The nematode Caenorhabditis elegans has a single gene, lev-11, encoding multiple tropomyosin isoforms. Two seventh exons are alternatively selected in the head and body regions of muscle cells and differentially affect muscle contractility in the respective regions.
Abstract
Tropomyosin, one of the major actin filament–binding proteins, regulates actin–myosin interaction and actin-filament stability. Multicellular organisms express a number of tropomyosin isoforms, but understanding of isoform-specific tropomyosin functions is incomplete. The nematode Caenorhabditis elegans has a single tropomyosin gene, lev-11, which has been reported to express four isoforms by using two separate promoters and alternative splicing. Here, we report a fifth tropomyosin isoform, LEV-11O, which is produced by alternative splicing that includes a newly identified seventh exon, exon 7a. By visualizing specific splicing events in vivo, we find that exon 7a is predominantly selected in a subset of the body wall muscles in the head, while exon 7b, which is the alternative to exon 7a, is utilized in the rest of the body. Point mutations in exon 7a and exon 7b cause resistance to levamisole-induced muscle contraction specifically in the head and the main body, respectively. Overexpression of LEV-11O, but not LEV-11A, in the main body results in weak levamisole resistance. These results demonstrate that specific tropomyosin isoforms are expressed in the head and body regions of the muscles and contribute differentially to the regulation of muscle contractility.
INTRODUCTION
Diverse functions of the actin cytoskeleton are supported by a variety of actin-regulatory proteins (Pollard and Cooper, 2009). Among them, tropomyosin serves as a major regulator of actin functions in fungi and metazoans (Lin et al., 1997; Perry, 2003; Dominguez, 2011; Gunning et al., 2015; Hitchcock-DeGregori and Barua, 2017). Tropomyosin is a coiled-coil dimer that binds along the side of actin filaments and influences actomyosin contractility and actin filament dynamics (Bailey, 1946; Maruyama, 1964; Smillie et al., 1980; Hitchcock-DeGregori et al., 1988; Rao et al., 2012; von der Ecken et al., 2015). It regulates actin–myosin interaction in an isoform-specific manner (Manstein and Mulvihill, 2016; Gateva et al., 2017) and stabilizes actin filaments by inhibiting actin depolymerizing factor/cofilin-mediated severing (Bernstein and Bamburg, 1982; Ono and Ono, 2002). While tropomyosin inhibits Arp2/3-dependent actin nucleation and branching (Blanchoin et al., 2001), it promotes formin-dependent actin elongation (Wawro et al., 2007; Skau et al., 2009; Ujfalusi et al., 2009, 2012; Alioto et al., 2016). Classically, tropomyosin has been well characterized as a regulator of muscle contraction. In striated muscles, tropomyosin mediates Ca2+-dependent conformational changes of the troponin complex to activate or inhibit actin–myosin interaction (Ebashi, 1984; Squire and Morris, 1998; Galinska-Rakoczy et al., 2008). In nonmuscle cells, tropomyosin is involved in a wide range of actin-dependent events including cell migration, cytokinesis, and morphogenesis (Gunning et al., 2015). In metazoan species, the presence of multiple tropomyosin isoforms with distinct functions is associated with the roles of tropomyosin in a variety of cellular processes (Gunning et al., 2005, 2015). Nevertheless, the functions and regulatory mechanisms of tropomyosin isoforms are not fully characterized.
Increased diversity of tropomyosin isoforms correlates with the evolution of complexity in cells and tissues (Gunning et al., 2015). The budding yeast Saccharomyces cerevisiae has two tropomyosin isoforms from separate genes (Drees et al., 1995) that give rise to different effects on actin–myosin interaction (Huckaba et al., 2006) and formin function (Alioto et al., 2016). Contrastingly, mammalian species have four tropomyosin genes that produce over 40 isoforms by alternative splicing (Schevzov et al., 2011; Geeves et al., 2015). In addition to the tissue-specific expression of tropomyosin isoforms, many isoforms are sorted to different subcellular regions (Temm-Grove et al., 1998; Bryce et al., 2003; Gallant et al., 2011). During stress fiber formation, at least four tropomyosin isoforms contribute to organizing the assembly of distinct compartments of stress fibers (Tojkander et al., 2011). In neurons, several tropomyosin isoforms localize to different actin-rich compartments and have unique effects on neuronal cell morphology (Schevzov et al., 2005; Curthoys et al., 2014). In striated muscle, major sarcomeric tropomyosins localize along the thin filaments, whereas the minor isoforms Tpm3.1 (Tm5NM1) and Tpm4.2 localize to narrow regions adjacent to the Z-lines (Schevzov et al., 2008; Vlahovich et al., 2008, 2009). Additionally, novel tropomyosin isoforms with unexpected functions were discovered in Drosophila: these functions include important roles in mitotic spindles (Goins and Mullins, 2015), microtubule-dependent mRNA localization (Veeranan-Karmegam et al., 2016), and intermediate filament-like cytoskeletal functioning (Cho et al., 2016). Notwithstanding our current understanding of isoform diversity, functions of many other tropomyosin isoforms remain unknown. Therefore, further studies on tropomyosin isoforms may reveal unexpected functions.
The nematode Caenorhabditis elegans has a single gene for tropomyosin, lev-11 (also known as tmy-1), which has been reported to produce four isoforms through the utilization of two separate promoters and alternative splicing (Kagawa et al., 1995; Anyanful et al., 2001). lev-11 mutants were originally isolated from a screen for levamisole-resistant strains (Lewis et al., 1980a, b). Levamisole, an agonist of the acetylcholine (ACh) receptor, induces contraction and paralysis in wild-type worms. However, lev-11 mutants remain motile in the presence of levamisole. Strong loss of function or null mutation of lev-11 causes severe paralysis and embryonic arrest at the two-fold stage (Pat phenotype) (Williams and Waterston, 1994). In the adult body wall muscle, which is obliquely striated muscle (Ono, 2014), RNA interference (RNAi) of lev-11 causes partial paralysis, disorganization of sarcomeres (Ono and Ono, 2002; Yu and Ono, 2006) and defective muscle arm development (Dixon and Roy, 2005). lev-11 is also important in the reproductive system through its role in regulating the contractility of male sex muscle (Gruninger et al., 2006) and hermaphroditic gonadal myoepithelial sheath (Ono and Ono, 2004). However, complex splicing patterns of lev-11 and the functional significance of the C. elegans tropomyosin isoforms remain elusive.
In this study, we cloned a novel C. elegans tropomyosin isoform, LEV-11O, containing a newly identified alternative lev-11 exon (Kuroyanagi et al., 2014) and found that this exon is predominantly utilized in a subset of head muscle cells. Mutational and transgenic analyses suggest that tropomyosin isoforms produced by alternative splicing differentially affect muscle contractility in the head and main-body regions. These results suggest that alternative splicing of the lev-11 tropomyosin gene is a key to differentiating the body wall muscle cells in the head and main-body regions.
RESULTS
A newly characterized alternative exon of lev-11 is included in a novel high-molecular-weight tropomyosin isoform
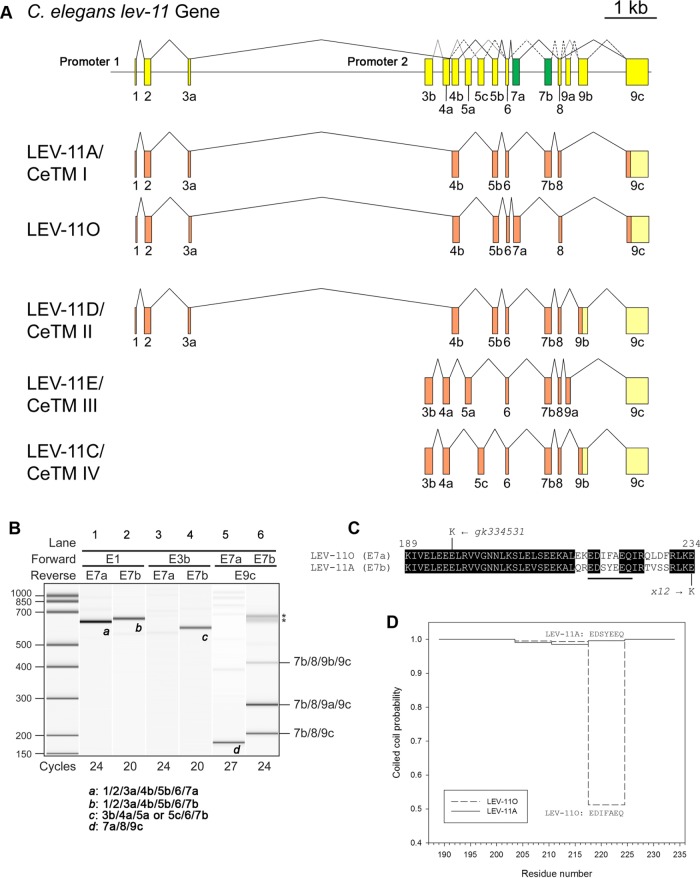
The structure of the C. elegans lev-11 gene (Figure 1A) is fairly complex. First, it has two promoters (Figure 1A): the upstream promoter (promoter 1) drives transcription from exon 1 (E1) to express high-molecular-weight isoforms, whereas the downstream promoter (promoter 2) drives transcription from exon 3b (E3b) to express low-molecular-weight isoforms (Kagawa et al., 1995; Anyanful et al., 2001). Second, the lev-11 gene contains multiple alternatively spliced exons 4 (a and b), 5 (a–c), and 9 (a–c) (Figure 1A) (Kagawa et al., 1995; Anyanful et al., 2001). Additionally, gene models in WormBase (www.wormbase.org) suggested that exon 7 is also duplicated (Figure 1A, shown in green). The sequence of an expressed sequence tag clone (yk783g10) and recent deep sequencing of poly(A)+ RNAs from synchronized L1 larvae confirmed the use of exon 7a (E7a) (Kuroyanagi et al., 2014), which is the same length (139 nt) as exon 7b (E7b; previously reported as exon 7) with 71% nucleotide sequence identity and is conserved in the genus Caenorhabditis (Supplemental Figure S1).
FIGURE 1:
Characterization of mutually exclusive seventh exons of the C. elegans lev-11 tropomyosin gene. (A) Structure of the lev-11 gene is shown schematically (top) with numbered boxes indicating exons. Recently characterized alternative exons 7a and 7b are shown in green. Below the gene structure are splicing patterns of LEV-11A/CeTMI, a newly identified isoform, LEV-11O, and three previously characterized isoforms, LEV-11D/CeTMII, LEV-11E/CeTMIII, and LEV-11C/CeTMIV. Coding and noncoding regions are shown in orange and light yellow, respectively. Note that exon 9c is used in all known isoforms either as a coding region (LEV-11A and LEV-11O) or as a noncoding region when 9a or 9b is used as a coding region (LEV-11C, LEV-11D, and LEV-11E). (B) Analysis of lev-11 mRNAs by RT-PCR. Total RNAs from synchronized wild-type L1 larvae were subjected to RT-PCR with indicated primer pairs and cycle numbers, and the PCR products were analyzed with a 2100 BioAnalyzer (Agilent). DNA size markers are shown on the left. Results are shown in gel-like presentations. Exon combinations of representative bands a–d are shown below. Multiple bands in E7b/E9c (lane 6) were cloned and sequenced, and the exon combinations are indicated on the right. Asterisks indicate artificial PCR products due to excessive cycles. (C) Alignment of amino acid sequences encoded by exons 7a and 7b. Identical residues are indicated with black backgrounds. Point mutations in lev-11(gk334531) and lev-11(x12) are shown at the top and bottom of the sequences, respectively. (D) Probability of coiled-coil formation (0–1) was calculated from the full-length sequences of LEV-11A and LEV-11O by COILS (Lupas et al., 1991), and plots of exon 7-coded regions are shown.
Splicing patterns in relation to the E7a/E7b selection were analyzed by reverse transcription-PCR (RT-PCR) (Figure 1B). With the E1/E7a and E1/E7b primer pairs, single bands were detected (Figure 1B, lanes 1 and 2), and both of them included E1/E2/E3a/E4b/E5b/E6, indicating that E7a and E7b are included in high-molecular-weight isoforms in a mutually exclusive manner. However, with the E3b primer, a band was detected for the E3b/E7b pair (Figure 1B, lane 4) but not for the E3b/E7a pair (Figure 1B, lane 3), indicating that E7a is not included in a low-molecular-weight isoform. With the E7a/E9c primer pair, only a single band, which excluded E9a and E9b (E7a/E8/E9c), was amplified (Figure 1B, lane 5). In contrast, three expected fragments corresponding to E7b/E8/E9c, E7b/E8/E9a/E9c, and E7b/E8/E9b/E9c were amplified with the E7b/E9c primer pair (Figure 1B, lane 6). With an effort to characterize a full-length cDNA containing E7a, we obtained clones for a novel isoform, LEV-11O (E1/E2/E3a/E4b/E5b/E6/E7a/E8/E9c) (Figure 1A), which was previously only predicted from a possible exon combination (GenBank accession # NM_001313528). LEV-11O is a high-molecular-weight tropomyosin isoform (284 amino acids) that is most similar to LEV-11A (E1/E2/E3a/E4b/E5b/E6/E7b/E8/E9c), with differences only in the E7a/E7b-encoded region (Figure 1C). Both E7a and E7b encode 46 residues, and their amino acid sequences show 78% identity (Figure 1C). These sequences are invariable among the examined Caenorhabditis species, suggesting their functional significance. The E7a- and E7b-encoded sequences are mostly different in the second half (Figure 1C) with notable substitutions of acidic (E215 and D228) and hydrophobic (I219 and F229) residues in E7a in place of polar residues (Q215, S219, S228, and S229) in E7b. Interestingly, prediction of coiled-coil formation by COILS (Lupas et al., 1991) suggests a low probability of coiled-coil formation at the E7a-encoded residues 218–224 of LEV-11O (Figure 1C underlined residues and 1D). Therefore, these isoforms may possess a conformational difference.
Alternative exons, E7a and E7b, of the lev-11 gene are differentially selected in the head and main-body regions of body wall muscles
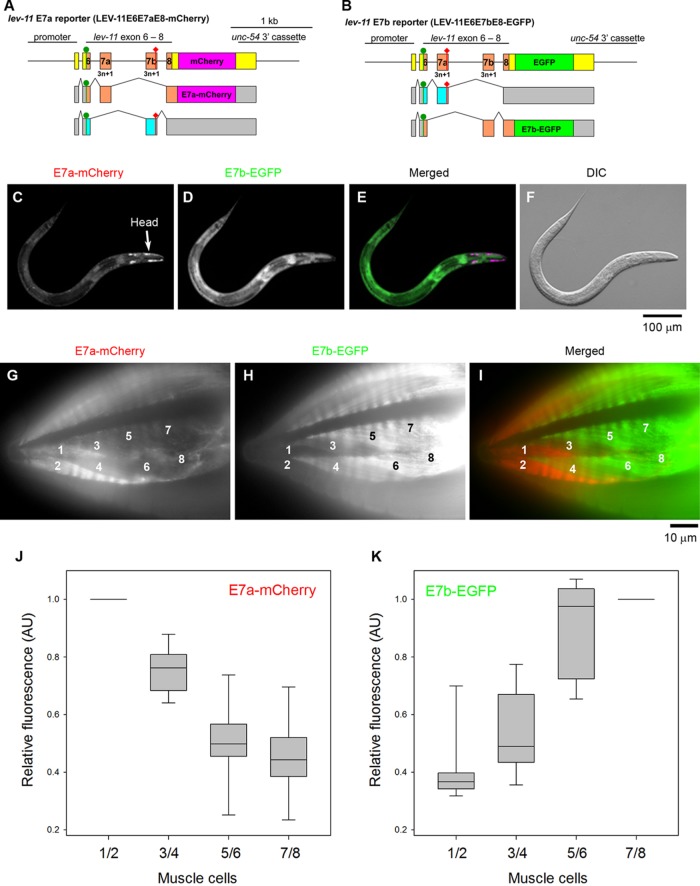
We utilized fluorescence reporter systems for alternative splicing to analyze the selection pattern for E7a/E7b in vivo. We constructed a pair of reporter minigenes in which a lev-11 genomic fragment spanning from E6 through E8 was connected in-frame to the cDNA for mCherry (E7a reporter) or EGFP (E7b reporter) (Figure 2, A and B; Kuroyanagi et al., 2010). A stop codon was introduced in E7b of the mCherry construct, such that only selection of E7a would result in mCherry expression (Figure 2A). Likewise, a stop codon in E7a of the EGFP construct would result in EGFP expression only when E7b was selected (Figure 2B). When these minigenes were ubiquitously expressed, only a small number of body wall muscle cells at the head predominantly expressed E7a-mCherry (Figure 2, C and E), whereas the other tissues expressed E7b-EGFP (Figure 2, D and E). To determine the E7a/E7b selections more clearly in the body wall muscle cells, these minigenes were expressed under the control of the myo-3 promoter (Figure 2, G–I), which is active in all body wall muscle cells (Okkema et al., 1993). These muscle-specific reporters showed that the two most anterior muscle cells in each muscle quadrant strongly expressed E7a-mCherry (Figure 2G, cells 1 and 2) but only weakly expressed E7b-EGFP (Figure 2H) (note that C. elegans body wall muscle cells are mononucleated and do not fuse). The two cells in the second row (Figure 2G, cells 3 and 4) weakly expressed both E7a-mCherry and E7b-EGFP. The rest of the body wall muscle cells (Figure 2, H and I) and vulval muscle cells (not shown in the figure) predominantly expressed E7b-EGFP. These cells also weakly expressed E7a-mCherry (Figure 2, G and I). The weak E7a-mCherry signals throughout the muscle indicate that selection of E7a from the reporter minigenes can occur as minor events even when selection of E7b is predominant. Quantitative analysis of the fluorescence signals further confirmed these differential selection patterns of E7a and E7b in the head region (Figure 2, J and K). Similar patterns were observed in muscle cells of multiple transgenic isolates with both ubiquitous and muscle-specific promoters (our unpublished data).
FIGURE 2:
Fluorescence splicing reporter analysis for lev-11 E7s. (A, B) Schematic illustration of reporter minigenes for lev-11 E7a (LEV-11-E6E7aE8-GGS6-mCherry) (A) and lev-11 E7b (LEV-11-E6E7bE8-GGS6-EGFP) (B). Structures of mRNA isoforms are indicated below each minigene. The cDNA cassettes and the predicted ORFs for mCherry and EGFP are colored in magenta and green, respectively. Green circles and red diamonds indicate the artificially introduced initiation and termination codons, respectively. Frames shown in cyan are interrupted by the artificial termination codons that prevent expression of the fluorescent reporters. (C–F) Fluorescent images show expression patterns of E7a-mCherry (C) and E7b-EGFP (D) under the control of the ubiquitous eef-1A.1 promoter. A merged image with mCherry in red and EGFP in green is shown in E. A DIC image is shown in F. Bar, 100 μm. (G–I) Expression patterns of E7a-mCherry (G) and E7b-EGFP (H) in the head region under the control of the myo-3 promoter were determined. A merged image with mCherry in red and EGFP in green is shown in I. Muscle cells in one of the four quadrants are numbered according to Hedgecock et al. (1987). Bar, 10 μm. (J, K) Relative fluorescence intensity of E7a-mCherry (J) and E7b-EGFP (K). Fluorescence intensity in the cytoplasm of indicated muscle cells were measured by ImageJ, and relative fluorescence intensity was calculated by setting the values of cells 1/2 for E7a-mCherry or 7/8 for E7b-EGFP as 1 (n = 12). Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles.
Missense mutations in exons 7a and 7b cause contractile phenotypes in different subsets of muscle cells
To determine the in vivo functions of tropomyosin isoforms containing E7a or E7b sequence, we examined phenotypes caused by a mutation in either E7a or E7b. lev-11(x12) is one of the original lev-11 mutant alleles (Lewis et al., 1980a) and has a missense mutation in E7b (Kagawa et al., 1995) resulting in E234K in the LEV-11A isoform (Figure 1C). We found that lev-11(gk334531), one of the alleles from the Million Mutation Project (Thompson et al., 2013), has a missense mutation in E7a that results in E196K in the LEV-11O isoform (Figure 1C). The strain that possesses lev-11(gk334531) was outcrossed six times with the wild type and used in the following experiments. E196 and E234 correspond to g and c positions in a heptad, which are exposed on the surface of a coiled coil (Brown et al., 2001; Minakata et al., 2008; Barua et al., 2011, 2013). Calculation of coiled-coil probability by COILS (Lupas et al., 1991) suggests that both of these missense mutations do not alter the coiled-coil formation of the tropomyosin molecule.
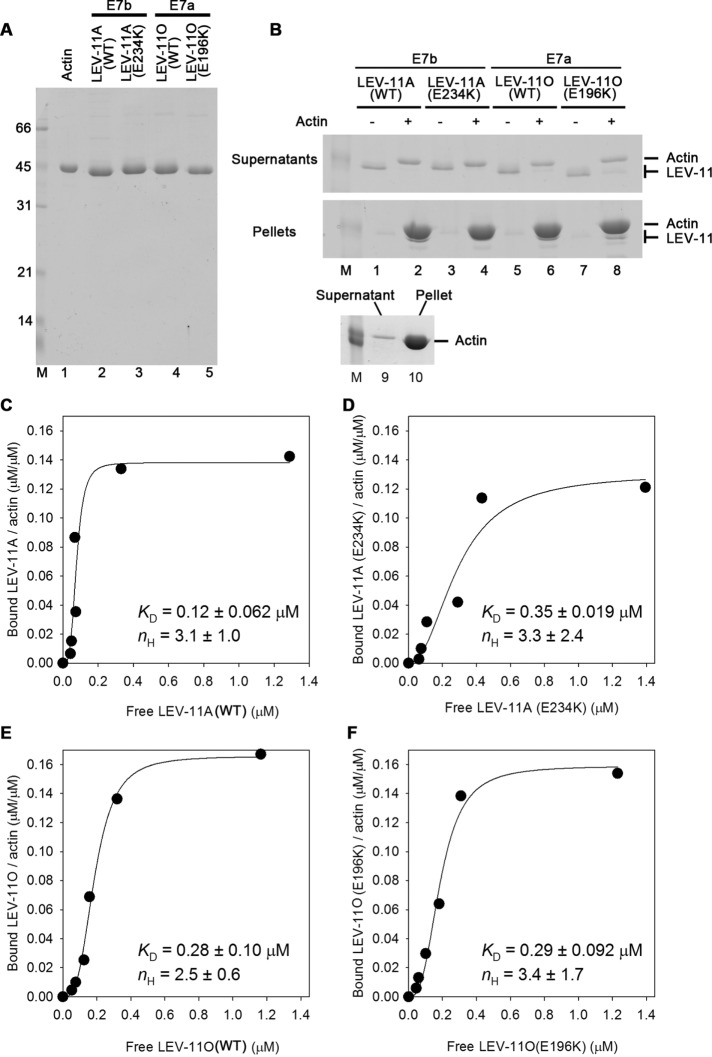
To determine biochemical properties of tropomyosin isoforms and effects of exon 7 mutations, we purified recombinant LEV-11A (with the E7b-encoded sequence) and LEV-11O (with the E7a-encoded sequence) in wild-type and mutant forms (Figure 3A) and examined their binding to actin. Their affinity to actin filaments was determined by actin cosedimentation assays (Figure 3, B–F). In the absence of actin filaments, all examined LEV-11 proteins remained in the supernatants after ultracentrifugation (Figure 3B, lanes 1, 3, 5, and 7). However, in the presence of actin filaments, all examined LEV-11 proteins coprecipitated with actin and were depleted from their respective supernatant (Figure 3, lanes 2, 4, 6, and 8). Using this cosedimentation assay, binding of LEV-11 proteins with actin was quantitatively analyzed (Figure 3, C–F). Wild-type LEV-11A bound to actin with high affinity (Figure 3C), while LEV-11A(E234K) showed an ∼3-fold decrease in the affinity to actin (Figure 3D). Wild-type LEV-11O showed a slightly lower affinity to actin than wild-type LEV-11A (Figure 3E), and LEV-11O(E196K) bound to actin in a similar manner to wild-type LEV-11O (Figure 3F). Despite the noticeable difference in dissociation constants (KD) between wild-type LEV-11A and LEV-11A(E234K), a statistical test using one-way analysis of variance (ANOVA) or Student’s t test gave p = 0.1 for comparison between wild-type LEV-11A and LEV-11A(E234K), suggesting that the difference was insignificant or very minor. All examined LEV-11 proteins showed saturable binding to actin at LEV-11/actin molar ratios of 0.14–0.16, suggesting a 1:6 or 1:7 binding of LEV-11 to actin with no detectable difference among isoforms and mutations. Hill coefficients (nH) for binding of LEV-11 proteins to actin were estimated to be 2.5–3.4 (Figure 3, C–F) suggesting that all examined LEV-11 proteins bound to actin with similar positive cooperativity. These results indicate that wild-type LEV-11A and wild-type LEV-11O bind to actin filaments in a manner similar to that for conventional high-molecular-weight tropomyosins, and that E234K but not E196K had a minor effect on the affinity to actin.
FIGURE 3:
Binding of LEV-11 isoforms and mutants to actin filaments. (A) Purified actin (0.8 μg) and recombinant LEV-11 proteins (1.0 μg each) were analyzed by SDS–PAGE. Molecular size markers in kDa (lane M) are shown on the left of the gel. (B) LEV-11 proteins (0.25 μM) were incubated without (–) or with (+) 5 μM F-actin for 1 h and ultracentrifuged. Supernatants (top panel) and pellets (bottom panel) were analyzed by SDS–PAGE (lanes 1–8). A control with actin alone is shown in lanes 9 and 10. All examined LEV-11 proteins remained in the supernatants in the absence of actin but precipitated in the presence of actin. Note that LEV-11A(E234K) could not be resolved from actin in the pellet but was clearly depleted from the supernatant (lane 4). Only size markers of 45 kDa (lane M) are included. (C–F) Quantitative analysis of actin binding of LEV-11 proteins was performed at various concentrations of LEV-11 proteins and a fixed concentration of actin (5 μM). Molar ratios of bound LEV-11 to actin were plotted as functions of free LEV-11 concentrations. The data were fitted to a Hill equation, and estimated dissociation constants (KD) and Hill constants (nH) are shown (means ± SD, n = 3). The graphs show representative results among three independent experiments.
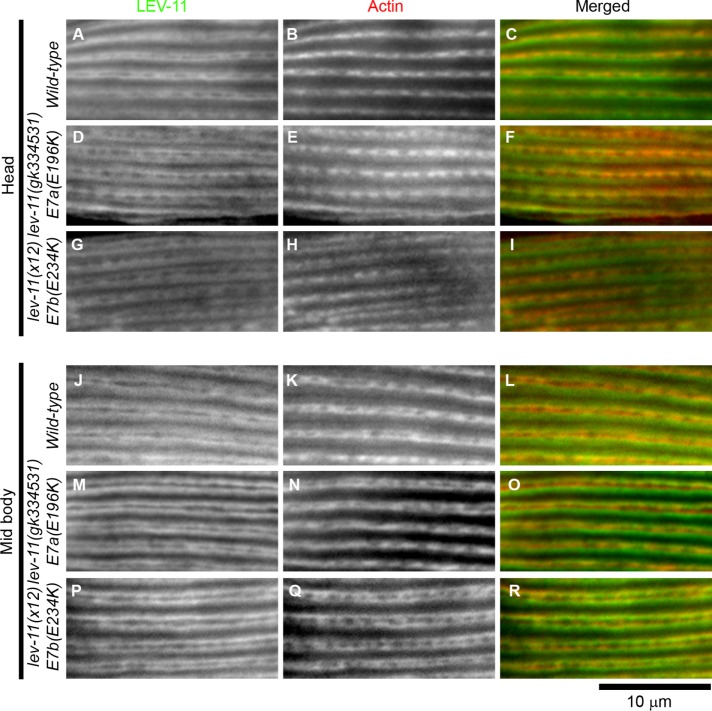
Expression and subcellular localization of LEV-11 proteins were examined in these strains by immunofluorescence microscopy (Figure 4). The anti-LEV-11 antibody that we have previously generated reacts with all known LEV-11 isoforms (Ono and Ono, 2002). In the wild type, the LEV-11 proteins colocalized with actin in sarcomeres in the body wall muscle cells of both the head (Figure 4, A–C) and main-body regions (Figure 4, J–L), suggesting that both E7a- and E7b-containing LEV-11 isoforms are components of the thin filaments in muscle sarcomeres. These patterns were not noticeably different in lev-11(gk334531)-E7a(E196K) or lev-11(x12)-E7b(E234K) in the head (Figure 4, D–I) or main-body regions (Figure 4, M–R), indicating that these missense mutations do not cause major changes in the protein levels or the localization patterns of LEV-11 protein isoforms.
FIGURE 4:
Localization of LEV-11 and actin in the body wall muscle. Wild-type (A–C, J–L), lev-11(gk334531)-E7a(E196K) (D–F, M–O), and lev-11(x12)-E7b(E234K) (G–I, P–R) adult worms were immunofluorescently stained for LEV-11 (left column) and actin (middle column), and the head (A–I) and midbody (J–R) regions were examined at high magnification. Merged images are shown in the right column (LEV-11 in green and actin in red). Bar, 10 μm.
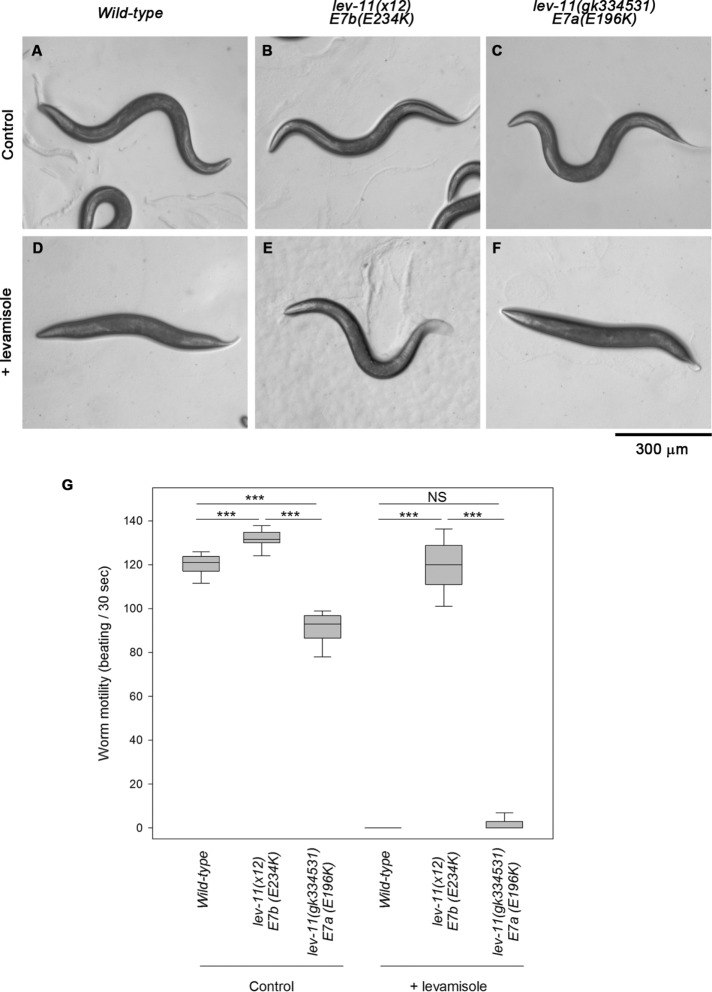
Motility examination of the lev-11 mutant worms in the absence or presence of levamisole, which is an agonist of the ACh receptor, demonstrated that both E7a and E7b are important for muscle contractility in the main body (Figure 5). Original lev-11 alleles, including lev-11(x12)-E7b(E234K), were isolated in a screen for mutants that were resistant to levamisole (Lewis et al., 1980a). Wild-type worms crawled in a sinusoidal pattern on control agar plates (Figure 5A); however, levamisole induced tonic body contraction and paralysis within a few minutes (Figure 5, D and G). lev-11(x12)-E7b(E234K) worms crawled in a similar manner to the wild type on control agar plates (Figure 5B), although they exhibited minor twitching of their bodies and moved slightly faster than the wild type (Figure 5G). Unlike the wild type, these mutant worms were resistant to levamisole and continued to move in the presence of levamisole (Figure 5, E and G) as reported by Lewis et al. (1980a). In contrast, lev-11(gk334531)-E7a(E196K) worms appeared to move normally on control agar plates (Figure 5C), but quantitative analysis showed that they moved slightly slower than the wild type (Figure 5G), indicating that the mutation in E7a partially impaired muscle contractility in the main body region. In the presence of levamisole, the worms with the E7a mutation were paralyzed, as well as the wild type (Figure 5, F and G).
FIGURE 5:
Differential responses to levamisole in the main body region caused by lev-11 exon 7 mutations. Wild-type (left), lev-11(x12)-E7b(E234K) (middle), and lev-11(gk334531)-E7a(E196K) (right) adult worms were treated with M9 buffer without (control) or with 500 μM levamisole (+ levamisole). (A–F) Entire worm bodies on agar plates are shown. The heads of the worms are oriented to the left. Bar, 300 μm. (G) Box plots of worm motility. Worm motility was quantified as beats per 30 s (n = 20). Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. NS, not significant. ***, p < 0.001.
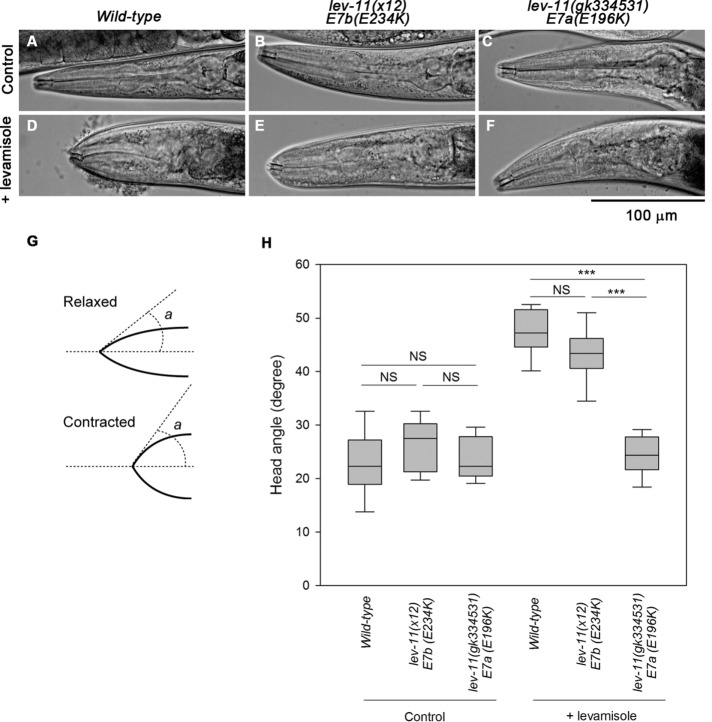
Interestingly, the lev-11 mutant worms exhibited different levamisole sensitivity in the head region, suggesting a functional difference between E7a and E7b in head muscle contractility (Figure 6). In wild-type worms, the head region became contracted in the presence of levamisole (Figure 6, A and D). Upon contraction, the head angle increased from ∼23° to ∼47° (Figure 6, G and H). The very tip of the head of the lev-11(x12)-E7b(E234K) worms also showed a contracted appearance in the presence of levamisole (Figure 6, B and E) to an extent similar to that in the wild type (Figure 6H), indicating that these worms are levamisole-sensitive only in the head. In contrast, the lev-11(gk334531)-E7a(E196K) worms exhibited a relaxed appearance in the head in the absence or presence of levamisole (Figure 6, C, F, and H). Therefore, lev-11(gk334531)-E7a(E196K) worms were resistant to levamisole in the head region but not in the main body. These results are consistent with the predominant selection of E7a in the head muscle cells (Figure 2), suggesting that a tropomyosin isoform containing the E7a-encoded sequence, that is, LEV-11O, plays a major role in the regulation of head-muscle contractility.
FIGURE 6:
Differential responses to levamisole in the head region caused by lev-11 exon 7 mutations. Wild-type (left), lev-11(x12)-E7b(E234K) (middle), and lev-11(gk334531)-E7a(E196K) (right) adult worms were treated with M9 buffer without (control) or with 500 μM levamisole (+ levamisole). (A–F) The head regions of mounted worms were imaged with DIC microscopy. Bar, 100 μm. (G) The head angle was defined as an angle (a) of a line drawn at the tip of a worm head from a midline. (H) Box plots of head angles. Head angles were measured using ImageJ (n = 10–22). Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. NS, not significant. ***, p < 0.001.
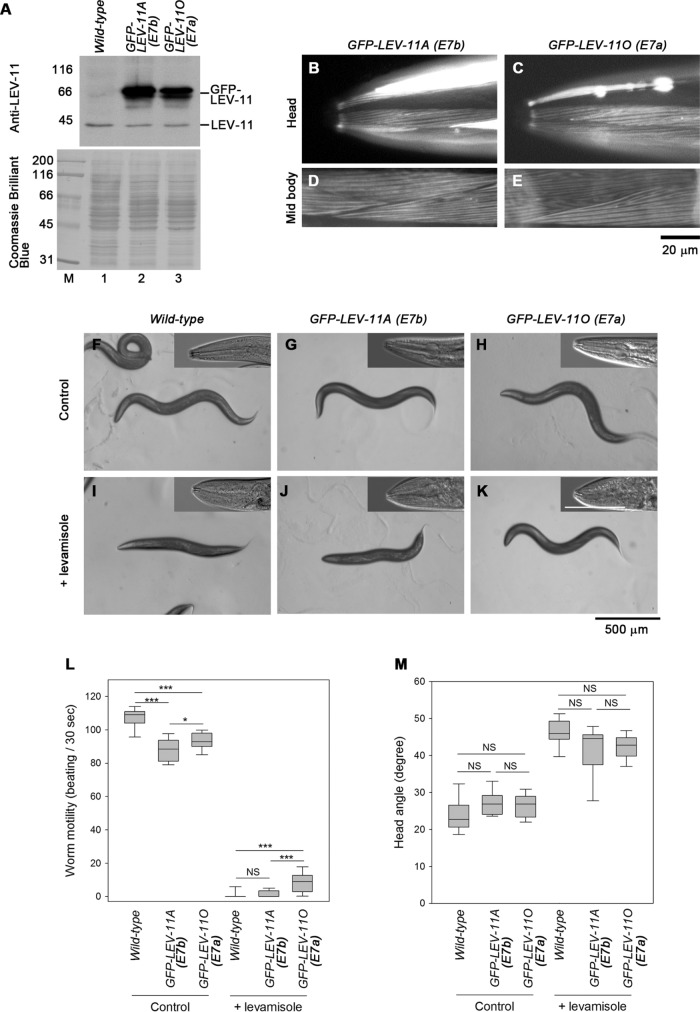
Compatibility of tropomyosin isoforms in muscle contractility regulation was further examined by altering their expression levels by transgenic overexpression. GFP-tagged LEV-11A (E7b) or LEV-11O (E7a) was expressed in wild-type background under the control of the myo-3 promoter, which is active in all body wall muscle cells (Okkema et al., 1993). Western blot showed that GFP-LEV-11A or GFP-LEV-11O was overexpressed in the transgenic strains (Figure 7A). GFP-LEV-11A or GFP-LEV-11O was expressed in both head and midbody regions and localized in sarcomeric patterns (Figure 7, B–E). Under normal (control) conditions, the worms expressing GFP-LEV-11A or GFP-LEV-11O moved slightly more slowly than the wild-type worms (Figure 7, F–H and L). In the presence of levamisole, the worms expressing GFP-LEV-11A were paralyzed (Figure 7, J and L) in a manner similar to that of wild-type worms (Figure 7, I and L). However, the worms expressing GFP-LEV-11O could maintain a curved appearance of the body (Figure 7K) and move very slowly (Figure 7L) even in the presence of levamisole, indicating that overexpression of GFP-LEV-11O in the body conferred partial levamisole resistance. In contrast, expression of GFP-LEV-11A or GFP-LEV-11O did not affect levamisole-induced contraction of the heads (Figure 7, I–K insets and M). Similar phenotypes were observed in at least one independent transgenic line for GFP-LEV-11A or GFP-LEV-11O (unpublished data). Therefore, contractility regulation in the main body region is sensitive to misexpression of LEV-11O (E7a), whereas that in the head muscle is tolerant to disturbance of tropomyosin isoforms.
FIGURE 7:
Effects of overexpression of LEV-11 isoforms on levamisole sensitivity. GFP-LEV-11A or GFP-LEV-11O was overexpressed in all muscle cells using the myo-3 promoter in the wild-type background. (A) Western blot analysis of GFP-tagged and endogenous LEV-11 proteins. Lysates of 15 adult worms from each strain were subjected to Western blot with anti-LEV-11 antibody (top). The blotted membrane was stained with Coomassie Brilliant Blue (bottom), which demonstrated equivalent loading of total proteins. Positions of GFP-LEV-11 and endogenous LEV-11 proteins are shown on the right. Molecular size markers (lane M) in kDa are indicated on the left. (B–E) Localization of GFP-LEV-11A (B, D) or GFP-LEV-11O (C, E) in the head (B, C) or midbody (D, E) regions in live worms. Bar, 20 μm. (F–K) Wild-type worms (F, I) or worms expressing GFP-LEV-11A (G, J) or GFP-LEV-11O (H, K) were treated with M9 buffer without (control) or with 500 μM levamisole (+ levamisole). Entire bodies on agar plates and head regions of mounted worms (insets) are shown. Black bar, 500 μm for images of entire bodies. White bar, 100 μm for insets. (L, M) Box plots of worm motility (beats per 30 s, n = 20) (L) and head angles (n = 7–13) (M) are shown. Boxes represent the range of the 25th and 75th percentiles, with the medians marked by solid horizontal lines, and whiskers indicate the 10th and 90th percentiles. NS, not significant. *, 0.01< p < 0.05. ***, p < 0.001.
unc-27 troponin I mediates levamisole resistance in the main bodies of lev-11 E7b mutant worms
To elucidate the mechanism of differential responses to levamisole in the head and main body, we examined the involvement of troponin I isoforms. The troponin complex, consisting of troponin T, troponin I, and troponin C, is a calcium-dependent regulator for muscle contraction (Ebashi, 1984). C. elegans has four genes encoding troponin I, a troponin component that cooperates with tropomyosin to inhibit actomyosin interaction (Burkeen et al., 2004; Ruksana et al., 2005; Obinata et al., 2010; Barnes et al., 2016). Among them, tni-3 is preferentially expressed in the head region, whereas unc-27 (also known as tni-2) is expressed as a major isoform in most of body wall muscle cells (Burkeen et al., 2004; Ruksana et al., 2005). Although expression patterns of tni-3 and unc-27 in the head have not been analyzed at single-cell levels, their expression patterns resemble the selection patterns or E7a (LEV-11O) and E7b (LEV-11A), suggesting isoform-specific compatibility for tropomyosin and troponin I.
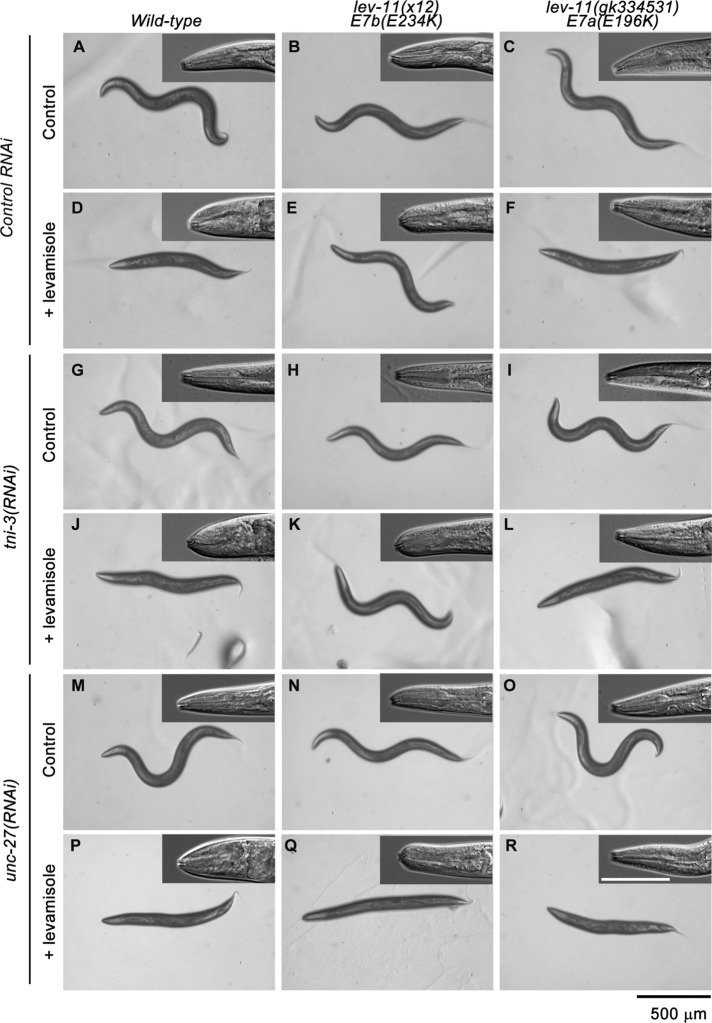
RNAi of tni-3 or unc-27 in lev-11 mutants revealed a compatible combination of troponin I and tropomyosin isoforms in the main body (Figure 8). Wild-type worms were levamisole-sensitive in the head and body in control RNAi, tni-3(RNAi), or unc-27(RNAi) (Figure 8 left column). lev-11(x12)-E7b(E234K) worms were resistant to levamisole in the body in control RNAi or tni-3(RNAi) (Figure 8, B, E, H, and K) but became sensitive to levamisole in unc-27(RNAi) (Figure 8, N and Q), indicating that the levamisole-resistant phenotype in the body of lev-11(x12)-E7b(E234K) was dependent on unc-27 troponin I. lev-11(x12)-E7b(E234K) worms remained levamisole-sensitive in the head in control RNAi, tni-3(RNAi), or unc-27(RNAi) (Figure 8, insets in the middle column). Levamisole sensitivity of lev-11(gk334531)-E7a(E196K) worms was not modified by the RNAi treatments (Figure 8, right column). Although lack of phenotypic modification by RNAi does not rule out functional significance in the genetic interaction between lev-11o and tni-3, these results, at least, show a close relationship between LEV-11A (E7b) tropomyosin and UNC-27 troponin I isoforms in the regulation of muscle contractility in the main-body region.
FIGURE 8:
Effects of RNAi of troponin I isoforms on levamisole sensitivity. Wild-type (left column), lev-11(x12)-E7b(E234K) (middle column), or lev-11(gk334531)-E7a(E196K) (right column) worms were treated with control RNAi (A–F), tni-3(RNAi) (G–L), or unc-27(RNAi) (M–R) and with M9 buffer without (control) or with 500 μM levamisole (+ levamisole). Entire bodies on agar plates and head regions of mounted worms (insets) are shown. The heads of the worms are oriented to the left. Black bars, 500 μm for images of entire bodies. White bars, 100 μm for insets.
DISCUSSION
In this study, we identified the LEV-11O tropomyosin isoform, which is the first isoform containing the E7a-encoded sequence. E7a was selected predominantly in the most anterior subset of the body wall muscle cells in the head, whereas E7b was selected in the rest of the muscle cells in the main body. Missense mutation in E7a or E7b caused levamisole-resistant phenotypes in the body regions where a corresponding exon is predominantly selected. Transgenic expression of LEV-11O in all muscle cells caused abnormal muscle contractility only in the main body. These results suggest that cell-specific alternative splicing of E7a and E7b is important to express appropriate tropomyosin isoforms for the regulation of muscle contraction.
LEV-11O (E7a) and LEV-11A (E7b) are different only by seven amino acids in the exon 7–encoded region (Figure 1C), which may confer different biophysical properties on the tropomyosin molecules. Tropomyosin is nearly entirely composed of coiled coil that is based on seven-residue (heptad) repeats (typically labeled as abcdefg). Residues a and d of a heptad repeat are often hydrophobic and contribute to formation of a hydrophobic core that stabilizes a tropomyosin dimer (Greenfield and Hitchcock-DeGregori, 1995). However, crystal structures of rat skeletal muscle α-tropomyosin fragments revealed several “broken cores” where the two polypeptide chains are separated and the coiled coil is locally unwound (Minakata et al., 2008). In particular, the charged side chain of E218 at residue a of the heptad at residues 218–224 causes destabilization of the coiled coil and makes the molecule locally flexible (Minakata et al., 2008). Interestingly, the corresponding heptads in LEV-11O and LEV-11A exhibit the greatest difference in coiled-coil probability (Figure 1D). Because bending (Brown et al., 2001) and local destabilization of the coiled coil (Singh and Hitchcock-DeGregori, 2003, 2006) are important to adapt the shape of tropomyosin to fit with the helical structure of actin filaments, a difference in the coiled-coil stability may alter the actin-regulatory functions of the tropomyosin isoforms. Although we did not detect a difference between LEV-11O and LEV-11A in the affinity with actin in vitro, there may be a significant difference in muscle thin filament regulation in the presence of troponin in vivo. In thin filaments, tropomyosin binds to actin only weakly, with a distance of ∼10 Å from the surface of actin to allow rapid troponin-regulated movement (Holmes and Lehman, 2008; El-Mezgueldi, 2014; Rynkiewicz et al., 2017). Therefore, the difference between a heptad of LEV-11O and LEV-11A may affect their dynamic biophysical properties, which may be of functional significance.
The specific selection of E7a in the limited head muscle cells suggests that the anteriormost muscle cells need to have different physiological properties than the rest of the muscle cells. The four most anterior muscle cells (cells 1–4) in each quadrant are innervated only by the nerve ring motor neurons, whereas all other muscle cells are innervated by the ventral nerve cord motor neurons (cells 5–8 receive innervation from both the nerve ring and the ventral nerve cord) (White et al., 1986). Because movement of the head is important in determining the direction of worm locomotion, the head muscle may have unique contractile properties in response to environmental cues to set a direction of movement. The roles of sensory neurons in the worm head during chemotaxis, thermotaxis, and mechanosensation have been extensively studied (Bargmann and Mori, 1997; Bargmann, 2006; Goodman, 2006). However, how the head muscle is regulated during these behavioral changes remains largely unknown. A recent single-cell transcriptional profiling study has revealed different gene expression profiles between anterior and posterior body wall muscle cells (Cao et al., 2017), indicating that the muscle cells in the head are qualitatively different from the rest of the muscle cells. In addition to these transcriptional regulation, head muscle–specific alternative splicing, such as E7a and E7b of lev-11, should further increase the complexity of differential gene products.
The RNAi experiments on troponin I isoforms suggest that isoform compatibility of tropomyosin and troponin may be involved in the regulation of differential muscle contractility between the head and body regions. Two troponin I genes, unc-27 and tni-3, show patterns of region-specific expression similar to those of E7a/E7b splicing: tni-3 is strongly expressed in the head region, whereas unc-27 is predominantly expressed in most of the body wall muscle cells (Burkeen et al., 2004; Ruksana et al., 2005). Our observation that unc-27(RNAi) suppresses the levamisole resistance of lev-11(x12)-E7b(E234K) suggests that UNC-27 troponin I and LEV-11A (E7b) tropomyosin are one of the compatible isoform combinations. Lack of phenotypic modifications by tni-3(RNAi) could be due to insufficient depletion of the TNI-3 protein. Ruksana et al. (2005) reported abnormal body morphology caused by tni-3(RNAi) by means of RNA microinjection, but we did not detect this phenotype in our tni-3(RNAi) treatment by the feeding method. Our recent optogenetic analysis showed that lev-11(x12)-E7b(E234K) worms fail to maintain a contracted state, while cholinergic neurons are active (Hwang et al., 2016). Additionally, unc-27 troponin I is crucial for inducing muscle relaxation in the main body (Burkeen et al., 2004). Together, these phenotypes suggest that the lev-11(x12)-E7b(E234K) mutation destabilizes the contracted state during excitation–contraction coupling. In contrast, LEV-11O (E7a) is involved in contractility regulation in both head and body regions. Nevertheless, overexpression of LEV-11O (E7a) in the body region caused levamisole resistance, independent of an E7a mutation. Therefore, having the right balance of tropomyosin–troponin isoform composition may be important for proper regulation of muscle contractility. In vertebrates, multiple isoforms of troponin components are expressed in a muscle type–specific manner, and a number of isoform-specific posttranslational modifications are known to regulate their functions (Gomes et al., 2002; Jin et al., 2008). However, isoform compatibility of tropomyosin and troponin in vitro and in vivo is poorly understood. As studies in the fruit fly Drosophila revealed nonequivalent functions of troponin C isoforms in muscle contractility (Eldred et al., 2014; Chechenova et al., 2017), C. elegans should also be a powerful model organism to address these questions. In addition to four troponin I isoforms, the C. elegans genome contains two genes for troponin C (Terami et al., 1999) and four genes for troponin T (Myers et al., 1996; Amin et al., 2007), which could potentially generate a large number of functionally distinct troponin–tropomyosin variants.
MATERIALS AND METHODS
Worm culture and microscopy
Worms were cultured following standard methods (Stiernagle, 2006). The following strains were obtained from the Caenorhabditis Genetics Center and used in this study: N2 wild type, ZZ12 lev-11(x12) (Lewis et al., 1980a), and VC20516 a Million Mutation Project strain containing lev-11(gk334531) (Thompson et al., 2013). VC20516 was outcrossed with wild type six times to isolate ON315 lev-11(gk334531) before phenotypic analysis. All mutant strains were analyzed as homozygotic hermaphrodites. Transgenic worms were generated as described previously (Kuroyanagi et al., 2010). Genotypes of strains are listed in Supplemental Table S1.
RT-PCR and sequencing
Total RNAs from synchronized L1-stage worms were extracted as described previously (Kuroyanagi et al., 2010, 2013b). RT-PCR was performed essentially as described previously (Kuroyanagi et al., 2010, 2013b).The sequences of the primers used in the RT-PCR experiments are available in Supplemental Table S2. RT-PCR products were analyzed by using a 2100 BioAnalyzer (Agilent Technologies). The sequences of the RT-PCR products were confirmed by direct sequencing or by cloning and sequencing.
Cloning of the lev-11o cDNA
First-strand cDNAs were reverse-transcribed from total RNAs from the N2 strain using oligo-dT by a Maxima H- First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). lev-11 cDNAs were amplified by PCR using a primer pair for E1 and E9c with Platinum Taq DNA polymerase (Thermo Fisher Scientific), purified by a PCR Cleanup kit (Qiagen), and digested by BstYI (New England BioLabs) that specifically recognized E7b. BstYI-resistant cDNAs were amplified by PCR using the same primer set and cloned in a pET-3d plasmid vector (EMD MilliporeA) by an In-Fusion HD Cloning kit (Takara Bio USA). Isolated cDNA clones were confirmed by DNA sequencing.
Construction of expression vectors for C. elegans genes and cDNAs
Fluorescence lev-11 splicing reporter minigenes were constructed essentially as described previously (Kuroyanagi et al., 2010, 2013a). Briefly, the lev-11 genomic DNA fragment was cloned into a Gateway pENTR-L1/R5 vector (Life Technologies). Mutagenesis in the exons were performed with PrimeStar GXL DNA polymerase (Takara Bio, Shiga, Japan). Expression vectors were constructed by homologous recombination between the genomic DNA cassette and a fluorescent protein cassette in the pENTR-L1/R5 vector and a destination vector pDEST-eef-1A.1p or pDEST-myo-3p (Kuroyanagi et al., 2006) with LR Clonase II Plus (Life Technologies). Expression vectors for GFP-LEV-11A and GFP-LEV-11O were constructed by inserting LEV-11A or LEV-11O cDNA at the EcoRI-NheI sites of pPD118.20 (provided by Andrew Fire, Stanford University) in-frame with the 3′-end of the GFP coding sequence. Briefly, the cDNA was amplified by PCR using Platinum Taq DNA polymerase High Fidelity (Thermo Fisher Scientific) and fused with pPD118.20 that had been cut with EcoRI and NheI using the In-fusion HD Cloning Kit (Takara Bio USA). The sequences of the primers used in the plasmid construction and mutagenesis are available in Supplemental Table S3. Genotypes of generated strains are listed in Supplemental Table S1. Sequence information on the reporter minigenes is available upon request to H.K.
Immunofluorescence staining
Adult hermaphrodites were fixed using a whole-mount method (Finney and Ruvkun, 1990) and stained with anti–C. elegans tropomyosin (LEV-11) guinea pig polyclonal antibody (Ono and Ono, 2002) and anti-actin mouse monoclonal antibody (C4, MP Biochemicals). The primary antibodies were further labeled with Alexa 488–conjugated goat anti-guinea pig immunoglobulin G (IgG) (Life Technologies) and Cy3-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories).
Fluorescence microscopy
Some of the images of fluorescence reporter worms were captured using a fluorescence compound microscope (DM6000B; Leica Microsystems, Wetzlar, Germany) equipped with a color-cooled CCD camera (DFC310FX; Leica Microsystems, Wetzlar, Germany). High-magnification images of fluorescence reporter worms and immunofluorescence staining were taken by a fluorescence inverted microscope (TE2000; Nikon Instruments, Tokyo, Japan) equipped with a SPOT RT monochrome CCD camera (Diagnostic Instruments). Images were processed with Photoshop CS3 (Adobe). The fluorescence intensity of mCherry (Figure 2J) or EGFP (Figure 2K) was measured with ImageJ. In a digital image (TIFF format), a straight line was drawn across a short axis at the center of each muscle cell, and the mean intensity was measured with the Measure tool of ImageJ. Then the relative fluorescence intensity was calculated by setting the values of cells 1/2 for E7a-mCherry or 7/8 for E7b-EGFP as 1.
Levamisole sensitivity assay
Adult worms were transferred into droplets of M9 buffer with or without 500 μM levamisole and incubated for 10 min on agar plates for observation of whole bodies or mounted immediately on 2% agarose pads for observation of head regions. Images of whole bodies were captured using a stereomicroscope (Stemi 2000C; Zeiss, Oberkochen, Germany) equipped with a digital camera (CoolPix 995; Nikon Corporation, Tokyo, Japan). Images of head regions were captured using an inverted microscope with differential interference contrast (DIC) optics (TE2000; Nikon Instruments, Tokyo, Japan) equipped with a SPOT RT monochrome CCD camera (Diagnostic Instruments). Worm motility was determined by counting swinging motions of worms in M9 buffer as described (Epstein and Thomson, 1974; Ono et al., 1999). Head angles were measured by the angle tool in ImageJ. The data were analyzed by one-way ANOVA using SigmaPlot 13 (Systat Software).
Preparation of proteins
Actin was prepared from rabbit skeletal muscle acetone powder (Pel-Freeze Biologicals) as described by Pardee and Spudich (1982). cDNAs for LEV-11A and LEV-11O were amplified by RT-PCR and cloned into a pET-3d vector as described above. The PCR primers included codons for Ala-Ser after the initiator Met to mimic the N-terminal acetylation state of tropomyosin (Monteiro et al., 1994; Supplemental Table S2). Expression vectors for LEV-11A(E234K) and LEV-11O(E196K) were generated by site-directed mutagenesis using a QuickChange mutagenesis kit (Agilent Technologies). All insert sequences were verified by DNA sequencing. All LEV-11 proteins were bacterially expressed and purified in essentially the same manner. Escherichia coli BL21(DE3) cells were transformed with the LEV-11 expression vectors, and protein expression was induced by 0.4 mM isopropyl β-d-1-thiogalactopyranoside for 3 h at 37°C. The cells were harvested by centrifugation at 5000 × g for 10 min at 4°C and homogenized by sonication in 0.1 M NaCl, 1 mM EDTA, 20 mM Tris-HCl, pH 8.0, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 0.2 mM dithiothreitol (DTT). The homogenates were cleared at 20,000 × g for 30 min at 4°C, mixed with 35% saturation of ammonium sulfate for 30 min with stirring, and centrifuged at 20,000 × g for 30 min at 4°C. Each supernatant was dialyzed against 0.1 M NaCl, 20 mM Tris-HCl, pH 6.8, applied to a DE53 anion exchange column, and eluted with a NaCl gradient from 0.1 to 0.6 M. Fractions containing LEV-11 proteins were applied to a Bio-Gel HTP hydroxyapatite column (Bio-Rad) and eluted with a gradient of phosphate from 0.01 to 0.3 M in the presence of 0.05 M KCl at pH 7.5. Fractions containing pure LEV-11 proteins were dialyzed against F-buffer (0.1 M KCl, 2 mM MgCl2, 20 mM HEPES-KOH, pH 7.5) containing 50% glycerol and stored at –20°C. Protein concentrations were determined by a BCA Protein Assay kit (Thermo Fisher Scientific). Molar concentrations of LEV-11 dimers were calculated using a molecular weight of 66,000.
Actin cosedimentation assay
Various concentrations of LEV-11 proteins were incubated with or without 5 μM F-actin in F-buffer for 1 h at room temperature and ultracentrifuged at 42,000 rpm for 20 min using a Beckman 42.2Ti rotor. Supernatants and pellets were separated, adjusted to the same volumes, and examined by SDS–PAGE (12% acrylamide gel). Protein markers (#29458-24; Nacalai USA) were used as molecular mass markers. The gels were stained with Coomassie Brilliant Blue R-250 (National Diagnostics) and scanned by an Epson Perfection V700 scanner at 300 dpi. Because LEV-11 proteins migrated too close to actin in the pellet fractions (see Figure 3B), actin-dependent depletion of LEV-11 proteins was densitometrically quantified by ImageJ from the supernatant fractions to estimate portions of LEV-11 proteins in the pellets. Actin-independent sedimentation of LEV-11 proteins was quantified from the experiments without actin and subtracted from the data. The data were fitted to the following Hill equation (three-parameter sigmoidal) using SigmaPlot 13 (Systat Software):
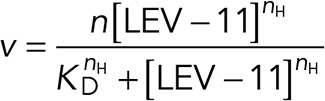 |
where v = bound LEV-11 relative to actin (μM/μM), n = maximal bound LEV-11 relative to actin (μM/ μM), nH = Hill coefficient, KD = dissociation constant, and [LEV-11] = [LEV-11]free.
Western blot
Fifteen adult worms were suspended in 15 μl of SDS lysis buffer (2% SDS, 80 mM Tris-HCl, 5% β-mercaptoethanol, 15% glycerol, 0.05% bromophenol blue, pH 6.8), heated at 97°C for 2 min, homogenized briefly by sonication, and subjected to SDS–PAGE (12% acrylamide gel). The proteins were transferred to a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membrane was blocked in 5% nonfat milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-T) and incubated for 1 h with anti-C. elegans tropomyosin (LEV-11) guinea pig polyclonal antibody (Ono and Ono, 2002). After being washed with PBS-T, the membrane was treated with horseradish peroxidase–conjugated goat anti-guinea pig IgG (MP Biomedicals) for 1 h. The reactivity was detected with Chemi-Lumi One Ultra (Nacalai USA) and exposure to x-ray films. Finally, the membrane was stained with 0.1% Coomassie Brilliant Blue R-250 (National Diagnostics) in 50% methanol and washed in a destaining solution containing 10% acetic acid and 50% ethanol to visualize total proteins (Welinder and Ekblad, 2011).
RNAi
RNAi experiments were performed by feeding with Escherichia coli HT115(DE3) expressing double-stranded RNA as described previously (Ono and Ono, 2002). All RNAi experiments were started by treating L4 larvae and observing phenotypes in the next generation. Control RNAi experiments were performed using the unmodified L4440 plasmid vector for feeding RNAi (kindly provided by Andrew Fire, Stanford University; Timmons et al., 2001). RNA clones for tni-3 (V-11A06) and unc-27 (X-4O16) were obtained from Source BioScience (Nottingham, United Kingdom).
Supplementary Material
Acknowledgments
We thank the members of Yuichiro Maeda’s laboratory (Nagoya University) for their expert support of our attempt to produce a recombinant C. elegans troponin complex. We also thank Masumi Saito and Marina Togo-Ohno for technical assistance. Some C. elegans strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by Grants-in-Aid for Scientific Research (KAKENHI, Grants JP26291003, JP15H01350, JP15H01467, JP15KK0252, JP17H03633, and JP17H05596) from the Japan Society for Promotion of Science (JSPS) to H.K. and a grant from the National Institutes of Health (AR048615) to S.O.
Abbreviations used:
- CCD
charge-coupled device
- DIC
differential interference contrast
- EGFP
enhanced green fluorescent protein
- ORFs
open reading frames
- RT-PCR
reverse transcription-PCR.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-03-0152) on February 26, 2018.
REFERENCES
- Alioto SL, Garabedian MV, Bellavance DR, Goode BL. (2016). Tropomyosin and profilin cooperate to promote formin-mediated actin nucleation and drive yeast actin cable assembly. Curr Biol , 3230–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin MZ, Bando T, Ruksana R, Anokye-Danso F, Takashima Y, Sakube Y, Kagawa H. (2007). Tissue-specific interactions of TNI isoforms with other TN subunits and tropomyosins in C. elegans: the role of the C- and N-terminal extensions. Biochim Biophys Acta , 456–465. [DOI] [PubMed] [Google Scholar]
- Anyanful A, Sakube Y, Takuwa K, Kagawa H. (2001). The third and fourth tropomyosin isoforms of Caenorhabditis elegans are expressed in the pharynx and intestines and are essential for development and morphology. J Mol Biol , 525–537. [DOI] [PubMed] [Google Scholar]
- Bailey K. (1946). Tropomyosin: a new asymmetric protein component of muscle. Nature , 368. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. (2006). Chemosensation in C. elegans. In: WormBook, ed. The C. elegans Research Community, /10.1895/wormbook.1.123.1. [DOI] [PMC free article] [PubMed]
- Bargmann CI, Mori I. (1997). Chemotaxis and thermotaxis. In: C. elegans II, ed. Riddle DL, Blumenthal T, Meyer BJ, Priess JR, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 717–737. [PubMed] [Google Scholar]
- Barnes DE, Hwang H, Ono K, Lu H, Ono S. (2016). Molecular evolution of troponin I and a role of its N-terminal extension in nematode locomotion. Cytoskeleton , 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B, Fagnant PM, Winkelmann DA, Trybus KM, Hitchcock-DeGregori SE. (2013). A periodic pattern of evolutionarily conserved basic and acidic residues constitutes the binding interface of actin–tropomyosin. J Biol Chem , 9602–9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua B, Pamula MC, Hitchcock-DeGregori SE. (2011). Evolutionarily conserved surface residues constitute actin binding sites of tropomyosin. Proc Natl Acad Sci USA , 10150–10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. (1982). Tropomyosin binding to F-actin protects the F-actin from disassembly by brain actin-depolymerizing factor (ADF). Cell Motil , 1–8. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. (2001). Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr Biol , 1300–1304. [DOI] [PubMed] [Google Scholar]
- Brown JH, Kim KH, Jun G, Greenfield NJ, Dominguez R, Volkmann N, Hitchcock-DeGregori SE, Cohen C. (2001). Deciphering the design of the tropomyosin molecule. Proc Natl Acad Sci USA , 8496–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce NS, Schevzov G, Ferguson V, Percival JM, Lin JJ, Matsumura F, Bamburg JR, Jeffrey PL, Hardeman EC, Gunning P, Weinberger RP. (2003). Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol Biol Cell , 1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkeen AK, Maday SL, Rybicka KK, Sulcove JA, Ward J, Huang MM, Barstead R, Franzini-Armstrong C, Allen TS. (2004). Disruption of Caenorhabditis elegans muscle structure and function caused by mutation of troponin I. Biophys J , 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, et al (2017). Comprehensive single-cell transcriptional profiling of a multicellular organism. Science , 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chechenova MB, Maes S, Oas ST, Nelson C, Kiani KG, Bryantsev AL, Cripps RM. (2017). Functional redundancy and nonredundancy between two Troponin C isoforms in Drosophila adult muscles. Mol Biol Cell , 760–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho A, Kato M, Whitwam T, Kim JH, Montell DJ. (2016). An atypical tropomyosin in Drosophila with intermediate filament-like properties. Cell Rep , 928–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curthoys NM, Freittag H, Connor A, Desouza M, Brettle M, Poljak A, Hall A, Hardeman E, Schevzov G, Gunning PW, Fath T. (2014). Tropomyosins induce neuritogenesis and determine neurite branching patterns in B35 neuroblastoma cells. Mol Cell Neurosci , 11–21. [DOI] [PubMed] [Google Scholar]
- Dixon SJ, Roy PJ. (2005). Muscle arm development in Caenorhabditis elegans. Development , 3079–3092. [DOI] [PubMed] [Google Scholar]
- Dominguez R. (2011). Tropomyosin: the gatekeeper’s view of the actin filament revealed. Biophys J , 797–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B, Brown C, Barrell BG, Bretscher A. (1995). Tropomyosin is essential in yeast, yet the TPM1 and TPM2 products perform distinct functions. J Cell Biol , 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebashi S. (1984). Ca2+ and the contractile proteins. J Mol Cell Cardiol , 129–136. [DOI] [PubMed] [Google Scholar]
- Eldred CC, Katzemich A, Patel M, Bullard B, Swank DM. (2014). The roles of troponin C isoforms in the mechanical function of Drosophila indirect flight muscle. J Muscle Res Cell Motil , 211–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mezgueldi M. (2014). Tropomyosin dynamics. J Muscle Res Cell Motil , 203–210. [DOI] [PubMed] [Google Scholar]
- Epstein HF, Thomson JN. (1974). Temperature-sensitive mutation affecting myofilament assembly in Caenorhabditis elegans. Nature , 579–580. [DOI] [PubMed] [Google Scholar]
- Finney M, Ruvkun G. (1990). The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell , 895–905. [DOI] [PubMed] [Google Scholar]
- Galinska-Rakoczy A, Engel P, Xu C, Jung H, Craig R, Tobacman LS, Lehman W. (2008). Structural basis for the regulation of muscle contraction by troponin and tropomyosin. J Mol Biol , 929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant C, Appel S, Graceffa P, Leavis P, Lin JJ, Gunning PW, Schevzov G, Chaponnier C, DeGnore J, Lehman W, Morgan KG. (2011). Tropomyosin variants describe distinct functional subcellular domains in differentiated vascular smooth muscle cells. Am J Physiol Cell Physiol , C1356–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateva G, Kremneva E, Reindl T, Kotila T, Kogan K, Gressin L, Gunning PW, Manstein DJ, Michelot A, Lappalainen P. (2017). Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr Biol , 705–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeves MA, Hitchcock-DeGregori SE, Gunning PW. (2015). A systematic nomenclature for mammalian tropomyosin isoforms. J Muscle Res Cell Motil , 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goins LM, Mullins RD. (2015). A novel tropomyosin isoform functions at the mitotic spindle and Golgi in Drosophila. Mol Biol Cell , 2491–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AV, Potter JD, Szczesna-Cordary D. (2002). The role of troponins in muscle contraction. IUBMB Life , 323–333. [DOI] [PubMed] [Google Scholar]
- Goodman MB. (2006). Mechanosensation. In: WormBook, ed. The C. elegans Research Community, /10.1895/wormbook.1.62.1.
- Greenfield NJ, Hitchcock-DeGregori SE. (1995). The stability of tropomyosin, a two-stranded coiled-coil protein, is primarily a function of the hydrophobicity of residues at the helix-helix interface. Biochemistry , 16797–16805. [DOI] [PubMed] [Google Scholar]
- Gruninger TR, Gualberto DG, LeBoeuf B, Garcia LR. (2006). Integration of male mating and feeding behaviors in Caenorhabditis elegans. J Neurosci , 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning PW, Hardeman EC, Lappalainen P, Mulvihill DP. (2015). Tropomyosin—master regulator of actin filament function in the cytoskeleton. J Cell Sci , 2965–2974. [DOI] [PubMed] [Google Scholar]
- Gunning PW, Schevzov G, Kee AJ, Hardeman EC. (2005). Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol , 333–341. [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Hall DH, Stern BD. (1987). Genetics of cell and axon migrations in Caenorhabditis elegans. Development , 365–382. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Barua B. (2017). Tropomyosin structure, function, and interactions: a dynamic regulator. Subcell Biochem , 253–284. [DOI] [PubMed] [Google Scholar]
- Hitchcock-DeGregori SE, Sampath P, Pollard TD. (1988). Tropomyosin inhibits the rate of actin polymerization by stabilizing actin filaments. Biochemistry , 9182–9185. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Lehman W. (2008). Gestalt-binding of tropomyosin to actin filaments. J Muscle Res Cell Motil , 213–219. [DOI] [PubMed] [Google Scholar]
- Huckaba TM, Lipkin T, Pon LA. (2006). Roles of type II myosin and a tropomyosin isoform in retrograde actin flow in budding yeast. J Cell Biol , 957–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Barnes DE, Matsunaga Y, Benian GM, Ono S, Lu H. (2016). Muscle contraction phenotypic analysis enabled by optogenetics reveals functional relationships of sarcomere components in Caenorhabditis elegans. Sci Rep , 19900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JP, Zhang Z, Bautista JA. (2008). Isoform diversity, regulation, and functional adaptation of troponin and calponin. Crit Rev Eukaryot Gene Expr , 93–124. [DOI] [PubMed] [Google Scholar]
- Kagawa H, Sugimoto K, Matsumoto H, Inoue T, Imadzu H, Takuwa K, Sakube Y. (1995). Genome structure, mapping and expression of the tropomyosin gene tmy-1 of Caenorhabditis elegans. J Mol Biol , 603–613. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. (2006). Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods , 909–915. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. (2010). Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc , 1495–1517. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi H, Takei S, Suzuki Y. (2014). Comprehensive analysis of mutually exclusive alternative splicing in C. elegans. Worm , e28459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Hagiwara M. (2013a). CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans. PLoS Genet , e1003337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroyanagi H, Watanabe Y, Suzuki Y, Hagiwara M. (2013b). Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans. Nucleic Acids Res , 4015–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Wu CH, Berg H, Levine JH. (1980a). The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics , 905–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Wu CH, Levine JH, Berg H. (1980b). Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience , 967–989. [DOI] [PubMed] [Google Scholar]
- Lin JJ, Warren KS, Wamboldt DD, Wang T, Lin JL. (1997). Tropomyosin isoforms in nonmuscle cells. Int Rev Cytol , 1–38. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. (1991). Predicting coiled coils from protein sequences. Science , 1162–1164. [DOI] [PubMed] [Google Scholar]
- Manstein DJ, Mulvihill DP. (2016). Tropomyosin-mediated regulation of cytoplasmic myosins. Traffic , 872–877. [DOI] [PubMed] [Google Scholar]
- Maruyama K. (1964). Interaction of tropomyosin with actin. A flow birefringence study. Arch Biochem Biophys , 142–150. [DOI] [PubMed] [Google Scholar]
- Minakata S, Maeda K, Oda N, Wakabayashi K, Nitanai Y, Maeda Y. (2008). Two-crystal structures of tropomyosin C-terminal fragment 176–273: exposure of the hydrophobic core to the solvent destabilizes the tropomyosin molecule. Biophys J , 710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro PB, Lataro RC, Ferro JA, Reinach Fde C. (1994). Functional alpha-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J Biol Chem , 10461–10466. [PubMed] [Google Scholar]
- Myers CD, Goh PY, Allen TS, Bucher EA, Bogaert T. (1996). Developmental genetic analysis of troponin T mutations in striated and nonstriated muscle cells of Caenorhabditis elegans. J Cell Biol , 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinata T, Ono K, Ono S. (2010). Troponin I controls ovulatory contraction of non-striated actomyosin networks in the C. elegans somatic gonad. J Cell Sci , 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. (1993). Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics , 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono K, Ono S. (2004). Tropomyosin and troponin are required for ovarian contraction in the Caenorhabditis elegans reproductive system. Mol Biol Cell , 2782–2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S. (2014). Regulation of structure and function of sarcomeric actin filaments in striated muscle of the nematode Caenorhabditis elegans. Anat Rec , 1548–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Baillie DL, Benian GM. (1999). UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J Cell Biol , 491–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S, Ono K. (2002). Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol , 1065–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. (1982). Purification of muscle actin. Methods Enzymol , 164–181. [DOI] [PubMed] [Google Scholar]
- Perry SV. (2003). What is the role of tropomyosin in the regulation of muscle contraction. J Muscle Res Cell Motil , 593–596. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA. (2009). Actin, a central player in cell shape and movement. Science , 1208–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao JN, Rivera-Santiago R, Li XE, Lehman W, Dominguez R. (2012). Structural analysis of smooth muscle tropomyosin α and β isoforms. J Biol Chem , 3165–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruksana R, Kuroda K, Terami H, Bando T, Kitaoka S, Takaya T, Sakube Y, Kagawa H. (2005). Tissue expression of four troponin I genes and their molecular interactions with two troponin C isoforms in Caenorhabditis elegans. Genes Cells , 261–276. [DOI] [PubMed] [Google Scholar]
- Rynkiewicz MJ, Prum T, Hollenberg S, Kiani FA, Fagnant PM, Marston SB, Trybus KM, Fischer S, Moore JR, Lehman W. (2017). Tropomyosin must interact weakly with actin to effectively regulate thin filament function. Biophys J , 2444–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Bryce NS, Almonte-Baldonado R, Joya J, Lin JJ, Hardeman E, Weinberger R, Gunning P. (2005). Specific features of neuronal size and shape are regulated by tropomyosin isoforms. Mol Biol Cell , 3425–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schevzov G, Fath T, Vrhovski B, Vlahovich N, Rajan S, Hook J, Joya JE, Lemckert F, Puttur F, Lin JJ, et al (2008). Divergent regulation of the sarcomere and the cytoskeleton. J Biol Chem , 275–283. [DOI] [PubMed] [Google Scholar]
- Schevzov G, Whittaker SP, Fath T, Lin JJ, Gunning PW. (2011). Tropomyosin isoforms and reagents. BioArchitecture , 135–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Hitchcock-DeGregori SE. (2003). Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry , 14114–14121. [DOI] [PubMed] [Google Scholar]
- Singh A, Hitchcock-DeGregori SE. (2006). Dual requirement for flexibility and specificity for binding of the coiled-coil tropomyosin to its target, actin. Structure , 43–50. [DOI] [PubMed] [Google Scholar]
- Skau CT, Neidt EM, Kovar DR. (2009). Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol Biol Cell , 2160–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie LB, Pato MD, Pearlstone JR, Mak AS. (1980). Periodicity of α-helical potential in tropomyosin sequence correlates with alternating actin binding sites. J Mol Biol , 199–202. [DOI] [PubMed] [Google Scholar]
- Squire JM, Morris EP. (1998). A new look at thin filament regulation in vertebrate skeletal muscle. FASEB J , 761–771. [DOI] [PubMed] [Google Scholar]
- Stiernagle T. (2006). Maintenance of C. elegans. In: WormBook, ed. The C. elegans Research Community, /10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed]
- Temm-Grove CJ, Jockusch BM, Weinberger RP, Schevzov G, Helfman DM. (1998). Distinct localizations of tropomyosin isoforms in LLC-PK1 epithelial cells suggests specialized function at cell–cell adhesions. Cell Motil Cytoskeleton , 393–407. [DOI] [PubMed] [Google Scholar]
- Terami H, Williams BD, Kitamura S, Sakube Y, Matsumoto S, Doi S, Obinata T, Kagawa H. (1999). Genomic organization, expression, and analysis of the troponin C gene pat-10 of Caenorhabditis elegans. J Cell Biol , 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, et al (2013). The million mutation project: a new approach to genetics in Caenorhabditis elegans. Genome Res , 1749–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene , 103–112. [DOI] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. (2011). A molecular pathway for myosin II recruitment to stress fibers. Curr Biol , 539–550. [DOI] [PubMed] [Google Scholar]
- Ujfalusi Z, Kovacs M, Nagy NT, Barko S, Hild G, Lukacs A, Nyitrai M, Bugyi B. (2012). Myosin and tropomyosin stabilize the conformation of formin-nucleated actin filaments. J Biol Chem , 31894–31904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujfalusi Z, Vig A, Hild G, Nyitrai M. (2009). Effect of tropomyosin on formin-bound actin filaments. Biophys J , 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeranan-Karmegam R, Boggupalli DP, Liu G, Gonsalvez GB. (2016). A new isoform of Drosophila non-muscle tropomyosin 1 interacts with kinesin-1 and functions in oskar mRNA localization. J Cell Sci , 4252–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahovich N, Kee AJ, Van der Poel C, Kettle E, Hernandez-Deviez D, Lucas C, Lynch GS, Parton RG, Gunning PW, Hardeman EC. (2009). Cytoskeletal tropomyosin Tm5NM1 is required for normal excitation-contraction coupling in skeletal muscle. Mol Biol Cell , 400–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlahovich N, Schevzov G, Nair-Shaliker V, Ilkovski B, Artap ST, Joya JE, Kee AJ, North KN, Gunning PW, Hardeman EC. (2008). Tropomyosin 4 defines novel filaments in skeletal muscle associated with muscle remodelling/regeneration in normal and diseased muscle. Cell Motil Cytoskeleton , 73–85. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Muller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. (2015). Structure of the F-actin–tropomyosin complex. Nature , 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. (2007). Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry , 8146–8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welinder C, Ekblad L. (2011). Coomassie staining as loading control in Western blot analysis. J Proteome Res , 1416–1419. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. (1986). The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci , 1–340. [DOI] [PubMed] [Google Scholar]
- Williams BD, Waterston RH. (1994). Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J Cell Biol , 475–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu R, Ono S. (2006). Dual roles of tropomyosin as an F-actin stabilizer and a regulator of muscle contraction in Caenorhabditis elegans body wall muscle. Cell Motil Cytoskeleton , 659–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.