This study analyzes the proteomes and phospho-proteomes of isogenic DLD-1 cancer cells differing in karyotypes and chromosome stability. Chromosome doubling is shown to trigger more extensive changes in (phospho-)proteomes than chromosome instability, and activation of mitotic pathways may explain differential responses to mitotic inhibitors.
Abstract
Although aneuploidy is poorly tolerated during embryogenesis, aneuploidy and whole chromosomal instability (CIN) are common hallmarks of cancer, raising the question of how cancer cells can thrive in spite of chromosome aberrations. Here we present a comprehensive and quantitative proteomics analysis of isogenic DLD-1 colorectal adenocarcinoma cells lines, aimed at identifying cellular responses to changes in ploidy and/or CIN. Specifically, we compared diploid (2N) and tetraploid (4N) cells with posttetraploid aneuploid (PTA) clones and engineered trisomic clones. Our study provides a comparative data set on the proteomes and phosphoproteomes of the above cell lines, comprising several thousand proteins and phosphopeptides. In comparison to the parental 2N line, we observed changes in proteins associated with stress responses and with interferon signaling. Although we did not detect a conspicuous protein signature associated with CIN, we observed many changes in phosphopeptides that relate to fundamental cellular processes, including mitotic progression and spindle function. Most importantly, we found that most changes detectable in PTA cells were already present in the 4N progenitor line. This suggests that activation of mitotic pathways through hyper-phosphorylation likely constitutes an important response to chromosomal burden. In line with this conclusion, cells with extensive chromosome gains showed differential sensitivity toward a number of inhibitors targeting cell cycle kinases, suggesting that the efficacy of anti-mitotic drugs may depend on the karyotype of cancer cells.
INTRODUCTION
Aneuploidy is a genomic state in which chromosome number is not a multiple of the haploid number. Constitutional aneuploidy originates during meiosis and is therefore present in all cells of an organism. In humans, most cases of constitutional aneuploidy cause embryonic lethality, with the exception of a few viable constellations such as trisomies 21, 13, or 18, which lead to Down, Patau, or Edwards syndrome, respectively. In contrast, most acquired somatic aneuploidies, as seen in a vast majority of all malignant human tumors, are nonclonal and generally reflect errors in chromosome segregation during mitosis (Santaguida and Amon, 2015a). Moreover, many human tumors display not just aneuploidy but also a constant chromosome missegregation phenotype known as chromosomal instability (CIN) (Lengauer et al., 1997; van Jaarsveld and Kops, 2016). This leads to states of aneuploidy that change in time and space, thereby generating extensive genome heterogeneity within tumor tissues. Therefore, CIN is thought to have important implications for both cancer development and therapy. Prominent causes of CIN include defects in DNA replication, chromatid cohesion or mitotic spindle function, particularly aberrant microtubule dynamics, erroneous microtubule–kinetochore attachments, spindle checkpoint dysfunctions, and/or deregulation of the centrosome duplication cycle (Holland and Cleveland, 2009; Thompson and Compton, 2010).
Studies carried out primarily in yeast and mammalian cell lines have shown that aneuploidy comes with a fitness cost. Aneuploid cells typically grow slower (McCoy et al., 1974; Torres et al., 2007; Williams et al., 2008; Tang et al., 2011; Siegel and Amon, 2012; Stingele et al., 2012) and suffer from replication stress that leads to DNA damage and gene mutation (Janssen et al., 2011; Crasta et al., 2012; Santaguida and Amon, 2015a; Passerini et al., 2016; Ly and Cleveland, 2017). Also, both in vitro engineered aneuploid cells and chromosomally unstable cancer cells display gene expression patterns (Sheltzer, 2013) reminiscent of stress responses first described in yeast (Gasch, 2007). Accordingly, aneuploid cells were found to show increased sensitivity toward compounds inducing energy stress and proteotoxic stress (Tang et al., 2011). In nontransformed cells, chromosome missegregation generally leads to p53-dependent cell cycle arrest and, ultimately, cell death (Li et al., 2010; Thompson and Compton, 2010; Uetake and Sluder, 2010; Janssen et al., 2011; Lambrus et al., 2016). Yet, despite this fitness cost, severe aneuploidy and CIN are hallmarks of human cancers (Hanahan and Weinberg, 2011; Holland and Cleveland, 2012; Funk et al., 2016; De Braekeleer et al., 2017). They contribute to increased transformative potential (Paulsson and Johansson, 2007; Weaver et al., 2007) and correlate with poor prognosis (McGranahan et al., 2012). To resolve this apparent conundrum, it is generally argued that aneuploidy and CIN result in deregulated gene expression, which then confers a selective advantage during the evolution of a tumor in a changing microenvironment (Baek et al., 2009; Pavelka et al., 2010; Kwon-Chung and Chang, 2012; Yona et al., 2012). As one example supporting this notion, DLD-1 cells engineered to carry single-chromosome aneuploidies were found to have a selective advantage over diploid control cells when cultured under nonstandard conditions, such as serum starvation, drug treatment, or hypoxia (Rutledge et al., 2016). Such observations, as well as data obtained in tumor models, strongly support the hypothesis that aneuploidy is not a by-product of cell transformation but, when present at appropriate levels, contributes to tumor development (Hanks et al., 2004; Holland and Cleveland, 2012; Davoli et al., 2013).
Aneuploidy in cancer cells may arise when diploid progenitors gain or lose individual chromosomes. However, chromosome loss is not well tolerated in diploid cells (Alvaro et al., 2006; Anders et al., 2009). Moreover, cancer cells often carry near-tetraploid chromosome numbers, indicative of whole genome duplication events (Zack et al., 2013). This suggests that aneuploid cancer cells often derive from tetraploid intermediates (Cowell and Wigley, 1980; Mayer and Aguilera, 1990; Storchova and Pellman, 2004; Storchova and Kuffer, 2008; Holland and Cleveland, 2012). Considering that tetraploidization creates redundancy in chromosome content, it is expected to protect descendant aneuploid cells from the negative effects of haploinsufficiency (Shackney et al., 1989; Storchova and Pellman, 2004; Ganem and Pellman, 2007; Thompson and Compton, 2010; Dewhurst et al., 2014).
Aneuploidy has traditionally been ascribed to defects in mitotic spindle organization and/or dysfunction of the spindle assembly checkpoint (Wang et al., 2007; Kops et al., 2005). However, although mutations in spindle checkpoint genes can indeed cause aneuploidy (Hanks et al., 2004; Yost et al., 2017), such mutations have not been commonly observed in cancers (Cahill et al., 1999; Haruki et al., 2001). Deregulated expression of essential regulators of chromosome segregation and cell division has been observed in cancers with high degrees of aneuploidy and, accordingly, a CIN marker signature (CIN70) was proposed (Carter et al., 2006). However, subsequent studies argued that this CIN signature reflects altered proliferation rate rather than chromosome missegregation (Venet et al., 2011; Sheltzer, 2013; Buccitelli et al., 2017). Thus, a specific cellular response to CIN has not yet been identified.
Here we established a set of transformed cancer cell lines of isogenic origin but differing in chromosome content and propensity to chromosome missegregation. To determine the effects of gains in chromosome mass versus CIN on protein expression and phosphorylation, we subjected the different cell lines to extensive proteomic and phosphoproteomic analyses. We found that proteomic changes in response to CIN are similar to those observed in response to tetraploidy and are more readily detectable at the level of protein phosphorylation than at the level of protein expression. Furthermore, our results indicate that large gains in chromosome number, as caused by tetraploidization, trigger widespread responses in protein expression and phosphorylation patterns, lending support to the notion that an initial genome doubling event can set the stage for survival and propagation of descendent aneuploid tumor cells.
RESULTS
Establishment of DLD-1–derived cell lines differing in ploidy and aneuploidy
Chromosome gains or losses result in massive changes in gene expression (Lyle et al., 2004; Upender et al., 2004; Stingele et al., 2012), and protein expression patterns in cancer cell lines are known to reflect tissue origin, a priori making it difficult to identify a proteomic signature attributable to CIN. This notwithstanding, we subjected a panel of human cell lines to a proteomic quantification based on multiplexed tandem mass tag (TMT) labeling, a method of choice for achieving high proteome coverage in multiple samples and within a reasonable time frame (Thompson et al., 2003; Ahrné et al., 2016) (Supplemental Figure S1A and Supplemental Table S1). This panel included seven karyotypically stable (nonCIN) and unstable (CIN) cancer cell lines originating from different tumor tissues (Gascoigne and Taylor, 2008) and the immortalized retinal cell line hTERT. In line with previous data (Gascoigne and Taylor, 2008), we found that differences in global protein expression patterns were too profound to allow a distinction between CIN and karyotypically stable (nonCIN) cell lines through hierarchical cluster analysis (Supplemental Figure S1B). Nevertheless, this pilot study showed that our proteomics approach allowed for reliable quantification of thousands of proteins in each cell line.
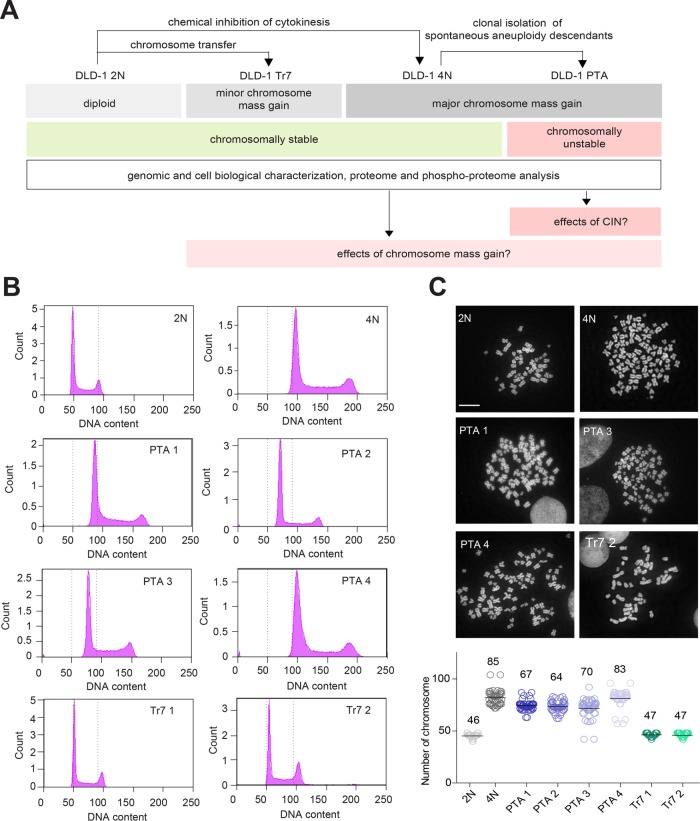
To reduce interline variation due to tissue origin, we next used the diploid colon cancer cell line DLD-1 to generate descendant lines differing in karyotype. DLD-1 cells show microsatellite instability (MIN) but proliferate in a near-diploid state (Lengauer et al., 1997). As DLD-1 cells are deficient in p53, tetraploid derivatives can readily be established through inhibition of cytokinesis (Drosopoulos et al., 2014). This afforded a syngeneic pair of stable diploid and tetraploid cells (Figure 1A). Starting with a culture of tetraploid DLD-1 cells, we then used single cell fluorescence-activated sorting (FACS) to isolate spontaneously arising aneuploid descendants. This provided us with four different PTA clones, specifically three near-triploid lines and one near-tetraploid line (Figure 1B). Finally, we applied microcell-mediated chromosome transfer (Stingele et al., 2012) to the parental diploid DLD-1 culture and obtained two viable trisomic clones carrying three copies of chromosome 7 (Tr 7) (Figure 1B). For all cell lines, DNA content was confirmed by chromosome counting (Figure 1C) and chromosome painting (Supplemental Figure S2A). This collection of isogeneic cell lines set the stage for analyzing chromosomally stable diploid, tetraploid, and trisomic cells as well as chromosomally unstable cells with complex aneuploidy (Figure 1A). We reasoned that this experimental approach might offer the possibility to dissect the effects of sheer gains in chromosome mass (represented by the tetraploid line) from those triggered by CIN (the PTA lines) or low-complexity aneuploidy (the trisomic lines).
FIGURE 1:
Establishment of DLD-1–derived lines differing in ploidy. (A) Schematic summarizing the generation, properties, and analysis of DLD-1 colon cancer cell lines. Starting from a diploid (2N) parental culture, tetraploid (4N) cells were obtained by inhibition of cytokinesis. The 4N cells were then used for clonal isolation of posttetraploid aneuploid (PTA) descendants. Clones harboring trisomies of chromosome 7 (Tr7) were generated from the diploid parental culture by microcell-mediated chromosome transfer. (B) Histograms document the DNA profiles of the cell lines used in this study. Cells were stained with propidium iodine and subjected to analysis by flow cytometry. Dotted lines indicate the G1 and G2/M peaks expected for the diploid culture. (C) Top panel: micrographs depict mitotic spreads of the indicated cell lines; chromosomes were stained with DAPI. Bottom panel: histograms show chromosome numbers for each cell line, with bars and numbers indicating mean values; at least 40 cells were counted for each line. Data represent results from three biological replicates. Scale bar denotes 10 μm.
Phenotypic characterization of DLD-1–derived cell lines
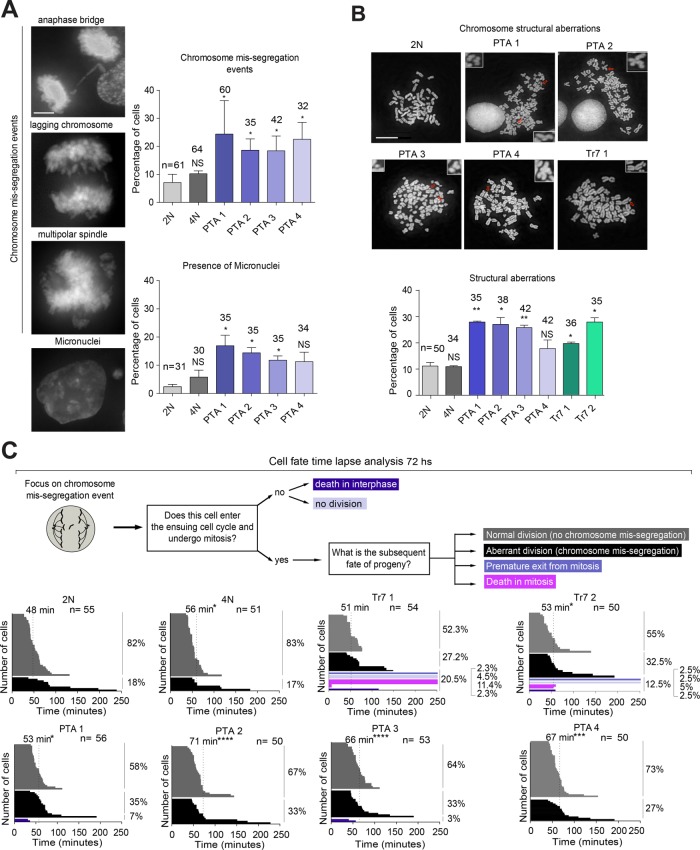
To further characterize the DLD-1 cell lines described above, we used microscopy to analyze chromosome segregation fidelity and mitotic duration. Compared to the diploid parental line, the frequencies of chromosome missegregation and micronuclei formation were significantly elevated in most PTA clones (Figure 2A) but not in the tetraploid line (Figure 2A). In agreement with previous work (Nicholson et al., 2015), the trisomic clones showed similar aberrations, albeit to a lesser extent (Supplemental Figure S2B). Furthermore, we observed an increase of structural aberrations in PTA lines and, consistent with earlier work (Kuznetsova et al., 2015; Passerini et al., 2016), also in trisomic clones (Figure 2B). In contrast, the frequencies of such aberrations were low in diploid and tetraploid cultures (Figure 2B). Mitotic spindle angle, which was determined as an indicator of proper spindle geometry, was not significantly altered in any of the cell lines (Supplemental Figure S2C), and a large majority of mitotic spindle poles in all cell lines contained the diploid equivalent of centrosomes and centrioles (Supplemental Figure S2D). Together, these findings support the notion that the unbalanced gain of chromosomes leads to an increase in chromosome segregation errors and genetic instability but that cells harboring these chromosomal aberrations are able to form largely normal mitotic spindles (Dodgson et al., 2016; Passerini et al., 2016).
FIGURE 2:
Mitotic properties of DLD-1–derived cells. (A) Analysis of chromosome segregation fidelity. Top panel: representative images illustrate chromosome missegregation events and micronucleation. Scale bar represents 5 μm. Right panel: histograms show the frequency of the above phenotypes in the indicated cell lines. (B) Left panel: micrographs show mitotic spreads of the indicated cell lines, with arrows pointing at structural chromosome aberrations (enlarged in insets). Scale bar represents 10 μm. Right panel: histogram shows the frequency of chromosome structural aberrations observed in the indicated cell lines. (C) Mitotic duration and cell fate in DLD-1–derived cells. Left panel: schematic summarizes cell fate analysis by time-lapse microscopy, using asynchronously growing cultures stably expressing GFP-tagged histone H2B. Dashed lines indicate mean mitotic duration. Frequencies of cell fates are shown to the right of each histogram. All fixed cells were stained with DAPI. Error bars in A and B show SD, and numbers of counted cells are indicated. Data represented in A and B result from three biological replicates; data in C from two biological replicates. Two-tailed t test: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Since supernumerary chromosomes are likely to prolong the time required for proper chromosome alignment on the mitotic spindle, and since chromosome missegregation can severely impair cell survival, we performed live cell imaging on cells transiently transfected with histone H2B-GFP. Specifically, we scored cells for the time spent in mitosis. Moreover, we focused on cell divisions displaying a spontaneous chromosome missegregation event and then analyzed the frequency of different fates after the completion of such a division. These fates included continued division with or without chromosome missegregation, premature mitotic exit/checkpoint slippage, or death in interphase or mitosis (Figure 2C). Interestingly, in the diploid culture, an occasional chromosome missegregation was often followed by an error-free division in the ensuing cell cycle, but in all PTA clones we observed an elevated rate of chromosome missegregation in the subsequent division, and we also measured a significant prolongation of mitotic duration (Figure 2C). In the tetraploid culture, mitotic length was also increased significantly, but this was not accompanied by an elevated rate of missegregation (Figure 2C). Trisomic clones responded to an initial chromosome missegregation event with a marginal (not statistically significant) prolongation of mitosis and continued chromosome missegregation; importantly, however, chromosome missegregation in these lines commonly triggered mitotic slippage and cell death (Figure 2C). Collectively, these data indicate that an increase in chromosome number provokes increased mitotic duration but not necessarily an increase of chromosome missegregation (as suggested by the different behaviors of PTA clones and tetraploid cells). Furthermore, in cells carrying an unbalanced genome (the PTAs and the trisomic clones), any spontaneous chromosome missegregation event is commonly followed by continued missegregation. However, while cells displaying complex aneuploidies (PTA) tolerate chromosome segregation errors, cells with low complexity aneuploidy (Tr7) often respond to such errors by cell death, thereby preserving the karyotype of the culture. On the basis of these findings, we classify the trisomic cultures as “chromosomally stable.”
Having characterized the different cell lines, we compared the karyotypically stable (diploid, trisomic, and tetraploid) clones with the karyotypically unstable (PTA) clones to investigate the effects of altered chromosome mass versus altered chromosome stability (CIN) on protein expression and protein phosphorylation (see also Figure 1A). Comparison of the doubling times or cell cycle profiles of the cell lines analyzed here revealed no significant differences. Moreover, we emphasize that all cells were synchronized in mitosis prior to analysis (see Materials and Methods).
Comparison of chromosome copy number and corresponding protein expression
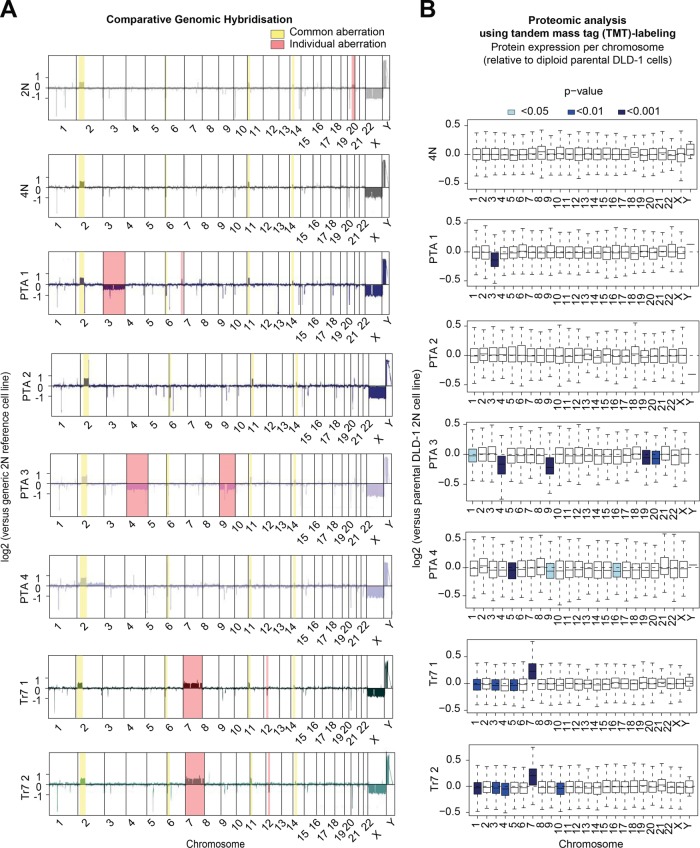
To test the impact of changes in chromosome copy numbers on protein levels, we submitted all cell lines to array comparative genomic hybridization (aCGH) and, in parallel, to quantitative proteome analysis using TMT labeling. Array hybridization assays showed that most small structural aberrations present in the parental DLD-1 line were propagated to the cells harboring tetraploid, PTA, and trisomic karyotypes (Figure 3A), and these minor aberrations were also detected in a DLD-1 line originating from a different laboratory (unpublished data; Upender et al., 2004). Second, prominent whole-chromosome copy number reductions were detected in two PTA clones, affecting chromosome 3 in PTA1 and chromosomes 4 and 9 in PTA3 (Figure 3A and Supplemental Figure S2A). We surmise that these chromosomes were lost early during the isolation of PTA1 and PTA3, while PTA2 and PTA4 apparently acquired near-triploid and near-tetraploid karyotypes through progressive chromosome missegregation, resulting in extensive chromosomal heterogeneity. Finally, the expected single chromosome gain could readily be observed in the trisomy 7 clones (Figure 3A and Supplemental Figure S2A).
FIGURE 3:
Comparison of chromosome copy number and protein expression. (A) Comparative genomic hybridization assay shows chromosome copy number changes for the indicated cell lines and chromosomes, relative to a generic diploid (2N) reference line. Copy number variations that remained largely unchanged in all cell cultures are highlighted in yellow, and individual variations are highlighted in red. (B) LC-MS/MS analysis using the TMT labeling approach. Box-whisker plots show protein abundance relative to the parental diploid (2N) DLD-1 culture for the indicated lines. Proteins are ordered by chromosome origin, and blue shading indicates the level of significance. Data in B are from three biological replicates.
A comparison of aCGH data with proteomic data revealed that chromosome copy number aberrations in PTA and trisomic clones showed a positive correlation with the average relative expression levels of the proteins encoded by the genes on the respective chromosomes, as compared with the 2N parental cell line (Figure 3B). These data confirm that alterations in gene dosage generally lead to corresponding changes in protein expression levels (Upender et al., 2004). Proteome analyses did not reveal significant chromosome-specific deregulations of protein expression in either the 4N culture or PTA2, likely reflecting the balanced karyotype (in case of 4N) or mild and clonally heterogeneous aneuploidy (in case of PTA2). Finally, in PTA3 and PTA4, as well as in both trisomic clones, we identified small but statistically significant reductions in protein expression patterns, even though the corresponding chromosomes that did not show obvious copy number deviations in the aCGH analysis (Figure 3B), illustrating the high sensitivity of the proteomic analysis.
Comparative proteomic analysis of DLD-1–derived cells
To provide information about relative protein expression at higher resolution, comparative proteomics studies were performed, in three biological replicates, using the 6- and 10-plex TMT labeling approach (Ahrné et al., 2016). This yielded information about expression levels for some 6000–7500 proteins across all DLD-1 cell lines described above (Supplemental Table S2). Compared to the 2N parental culture, the 4N clone showed a similar frequency of up- and down-regulations, while the PTA and trisomic clones showed a slight predominance of down-regulations. Previous studies had suggested that aneuploidy commonly induces the activation of stress response pathways, resulting in deregulated expression of genes associated with proteolysis, protein folding, autophagy, DNA damage, and oxidative stress (Sheltzer et al., 2012; Sheltzer, 2013; Stingele et al., 2012; Dephoure et al., 2014; Durrbaum et al., 2014; Ohashi et al., 2015; Santaguida and Amon, 2015a, b). Confirming and extending these observations, we found that proteins associated with aneuploidy-induced stress responses showed a trend toward up-regulation in all cell lines that had undergone major chromosome gains, notably the 4N clone and the PTA clones (Supplemental Figure S3A). Prominent examples of proteins up-regulated by >1.5 fold are summarized in Supplemental Table S3.
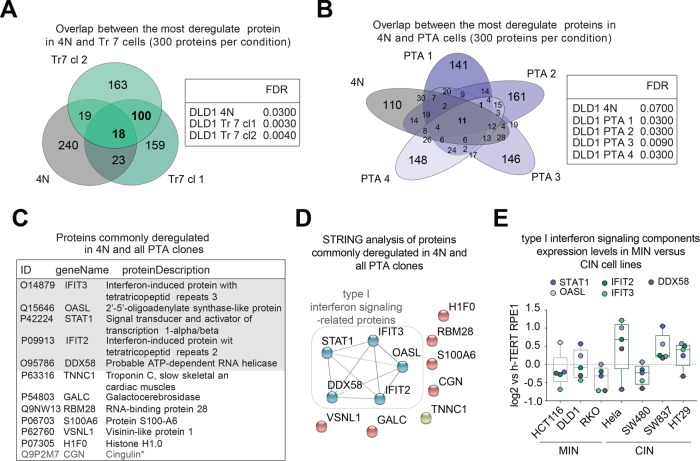
Next we asked whether deregulated expression of any particular set of proteins could be correlated with either chromosome mass gains per se (in 4N, PTA, and trisomic clones) and/or the genetic instability present in CIN cells (in PTA clones). To this end, we first examined our proteome data sets by comparing the 300 most significantly deregulated proteins in the 4N line and the two trisomic clones, standardized against the 2N parental line (Figure 4A). Although this led to the identification of 18 commonly deregulated proteins (Supplemental Table S2), these showed no obvious functional relationship by search tool for the retrieval of interacting genes/proteins (STRING) analysis (unpublished data). A similar analysis was then carried out comparing the 4N line and each of the PTA samples (Figure 4B). Surprisingly, only 12 proteins were consistently deregulated in all four PTA lines, and 11 of these (except for CGN; Figure 4D) were deregulated also in response to a doubling of chromosome mass in the 4N line (Figure 4B and Supplemental Table S2). This suggests that deregulation of these proteins reflected a response to extra chromosomes mass rather than CIN. These 11 deregulated proteins comprised five gene products (IFIT2, IFIT3, OASL, STAT1, and DDX58) whose annotation suggests an involvement in the regulation of interferon signaling (Figure 4C and Supplemental Figure S3B), and these five proteins could also be functionally linked through STRING analysis (Figure 4D). This observation is reminiscent of several recent studies demonstrating a link between aneuploidy and activation of the innate immune response, which in turn has been related to the presence of cytoplasmic DNA (Weichselbaum et al., 2008; Stingele et al., 2012; Durrbaum et al., 2014; Shen et al., 2015; Ho et al., 2016; Erdal et al., 2017). In line with this argumentation, BrdU staining revealed cytoplasmic signals consistent with the presence of cytoplasmic DNA in cells harboring excess chromosomes (Supplemental Figure S3C).
FIGURE 4:
Comparative proteomic analysis of DLD-1–derived cells. (A, B) Left panels: Venn diagrams represent the numbers of shared protein deregulated across the indicated cell lines. Results were obtained by selecting the 300 most deregulated proteins per cell line (based on a false discovery rate [FDR] of <10%). Right panels: tables listing the FDR for each cell line. (C) Listing of the 11 proteins commonly deregulated across tetraploid and PTA clones, as shown in B. Asterisk demarks the single protein found to be deregulated specifically to PTA clones only, as shown in B. Shaded area highlights proteins involved in type I interferon signaling. (D) STRING functional network analysis of the 11 proteins commonly deregulated in 4N and PTA clones, as shown in B and C. Nodal connections are based on a confidence value of 0.9, using experimental and database evidence. (E) Box-whisker plots show the relative abundance of proteins involved in type I interferon signaling across microsatellite instable (MIN) and chromosomally instable (CIN) cell lines. The graph is based on experimental data shown in Supplemental Figure S1, A and B, and Supplemental Table S1. Data in E are from a single biological replicate (pilot experiment).
To corroborate the conclusion that complex aneuploidy correlates with deregulation of interferon signaling, we reexamined the data set from our pilot study, comparing MIN and CIN cell lines (Supplemental Figure S1 and Supplemental Table S1). While none of the diploid MIN cell lines (HCT116, RKO, DLD-1) showed up-regulated expression of IFIT2, IFIT3, OASL, STAT1, or DDX58, three of four tested CIN cell lines (HeLa, HT29, SW837) showed elevated levels of these proteins (Figure 4E).
Targeted analysis of protein expression in DLD-1–derived cells
To determine whether increases in chromosome mass and/or missegregation rate lead to deregulation of proteins previously implicated in malignant transformation, cell-cycle progression, or mitotic functions, we designed protein lists for targeted data analysis. A first inclusion list featured 550 tumor suppressor genes (TSG) and oncogenes (OG) (Davoli et al., 2017), encompassing 14 signaling pathways commonly deregulated in cancer (TSG/OG inclusion list; Supplemental Table S4). A second list, comprising 737 proteins, was compiled by combining gene products alleged to represent a CIN signature (Carter et al., 2006), gene products associated with cell division through the MitoCheck project (Neumann et al., 2010), and products of genes with assigned gene ontology (GO) terms referring to kinetochore-, centrosome-, microtubule-, cell-cycle checkpoint–, cell division–, and chromosome segregation–related processes (CIN/cell division inclusion list; Supplemental Table S4). Third, we generated several smaller inclusion lists covering pathways and protein complexes previously linked to chromosome missegregation and genetic instability (Babu et al., 2003; Weaver and Cleveland, 2006; Kabeche and Compton, 2012), including key proteins with functions in the spindle assembly checkpoint, the anaphase-promoting complex/cyclosome (APC/C), the kinetochore-microtubule interface, as well as DNA replication, notably minichromosome maintenance (MCM) and origin recognition complex (ORC) proteins.
Analysis of the 300 most deregulated proteins in each cell line with help of the TSG/OG inclusion list did not identify any significant enrichments in 4N, PTA, or trisomic samples (Supplemental Figure S4A), possibly reflecting the fact that all these cells were descendant from a diploid colon cancer cell line that had already been transformed. Similarly, none of the most deregulated proteins showed a significant enrichment of CIN- or cell division–related gene products (Supplemental Figure S4B), and no consistent correlation could be detected between gains in chromosome mass, or the presence of CIN, and proteins implicated in the spindle assembly checkpoint, the APC/C complex, or the kinetochore–microtubule interface (Supplemental Figure S4C). Regarding proteins implicated in DNA replication, we examined the levels of DNA replication-licensing MCM proteins, for which low expression levels had previously been correlated with genetic instability in p53-proficient cells displaying low-complexity aneuploidy (Passerini et al., 2016). In our data sets, obtained with p53-deficient cells, we observed down-regulation of MCM subunits in only one PTA clone but not in any of the other clones (Supplemental Figure S4D, top panel). Similarly, we could not detect consistent deregulation of any origin of replication complex (ORC) subunits (Supplemental Figure S4D, bottom panel).
Our inability to detect consistent patterns of deregulation of cell-cycle proteins in response to increased chromosome mass and/or CIN suggests that cells can react to chromosome aberrations in different ways, perhaps by optimizing combinatorial interactions between many components. Moreover, any minor changes in protein expression may have been masked by clonal heterogeneity within the aneuploid cultures. Alternatively, it is possible that the bulk of cellular adaptation to chromosome aberrations relies on posttranslational modifications rather than changes in protein expression. To investigate the latter possibility, we subjected all cell lines to quantitative phosphoproteome analyses, with the aim of correlating deregulated phosphorylation with a gain of chromosome mass and/or CIN.
Comparative phosphoproteomic analysis of DLD-1–derived cells
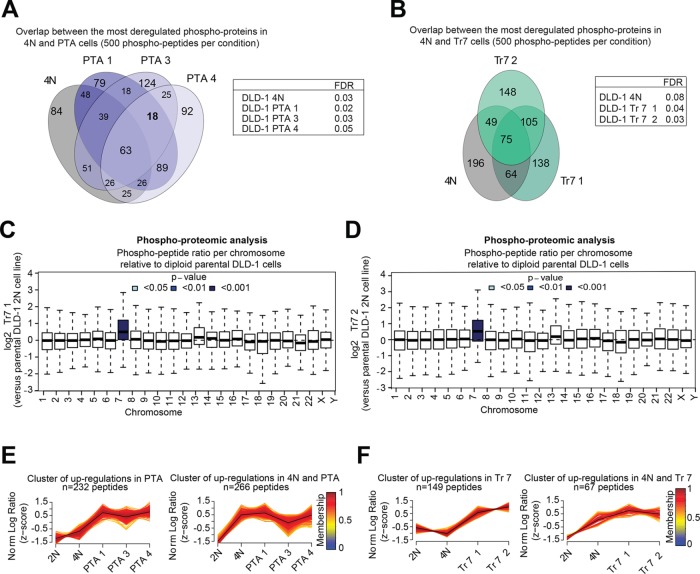
Biological triplicates of all cell lines were subjected to phosphopeptide enrichment through TiO2, followed by high-performance liquid chromatography mass spectometry (HPLC-MS/MS). This resulted in the reproducible identification and quantification of 15–300 phospho-peptides from 3 to 192 different proteins of the 2N and 4N lines and three of the PTA lines (Supplemental Table S5); data for PTA2 were not considered for further analysis, due to an unexpectedly large variance of phosphopeptide abundances across replicate measurements. In parallel, the same approach was applied to the trisomic cell lines, resulting in identification of 8–960 phospho-peptides from 2–553 different proteins (Supplemental Table S5). To identify changes in protein phosphorylation that might correlate with gains in chromosome mass, we compared the 500 most deregulated phosphopeptides (based on a false discovery rate (FDR) of <10%, comprising up- as well as down-regulations) in 4N, PTA, and trisomic clones (Figure 5, A and B). We identified 63 proteins showing significantly deregulated phosphorylation across 4N and PTA clones (Figure 5A and Supplemental Table S6) and 75 proteins across 4N and trisomic clones (Figure 5B and Supplemental Table S6). This demonstrates that changes in chromosome number affect not only protein expression but also protein phosphorylation. However, only few proteins showed deregulated phosphorylation exclusively in all PTA clones, which might have suggested a correlation with CIN (Figure 5A and Supplemental Table S6). Instead, most of these phosphopeptides were deregulated also in 4N clones, suggesting that a major gain in chromosome mass is a more important determinant of deregulated protein phosphorylation than frequent chromosome missegregation. Finally, we emphasize that proteins encoded on chromosome 7 were not only expressed at higher levels in the two trisomy 7 clones (Figure 3B) but also phosphorylated at higher levels, with virtually indistinguishable results obtained for both clones (Figures 5, C and D).
FIGURE 5:
Comparative phosphoproteomic analysis of DLD-1–derived cells. (A, B) Left panel: Venn diagrams represent the number of shared protein deregulations across the indicated cell lines (with each protein showing at least one deregulated phosphopeptide). Results were obtained by selecting the 500 most deregulated phosphopeptides per cell line (FDR of <10%, yielding a total of 1410 phosphopeptides from 807 proteins). Right panel: table listing the FDR for analyzed cell lines. (C, D) The box-whisker plots show the distribution of phosphopeptide abundances of the two trisomy 7 clones relative to the parental diploid (2N) DLD-1, per chromosome. Each box spans the interquartile range. The notches extend to the most extreme data point, which is no more than 1.5 times the interquartile range from the box. The thick horizontal line in each box show the median. The color coding indicates two-sided Student’s t test p value significance, testing the probability of observing a mean log2 ratio at least large as the one observed by chance alone. Data are from three biological replicates (Supplemental Table S4, gray columns). (E, F) Cluster analysis of the data presented in A and B using the fuzzy C-means algorithm “MFuzz.” Depicted clusters show phosphopeptide up-regulations common to PTA (left graph) or 4N and PTA (right graph) clones. Log2 ratios were normalized to yield a SD of 1 and a mean of 0 (z-score). Black lines indicate the optimal membership of 1; color-coding represents cluster membership values. Note that clusters were formed based on peptides that showed significant deregulation in at least one condition; statistical significance for all peptides is given in Supplemental Table S6.
To detect regulation patterns that might reveal a common response to aberrant karyotypes, we grouped the phosphopeptide data using fuzzy C-means clustering (Futschik and Carlisle, 2005). In cells harboring large increases in chromosome numbers (the 4N and PTA clones), we predominantly observed patterns of phosphopeptide up-regulation (Figure 5E and Supplemental Table S6), while the gain of a single chromosome in the trisomic clones revealed phosphopeptide up- as well as down-regulations (Figure 5F and Supplemental Table S7). Next we submitted the phosphopeptide lists obtained for each cluster to GO-term enrichment analysis. The 20 most significantly enriched GO terms per cluster included references to cell-cycle–, cytoskeleton-, as well as RNA-related processes (Supplemental Figure S5, A and B). Remarkably, this observation held true for all cell lines that had undergone any kind of chromosome gain, the 4N and PTA clones (Supplemental Figure S5A), as well as the trisomic clones (Supplemental Figure S5B). Additionally, the clones that had acquired major chromosome gains (4N and PTA) revealed enrichments of GO terms related to nuclear pore organization (Supplemental Figure S5A). Only few processes, including protein sumoylation and G-protein coupled receptor signaling, showed highly significant GO-term enrichments in clones displaying CIN (Supplemental Figure S5A).
Similar results were obtained when clusters of phosphopeptide up-regulations were examined by STRING functional network analysis. First, we observed a greater number of functional clusters in cells that had undergone major chromosome gains (4N and PTA clones) (Supplemental Figure S6, A and B) than in the trisomic clones (Supplemental Figure S6, C and D). Second, the majority of the networks identified in clones harboring both major and minor chromosome gains were related to similar processes: replication, transcription, and translation; transport through nuclear pores; DNA damage response; chromatin organization; as well as microtubule and centrosome regulation (Supplemental Figure S6, A–D). However, while networks for DNA- and/or RNA-related processes were identified in all clones, networks of proteins implicated in the mitotic apparatus and DNA damage responses were enriched primarily in cells carrying a strongly increased chromosome mass (4N and PTA clones) (Supplemental Figure S6, A and B). These observations are reminiscent of a genomic analysis of polyploidy in yeast (Storchová et al., 2006).
Collectively, the above observations suggest that the most prominent changes in protein phosphorylation correlate with a gain in chromosome number rather than the presence of CIN. Furthermore, we conclude that all chromosome aberrations trigger responses related to cell-cycle, cytoskeleton, and RNA metabolism. Additionally, a strong increase in chromosome mass imposes a stress on mitotic spindle organization and provokes a DNA damage response, while a gain of a single chromosome appears to predominantly elicit adaptations of DNA- and RNA-related transactions.
Targeted analysis of protein phosphorylation in DLD-1–derived cells
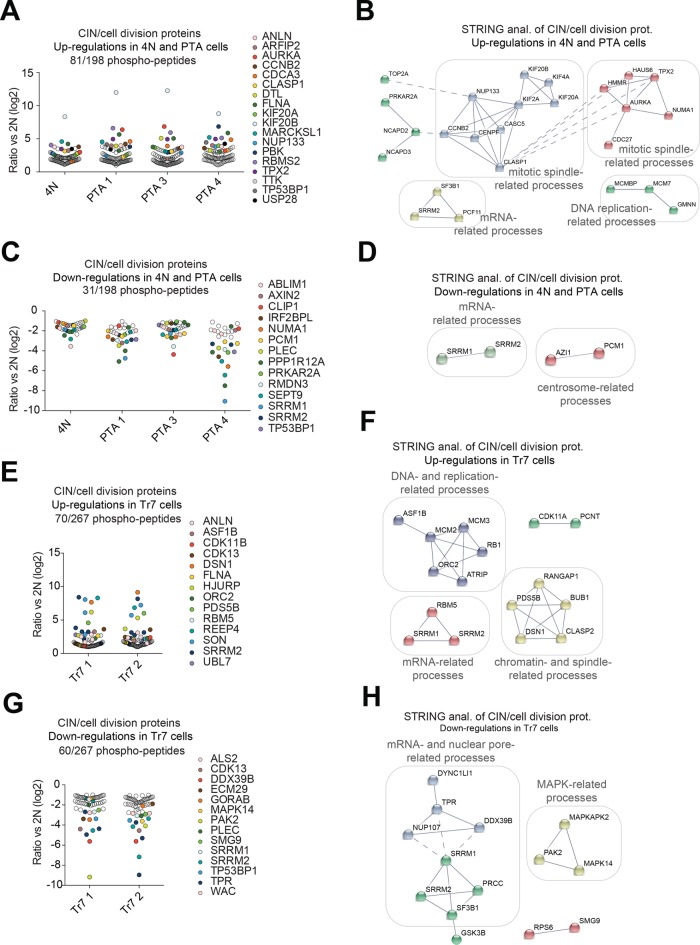
As done above for protein abundance analyses, we complemented the unbiased analysis of the phospho-proteome data set by targeted enrichment analysis, using the CIN/cell division inclusion list (Supplemental Table S4). This allowed us to identify enrichments for phospho-peptides that were significantly up-regulated in 4N and PTA clones, as well as in one trisomic clone, but only few enrichments for significantly down-regulated phosphopeptides could be seen in any clones (Supplemental Figure S7A). For closer inspection, all phosphopeptides deregulated by at least twofold were filtered using the CIN/cell division inclusion list (Supplemental Table S5), but this revealed no proteins showing consistently altered phosphorylation across all PTA clones (unpublished data). In contrast, several proteins related to mitotic spindle function and chromosome segregation were identified whose phosphorylation was commonly altered in both 4N and PTA clones (Figure 6A) and, to a lesser extent, in trisomic clones (Figure 6B). These hits included regulators of microtubule dynamics, kinetochore–microtubule interactions, centrosome function as well as spindle assembly checkpoint signaling (Figure 6, A and B). STRING network analysis of commonly deregulated phosphopeptides also identified mitotic spindle- and chromatin-related processes and processes related to DNA replication and transcription (Figure 6, C–F).
FIGURE 6:
Targeted analysis of protein phosphorylation in DLD-1–derived cells. (A, C, E, G) Dot plots showing the phosphopeptide ratios (versus parental 2N cells) of detected proteins belonging to the CIN/cell division inclusion list and showing at least twofold deregulation. Dots represent significant (p ≤ 0.05) phosphopeptide log2 ratios. Proteins related to mitotic spindle regulation and chromosome segregation are shown in bold. (B, D, F, H) STRING functional network analysis of the data shown in A, C, E, and G. Nodal connections are based on a confidence value of 0.9 using experimental and database evidence. Solid lines indicate intranetwork and dashed lines internetwork connections.
We conclude that a strong increase in chromosome number leads to changes in the phosphoproteome related to spindle function and mitotic regulatory pathways, possibly reflecting stress conditions triggered by the need to segregate large numbers of chromosomes. In agreement with this conclusion, changes were observed predominantly in both 4N and PTA clones and only to a lesser extent in trisomic cells.
Drug sensitivity assays in cultures of DLD-1–derived cells
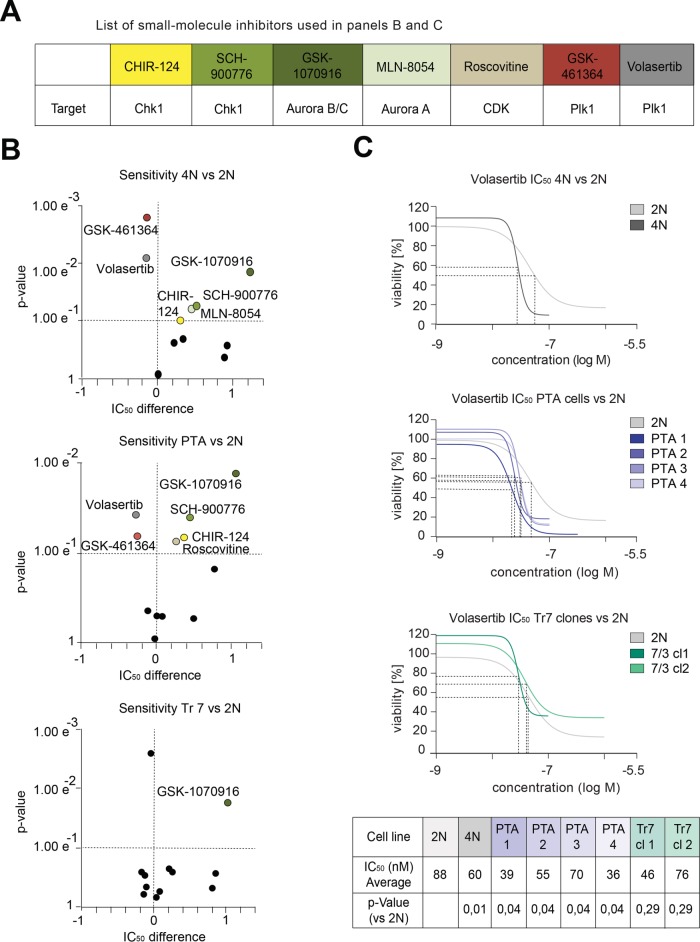
The above observations prompted us to ask whether adaptations of mitotic functions to large gains of chromosomes might translate into differential sensitivities to pharmacological inhibitors of spindle and cell-cycle regulatory kinases. A collection of 38 well-annotated anti-cancer agents, mostly targeting cell division–related protein kinases, was used to perform drug sensitivity assays on all DLD-1–derived cell lines (Figure 7A and Supplemental Table S8). Compounds were tested in a concentration range of 0.32–32 μM, and intracellular ATP content was measured as an indirect readout for cell survival (Kuznetsova et al., 2015). Most observed changes in drug sensitivity were common to cells harboring large gains in chromosome mass (Figure 7B and Supplemental Table S8). Specifically, both 4N and PTA cultures showed increased resistance toward inhibitors of the mitotic spindle kinase Aurora A (GSK-1070916) (Adams et al., 2010) and the DNA-damage checkpoint kinase Chk1 (CHIR-124 and SCH-900776) (Tse et al., 2007; Karp et al., 2012); additionally, the PTA cultures were resistant to Roscovitine, an inhibitor of cyclin-dependent kinases (Meijer and Raymond, 2003) (Figure 7B and Supplemental Table S8). This trend is reminiscent of previous reports on multidrug resistance in response to aneuploidy (Lee et al., 2011; Kuznetsova et al., 2015). However, we emphasize that both 4N and PTA cultures showed increased sensitivity for two distinct compounds targeting Plk1 (GSK-461364 and volasertib) (Shin et al., 2015) (Figure 7B and C, Supplemental Table S8), arguing that multidrug resistance is unlikely to fully explain the resistance to Aurora A, Chk1, and Cdk1 inhibitors. Likewise, we note that the phosphoproteome analyses of 4N and PTA cells revealed a tendency for increased phosphorylation at both the Aurora A T-loop site (Eyers et al., 2003) (Supplemental Figure S7 and Supplemental Table S5) and the Plk1 T-loop site (T210) (Jang et al., 2002; Macůrek et al., 2008) (Supplemental Figure S7), while no alterations at Chk1 or Cdk1 regulatory sites were observed (Smits and Gillespie, 2015; Malumbres, 2014). This argues that major chromosome aberrations trigger changes in the activities of Aurora A and Plk1 that, through mechanisms not yet understood, confer altered sensitivities for the corresponding inhibitors.
FIGURE 7:
Drug sensitivity assays in cultures of DLD-1–derived cells. (A) Table shows the small molecule inhibitors and their kinase targets, used in B and C. (B) Dot plots show the sensitivities (significance determined by unpaired two-tailed t test) on the y-axes and the IC50 differences (log ratios relative to the parental 2N DLD-1 cell line) on the x-axes, as observed after adding compounds to the indicated cell cultures. Dashed lines demark the quadrants of significant increases in sensitivity (top left), significant decreases in sensitivity (top right), and insignificant changes of IC50 values (bottom quadrants). Data in B are from three biological replicates. (C) Graphs show dose–response curves using the Plk1 inhibitor Volasertib on the indicated cell cultures. Dashed lines indicate IC50 values. Table lists IC50 averages from three biological replicates (see Supplemental Table S7) and p values for the indicated cell lines (unpaired two-tailed t test).
DISCUSSION
In this study, we have used proteomics approaches to analyze the effects of chromosome gains and/or CIN on protein levels and protein phosphorylation. To this end, we have generated and characterized a syngeneic set of cell lines derived from DLD-1, a well-established MIN colon cancer line. Specifically, our set of cells comprised the parental diploid DLD-1 line (2N) and descendant trisomic (for chromosome 7; Tr 7), 4N, as well as four distinct PTA lines. As shown by both genomic and proteomic analyses, as well as extensive cell biological characterizations, the 4N line exhibited a low level of chromosome missegregation, comparable to the 2N line, while all four PTA clones displayed elevated rates of missegregation and hence a CIN phenotype. The two trisomic clones also showed elevated levels of chromosome missegregation, but because these errors frequently resulted in cell death, the trisomic cultures maintained an essentially stable karyotype.
Broadly speaking, we found that changes in chromosome copy number exerted proportional effects on the expression of the proteins encoded by the respective chromosomes, confirming that human cells lack a global dosage compensation system for autosomal aneuploidy. Furthermore, we found that the presence of extra chromosomes in the 4N and PTA clones led to a deregulation of proteins involved in protein degradation and folding, autophagy, DNA damage, and oxidative stress response. So far, none of the analysis tools applied to our data sets revealed a unifying protein or phosphoprotein signature that could be considered specific for CIN. This does not exclude the existence of such signatures, but it suggests that their detection is difficult. One complicating factor is that changes in proteins and phosphoproteins may exert combinatorial effects. Thus, a given phenotype, that is, resistance to CIN, may a priori be brought about by deregulation of different sets of (phospho-)proteins in different PTA clones, provided that these achieve a similar physiological outcome. Another problem relates to the clonal heterogeneity of CIN cultures, which makes detection of quantitatively minor changes in protein expression and/or phosphorylation patterns difficult.
Importantly, our inclusion of a tetraploid line in all comparative analyses led us to discover that both the proteomic and the phosphoproteomic signatures of 4N cells closely resembled those seen in the four PTA lines. This key observation suggests that major changes in ploidy already trigger a general proteomic and phosphoproteomic response, regardless of the degree of CIN. We emphasize that these data on the effects of genome doubling and aneuploidy fall in line with a recent comprehensive genomic study on non–small cell lung cancer patients (Jamal-Hanjani et al., 2017), strongly suggesting that genome doubling is an early clonal event associated with frequent subclonal mutations and copy number alterations in a clinically relevant context. While we propose that the (phospho-)proteome changes observed in our study reflect primarily a response to mitotic stresses imposed by increased chromosome burden (see also Storchová et al., 2006), it is conceivable that they also set the stage for increased tolerance toward chromosome missegregation (Dewhurst et al., 2014; Kuznetsova et al., 2015). In particular, initial changes triggered by increased chromosome numbers may confer increased robustness to the spindle, which may then confer tolerance to CIN and facilitate the survival of emerging clones. Because CIN might conceivably be triggered by a myriad of distinct mechanisms, the identification of proteomic CIN signatures remains a daunting task.
Most intriguingly, we observed changes in the protein levels of type I interferon-signaling components in both 4N and PTA cells (Figure 4, C and D, and Supplemental Figure S3B). To some extent, this could be correlated with an increased presence of cytoplasmic DNA (Supplemental Figure S3C) and increased phosphorylation of DNA damage response proteins (Supplemental Figure S6, A and B). These observations lend support to several recent studies that establish a link between aneuploidy-related chromosome lesions and increased abundance of cytoplasmic DNA fragments, which then trigger a DNA damage response and induction of type I interferon signaling (Shen et al., 2015; Ho et al., 2016; Erdal et al., 2017). Moreover, resistance to DNA-damaging cancer therapy has been linked to an interferon-related DNA damage response (Weichselbaum et al., 2008). However, although we observed a general up-regulation of interferon levels in 4N and PTA cells, STAT1 expression was mostly down-regulated (Supplemental Figure S3B). Regardless of the exact mechanisms underlying these changes, our data suggest that elevated chromosome numbers will influence the immune response to transformed cancer cells, regardless of the presence of CIN1. This conclusion falls in line with a recent gene expression analysis showing that hTERT-RPE1–derived cells with complex karyotypes produce pro-inflammatory cytokines, which was proposed to stimulate their clearance by the immune system (Santaguida et al., 2017).
Regarding the changes in protein phosphorylation that could be observed in 4N and PTA cells, these affected primarily the regulation of the mitotic spindle, transcription, translation, nuclear pore-dependent transport, and DNA damage responses. A single chromosome gain in trisomic cells (chromosome 7) showed similar, albeit less pronounced responses. Of particular interest, we observed altered phosphorylation of several proteins associated with cell division (Figure 6 and Supplemental Figure S7A). For instance, we observed strongly increased phosphorylation of the mitotic proteins KIF20B and TPX2 (Figure 6, A and B) as well as the T-loops of the mitotic kinases Aurora A (AURKA) and Plk1 (Supplemental Figure S7B). KIF20B is a kinesin-related motor important for cytokinesis and was previously found to be highly phosphorylated in M phase (Abaza et al., 2003). TPX2, an activator of Aurora A, is frequently deregulated in aneuploid cancer cells (Perez de Castro and Malumbres, 2013), and its overexpression was reported to correlate with CIN (Carter et al., 2006). Aurora-A, in turn, is required for the activation of Plk1 (Jang et al., 2002; Macu˚rek et al., 2008). Together, these data are consistent with the notion that increased numbers of chromosomes require increased activity of mitotic kinases to bring about chromosome segregation during mitosis. This in turn may explain why 4N and PTA clones showed distinct sensitivities for inhibitors of these kinases when compared with the parental 2N clone (Figure 7). Although further research is needed, these data raise the intriguing possibility that polyploid and extensively hyperdiploid aneuploid cells might preferentially be targeted by inhibitors that interfere with spindle regulatory enzymes. From a more general perspective, it seems legitimate to hope that continued study of the phosphorylation dynamics of the mitotic spindle phosphoproteome might eventually translate into better clinical use of anti-cancer drugs.
MATERIALS AND METHODS
Cell culture
Colon carcinoma lines HCT116, RKO, HT29, SW480, and SW837 (a gift from Stephen Taylor, University of Manchester, UK) were cultured as previously described (Gascoigne and Taylor, 2008). HeLa S3 cells were grown in DMEM-Glutamax (Invitrogen, CA) supplemented with 10% heat-inactivated fetal calf serum (FCS) (PAN Biotech, Aidenbach, Germany) and penicillin-streptomycin (Pen-Strep; 100 IU/ml and 100 mg/ml; Life Technologies, Zug, Switzerland). hTERT-RPE1 cells were cultured in F12 DMEM (Sigma Aldrich, MO) supplemented with 10% heat-inactivated FCS, l-glutamine (2 mM; PAN Biotech, Aidenbach, Germany), sodium bicarbonate (0.35%; Sigma-Aldrich, MO), and Pen-Strep. DLD-1 cells (2N and 4N) were kindly provided by Spiros Linardopoulos (ICR, London, UK) and maintained as described (Drosopoulos et al., 2014). H2B-GFP cultures were generated by retrovirus transduction, using a pLPCX- based plasmid (Gascoigne and Taylor, 2008), selected in the presence of 2 μg/ml puromycin for 72 h and subsequently maintained in the presence of 0.5 μg/ml puromycin. All lines were grown at 37°C in a humidified 5% CO2 incubator. DLD-1–derived PTA and trisomic clones were generated as described below.
Generation of trisomic and PTA clones
To generate DLD-1 cells containing an additional chromosome 7, microcell fusion was performed by microcell-mediated chromosome transfer, as previously described (Stingele et al., 2012). Clonal populations arising from single cells after chromosome transfer were isolated and further expanded in presence of 2 μg/ml puromycin and G418 0.2 mg/ml.
Spontaneously arising posttetraploid aneuploidy (PTA) clones were derived from a tetraploid DLD-1 parental culture (Drosopoulos et al., 2014) by sorting according to DNA content, using a BD FACS Aria cell sorter. 4N cells were harvested in trypsin, washed in phosphate-buffered saline (PBS), and resuspended for 30 min at 37° in 50 mg/ml RNase A and 1 mg/ml propidium iodine. After sorting, single cells were placed into three 96-well plates and cultured in medium without antibiotics. After visual inspection to ensure the presence of single cells and 1 mo of clonal expansion, multiple clones were obtained and four could be validated, by FACS, aCGH, and chromosome spreads, as PTA clones.
Chromosome spreads
Cells were treated with 50 ng/ml colchicine (Sigma-Aldrich) for 5 h and submitted to hypotonic swelling in 75 mM KCl at 37°C for 15 min. They were fixed by dropwise addition of ice-cold Carnoy solution (75% methanol and 25% acetic acid) and spread on a glass slide. Slides were dried at 42°C and stained with 4′, 6-diamidino-2-phenylindole (DAPI) (Life Technologies) or prepared for whole chromosome fluorescence in situ hybridization (FISH).
Whole chromosome FISH
Multicolor FISH was performed using a DNA probe mixture, according to manufacturer’s instructions (Chromosome specific painting probe kit; ChromBios GmbH, Nussdorf, Germany). We used probes directly labeled with red fluorochrome for chromosomes 3 and 5 and probes labeled with digoxin for chromosomes 4 and 7. In brief, chromosome spreads were incubated with probe mixture (1 μl of each probe, adjusted to 10 μl with HybMix buffer). After denaturation at 72° for 6 min, slides were kept at 37° in a humid chamber overnight. Slides were washed for 5 min in 2X saline sodium citrate (SSC) solution and then for 1 min in prewarmed 70° 0.4X SSC, 0.1% Tween solution, and, finally, in 4X SSC, 0.1% Tween solution for 5 min at room temperature. Then slides were incubated for 30 min at 37° with 100 μl fluorescein isothiocyanate (FITC) mouse anti-digoxin (Jackson ImmunoResearch) solution (1:300 in 4X SSC/0.1% Tween) and washed twice in 45° prewarmed 4X SSC/0.1% Tween solution for 5–10 min. Finally, DAPI staining was performed and microscopic analysis was carried out using Fiji for visual inspection of the images, using a Deltavision instrument (see below).
Fluorescence microscopy and image processing
Cells were grown on coverslips and fixed in PTEMF buffer (20 mM PIPES, pH 6.8, 0.2% Triton X-100, 10 mM egtazic acid [EGTA], 1 mM MgCl2, 4% formaldehyde). Z-stacks of randomly selected cells were acquired using a DeltaVision microscope (GE Healthcare) on an Olympus IX71 base (Applied Precision, WA), equipped with a Plan Apochromat N 60×/NA1.42 oil immersion objective (Olympus) and a CoolSNAP HQ2 camera (Photometrics). Deconvolution and projection were done using SoftWorx software (GE Healthcare). Statistical analysis was performed on two to three independent experiments and GraphPad Prism software was used for parametric two-tailed t tests.
For time-lapse imaging, cells were monitored using a Nikon ECLIPSE Ti microscope equipped with a CoolLED pE-1 illumination system and a 20×/NA0.75 air Plan Apochromat objective (Nikon) in a climate-controlled environment. Images were acquired every 9 min for 72 h. MetaMorph 7.7 software (MDS Analytical Technologies, Sunnyvale, CA) was used for acquisition and processing of data.
Array-comparative genomic hybridization
aCGH was performed on DLD-1 cell lines (2N, 4N, PTA, and trisomic clones) as previously described (Ruiz et al., 2011; Juskevicius et al., 2016), with minor modifications. PTA clones were analyzed two passages after establishing the lines. In brief, 1 μg of sample DNA and equal amounts of female reference genomic DNA (Promega 46/XX, Madison, WI) were digested with DNaseI to a size range of 200–500 base pairs. Subsequent labeling of sample and reference DNA with Cy3-dUTP and Cy5-dUTP, respectively, was performed with the BioPrime Array CGH Genomic Labeling System (Invitrogen, Carlsbad, CA). Labeling efficiency was quantified by measuring the specific activity of the incorporated dyes with a Nanodrop (Thermo Fischer Scientific, Waltham, MA). Reference and sample DNA were mixed and hybridized to 180k CGH arrays (Agilent Technologies, Santa Clara, CA) for 24 h in a rotating oven at 67°C. Microarray slides were scanned with the Agilent 2565C DNA scanner and images were analyzed with Agilent’s Feature Extraction using default settings. Feature extracted array CGH data were evaluated using Agilent’s CytoGenomics software v3.0.1.1. Aberrations were called with the aberration detection algorithm ADM2 set to a threshold of 12.0, with Fuzzy Zero and GC-content (window size: 2kb) correction. A minimum of three probes was necessary to call an aberration.
Cell proliferation assay
Cells were dispensed in 384-well plates at optimal density and cultured for 24 h. Compound dilution series (log10) were performed in duplicate, using a Biomek FX Lab Automation Workstation. Compounds were diluted from stock solutions (100% dimethyl sulfoxide [DMSO]) into 20 mM HEPES, such that 5 μl of compound dilution could be added to 45 μl of cell suspension (resulting in final DMSO concentrations of 0.4%). After 72 h, 24 μl of ATPlite 1Step (PerkinElmer, Groningen, The Netherlands) solution was added to each well before plates were shaken for 2 min and incubated for 5 min in the dark. Luminescence read out was performed on an Envision multimode reader (PerkinElmer, Waltham, MA). For each cell line, the maximum luminescence was recorded without compound (in the presence of 0.4% DMSO) after incubation until t = 72 h or 120 h (see Supplemental Table S8). Half-maximal inhibitory concentrations (IC50s) were fitted by nonlinear regression using XLfit5 (four-parameter method). A two-tailed Student’s t test was performed to determine whether differences in sensitivity (ΔpIC50) were statistically significant (p value < 0.1). Monitoring the influence of drugs on doubling times revealed effects falling between 0.5 and 2 times the doubling times measured in untreated cells.
Sample preparation and tandem mass tag labeling
Cells were cultured as described above and synchronized in G2/M phase by incubating them for 24 h with 2 mM thymidine and, subsequently, for 12 h with STLC 10 μM. From each culture, 106 cells were collected by mitotic shake off and centrifuged, and pellets were washed twice with PBS. Cells were lysed in 200 μl lysis buffer (2% sodium deoxycholate [DOC]), 0.1 M ammoniumbicarbonate) using strong ultrasonication (two cycles of sonication S3 for 10 s, Hielscher Ultrasonicator). Protein concentration was determined by bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) using a small sample aliquot. Proteins (50 μg) were digested as described previously (Ahrné et al., 2016), reduced with 5 mM Tris 2-carboxyethylphosphine (TCEP) for 15 min at 95°C, and alkylated with 10 mM iodoacetamide for 30 min in the dark at 25°C. After diluting samples with 100 mM ammonium bicarbonate buffer to a final DOC concentration of 1%, proteins were digested by incubation with sequencing-grade modified trypsin (1/50, wt/wt; Promega, Madison, WI) overnight at 37°C. Then, the samples were acidified with 2 M HCl to a final concentration of 50 mM and incubated for 15 min at 37°C, and the precipitated detergent removed by centrifugation at 10,000 × g for 15 min. Subsequently, peptides were desalted on C18 reversed-phase spin columns according to the manufacturer’s instructions (Microspin; Harvard Apparatus) and dried under vacuum. The dried peptide samples were subsequently labeled with isobaric tags (TMT 10-plex, Thermo Fisher Scientific) according to the manufacturer’s instructions. Owing to cofragmentation of coeluting peptide species TMT quantification tends to underestimate the magnitude of protein abundance changes, but, as shown previously (Ahrné et al., 2016), this does not compromise identification of deregulated proteins. After pooling the TMT labeled peptide samples, peptides were again desalted on C18 reversed-phase spin columns according to the manufacturer’s instructions (Macrospin; Harvard Apparatus) and dried under vacuum. TMT-labeled peptides were fractionated by high-pH reversed-phase separation using a XBridge Peptide BEH C18 column (3.5 µm, 130 Å, 1 mm × 150 mm; Waters) on an Agilent 1260 Infinity HPLC system. Peptides were loaded onto the column in buffer A (ammonium formate [20 mM, pH 10] in water) and eluted using a two-step linear gradient starting from 2%–10% in 5 min and then to 50% (vol/vol) buffer B (90% acetonitrile/10% ammonium formate [20 mM, pH 10]) over 55 min at a flow rate of 42 µl/min. Elution of peptides was monitored with a UV detector (215 nm, 254 nm). A total of 36 fractions were collected, pooled into 12 fractions using a postconcatenation strategy as previously described (Wang et al., 2011), dried under vacuum and subjected to liquid chromatography (LC)-MS/MS analysis.
Mass-spectrometric analysis
The setup of the μ reversed-phase liquid chromatography-MS system was as described previously (Ahrné et al., 2016). Chromatographic separation of peptides was carried out using an EASY nano-LC 1000 system (Thermo Fisher Scientific), equipped with a heated reversed-phase-HPLC column (75 μm × 37 cm) packed in-house with 1.9 μm C18 resin (Reprosil-AQ Pur, Dr. Maisch). Aliquots of 1 μg total peptides were analyzed per LC-MS/MS run, using a linear gradient ranging from 95% solvent A (0.15% formic acid, 2% acetonitrile) and 5% solvent B (98% acetonitrile, 2% water, 0.15% formic acid) to 30% solvent B over 90 min at a flow rate of 200 nl/min. Mass spectrometry analysis was performed on a Q-Exactive HF mass spectrometer equipped with a nanoelectrospray ion source (both Thermo Fisher Scientific). Each MS1 scan was followed by high-collision dissociation of the 10 most abundant precursor ions with dynamic exclusion for 20 s. Total cycle time was ∼1 s. For MS1, 3e6 ions were accumulated in the Orbitrap cell over a maximum time of 100 ms and scanned at a resolution of 120,000 full width at half maximum (FWHM) (at 200 m/z). MS2 scans were acquired at a target setting of 1e5 ions, accumulation time of 100 ms, and a resolution of 30,000 FWHM (at 200 m/z). Singly charged ions and ions with unassigned charge state were excluded from triggering MS2 events. The normalized collision energy was set to 35%, the mass isolation window was set to 1.1 m/z, and one microscan was acquired for each spectrum.
Database searching and protein quantification
The acquired raw files were converted to the mascot generic file (mgf) format using the msconvert tool (part of ProteoWizard, version 3.0.4624 [2013-6-3]). Using the MASCOT algorithm (Matrix Science, Version 2.4.1), the mgf files were searched against a decoy database containing normal and reverse sequences of the predicted SwissProt entries of Homo sapiens (www.ebi.ac.uk), the six calibration mix proteins (Ahrné et al., 2016), and commonly observed contaminants (in total 84,610 sequences for Homo sapiens) generated using the SequenceReverser tool from the MaxQuant software (version 1.0.13.13). The precursor ion tolerance was set to 15 ppm, and fragment ion tolerance was set to 0.02 Da. The search criteria were set as follows: full tryptic specificity was required (cleavage after lysine or arginine residues unless followed by proline), three missed cleavages were allowed, and carbamidomethylation (C) and TMT6plex (K and peptide n-terminus) were set as fixed modification and oxidation (M) as a variable modification. Next, the database search results were imported to the Scaffold Q+ software (version 4.3.2; Proteome Software, Portland, OR) and the protein false identification rate was set to 1% based on the number of decoy hits. Specifically, peptide identifications were accepted if they could be established at greater than 96.0% probability to achieve an FDR less than 1.0% by the scaffold local FDR algorithm. Protein identifications were accepted if they could be established at greater than 77.0% probability to achieve an FDR less than 1.0% and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet program (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Acquired reporter ion intensities in the experiments were employed for automated quantification and statically analysis using a modified version of our in-house developed SafeQuant R script (Ahrné et al., 2016). In a first step, missing values were replaced by half of the global minimum among TMT intensities or label free quantification MS1 intensities (Webb-Robertson et al., 2015). In the TMT experiments the spectrum level quantification matrix included approximately ∼2% missing values per sample. In the phospho label-free quantification experiments, each run had ∼7% missing values. The subsequent analysis included adjustment of reporter ion intensities, global data normalization by equalizing the total reporter ion intensity across all channels, summation of reporter ion intensities per protein and channel, calculation of protein abundance ratios, and testing for differential abundance using empirical Bayes moderated t statistics. Finally, the calculated p values were corrected for multiple testing using the Benjamini−Hochberg method.
Phosphoproteome analysis
Cells, synchronized as described above, were collected by mitotic shake off and lysed in 8 M urea (Sigma) and 0.1 M ammonium bicarbonate in the presence of phosphatase inhibitors (Sigma P5726 and P0044). Protein lysate (2 mg) was digested with trypsin, cleaned up using an C18 column, and enriched for phosphorylated peptides using titanium dioxide beads as previously described (Schmutz et al., 2013). After C18 cleanup, peptides (1 μg) were LC-MS analyzed as described above. The acquired raw files were imported into the Progenesis QI software (v2.0; Nonlinear Dynamics Limited), which was used to extract peptide precursor ion intensities across all samples applying the default parameters. The generated mgf-files were searched using MASCOT as described above, using the following search criteria: full tryptic specificity was required (cleavage after lysine or arginine residues, unless followed by proline); three missed cleavages were allowed; carbamidomethylation (C) was set as fixed modification; oxidation (M) and phosphorylation (STY) were applied as variable modifications; mass tolerance of 10 ppm (precursor) and 0.02 Da (fragments). The database search results were filtered using the ion score to set the FDR to 1% on the peptide and protein level, respectively, based on the number of reverse protein sequence hits in the data sets. The relative quantitative data obtained were normalized and statistically analyzed using our in-house script as described above (Ahrné et al., 2016).
Enrichment analysis
For all relative protein quantifications, the DLD-1 2N line was used as a standard. The quantified proteins were sorted by increasing p value and the 300 most significantly deregulated proteins, as well as the 500 most deregulated phosphopeptides for each cell line, were subjected to functional enrichment analysis. To this end, Biological Process Gene Ontology (GO) (Ashburner et al., 2000) annotations were mapped to all identified proteins using the R package GO.db v.3.4.1. Next, GO-term enrichment was investigated for each set of 300 deregulated proteins with respect to the set of nonderegulated proteins. Here the R package topGO was used, setting the nodeSize filter to 10 and calculating enrichment p values for each GO term using a one-sided Fisher’s exact test (Alexa et al., 2006). Similarly, enrichment of CIN/cell cycle and OG/TSG inclusion lists among deregulated proteins, with respect to the set of nonderegulated proteins, was assessed using a one-sided Fisher’s exact test. The CIN/cell-cycle inclusion list was created by compiling hits identified from previous studies. In particular, this list included the identifiers from CIN70 gene expression data (Carter et al., 2006); 572 validated mitotic genes from the Mitocheck consortium www.mitocheck.org (Neumann et al., 2010); and all the identifiers corresponding to mitotic and cell cycle related GO terms (included GO identifiers: GO:0007094, GO:0007051, GO:0005828, GO:0005813, GO:0007059, GO:0005813, GO:0007049, and GO:1905115. Annotation source www.uniprot.org). OGs and TSGs list was previously published in Davoli et al. (2017) (http://science.sciencemag.org/content/355/6322/eaaf8399.full).
Supplementary Material
Acknowledgments
We thank Stephen Taylor (University of Manchester) for generously providing colon carcinoma lines HCT116, RKO, HT29, SW480, and SW837 and Spiros Linardopoulos (Institute for Cancer Research, London) for kindly providing 2N and 4N DLD-1 colon cancer cell lines. We also thank Janine Bögli of the FACS Core Facility (Biozentrum, Basel) for help with cell sorting and Aline Sewo-Pires de Campos for excellent technical support with chromosome transfer. Work in E.A.N.’s laboratory was supported by the University of Basel and the Swiss National Science Foundation (310030B_149641). Z.S. acknowledges support by DFG STO918/2-2. This work was supported by the Marie Curie Network PloidyNet, funded by the European Union Seventh Framework Programme (FP7/2007–2013) under Grant Agreement no. 316964 to C.V., C.v.S., G.J.R.Z., Z.S., and E.A.N.
Abbreviations used:
- aCGH
array comparative genomic hybridization
- CIN
chromosome instability
- MIN
microsatellite instability
- PTA
posttetraploid aneuploid
- TMT
tandem mass tag
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-10-0577) on February 26, 2018.
REFERENCES
- Abaza A, Soleilhac J-M, Westendorf J, Piel M, Crevel I, Roux A, Pirollet F. (2003). M phase phosphoprotein 1 is a human plus-end-directed kinesin-related protein required for cytokinesis. J Biol Chem , 27844–27852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams ND, Adams JL, Burgess JL, Chaudhari AM, Copeland RA, Donatelli CA, Drewry DH, Fisher KE, Hamajima T, Hardwicke MA, et al (2010). Discovery of GSK1070916, a potent and selective inhibitor of aurora B/C kinase. J Med Chem , 3973–4001. [DOI] [PubMed] [Google Scholar]
- Ahrné E, Glatter T, Viganó C, Schubert CV, Nigg EA, Schmidt A. (2016). Evaluation and improvement of quantification accuracy in isobaric mass tag-based protein quantification experiments. J Proteome Res , 2537–2547. [DOI] [PubMed] [Google Scholar]
- Alexa A, Rahnenfuhrer J, Lengauer T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics , 1600–1607. [DOI] [PubMed] [Google Scholar]
- Alvaro D, Sunjevaric I, Reid RJD, Lisby M, Stillman DJ, Rothstein R. (2006). Systematic hybrid LOH: a new method to reduce false positives and negatives during screening of yeast gene deletion libraries. Yeast , 1097–1106. [DOI] [PubMed] [Google Scholar]
- Anders KR, Kudrna JR, Keller KE, Kinghorn B, Miller EM, Pauw D, Peck AT, Shellooe CE, Strong IJ. (2009). A strategy for constructing aneuploid yeast strains by transient nondisjunction of a target chromosome. BMC Genet , 36–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al (2000). Gene ontology: tool for the unification of biology. Nat Genet , 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. (2003). Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol , 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K-H, Zaslavsky A, Lynch RC, Britt C, Okada Y, Siarey RJ, Lensch MW, Park I-H, Yoon SS, Minami T, et al (2009). Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature , 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccitelli C, Salgueiro L, Rowald K, Sotillo R, Mardin BR, Korbel JO. (2017). Pan-cancer analysis distinguishes transcriptional changes of aneuploidy from proliferation. Genome Res , 501–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, da Costa LT, Carson-Walter EB, Kinzler KW, Vogelstein B, Lengauer C. (1999). Characterization of MAD2B and other mitotic spindle checkpoint genes. Genomics , 181–187. [DOI] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. (2006). A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet , 1043–1048. [DOI] [PubMed] [Google Scholar]
- Cowell JK, Wigley CB. (1980). Changes in DNA content during in vitro transformation of mouse salivary gland epithelium. J Natl Cancer Inst , 1443–1449. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. (2012). DNA breaks and chromosome pulverization from errors in mitosis. Nature , 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Uno H, Wooten EC, Elledge SJ. (2017). Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science , eaaf8399–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. (2013). Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell , 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Braekeleer E, Huret J-L, Mossafa H, Dessen P. (2017). Cytogenetic resources and information. Methods Mol Biol , 311–331. [DOI] [PubMed] [Google Scholar]
- Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, Torres EM. (2014). Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. eLife , 36–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al (2014). Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov , 175–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson SE, Kim S, Costanzo M, Baryshnikova A, Morse DL, Kaiser CA, Boone C, Amon A. (2016). Chromosome-specific and global effects of aneuploidy in Saccharomyces cerevisiae. Genetics , 1395–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosopoulos K, Tang C, Chao WCH, Linardopoulos S. (2014). APC/C is an essential regulator of centrosome clustering. Nat Commun , 3686. [DOI] [PubMed] [Google Scholar]
- Durrbaum M, Kuznetsova AY, Passerini V, Stingele S, Stoehr G, Storchova Z. (2014). Unique features of the transcriptional response to model aneuploidy in human cells. BMC Genomics , 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdal E, Haider S, Rehwinkel J, Harris AL, McHugh PJ. (2017). A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev , 353–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers PA, Erikson E, Chen LG, Maller JL. (2003). A novel mechanism for activation of the protein kinase aurora A. Curr Biol , 691–697. [DOI] [PubMed] [Google Scholar]
- Funk LC, Zasadil LM, Weaver BA. (2016). Living in CIN: mitotic infidelity and its consequences for tumor promotion and suppression. Dev Cell , 638–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futschik ME, Carlisle B. (2005). Noise-robust soft clustering of gene expression time-course data. J Bioinform Comput Biol , 965–988. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. (2007). Limiting the proliferation of polyploid cells. Cell , 437–440. [DOI] [PubMed] [Google Scholar]
- Gasch AP. (2007). Comparative genomics of the environmental stress response in ascomycete fungi. Yeast , 961–976. [DOI] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS. (2008). Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell , 111–122. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. (2011). Hallmarks of cancer: the next generation. Cell , 646–674. [DOI] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, FitzPatrick D, Kidd A, Méhes K, Nash R, Robin N, et al (2004). Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet , 1159–1161. [DOI] [PubMed] [Google Scholar]
- Haruki N, Harano T, Masuda A, Kiyono T, Takahashi T, Tatematsu Y, Shimizu S, Mitsudomi T, Konishi H, Osada H, et al (2001). Persistent increase in chromosome instability in lung cancer. Am J Pathol , 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SSW, Zhang WYL, Tan NYJ, Khatoo M, Suter MA, Tripathi S, Cheung FSG, Lim WK, Tan PH, Ngeow J, Gasser S. (2016). The DNA structure-specific endonuclease MUS81 mediates DNA sensor STING-dependent host rejection of prostate cancer cells. Immunity , 1177–1189. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. (2009). Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol , 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. (2012). Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep , 501–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, Shafi S, Johnson DH, Mitter R, Rosenthal R, et al (2017). Tracking the evolution of non–small-cell lung cancer. N Engl J Med , 2109–2121. [DOI] [PubMed] [Google Scholar]
- Jang Y-J, Lin CY, Ma S, Erikson RL. (2002). Functional studies on the role of the C-terminal domain of mammalian polo-like kinase. Proc Natl Acad Sci USA , 1984–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJPL, Medema RH. (2011). Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science , 1895–1898. [DOI] [PubMed] [Google Scholar]
- Juskevicius D, Lorber T, Gsponer J, Perrina V, Ruiz C, Stenner-Liewen F, Dirnhofer S, Tzankov A. (2016). Distinct genetic evolution patterns of relapsing diffuse large B-cell lymphoma revealed by genome-wide copy number aberration and targeted sequencing analysis. Leukemia , 2385–2395. [DOI] [PubMed] [Google Scholar]
- Kabeche L, Compton DA. (2012). Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in H human cells. Curr Biol , 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp JE, Thomas BM, Greer JM, Sorge C, Gore SD, Pratz KW, Smith BD, Flatten KS, Peterson K, Schneider P, et al (2012). Phase I and pharmacologic trial of cytosine arabinoside with the selective checkpoint 1 inhibitor Sch 900776 in refractory acute leukemias. Clin Cancer Res , 6723–6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJPL, Weaver BAA, Cleveland DW. (2005). On the road to cancer: aneuploidy and the mitotic checkpoint. Nat Rev Cancer , 773–785. [DOI] [PubMed] [Google Scholar]
- Kuznetsova AY, Seget K, Moeller GK, de Pagter MS, de Roos JADM, Dürrbaum M, Kuffer C, Müller S, Zaman GJR, Kloosterman WP, Storchová Z. (2015). Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle , 2810–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Chang YC. (2012). Aneuploidy and drug resistance in pathogenic fungi. PLoS Pathog , e1003022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrus BG, Daggubati V, Uetake Y, Scott PM, Clutario KM, Sluder G, Holland AJ. (2016). A USP28–53BP1–p53–p21 signaling axis arrests growth after centrosome loss or prolonged mitosis. J Cell Biol , 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJX, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, Downward J, Szallasi Z, Tomlinson IPM, Howell M, et al (2011). Chromosomal instability confers intrinsic multidrug resistance. Cancer Res , 1858–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. (1997). Genetic instability in colorectal cancers. Nature , 623–627. [DOI] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P. (2010). The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci USA , 14188–14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly P, Cleveland DW. (2017). Interrogating cell division errors using random and chromosome-specific missegregation approaches. Cell Cycle , 1252–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyle R, Gehrig C, Neergaard-Henrichsen C, Deutsch S, Antonarakis SE. (2004). Gene expression from the aneuploid chromosome in a trisomy mouse model of down syndrome. Genome Res , 1268–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macu˚rek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, Clouin C, Taylor SS, Yaffe MB, Medema RH. (2008). Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature , 119–123. [DOI] [PubMed] [Google Scholar]
- Malumbres M. (2014). Cyclin-dependent kinases. Genome Biol , 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer VW, Aguilera A. (1990). High levels of chromosome instability in polyploids of Saccharomyces cerevisiae. Mutat Res , 177–186. [DOI] [PubMed] [Google Scholar]
- McCoy EE, Segal DJ, Bayer SM, Strynadka KD. (1974). Decreased ATPase and increased sodiun content of platelets in Down’s syndrome. Relation to decreased serotonin content. N Engl J Med , 950–953. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Burrell RA, Endesfelder D, Novelli MR, Swanton C. (2012). Cancer chromosomal instability: therapeutic and diagnostic challenges. EMBO Rep , 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer L, Raymond E. (2003). Roscovitine and other purines as kinase inhibitors: from starfish oocytes to clinical trials. Acc Chem Res , 417–425. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii Al, Keller A, Kolker E, Aebersold R. (2003). A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem , 4646–4658. [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Hériché J-K, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al (2010). Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature , 721–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JM, Macedo JC, Mattingly AJ, DarawaleeWangsa Jps, Lima V, Gomes AM, Do ria S, Ried T, Logarinho E, Cimini D. (2015). Chromosome mis-segregation and cytokinesis failure in trisomic human cells. eLife , 05068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A, Ohori M, Iwai K, Nakayama Y, Nambu T, Morishita D, Kawamoto T, Miyamoto M, Hirayama T, Okaniwa M, et al (2015). Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat Commun , 7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, Kerem B, Aacute ZS. (2016). The presence of extra chromosomes leads to genomic instability. Nat Commun , 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson K, Johansson B. (2007). Trisomy 8 as the sole chromosomal aberration in acute myeloid leukemia and myelodysplastic syndromes. Pathol Biol , 37–48. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R. (2010). Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature , 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Castro I, Malumbres M. (2013). Mitotic stress and chromosomal instability in cancer: the case for TPX2. Genes Cancer , 721–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz C, Lenkiewicz E, Evers L, Holley T, Robeson A, Kiefer J, Demeure MJ, Hollingsworth MA, Shen M, Prunkard D, et al (2011). Advancing a clinically relevant perspective of the clonal nature of cancer. Proc Natl Acad Sci USA , 12054–12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge SD, Douglas TA, Nicholson JM, Vila-Casadesús M, Kantzler CL, Wangsa D, Barroso-Vilares M, Kale SD, Logarinho E, Cimini D. (2016). Selective advantage of trisomic human cells cultured in non-standard conditions. Sci Rep , 22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. (2015a). Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol , 473–485. [DOI] [PubMed] [Google Scholar]
- Santaguida S, Amon A. (2015b). Aneuploidy triggers a TFEB-mediated lysosomal stress response. Autophagy , 2383–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Richardson A, Iyer DR, M’Saad O, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A, Amon A. (2017). Chromosome mis-segregation generates cell-cycle-arrested cells with complex karyotypes that are eliminated by the immune system. Dev Cell , 638–651.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz C, Ahrné E, Kasper CA, Tschon T, Sorg I, Dreier RF, Schmidt A, Arrieumerlou C. (2013). Systems-level overview of host protein phosphorylation during Shigella flexneri infection revealed by phosphoproteomics. Mol Cell Proteom , 2952–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackney SE, Smith CA, Miller BW, Burholt DR, Murtha K, Giles HR, Ketterer DM, Pollice AA. (1989). Model for the genetic evolution of human solid tumors. Cancer Res , 3344–3354. [PubMed] [Google Scholar]
- Sheltzer JM. (2013). A transcriptional and metabolic signature of primary aneuploidy is present in chromosomally unstable cancer cells and informs clinical prognosis. Cancer Res , 6401–6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. (2012). Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA , 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YJ, Le Bert N, Chitre AA, Koo CX, Nga XH, Ho SSW, Khatoo M, Tan NY, Ishii KJ, Gasser S. (2015). Genome-derived cytosolic DNA mediates Type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep , 460–473. [DOI] [PubMed] [Google Scholar]
- Shin S-B, Woo S-U, Yim H. (2015). Differential cellular effects of Plk1 inhibitors targeting the ATP-binding domain or polo-box domain. J Cell Physiol , 3057–3067. [DOI] [PubMed] [Google Scholar]
- Siegel JJ, Amon A. (2012). New insights into the troubles of aneuploidy. Annu Rev Cell Dev Biol , 189–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits VAJ, Gillespie DA. (2015). DNA damage control: regulation and functions of checkpoint kinase 1. FEBS J , 3681–3692. [DOI] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchová Z. (2012). Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol , 608–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z, Breneman A, Cande J, Dunn J, Burbank K, O’Toole E, Pellman D. (2006). Genome-wide genetic analysis of polyploidy in yeast. Nature , 541–547. [DOI] [PubMed] [Google Scholar]
- Storchova Z, Kuffer C. (2008). The consequences of tetraploidy and aneuploidy. J Cell Sci , 3859–3866. [DOI] [PubMed] [Google Scholar]
- Storchová Z, Pellman D. (2004). From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol , 45–54. [DOI] [PubMed] [Google Scholar]
- Tang Y-C, Williams BR, Siegel JJ, Amon A. (2011). Identification of aneuploidy-selective antiproliferation compounds. Cell , 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Schäfer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Hamon C. (2003). Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem , 1895–1904. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. (2010). Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol , 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. (2007). Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science , 916–924. [DOI] [PubMed] [Google Scholar]
- Tse AN, Rendahl KG, Sheikh T, Cheema H, Aardalen K, Embry M, Ma S, Moler EJ, Ni ZJ, Lopes de Menezes DE, et al (2007). CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res , 591–602. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. (2010). Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol , 1666–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upender MB, Habermann JK, McShane LM, Korn EL, Barrett JC, Difilippantonio MJ, Ried T. (2004). Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res , 6941–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Jaarsveld RH, Kops GJPL. (2016). Difference makers: chromosomal instability versus aneuploidy in cancer. Trends Cancer , 561–571. [DOI] [PubMed] [Google Scholar]
- Venet D, Dumont JE, Detours V. (2011). Most random gene expression signatures are significantly associated with breast cancer outcome. PLoS Comput Biol , e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH-C, Schwarzbraun T, Speicher MR, Nigg EA. (2007). Persistence of DNA threads in human anaphase cells suggests late completion of sister chromatid decatenation. Chromosoma , 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, et al (2011). Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics , 2019–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. (2006). Does aneuploidy cause cancer?. Curr Opin Cell Biol , 658–667. [DOI] [PubMed] [Google Scholar]
- Weaver BAA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. (2007). Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell , 25–36. [DOI] [PubMed] [Google Scholar]
- Webb-Robertson BJ, Wiberg HK, Matzke MM, Brown JN, Wang J, McDermott JE, Smith RD, Rodland KD, Metz TO, Pounds JG, Waters KM. (2015). Review, evaluation, and discussion of the challenges of missing value imputation for mass spectrometry-based label-free global proteomics. J Proteome Res , 1993–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum RR, Ishwaran H, Yoon T, Nuyten DSA, Baker SW, Khodarev N, Su AW, Shaikh AY, Roach P, Kreike B, et al (2008). An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci USA , 18490–18495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A. (2008). Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science , 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. (2012). Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci USA , 21010–21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost S, de Wolf B, Hanks S, Zachariou A, Marcozzi C, Clarke M, de Voer RM, Etemad B, Uijttewaal E, Ramsay E, et al (2017). Biallelic TRIP13 mutations predispose to Wilms tumor and chromosome missegregation. Nat Genet , 1148–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang C-Z, Wala J, Mermel CH, et al (2013). Pan-cancer patterns of somatic copy number alteration. Nat Genet , 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.