Plant Cell 24: 4465–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnholt N., Li B., D’Alvise J., Janfelt C. (2014) Mass spectrometry imaging of plant metabolites—principles and possibilities. Nat. Prod. Rep. 31: 818–837. [DOI] [PubMed] [Google Scholar]
- Boonchird C., Flegel T.W. (1982) In vitro antifungal activity of eugenol and vanillin against Candida albicans and Cryptococcus neoformans. Can. J. Microbiol. 28: 1235–1241. [DOI] [PubMed] [Google Scholar]
- Brillouet J.M., Romieu C., Schoefs B., Solymosi K., Cheynier V., Fulcrand H., et al. (2013) The tannosome is an organelle forming condensed tannins in the chlorophyllous organs of Tracheophyta. Ann. Bot. 112: 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brillouet J.M., Verdeil J.L., Odoux E., Lartaud M., Grisoni M., Conejero G. (2014) Phenol homeostasis is ensured in vanilla fruit by storage under solid form in a new chloroplast-derived organelle, the phenyloplast. J. Exp. Bot. 65: 2427–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce B.D. (2000) Chloroplast transit peptides: structure, function and evolution. Trends Cell Biol. 10: 440–447. [DOI] [PubMed] [Google Scholar]
- Burri J., Graf M., Lambelet P., Lolige R.J. (1989) Vanillin—more than a flavoring agent—a potent antioxidant. J. Sci. Food Agric. 48: 49–56. [Google Scholar]
- Camara B., Bardat F., Moneger R. (1982) Sites of carotenoid biosynthesis in pepper (Capsicum annuum L) fruit chromoplasts. C.R. Acad. Sci. IIII Vie 294: 649–652. [Google Scholar]
- Cambra I., Hernandez D., Diaz I., Martinez M. (2012) Structural basis for specificity of propeptide–enzyme interaction in barley C1A cysteine peptidases. PLoS One 7: e37234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron K.M. (2004) Utility of plastid psaB gene sequences for investigating intrafamilial relationships within Orchidaceae. Mol. Phylogenet. Evol. 31: 1157–1180. [DOI] [PubMed] [Google Scholar]
- Cameron K.M., Chase M.W., Whitten W.M., Kores P.J., Jarrell D.C., Albert V.A., et al. (1999) A phylogenetic analysis of the Orchidaceae: evidence from rbcL nucleotide sequences. Amer. J. Bot. 86: 208–224. [PubMed] [Google Scholar]
- Cameron K.M., Molina M.C. (2006) Photosystem II gene sequences of psbB and psbC clarify the phylogenetic position of Vanilla (Vanilloideae, Orchidaceae). Cladistics 22: 239–248. [Google Scholar]
- Chee M.J.Y., Lycett G.W., Khoo T.J., Chin C.F. (2017) Bioengineering of the plant culture of Capsicum frutescens with vanillin synthase gene for the production of vanillin. Mol. Biotechnol. 59: 1–8. [DOI] [PubMed] [Google Scholar]
- Dellapenna D., Pogson B.J. (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57: 711–738. [DOI] [PubMed] [Google Scholar]
- Egea I., Barsan C., Bian W.P., Purgatto E., Latche A., Chervin C., et al. (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol. 51: 1601–1611. [DOI] [PubMed] [Google Scholar]
- Gachon C.M., Langlois-Meurinne M., Saindrenan P. (2005) Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10: 542–549. [DOI] [PubMed] [Google Scholar]
- Gallage N.J., Hansen E.H., Kannangara R., Olsen C.E., Motawia M.S., JØrgensen K., et al. (2014) Vanillin formation from ferulic acid in Vanilla planifolia is catalysed by a single enzyme. Nat Commun. 5: 4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallage N.J., MØller B.L. (2015) Vanillin—bioconversion and bioengineering of the most popular plant flavor and its de novo biosynthesis in the vanilla orchid. Mol. Plant 8: 40–57. [DOI] [PubMed] [Google Scholar]
- Gallage N.J., MØller B.L., Hansen E.H., Hansen J. (2014) Vanillin synthase. Patent no. PCT/DK2013/050357.
- Gnanasekaran T., Karcher D., Nielsen A.Z., Martens H.J., Ruf S., Kroop X., et al. (2016) Transfer of the cytochrome P450-dependent dhurrin pathway from Sorghum bicolor into Nicotiana tabacum chloroplasts for light-driven synthesis. J. Exp. Bot. 67: 2495–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzonka Z., Jankowska E., Kasprzykowski F., Kasprzykowska R., Lamkiewicz L., Wiczk W., et al. (2001) Structural studies of cysteine proteases and their inhibitors. Acta Biochim. Pol. 48: 1–20. [PubMed] [Google Scholar]
- Haldrup A., Lunde C., Scheller H.V. (2003) Arabidopsis thaliana plants lacking the PSI-D subunit of photosystem I suffer severe photoinhibition, have unstable photosystem I complexes, and altered redox homeostasis in the chloroplast stroma. J. Biol. Chem. 278: 33276–33283. [DOI] [PubMed] [Google Scholar]
- Halkier B.A., MØller B.L. (1989) Biosynthesis of the cyanogenic glucoside dhurrin in seedlings of Sorghum bicolor (L.) Moench and partial purification of the enzyme system involved. Plant Physiol. 90: 1552–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havkin-Frenkel D., Podstolski A. (2007) Vanillin production. USA patent application, US 20070231864 A1.
- Havkin-Frenkel D., Zylstra G., Frenkel C., Belanger F. (2006) Production of vanillin in microbial cells. USA patent application, US 10/532,464.
- Joel D.M., French J.C., Graft N., Kourteva G., Dixon R.A., Havkin-Frenkel D. (2003) A hairy tissue produces vanillin. Isr. J. Plant Sci. 51: 157–159. [Google Scholar]
- Joyard J., Ferro M., Masselon C., Seigneurin-Berny D., Salvi D., Garin J., et al. (2009) Chloroplast proteomics and the compartmentation of plastidial isoprenoid biosynthetic pathways. Mol. Plant 2: 1154–1180. [DOI] [PubMed] [Google Scholar]
- Kannangara R., Motawia M.S., Hansen N.K., Paquette S.M., Olsen C.E., MØller B.L., et al. (2011) Characterization and expression profile of two UDP-glucosyltransferases, UGT85K4 and UGT85K5, catalyzing the last step in cyanogenic glucoside biosynthesis in cassava. Plant J. 68: 287–301. [DOI] [PubMed] [Google Scholar]
- Kiessling J., Kruse S., Rensing S.A., Harter K., Decker E.L., Reski R.. 2000. Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J. Cell Biol. 151: 945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk P.R., Leech R.M. (1972) Amino acid biosynthesis by isolated chloroplasts during photosynthesis. Plant Physiol. 50: 228–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee H.J., Giovannoni J.J. (2011) Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45: 41–59. [DOI] [PubMed] [Google Scholar]
- Klinkert I., Chughtai K., Ellis S.R., Heeren R.M.A. (2014) Methods for full resolution data exploration and visualization for large 2D and 3D mass spectrometry imaging datasets, International. Journal of Mass Spectrometry, 2014. 362: 40–47. [Google Scholar]
- Kutik J. (1998) The development of chloroplast structure during leaf ontogeny. Photosynthetica 35: 481–505. [Google Scholar]
- Lassen L.M., Nielsen A.Z., Ziersen B., Gnanasekaran T., Moller B.L., Jensen P.E. (2014) Redirecting photosynthetic electron flow into light-driven synthesis of alternative products including high-value bioactive natural compounds. ACS Synth. Biol. 3: 1–12. [DOI] [PubMed] [Google Scholar]
- Leyon H. (1953) The structure of chloroplasts. 2. The 1st differentiation of the chloroplast structure in Vallota and Taraxacum studied by means of electron microscopy. Exp. Cell Res. 5: 520–529. [DOI] [PubMed] [Google Scholar]
- Lippold F., Vom Dorp K., Abraham M., Holzl G., Wewer V., Yilmaz J.L., et al. (2012) Fatty acid phytyl ester synthesis in chloroplasts of Arabidopsis. Plant Cell 24: 2001–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gao L., Xia T., Zhao L. (2009) Investigation of the site-specific accumulation of catechins in the tea plant (Camellia sinensis (L.) O. Kuntze) via vanillin-HCl staining. J. Agric. Food Chem. 57: 10371–10376. [DOI] [PubMed] [Google Scholar]
- Llorente B., D’Andrea L., Rodriguez-concepcion M. (2016) Evolutionary recycling of light signaling components in fleshy fruits: new insights on the role of pigments to monitor ripening. Front. Plant Sci. 7: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B., Martinez-Garcia J.F., Stange C., Rodriguez-Concepcion M. (2017) Illuminating colors: regulation of carotenoid biosynthesis and accumulation by light. Curr. Opin. Plant Biol. 37: 49–55. [DOI] [PubMed] [Google Scholar]
- Lopezmalo A., Alzamora S.M., Argaiz A. (1995) Effect of natural vanillin on germination time and radial growth of molds in fruit-based agar systems. Food Microbiol. 12: 213–219. [Google Scholar]
- Mackenzie S.H., Clark A.C. (2012) Death by caspase dimerization. Adv. Exp. Med. Biol. 747: 55–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marianayagam N.J., Sunde M., Matthews J. M. (2004) The power of two: protein dimerization in biology. Trends Biochem. Sci. 29: 618–6S25. [DOI] [PubMed] [Google Scholar]
- McAndrew R.S., Froehlich J.E., Vitha S., Stokes K.D., Osteryoung K.W. (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol. 127: 1656–1666. [PMC free article] [PubMed] [Google Scholar]
- Metraux J.P. (2002) Recent breakthroughs in the study of salicylic acid biosynthesis. Trends Plant Sci. 7: 332–334. [DOI] [PubMed] [Google Scholar]
- MØller B.L. (2014) Disruptive innovation: channeling photosynthetic electron flow into light-driven synthesis of high-value products InSynthetic Biology. Edited by Ryadnov B., Suga pp. 330–354. The Royal Society of Chemistry, Cambridge. [Google Scholar]
- MØller B.L. (2017) Solar-driven synthesis of bioactive natural products. Chem. Eng. Prog. 113: 46–54. [Google Scholar]
- Negishi O., Sugiura K., Negishi Y. (2009) Biosynthesis of vanillin via ferulic acid in Vanilla planifolia. J. Agric. Food Chem. 57: 9956–9961. [DOI] [PubMed] [Google Scholar]
- Nielsen V.S., Mant A., Knoetzel J., Moller B.L., Robinson C. (1994) Import of barley photosystem-I subunit-N into the thylakoid lumen is mediated by a bipartite presequence lacking an intermediate processing site. Role of the delta-pH in translocation across the thylakoid membrane. J. Biol. Chem. 269: 3762–3766. [PubMed] [Google Scholar]
- Odoux E. (2005) Glucosylated aroma precursors and glucosidase(s) in vanilla bean (Vanilla planifolia G. Jackson). Fruits 61:171–184. [Google Scholar]
- Odoux E., Brillouet J.M. (2009) Anatomy, histochemistry and biochemistry of glucovanillin, oleoresin and mucilage accumulation sites in green mature vanilla pod (Vanilla planifolia; Orchidaceae): a comprehensive and critical reexamination. Fruits 64: 221–241. [Google Scholar]
- Odoux E., Chauwin A., Brillouet J.M. (2003a) Purification and characterization of vanilla bean (Vanilla planifolia Andrews) beta-d-glucosidase. J. Agric. Food Chem. 51: 3168–3173. [DOI] [PubMed] [Google Scholar]
- Odoux E., Escoute J., Verdeil J.L. (2006) The relation between glucovanillin, beta-d-glucosidase activity and cellular compartmentation during the senescence, freezing and traditional curing of vanilla beans. Ann. Appl. Biol. 149: 43–52. [Google Scholar]
- Odoux E., Escoute J., Verdiel J.L., Brillouet J.M. (2003b) Localization of beta-d-glucosidase activity and glucovanillin in vanilla bean (Vanilla planifolia Andrews). Ann. Bot. 92: 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odoux E., Grisoni M. (2010) Vanilla (Medicinal and Aromatic Plants—Industrial profiles). CRC Press, Boca Raton, FL. [Google Scholar]
- Okazaki Y., Shimojima M., Sawada Y., Toyooka K., Narisawa T., Mochida K., et al. (2009) A chloroplastic UDP-glucose pyrophosphorylase from Arabidopsis is the committed enzyme for the first step of sulfolipid biosynthesis. Plant Cell 21: 892–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J.G., Dagil R., Niclasen L.M., Sorensen O.E., Kragelund B.B. (2009) Structure of the mature streptococcal cysteine protease exotoxin mSpeB in its active dimeric form. J. Mol. Biol. 393: 693–703. [DOI] [PubMed] [Google Scholar]
- Otto H.H., Schirmeister T. (1997) Cysteine proteases and their inhibitors. Chem. Rev. 97: 133–171. [DOI] [PubMed] [Google Scholar]
- Palama T.L., Khatib A., Choi Y.H., Payet B., Fock I., Verpoorte R., et al. (2009) Metabolic changes in different developmental stages of Vanilla planifolia pods. J. Agric. Food. Chem. 57: 7651–7658. [DOI] [PubMed] [Google Scholar]
- Roberts E.A. (1946) Electron microscope studies of the structure of chloroplasts, chromoplasts, leucoplasts showing their relationship to cellulose particles, carotin crystals and colloidal carbon. J. Appl. Phys. 17: 68–68. [Google Scholar]
- Robinson C., Mant A. (2002) Molecular Plant Biology. Oxford University Press, Oxford. [Google Scholar]
- Sainders J.A., McClure J.W. (1975) Phytochrome controlled phenylalanine ammonia lyase in Hordeum vulgare plastids. Phytochemistry 14: 1285–1289. [Google Scholar]
- Sanchez-Perez R., Belmonte F.S., Borch J., Dicenta F., Moller B.L., Jorgensen K. (2012) Prunasin hydrolases during fruit development in sweet and bitter almonds. Plant Physiol. 158: 1916–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M. (1966) Metabolism of phenolic substances by the chloroplasts-II: Conversion by the isolated chloroplasts of p-coumaric acid to caffeic acid. Phytochemistry 5: 385–389. [Google Scholar]
- Saunders J.A., Conn E.E. (1977) Subcellular localization of the cyanogenic glucoside of sorghum by autoradiography. Plant Physiol. 59: 647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders J.A., McClure J.W. (1976) The distribution of flavonoids in chloroplasts of twenty five species of vascular plants. Phytochemistry 15: 809–810. [Google Scholar]
- Schramm T., Hester A., Klinkert I., Both J.P., Heeren R.M., Brunelle A., et al. (2012) imzML—a common data format for the flexible exchange and processing of mass spectrometry imaging data. J. Proteomics 75: 5106–5110. [DOI] [PubMed] [Google Scholar]
- Shindo T., Van der Hoorn R.A.L. (2008) Papain-like cysteine proteases: key players at molecular battlefields employed by both plants and their invaders. Mol. Plant Pathol. 9: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A.K., Sharma U.K., Sharma N. (2008) A comprehensive review on vanilla flavor: extraction, isolation and quantification of vanillin and others constituents. Int. J. Food Sci. Nutr. 59: 299–326. [DOI] [PubMed] [Google Scholar]
- Sitte P. (1977) Chromoplasten—bunte Objekte der modernen Zellbiologie. Biologie in unserer Zeit. Wiley Subscription Services, Inc., A Wiley Company. [Google Scholar]
- Smolarski N. (2012) High-Value Opportunities for Lignin: Unlocking its Potential. Frost and Sullivan. Available at: http://www.greenmaterials.fr/wp-content/uploads/2013/01/High-value-Opportunities-for-Lignin-Unlocking-its-Potential-Market-Insights.pdf.
- Storer A.C., Menard R. (1996) Recent insights into cysteine protease specificity: lessons for drug design. Perspect. Drug Des. 6: 33–46. [Google Scholar]
- Thunig J., Hansen S.H., Janfelt C. (2011) Analysis of secondary plant metabolites by indirect desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 83: 3256–3259. [DOI] [PubMed] [Google Scholar]
- Turk V., Stoka V., Vasiljeva O., Renko M., Sun T., Turk B., et al. (2012) Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim. Biophys. Acta 1824: 68–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent J.L., Brewin N.J. (2000) Immunolocalization of a cysteine protease in vacuoles, vesicles, and symbiosomes of pea nodule cells. Plant Physiol. 123: 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Tai F., Chen S. (2008) Optimizing protein extraction from plant tissues for enhanced proteomics analysis. J. Sep. Sci, 31: 2032–2039. [DOI] [PubMed] [Google Scholar]
- Wang Z., Benning C. (2012) Chloroplast lipid synthesis and lipid trafficking through ER–plastid membrane contact sites. Biochem. Soc. Trans. 40: 457–463. [DOI] [PubMed] [Google Scholar]
- Weier E. (1936) The structure of the non-starch-containing beet chloroplast. Amer. J. Bot. 23: 645–652. [Google Scholar]
- Wiederanders B., Kaulmann G., Schilling K. (2003) Functions of propeptide parts in cysteine proteases. Curr. Protein Pept. Sci. 4: 309–326. [DOI] [PubMed] [Google Scholar]
- Yang H., Barros-Rios J., Kourteva G., Rao X., Chen F., Shen H., et al. (2017) A re-evaluation of the final step of vanillin biosynthesis in the orchid Vanilla planifolia. Phytochemistry 139: 33–46. [DOI] [PubMed] [Google Scholar]
- Zaprometov M.N., Nikolaeva T.N. (2003) Chloroplasts isolated from kidney bean leaves are capable of phenolic compound biosynthesis. Russ. J. Plant Physiol. 50: 623–626. [Google Scholar]
- Zenk M.H. (1965) Biosynthese von vanillin in Vanilla planifolia Andr. Z. Pflanzenphysiol. 53: 404–414. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.