The nuclear pore complex is an ancient component of the eukaryotic cell. We show here that an FG nucleoporin, TbNup53b, in trypanosomes has an association with the splicing machinery and roles in gene expression, indicating that moonlighting roles for nucleoporins are highly ancient and present in the earliest eukaryotes.
Abstract
Components of the nuclear periphery coordinate a multitude of activities, including macromolecular transport, cell-cycle progression, and chromatin organization. Nuclear pore complexes (NPCs) mediate nucleocytoplasmic transport, mRNA processing, and transcriptional regulation, and NPC components can define regions of high transcriptional activity in some organisms at the nuclear periphery and nucleoplasm. Lineage-specific features underpin several core nuclear functions and in trypanosomatids, which branched very early from other eukaryotes, unique protein components constitute the lamina, kinetochores, and parts of the NPCs. Here we describe a phenylalanine-glycine (FG)-repeat nucleoporin, TbNup53b, that has dual localizations within the nucleoplasm and NPC. In addition to association with nucleoporins, TbNup53b interacts with a known trans-splicing component, TSR1, and has a role in controlling expression of surface proteins including the nucleolar periphery-located, procyclin genes. Significantly, while several nucleoporins are implicated in intranuclear transcriptional regulation in metazoa, TbNup53b appears orthologous to components of the yeast/human Nup49/Nup58 complex, for which no transcriptional functions are known. These data suggest that FG-Nups are frequently co-opted to transcriptional functions during evolution and extend the presence of FG-repeat nucleoporin control of gene expression to trypanosomes, suggesting that this is a widespread and ancient eukaryotic feature, as well as underscoring once more flexibility within nucleoporin function.
INTRODUCTION
The nucleus mediates transcription, DNA replication, DNA repair, DNA modifications, ribosome biogenesis, and other functions and is compartmentalized into several discrete subdomains, including the nucleolus, nuclear bodies, nuclear periphery, and chromosomal territories to facilitate these roles. At the nuclear periphery embedded within the nuclear envelope (NE) are nuclear pore complexes (NPCs), elaborate structures allowing exchange of macromolecules between the nucleus and cytoplasm. NPCs comprise ∼25–30 different proteins termed nucleoporins (Rout et al., 2000; Cronshaw et al., 2002). At the nuclear face of the NPC, the nuclear basket projects into the nucleoplasm, forming a molecular platform that interacts with the nuclear lamina, DNA repair, and cell-cycle checkpoint machinery, transcription factors, and various mRNA processing and transport factors (Galy et al., 2000 , 2004; Iouk et al., 2002; de Souza et al., 2009; Niepel et al., 2013). In addition to facilitating heterochromatin exclusion from the immediate vicinity of the NPC in mammalian cells and yeast (Ishii et al., 2002; Krull et al., 2010), the nuclear basket likely facilitates transcription and efficient processing of mRNAs for subsequent export (Casolari et al., 2004; Dieppois et al., 2006; Kurshakova et al., 2007; Ahmed et al., 2010; Dilworth et al., 2005; Vaquerizas et al., 2010; Light et al., 2013).
Significantly, several nucleoporins, particularly those located toward the nucleoplasm, are implicated in regulating genome architecture, gene expression, and cell-cycle progression. In animals and fungi, several nucleoporins and NPC-associated proteins define a robust boundary activity facilitating tethering of actively transcribed genes in proximity to the NPC (Ishii et al., 2002; Casolari et al., 2004; Dieppois et al., 2006; Kurshakova et al., 2007; Ahmed et al., 2010). Moreover, some phenylalanine-glycine (FG)-repeat–containing nucleoporins (FG-Nups), which are primarily responsible for the gating and transport functions of the NPC, are implicated in activation of developmentally regulated loci within the nuclear interior (Kasper et al., 1999; Capelson et al., 2010; Kalverda et al., 2010; Liang et al., 2013), while nuclear basket components also interact with the mitotic spindle. These findings indicate that the NPC mediates transcription and mRNA processing together with transport and that several nucleoporins have additional roles distinct beyond NPC functions.
The African trypanosome Trypanosoma brucei is an evolutionary divergent protozoan parasite and an important model for a vast group of organisms of both health and agricultural importance. The NPC in trypanosomes has a well-conserved core compared with yeast and vertebrates (DeGrasse et al., 2009), but peripheral regions are more divergent (duBois et al., 2012; Holden et al., 2014). This likely reflects both diversity in the client proteins with which the NPC interacts and potentially divergent functions. For example, the transcription and export complex 2 (TREX-2) complex, which interacts with the nuclear face of the animal/fungal NPC, appears poorly conserved in trypanosomatids, while the mRNA export system is radically different (Holden et al., 2014). In line with this, the requirement for rapid adaptation to multiple host environments (Palenchar and Bellofatto, 2006; Daniels et al., 2010) necessitates coordinated alterations to gene expression profiles to fulfill needs for survival within each host.
The genome of T. brucei contains ∼8,000 protein coding genes, mostly arranged into large polycistronic transcription units (PTUs). With the exception of the RNA polymerase I-transcribed (Pol I) developmentally regulated surface antigen genes encoding the variant surface glycoprotein (VSG) and procyclin, expressed in mammalian and insect forms, respectively (Navarro and Gull, 2001; Landeira and Navarro, 2007), RNA polymerase II (Pol II) is responsible for the vast majority of PTU transcription (Ouellette and Papadopoulou, 2009). The absence of promoters associated with individual genes indicates that regulation of gene expression is predominantly posttranscriptional (Clayton, 2002). In yeast and animals, mRNA degradation is initiated by poly-A tail removal, and recently three deadenylation complexes in trypanosomes have been described (Schwede et al., 2008, 2009; Utter et al., 2011), indicating that mRNA degradation pathways regulating trypanosome gene expression are present, although the precise signals required remain ill defined. Additional mechanisms controlling mRNA abundance have also been uncovered and include a role for a specific RNA-binding protein 6 (RBP6) in recapitulating the entire insect stage portion of the life cycle, while evidence supporting the position within the PTU as a contributory factor in control of mRNA levels has also emerged (Kolev et al., 2012; Kramer et al., 2013). Overall, these data suggest complex control of gene expression in trypanosomes and multiple mechanisms contributing to attaining steady-state levels of mRNA species.
To determine whether nucleoporins have additional functions in trypanosomes beyond being NPC components, we screened nucleoporins for intranuclear location, suggestive of non–NPC-associated activity. We find that TbNup53b, an FG-Nup, localizes both at the NPC and nucleoplasm and that TbNup53b contributes to mRNA processing. Interestingly, the opisthokont orthologues have not been associated with any such role, suggesting that nucleoporins can be recruited in a lineage-specific manner for intranuclear function.
RESULTS
TbNup53b is present at the NPC and nuclear matrix throughout the cell cycle
In common with mammalian cells and yeast, the majority of T. brucei nucleoporins have an almost exclusive localization at the NPC, but a number are also observed elsewhere within the nucleus (deGrasse et al., 2009). We previously described one of this group, a nuclear basket nucleoporin, TbNup92, which associates with the mitotic spindle and spindle anchor at mitosis (Holden et al., 2014). We analyzed GFP-tagged nucleoporins in both interphase and mitosis to ascertain which, if any, trypanosome Nups displayed non–NPC locations. We found evidence for several, but one of these, an FG-Nup TbNup53b (Tb927.3.3540), was the most prominent (Supplemental Figure S1). TbNup53b and TbNup98 were genomically tagged at the C-terminus with GFP (deGrasse et al., 2009) and their locations throughout the cell cycle determined by confocal microscopy (Figure 1).
FIGURE 1:
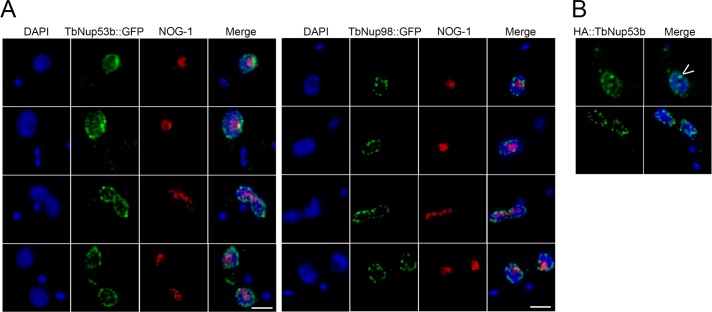
TbNup53b is localized within the nucleoplasm and nucleolar periphery. (A) TbNup53b::GFP-tagged PCF cells were fixed and stained with anti-GFP and antibody to the nucleolar protein NOG-1 to visualize TbNup53::GFP (green) and the nucleolus (red), respectively, by confocal immunofluorescence microscopy. Central slices from z-stacks are displayed. In addition to localizing at the NE, TbNup53b::GFP displays a speckled-like appearance within the nucleoplasm and is additionally localized at a single punctum at the periphery of the nucleolus. (B) BSF cells expressing 12HA::TbNup53b were fixed and stained with anti-HA antibody to visualize HA::TbNup53b imaged by confocal microscopy. The distribution of 12HA::TbNup53b in BSFs is indistinguishable from the localization of TbNup53b::GFP in PCFs. Arrowhead indicates a punctum of TbNup53B at the nucleolar periphery. For all images, DAPI was used to visualize DNA (blue). Scale bars: 2 μm.
As well as maintaining a distinct presence at the nuclear periphery, corresponding to a role as an NPC component, TbNup53b::GFP was distributed within the nucleoplasm as discrete puncta (Figure 1A, top). This localization is clearly distinct from a second FG-Nup, TbNup98, which is exclusively localized to the nuclear periphery, as are the other T. brucei FG-Nups documented to date (Figure 1A and Supplemental Figure S1). In addition, a small, but significant punctum of TbNup53b::GFP was present close to the interphase nucleolus periphery (Figure 1A, red), highlighted using an antibody to the nucleolar protein NOG-1, but was notably absent from mitotic cells.
To test whether this unique localization of TbNup53b was stage specific, the N-terminus of a single TbNup53b allele was tagged with 12xHA epitopes in bloodstream forms (BSF) cells. The distribution of 12HA::TbNup53b was indistinguishable from procyclic culture form (PCF) cells, indicating that intranuclear TbNup53b is not stage dependent (Figure 1B). Further, these data suggest that TbNup53b localization was not influenced by the epitope tag, as both C-terminal GFP and N-terminal HA provided similar locations.
TbNup53b is orthologous to HsNup58/45/ScNup49
TbNup53b consists of a central coiled-coil region flanked at N- and C-termini by small, unstructured, FG-repeat–containing units (deGrasse et al., 2009). To identify possible orthologues, we first sampled orthologous protein sequences from additional kinetoplastid genomes (Supplemental Figure S1). Using an alignment omitting the unstructured FG-containing N- and C-terminal regions, we created a profile for a hidden Markov model (HMM) in HMMER to search the UniProtKB database. With an e-value threshold of 0.01, a jackhmmer iterative search identified both HsNup58/45/ScNup49 and HsNup62/ScNsp1 as significant. However, only HsNup58/45/ScNup49 and orthologues in other eukaryotes were found with more stringent e-value thresholds, suggesting that TbNup53b is orthologous to HsNup58/45/ScNup49 and more distantly related to HsNup62/ScNsp1 (Figure 2). Interactome analysis indicates that NPC-associated TbNup53b is a component of the inner ring (Obado et al., 2016).
FIGURE 2:
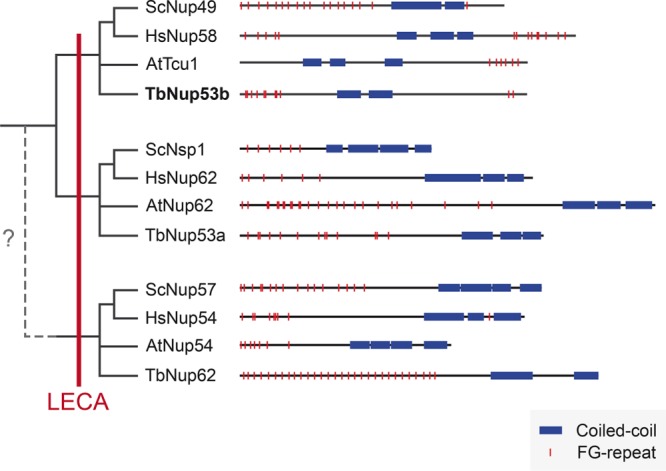
Domain architecture and evolutionary origins of the subunits of the Nup62 subcomplex. The distribution of FG-repeats (red) and coiled-coil regions (blue) is shown for proteins of representatives of distinct eukaryotic taxa: Arabidopsis thaliana (At), Saccharomyces cerevisiae (Sc), Homo sapiens (Hs), and Trypanosoma brucei (Tb). Iterative hmmer searches identified HsNup58/45/ScNup49/AtTcr1/TbNup53b as paralogues of HsNup62/ScNsp1/AtNup62/TbNup53a and vice versa, but neither demonstrated significant homology with HsNup54/ScNup57/AtNup54/TbNup62, although the shared domain architecture suggests possible common ancestry. The phylogenetic distribution suggests that all three subunits were present in the LECA.
Two other trypanosome FG Nups contain coiled-coils, TbNup53a and TbNup62 (deGrasse et al., 2009). Similarly to Nup53b, the HMM profile of kinetoplastid Nup53a identified both HsNup58/45/ScNup49 and HsNup62/ScNsp1 as significant using default settings, but only HsNup62/ScNsp1 were identified using a more stringent cutoff. Phylogenetic analysis of TbNup53b and TbNup53a homologues corroborated their phylogenetic affinity to HsNup58/45/ScNup49 and HsNup62/ScNsp1, respectively, and which are also components of the same inner ring complex in yeast (Obado et al., 2016) (Supplemental Figure S2). The phylogenetic distribution of these three Nup62 subcomplex subunits indicates that all three were present in the last eukaryotic common ancestor (LECA) (Figure 2). Given these complex evolutionary patterns of FG Nups, while we consider these assignments tentative, they indicate potential affinity between TbNup53b and HsNup58/45/ScNup49, consistent with assignment to the inner ring complex.
TbNup53b interacts with the NPC and the splicing component TSR1
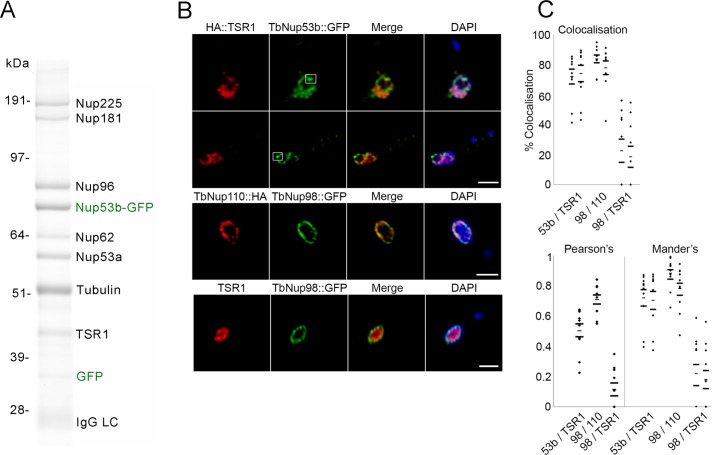
Given the extensive nuclear distribution of TbNup53b::GFP, we examined protein interactions made by TbNup53b using cryomilling/affinity isolation followed by mass spectrometry. As expected, extensive interactions between TbNup53b and TbNup225, 181, 96, 62, and 53a were revealed (Figure 3A), confirming assignment as a bona fide nucleoporin.
FIGURE 3:
TbNup53b interacts with the NPC and a splicing component TSR1. (A) Immunoisolation of TbNup53b complexes was performed using TbNup53b::GFP as the bait. Affinity isolated proteins were fractionated by SDS–PAGE, and the bands were identified by mass spectrometry. TbNup53b::GFP interacts with numerous core components of the NPC in addition to the splicing component TSR1. (B) Cells expressing 12HA::TSR1 and TbNup53b::GFP were fixed and stained with anti-GFP (green) and anti-HA. Shown are central slices along the z-axis imaged by confocal microscopy. 12HA::TSR1 (red) and TbNup53b::GFP (green) signals closely overlap within the nucleoplasm. White squares highlight the localization of TbNup53b::GFP within the vicinity of the nucleolar periphery. DAPI was used to visualize DNA (blue). Scale bars: 2 μm. (C) Following image acquisition using confocal microscopy, ImageJ software was used to calculate the percentage colocalization (top), Pearson’s correlation coefficients (bottom), and Mander’s correlation coefficients (bottom) for the following: TbNup53b::GFP and 12HA::TSR1; TbNup98::GFP and 12HA::TSR1; and TbNup98::GFP and TbNup110::HA. Twenty cells were analyzed for each condition. From left to right, the dot plots display the percentage localization and Manders coefficients for TbNup53b on TSR1, TSR1 on TbNup53b, TbNup98 on TbNup110, TbNup110 on TbNup98, TbNup98 on TSR1, and, finally, TSR1 on TbNup98. The high percentage colocalization and positive correlation coefficients suggest that TbNup53b and TSR1 signals exhibit a degree of overlap within the nucleoplasm.
Additionally, a strong interaction with a serine-arginine–rich protein, Trypanosoma SR-1 (TSR1) was detected. TSR1 is orthologous to yeast cis-spliceosomal SR proteins, and yeast three-hybrid demonstrates interactions between TSR1 and the spliced leader (SL) RNA of trypanosomes (Ismaïli et al., 1999), implying a role for TSR1 in splicing. In mammals, SR proteins are components of the splicing machinery, responsible for recruitment of spliceosomal components to mRNA 3′ splice sites (Gravely, 2000). SR proteins are typically present in multiple phosphorylation states that dictate functionality within the nucleoplasm; TSR1 is present as a 30- to 32-kDa doublet in bloodstream form cells and is additionally found as a 43-kDa protein present in all developmental stages, suggestive of extensive post-translational modification (Ismaïli et al., 1999; Gupta et al., 2014). The presence of a single 43-kDa band corresponding to TSR1 in the TbNup53b pull down suggests that TbNup53b interacts with a restricted TSR1 isoform subset. Confocal microscopy revealed that TSR1 displays a speckled appearance closely overlapping with TbNup53b::GFP (green) within the nucleoplasm (Figure 3B). However, distinct from TbNup53b, TSR1 was absent from both the nucleolar and nuclear periphery, indicating that their interaction is spatially restricted.
As the nucleoplasmic distribution of TbNup53b::GFP and TSR1 was quite extensive, we quantified colocalization to exclude stochastic overlap from random juxtaposition. We calculated the colocalization of TbNup53b and TSR-1 in confocal z-slices of 20 trypanosome nuclei (Figure 3C, top). As positive and negative controls, TbNup98::GFP/TbNup110::HA12 and HA::TSR1/TbNup98::GFP were compared; TbNup98 is exclusively located at the NPC, similarly to the nuclear basket nucleoporin TbNup110 (deGrasse et al., 2009). For each pair the proportion of colocalization of one signal was tested independently against the other. Colocalization between TbNup53b and TSR1 was 70–90%, suggesting that these two proteins closely overlap; by comparison, TbNup98::GFP/TbNup110::HA12 and HA::TSR1/TbNup98::GFP scored 70–95% and ∼20%, respectively. Object Pearson’s and Mander’s correlation coefficients were used to quantify the degree of colocalization (Figure 3C, bottom); both Pearson’s and Mander’s scores indicate high correlation for TbNup53b/TSR1 and TbNup98/TbNup110 and much lower for TbNup98 (Figure 3C, bottom). Taken together, the demonstrable protein–protein interaction between TSR1 and TbNup53b and the localization of TbNup53b::GFP and HA::TSR1 suggests that these proteins interact within the nucleoplasm. Multiple attempts to identify TbNup53b in a TSR pullout were unsuccessful, although this did identify multiple splicing and mRNA processing factors (Supplemental Table S2). In further support of the interaction, we also note that TSR was captured by pullout of TbNup96, a second component of the NPC inner ring and which interacts with TbNup53b (Obado et al., 2016).
TbNup53b is required for normal cell division
RNA interference (RNAi)-mediated knockdown was used to investigate TbNup53b function. The specificity and efficacy of knockdown was verified by quantitative reverse transcription–PCR (qRT–PCR), with ∼90 and 40% reduction to TbNup53b mRNA levels 24 h postinduction in PCFs and BSFs, respectively (Supplemental Figure S3A). Consistent with RNAi-target sequencing (RITSeq) (Alsford et al., 2011), a significant proliferative defect was observed in BSF but not PCF cells following TbNup53b depletion (Supplemental Figure S3B), and the BSF population failed to recover following 7 d induction. This is significant, as TbNup53b has a similar location in PCF and BSF cells and is constitutively expressed at the RNA level, based on multiple transcriptomes. Further, cell-cycle progression was altered in BSF but not PCF (Supplemental Figure S3C). In BSF cells, a significant decrease in cells where the kinetoplast (K) had divided, but not the nucleus (N) (2K1N), were recorded 72 h postinduction (Supplemental Figure S3C), and an increase in abnormal cell types was observed (1K2N, 0K1N, >2K2N), suggesting that TbNup53b is required for normal cytokinesis and positioning of the cytokinesis furrow following nuclear division. This is, however, most likely a secondary effect of disruption of nuclear function.
Using anti–NOG-1 and anti–β-tubulin (KMX) antibodies to highlight the nucleolus and mitotic spindle respectively, it was observed that replication and segregation of the nucleolus was perturbed in BSF cells, with two NOG-1 puncta often seen in knockdown interphase nuclei (Figure 4A). In addition, a significant increase in the frequency of mitotic knockdown cells displaying >2 nucleoli was observed, and the spindle was often undetectable in these cells with the KMX antibody (Figure 4B). By contrast, neither NOG-1 distribution nor spindle formation was affected in PCF knockdown cells (Supplemental Figure S4, A and B).
FIGURE 4:
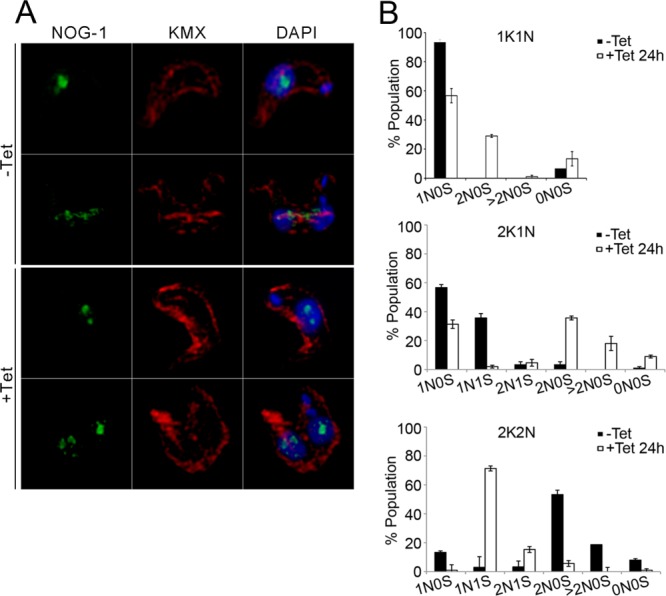
TbNup53b does not influence mitosis in PCFs. TbNup53b RNAi was induced in PCF cells. (A) Uninduced and induced cells were fixed and probed with anti–β-tubulin antibody (KMX) (red) and anti–NOG-1 antibody (green) to highlight the spindle microtubules and the nucleolar protein NOG-1, respectively. DAPI was used to visualize DNA (blue). Scale bar: 2 μm. (B) The number of nucleoli (NO) and presence of the mitotic spindle (S) were recorded in 1K1N (interphase), 2K1N (early mitosis), and 2K2N (post-mitotic) cells. Mean scores from triplicate experiments are shown (n = 300), with the nucleolus (green) and spindle (red) shown in the diagrams at right. The frequency of mitotic spindles and nucleolar division are indistinguishable throughout the cell cycle for uninduced and induced cells.
Together, these data suggest that TbNup53b depletion leads to errors in preparing for, and executing, mitosis and correct nucleolar partitioning in BSF cells, but that remarkably such a role is not readily apparent in the PCF, highlighting developmental variation in TbNup53b function as well as possible differences between life-stage mitotic mechanisms. Owing to the apparent complex roles of TbNup53b, both as a nucleoporin involved in nucleocytoplasmic transport and as an interactor with TSR1, it is not possible to decouple which of these processes is the primary mediator of the cell-cycle/mitosis defect in BSF cells. In both life-cycle stages, TSR1 maintains a speckled distribution when visualized with an anti-TSR1 antibody (kind gift of Etienne Pays, Université libre de Bruxelles, Bruxelles, Belgium) within the nucleoplasm and was indistinguishable between uninduced and TbNup53b knockdown cells (Supplemental Figure S5), indicating that TSR1 localization is not reliant on TbNup53b.
TbNup53b modulates mRNA processing
Monoallelic expression of VSG in BSFs is achieved by silencing all but one subtelomeric expression site (ES) locus (Navarro and Gull, 2001). In contrast, the genes encoding the PCF procyclin surface coat are transcribed at the nucleolar periphery and located within the core of megabase chromosomes 6 and 10 (Landeira and Navarro, 2007). The distinct localization of TbNup53b at both nuclear and nucleolar peripheries led us to ask if TbNup53b was present at the procylin locus in PCF cells. TbNup53b::HA distribution was recorded in three independent clones expressing GFP-LacI bound to the lac operator sequence inserted into the procyclin locus (Figure 5). Between 80% and 100% of cells showed colocalization between the procyclin locus and TbNup53b (Figure 5B); similarly, Mander’s correlation analyses revealed a positive overlap of the TbNup53b::HA signal with the lac operator-tagged procyclin locus in PCFs, suggesting that at least some of the intranuclear population of TbNup53b is associated with, or in the vicinity of, the active procyclin locus and hence a region of high transcriptional activity (Figure 5C).
FIGURE 5:
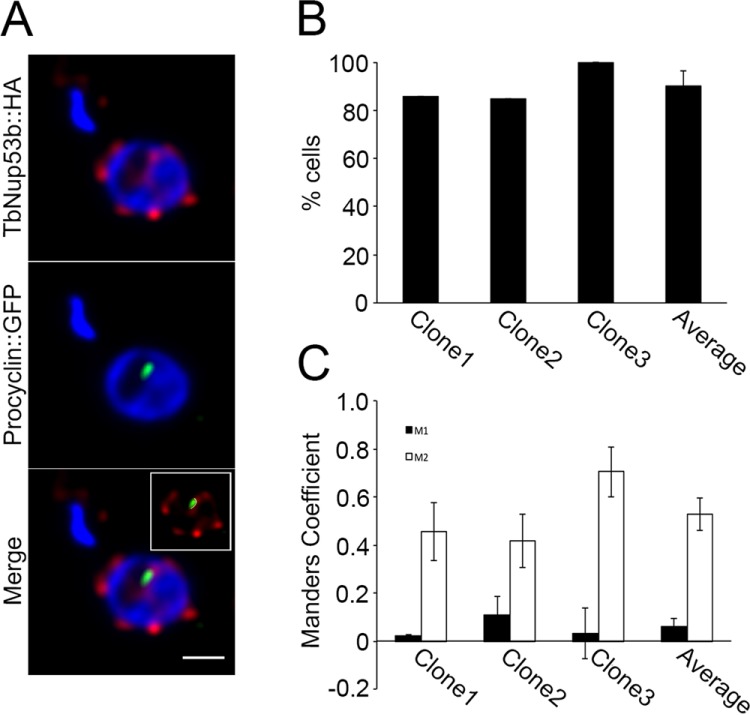
TbNup53b colocalizes with the procyclin locus at the nucleolar periphery. (A) Confocal immunofluorescence microscopy was used to visualize TbNup53b::HA12 (red) in cells expressing GFP-LacI bound to the lac operator sequence (green) inserted into the procyclin locus (34). DAPI was used to visualize DNA. Scale bar: 0.5 μm. (B) Percentage colocalization between TbNup53b::HA and procyclin loci signals at the nucleolar periphery were recorded in three independent clones using confocal immunofluorescence microscopy. Co-occurance of the two signals at the nucleolar periphery was in the region of ∼80–100% cells tested. (C) Manders correlation coefficients were used to measure the overlap of the two signals within the nucleoplasm. M1/M2 denotes the overlap of TbNup53b::HA12 signal on the GFP-procyclin and GFP-procyclin on TbNup53b::HA12, respectively. The high percentage colocalization and positive Manders correlation scores suggest that TbNup53b colocalizes with the procyclin locus at the nucleolar periphery.
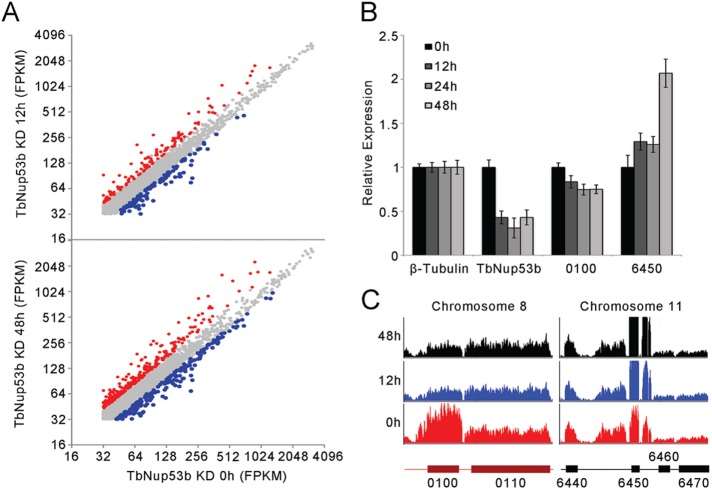
On the basis of TbNup53b and TSR1 location and their physical interaction, together with proximity to the procyclin locus, we considered that TbNup53b may play a role in regulating localized transcription and compared genome wide transcript levels between uninduced and knockdown cells using RNA-seq at 12 and 48 h postinduction (Supplemental Table S3). TbNup53b knockdown led to significant alterations to mRNA abundance (Figures 6A and 7 and Supplemental Table S3). Two of the most prominently altered transcripts, Tb927.8.6450 (inhibitor of cysteine peptidase, implicated in sensitivity to human serum lysis [Alsford et al., 2011]) and Tb927.8.0100 (TbMCP, a ClanMA(E) cowrin family metallopeptidase), were verified by qRT–PCR (Figure 6B). These were further validated by inspection of the raw reads, where specific changes to these two ORFs were observed. Significantly, adjacent ORFs were unaffected, suggesting an impact on mRNA stability/processing but not transcription. There is currently no evidence for a functional connection between these two genes or their products, and the former is lysosomal and the latter cytosolic.
FIGURE 6:
TbNup53b does not affect mRNA levels of entire PTUs. (A) Transcript abundances of individual genes in uninduced and TbNup53b knockdown cells based on FKPM. All FPKM’s of annotated transcripts were normalized by quantile normalization. Of 4824 genes, 163 genes were significantly up-regulated (red), and 144 genes were significantly down-regulated (blue) at 12 h postinduction. At 48 h postinduction, an additional 228 transcripts were down-regulated (blue) or 334 up-regulated (red) in expression. (B) The relative expression of the up-regulated (Tb427.08.6450) and down-regulated (Tb427tmp.02.0100; Tb427.03.3540) genes identified by RNA-seq were verified by qRT–PCR. Expression levels were normalized to β-tubulin. (C) RNAseq profiles of the up (Tb427.08.6450, designated 6450) or down-regulated (Tb427tmp.02.0100; Tb427.03.3540, designated 0100) genes in uninduced cells (red) and TbNup53b knockdown cells (12 h, blue; 48 h, black).
FIGURE 7:
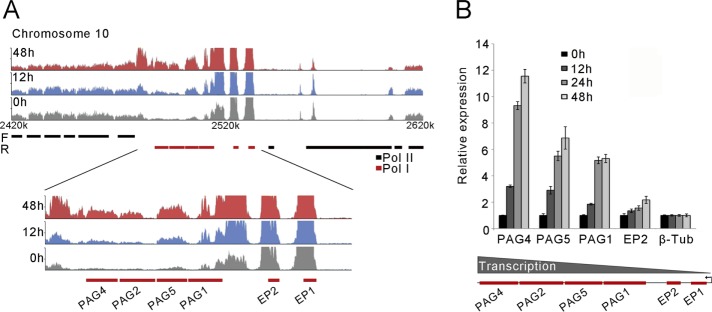
TbNup53b increases mRNA levels for genes transcribed from the procyclin locus. (A) RNA-seq profiles of uninduced (gray), induced 12 h (blue), and induced 48 h (red) at the procyclin region on chromosome 10 (top). Bottom: the expression profiles of genes contained within the pol I transcribed procyclin locus are significantly up-regulated following TbNup53b knockdown. (B) The relative expression of genes contained within the procyclin locus following TbNup53b knockdown were validated by qRT–PCR. The abundance of individual mRNAs appears to increase with distance from the promoter. Expression levels were normalized to β-tubulin.
TbNup53b knockdown impacts RNA PolI-transcribed surface gene cohorts
We rank-ordered the RNAseq data set by fold change at 48 h but considered only transcripts where there was also a change observed at 12 h and of the same sign. We considered a threefold up-regulation (85 genes) and 0.5-fold (180 genes) down-regulation as potentially significant changes and subjected these genes to annotation using the Basic Local Alignment Search Tool (BLAST)2Go.
Among the annotated set of genes within the up-regulated cohort were found VSG expression site associated genes (ESAGs) 1, 2, 3, 8 (annotated as LRRP protein), and 11; VSG itself; as well as invariant surface glycoproteins 64 and 65, together with procyclin-associated genes PAG1, 2, 4, 5, and EP procyclin itself (Supplemental Table S3). Most other hits were primarily metabolic enzymes with no obvious coherent pathway affiliation.
We also chose to investigate the procyclin locus present on chromosome 10, as reads from this locus can be uniquely mapped, unlike the ESAGs and VSGs. Four VSG-related procyclin-associated genes (PAGs) displayed highly significant increases in transcript level following TbNup53b knockdown (Figure 7A). We tested the expression levels of genes within the procyclin locus independently by qRT–PCR (Figure 7B) and found the number of transcripts increased with distance from the promoter and time of knockdown (Figure 7B, bottom). In contrast, mRNA levels of glu-pro (EP) procyclins and associated PAGs were not increase following knockdown in BSF cells (Supplemental Figure S8), indicating a further specific role for TbNup53b in PCF cells and fully consistent with the RNAseq data. Once more, the effect on the EP locus was highly specific, as similar changes were not detected with the gly-pro-glu-glu-thr (GPEET)/EP locus on chromosome 6 or for tandem repeat or single copy Pol II transcripts (Supplemental Figure S9). Western analysis indicated essentially no significant changes to procyclin protein levels (Supplemental Figure S10). It is likely that quality control mechanisms prevent additional procyclin expression to any great degree at the parasite surface (Engstler and Boshart, 2004).
Of the down-regulated cohort, the only obvious association was with small nucleolar RNAs (snoRNAs) and tRNAs, where a large cohort of each was identified. Both C/D and H/ACA snoRNAs were impacted. Little is known on snoRNA function in trypanosomes, although a connection to rRNA processing/modification has been described (Barth et al., 2008; Chikne et al., 2016), providing additional connections between the nucleolus and TbNup53B. Interestingly, snoRNAs are down-regulated in the procyclic stage compared with the mammalian stage, which correlates with the level of modification of the rRNA, and also snoRNA down-regulation correlates with transcriptional activation of the procyclin locus but not of ES genes.
Finally, the nucleolar periphery localization of a subpopulation of TbNup53b and possible impact of TbNup53b on this organelle led us to investigate a potential role for TbNup53b in SL-RNA transcription. SL-RNA is transcribed at the nucleolar periphery (Das et al., 2005; Alsford and Horn, 2011), and, using SNAP-42::cMyc as a marker to highlight this site, we observed no differences in the localization of SL-RNA transcription between uninduced and TbNup53b knockdown cells (Supplemental Figure S7). This, combined with no obvious alteration in the relative read counts for the 5′- or 3′-UTRs (unpubished data and Supplemental Figure S10), suggests that gross effects on RNA processing are not present.
DISCUSSION
The majority of nucleoporins remain anchored at the NPC throughout the cell cycle, reflecting roles in maintaining NPC structure and nucleocytoplasmic trafficking (Bapteste et al., 2005; deGrasse et al., 2009; Neumann et al., 2010). However, in common with metazoa and yeast, several T. brucei nucleoporins are also present within the nucleoplasm. We previously reported that TbNup92, a nuclear basket nucleoporin functionally similar to Mlp/Tpr, is recruited to the spindle, providing the first example of a moonlighting trypanosome nucleoporin (deGrasse et al., 2009). Given the divergence in transcriptional mechanisms, chromosomal segregation, and nuclear organization among trypanosomes, mammals, and yeast, it was unclear a priori if additional moonlighting functions exist or if they such functions are mediated by orthologous nucleoporins (duBois et al., 2012; Akiyoshi and Gull, 2014). In trypanosomes, control of mRNA level is complex but does not depend greatly on promoter activity. Multiple mechanisms, including turnover mediated by sequence elements within the 3′ untranslated region, recognition by specific RNA-binding proteins, and even position within a transcription unit, have all been shown to influence expression (Schwede et al., 2008, 2009; Utter et al., 2011). Additional mechanisms likely remain to be uncovered, and certainly the contribution that nuclear organization plays is only just beginning to emerge. Significantly, we recently reported that the later stages of RNA processing/export associated with the cytoplasmic face of the NPC in animals and fungi are absent or minimized in trypanosomes, which may be a reflection of reliance on essentially a single mode of trans-splicing, as well as the absence of an obvious barrier function for the nuclear basket.
In animals and fungi, several nucleoporins are implicated in transcriptional regulation. While a number of these apparently influence transcriptional processes from their location at the NE (Menon et al., 2005; Ahmed et al., 2010; Tous et al., 2011; Liang et al., 2013; Light et al., 2013), several FG-repeat nucleoporins, including Nup50, 98, and 153, are highly mobile within the nucleus with locations dependent on ongoing transcription (Griffis et al., 2002; Buchwalter et al., 2014). Nup50 also interacts with karyopherins and Ran (Buchwalter et al., 2014), and based on these observations it was suggested that these nucleoporins act to escort or ferry transcripts to the NPC. More recently, evidence emerged for a direct interaction between chromatin and additional FG-repeat nucleoporins, together with transcriptional regulation and potential formation of a sub-NPC complex (Morchoisne-Bolhy et al., 2015). Nup98 interacts with a large cohort of proteins, and specifically DHX9, an RNA helicase, where Nup98 expression is required for correct DHX9 targeting (Capitanio et al., 2017) Moreover, recent studies have indicated that Nup98 is a major player in modulation of transcription of developmental Hox genes and super-enhancer elements, and especially when fused to chromatin-remodelling genes as occurs in many cancer-related chromosomal translocations (Ibarra et al., 2016; Xu et al., 2016). Combined, these studies suggest very specific and high-order functions for FG Nups in control of cell fate. Here we investigated whether such processes were present in trypanosomes.
TbNup53b, a trypanosome FG-nucleoporin, has multiple locations, specifically the NPC, nucleoplasm, and nucleolar periphery, where it colocalizes with the highly active procyclin locus. At the NPC TbNup53B is a component of the central scaffold and has extensive interactions with additional Nups; it is unclear whether the nucleocytoplasmic and NPC pools are distinct or exchange with each other, and attempts to identify nuclear TbNup53B using chromatin immunoprecipitation provided nonspecific association with DNA. However, within the nucleoplasm TbNup53b interacts with the splicing component TSR1; a significant cooccurrence of both TbNup53b::GFP and 12HA::TSR1 signals within the nucleoplasm suggests that the two likely associate within the nuclear interior.
The splicing machinery of trypanosomes is organized into subnuclear compartments reminiscent of metazoan cells. Several trypanosome splicing components, including TSR1, TSR1P, RRM1, and Prp31, are organized into nuclear speckles (Manger and Boothroyd, 1998; Ismaïli et al., 2000; Liang et al., 2006). Consistent with a trans-splicing role for TSR1, yeast-three-hybrid demonstrates interactions between TSR1 and the spliced leader RNA (Ismaïli et al., 2000). Association of TSR1 and TSR1IP with nucleolar proteins such as NOG1, snoRNPs, and now TbNup53b combined with transcriptomic analyses provide support for a role of TSR1 and TSR1IP in RNA metabolism (Gupta et al., 2014). Significantly, evidence suggests an interaction with both NOG1 and snoRNAs for TbNup53b based on gene silencing, as both nucleolar segregation and steady-state levels of snoRNAs are impacted. How this is mediated precisely is unclear, but there is a clear interdependency among NOG1, snoRNAs, and rRNA processing, the latter of which is crucial to nucleolar structure. Further, how precisely this impacts the nucleolar peripheral procyclin locus remains incompletely understood. Regardless, it appears that TbNup53 is involved in control of important developmental genes, specifically the procyclin locus, and an interesting parallel with Nup98 in metazoa.
While in higher eukaryotes nucleoporins are clearly involved in transcriptional activation of extensive regions of the genome (Vaquerizas et al., 2010; Niepel et al., 2013), with the exception of the EP procyclin locus on chromosome 10 there was no evidence to support a role for TbNup53b in transcription of entire PTUs. These data are consistent with an apparent absence of regulation at the level of transcription initiation and with primary regulation residing at the posttranscriptional level in T. brucei. The mRNA abundance of genes contained within this locus increased with distance from the promoter, a finding remarkably similar to knockdown of the DNA binding proteins TbISWI and TbRAP1 that favor a compact chromatin structure and prevent elongation by RNA Pol I (Yang et al., 2009; Stanne et al., 2011). However, the finding that TbNup53b knockdown did not result in significant up-regulation of the normally silent GPEET procyclins (Vassella et al., 2000, Acosta-Serrano et al., 2001) suggests significant specificity within the control of procyclin loci. While the mechanism by which TbNup53b mediates these alterations to transcript abundance remains to be determined, one possibility is that TbNup53b functions in escorting nascent transcripts to the NPC for export or binding to cis-acting elements, both roles proposed for Nup98 in mammalian cells, and consistent with the extensive nuclear matrix distribution of the trypanosome protein (Griffis et al., 2002; Liang et al., 2013; Light et al., 2013).
In summary, we demonstrate that TbNup53b, in addition to being a bona fide nucleoporin, has distinct localization at the nucleolar periphery, coincident with the active procyclin locus. Significantly, these observations mirror various studies in metazoan cells, where roles for FG-repeat nucleoporins are also implicated, and where there is extensive distribution through the nucleus dependent on transcription. However, our data are distinct in that only a single FG-repeat nucleoprotein is involved, which is nonorthologous to the transcriptional mediating FG-repeat nucleoporins of mammals. These data indicate considerable evolutionary flexibility in the co-option of FG-repeat nucleoporins to other functions.
MATERIALS AND METHODS
Bioinformatics
The orthologues of TbNup53b and the other two trypanosome FG-nuceloporins that contain a coiled-coil region (TbNup53a and TbNup62) were identified in other kinetoplastids by BLAST using predicted protein and nucleotide databases (http://tritrypdb.org/tritrypdb/, www.genedb.org, NCBI GenBank, and an in-house database of kinetoplastid transcriptomes). The identification of orthologues in other eukaryotes was performed by HMMER (Finn et al., 2011) in the UniProtKB database using kinetoplastid sequence alignments as queries (Supplemental Table S1). Coiled-coil regions were predicted by PCOILS (http://toolkit.tuebingen.mpg.de/pcoils/) using protein alignments of related taxa. Protein sequences were aligned using Mafft (Katoh et al., 2005). The phylogenetic tree was constructed in PhyML 3.1 (Guindon et al., 2010) using the default settings and the robustness of individual branches evaluated by SH-like approximated likelihood ratio test and bootstrap after 100 iterations.
Cell culture
PCF T. brucei brucei MITat 1.2 (Lister 427) was grown as previously described (Brun and Schonenberger, 1979; Hirumi and Hirumi, 1994). Single marker bloodstream and procyclic culture form cell lines were used for expression of tetracycline-inducible constructs (Bastin et al., 1999; Wirtz et al., 1999). Expression of plasmid constructs was maintained using antibiotic selection at the following concentrations: G418 and hygromycin B at 1 µg/ml and phleomycin at 0.1 µg/ml for BSFs, G418 at 20 µg/ml, blasticidin at 10 µg/ml, phleomycin at 5 µg/ml, and hygromycin B at 25 µg/ml for PCFs. Cells were induced with tetracycline at a concentration of 1 µg/ml.
In situ genomic tagging
The TbNup53b, TbNup110, and TbNup98 open reading frames (ORF) were tagged using the pMOTag4G and pMOTag3H (Oberholzer et al., 2006) tagging vectors as a template. The following primers were used (all primer sequences are given in the 5′ to 3′ direction throughout): TbNup53bFOR: AATCGGAGGCGCCACCATATTTGGTGCGGGGACCTCTGCTGCTGACCCAAGGAAGACCCTGAACAAAACCGGTCGCTCAGGTACCGGGCCCCCCCTCGAG; TbNup53bREV: CAGGGGGACGCCGGCGGCGGAATCAAAGCAACCCTGCACACATGCACCGTTCGAATACCCTTGCGACGGAGGTATGGCGGCCGCTCTAGAACTAGTGGAT; TbNup98FOR: TGGGAATGCTTCAGCAAGTGGTGAAAAGAACAATGCTCCACGGAATCCCTTCTCATTTGGTGCCTCTTCTGGGAATGCTGGTACCGGGCCCCCCCTCGAG; TbNup98REV: ACTAAAGAAGGGTAGAAAACAAAGAAAACACCAAATAAGGTACCTGACGCAGCGGCAACACCACGTCGACTTGCTGGCGGCCGCTCTAGAACTAGTGGAT; TbNup110FOR: GAAAAGGCGATGCGACTACTGCACGTCAACAAGCAACTTGTGGAGAGAGTCAAAACCAGTCGAACTGAAGGAGAATCCCAGTCCAGTGGTACCGGGCCCCCCCTCGAG; TbNup110REV: GAGCATACGTACACGTACACGTACACGTACACGAATTGTCATACAACCTGACTAGCAGACGTAAGGCGCTACGAACCTTTACTGTGGTTCAAACAAAAATGGCGGCCGCTCTAGAACTAGTGGAT. For tagging the N-terminus of TbNup53b and TSR1, the first 500 bases of the ORF were PCR amplified using the following primers: TbNup53b_NtermFOR: CCGGGATCCATGATGTCAACTGCCCCAACG, TbNup53b_NtermREV: CCGGGTACCGTGTTCCGCTTGTATGAAGTT; TSR1NtermFOR: CGCGGATCCATGGATTCCAGAGACGGGAGTG, TSR1NtermREV: CGCGGTACCCTCCCGCGCTGTTGTTGTAGCC and cloned into a HindIII and BamHI restricted p2929 tagging vector (Kelly et al., 2007). p2929::53b and p2929::TSR1 were linearized with the unique restriction sites HpaI and BsmI, respectively, before transfection into PCFs. pNAT::SNAP42::12xcMyc (Alsford and Horn, 2011) was used to genomically tag the snRNA-activating protein complex subunit (SNAP42) ORF at the C-terminus (kind gift from Sam Alsford, London School of Tropical Medicine and Hygiene, London, UK).
Immunofluorescence microscopy
Antibodies were used at the following concentrations: rabbit anti-GFP 1:3000, rabbit anti–NOG-1 1:2000 (Park et al., 2001), goat anti-rabbit immunoglobulin G (IgG) Alexa Fluor 488 (Molecular Probes) 1:1000, mouse anti-HA (Santa Cruz Biotechnology) 1:1000, mouse anti-tubulin clone KMX-1 (Millipore) 1:3000, mouse anti-cMyc (Sigma) 1:1500, goat anti-mouse IgG Alexa Fluor 568 (Molecular Probes) 1:1000, and goat anti-mouse IgG Alexa Fluor 594 (Molecular Probes) 1:1500. Confocal images were acquired with a Leica TCS-NT confocal microscope with a 100×/1.4 numerical aperture objective and deconvolved using Huygens deconvolution software (Scientific Volume Imaging). A Nikon Eclipse E600 epifluorescence microscope equipped with a Hamamatsu ORCA charge-coupled device camera was used to acquire wide-field epifluorescence images and data captured using Metamorph (Universal Imaging Corp.). ImageJ (Rasband and Bright, 1995) and Fiji (Schindelin et al., 2012) software was used to calculate the percentage colocalization, object Pearson’s, and Mander’s correlation coefficients. The Object Pearson’s coefficient measures linear dependencies between two channels (excluding background) giving a value in the range of -1 to +1 whereby +1 represents a perfect correlation and -1 is a perfect negative correlation. The Mander’s coefficient was used to define the co-occurrence of the two signals, a score of 1 defining a complete overlap of signal and 0 denoting no signal overlap. Each signal was tested independently against the other. Images were further processed using Adobe Photoshop (Adobe Systems).
Real-time quantitative reverse transcriptase PCR
RNA was purified from cell lysates using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. Superscript III Reverse Transcriptase (Invitrogen) accompanied with RNase OUT (Invitrogen) was used to synthesize cDNA from 1 μg RNA. qRT–PCR using cDNA templates was carried out using iQ-SYBRGreen Supermix and a MiniOpticon Real-Time PCR Detection System (Bio-Rad). Results were analyzed using MiniOpticon software (Bio-Rad) with β-tubulin as a housekeeping gene to normalize RNA input. The following primer pairs were used: TbNup53bRNAiFOR: GCTAAGCAGGTTGTTGAGGC, TbNup53bRNAiREV: TTTATCCGAGAAACGGGATG; Tb427tmp.02.0100FOR: GAAAGCAGACCTCGACGTAA, Tb427tmp.02.0100REV: AAGTGCTCTCGGTGGTATTG; Tb427.07.190FOR: CCAAGGTGTTTGCTCTTGAT, Tb427.07.190REV: CGAGGAAGTTTCTCAGCATC; Tb427.08.6450FOR: AACCACAAGAACCACAGACG, Tb427.08.6450REV: TCGACGTGAATGTTGTAACG; Tb427.10.10210FOR: GTTGAGGATGCACTGGAAAG, Tb427.10.10210REV: TTGTCGCTTGTGTGTGAAAT; Tb427.10.10230FOR: AGCAAATTCTGCCGTTGATA, Tb427.10.10230REV: ACCCCACAATCCTTCAACTC; Tb427.10.10240FOR: ACACTGGAACTGGGAATACG, Tb427.10.10240REV: AACAGCTTCTCCTCTTGCAC; Tb427.10.10250FOR: CAGGACGAAGTTGAGCCTG, Tb427.10.10250REV: TGCAAGTGTCCTGTCGCC TbBetaTubulinFOR: CAAGATGGCTGTCACCTTCA; TbBetaTubulinREV: GCCAGTGTACCAGTGCAAGA.
Nucleic acid sequencing
For RNA-seq, RNA was extracted from PTT::RNAi53b cells following induction with tetracycline for 12 and 48 h, and cDNA was synthesized as described above. Sheared cDNA was sequenced by 76–base-pair paired-end Ilumina sequencing. Paired-end reads were mapped to the T. brucei 427 strain reference genome sequence using the Burrows-Wheeler Aligner. Mapped reads were further filtered by SAMtools (Li et al., 2009), allowing that the read is mapped in proper pair by a minimum 20 MAPQ (MAPing Quality). After filtering, the paired reads were aligned to annotated transcripts. Transcript abundances were calculated based on fragments per kilobase of transcript per million mapped reads (FPKM) (Trapnell et al., 2010). All FPKMs of annotated transcripts were normalized by quantile normalization. Low FPKM values (less than the median of FPKM values for all annotated transcripts) were removed. Transcripts that are annotated as rRNA or tRNA were excluded. To calculate the statistical significance for transcript level changes, the observed log-fold expression changes (FPKMtreat/FPKMwt) were tested against an empirical null distribution using the t test. The empirical null distribution of the log-fold changes was computed from all possible permutations of the samples as described (Budhraja et al., 2003). A p value for the comparison of the distributions of the positions of all genes and differentially expressed genes relative to the cistron transcription start sites were calculated based on the two-sample Kolmogorov–Smirnov test using the kstest2 MATLAB routine.
Protein interactome analysis
Immunoisolation of TbNup53b following cryomilling of parasites was used to determine the interactions between TbNup53b and other proteins. Full details of the methodology are published (Obado et al., 2016). Briefly, ∼1010 TbNup53b::GFP-tagged PCF cells were lysed by mechanical milling in a Retsch Planetary Ball Mill PM200 using liquid nitrogen cooling (Retsch, UK). Aliquots of powder were thawed in the presence of solubilization buffer (20 mM HEPES, pH 7.4, 250 mM NaCl, 2 mM MgCl2, 0.5% Triton, 0.5% deoxy Big CHAP), clarified by centrifugation, and TbNup53b::GFP was isolated using polyclonal llama anti-GFP antibodies coupled to Dynabeads (Invitrogen). Prior to fractionation by SDS–PAGE, affinity isolates of TbNup53b were reduced in 50 mM dithiothreitol and alkylated using 75 mM iodoacetamide. Following staining using GelCode Blue (Thermo Scientific), the protein bands were excised and identified using a SCIEX proTOF 2000 MALDI-TOF Proteomics Mass Spectrometer (Perkin Elmer). Peak lists were submitted to ProFound and searched against an in-house curated T. brucei database using data from GeneDB.
Supplementary Material
Acknowledgments
This work was supported by the Wellcome Trust (program grant 090007 to M.C.F.), the Medical Research Council (studentship to J.M.H.), and the National Institutes of Health (R21 AI096069, U54 GM103511 to B.T.C., J.D.A., and M.P.R.; U01 GM098256 to M.P.R.; P50 GM076547 to J.D.A.; P41 GM109824 to M.P.R., J.A., and B.T.C.; and P41 GM103314 to B.T.C.). M.C.F. is a Wellcome Trust Investigator.
Abbreviations used:
- BLAST
Basic Local Alignment Search Tool
- FG
phenylalanine–glycine
- NE
nuclear envelope
- NPC
nuclear pore complex
- qRT–PCR
quantitative reverse transcription PCR
- snoRNAs
small nucleolar RNAs.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0430) on February 26, 2018.
REFERENCES
- Acosta-Serrano A, Vassella E, Liniger M, Kunz Renggli C, Brun R, Roditi I, Englund PT. (2001). The surface coat of procyclic Trypanosoma brucei: programmed expression and proteolytic cleavage of procyclin in the tsetse fly. Proc Natl Acad Sci USA , 1513–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Brickner JH. (2010). DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol , 111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B, Gull K. (2014). Discovery of unconventional kinetochores in kinetoplastids. Cell , 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Horn D. (2011). Elongator protein 3b negatively regulates ribosomal DNA transcription in African trypanosomes. Mol Cell Biol , 1822–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsford S, Turner DJ, Obado SO, Sanchez-Flores A, Glover L, Berriman M, Hertz-Fowler C, Horn D. (2011). High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res , 915–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bapteste E, Charlebois RL, MacLeod D, Brochier C. (2005). The two tempos of nuclear pore complex evolution: highly adapting proteins in an ancient frozen structure. Genome Biol , R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Shalem B, Hury A, Tkacz ID, Liang X-H, Uliel S, Myslyuk I, Doniger T, Salmon-Divon M, Unger R, Michaeli S. (2008). Elucidating the Role of C/D snoRNA in rRNA Processing and Modification in Trypanosoma brucei. Eukaryotic Cell , 86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin P, MacRae TH, Francis SB, Matthews KR, Gull K. (1999). Flagellar morphogenesis: protein targeting and assembly in the paraflagellar rod of trypanosomes. Mol Cell Biol , 8191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Schonenberger M. (1979). Cultivation and in-votro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop , 289–292. [PubMed] [Google Scholar]
- Buchwalter AL, Liang Y, Hetzer MW. (2014). Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol Biol Cell , 2472–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budhraja V, Spitznagel E, Schaiff WT, Sadovsky Y. (2003). Incorporation of gene-specific variability improves expression analysis using high-density DNA microarrays. BMC Biol , 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. (2010). Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell , 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio JS, Montpetit B, Wozniak RW. (2017). Human Nup98 regulates the localization and activity of DExH/D-box helicase DHX9. Elife , e18825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. (2004). Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell , 427–439. [DOI] [PubMed] [Google Scholar]
- Chikne V, Doniger T, Rajan KS, Bartok O, Eliaz D, Cohen-Chalamish S, Tschudi C, Unger R, Hashem Y, Kadener S, Michaeli S. (2016). A pseudouridylation switch in rRNA is implicated in ribosome function during the life cycle of Trypanosoma brucei. Sci Rep , 25296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton CE. (2002). Life without transcriptional control? From fly to man and back again. EMBO J , 1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. (2002). Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol , 915–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels JP, Gull K, Wickstead B. (2010). Cell biology of the trypanosome genome. Microbiol Mol Biol Rev , 552–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Zhang Q, Palenchar JB, Chatterjee B, Cross GA, Bellofatto V. (2005). Trypanosomal TBP functions with the multisubunit transcription factor tSNAP to direct spliced-leader RNA gene expression. Mol Cell Biol , 7314–7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza CP, Hashmi SB, Nayak T, Oakley B, Osmani SA. (2009). Mlp1 acts as a mitotic scaffold to spatially regulate spindle assembly checkpoint proteins in Aspergillus nidulans. Mol Biol Cell , 2146–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. (2009). Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics , 2119–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. (2006). Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol , 7858–7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF Chait BT, Wozniak RW, Aitchison JD. (2005). The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol , 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- duBois KN, Alsford S, Holden JM, Buisson J, Swiderski M, Bart JM, Ratushny AV, Wan Y, Bastin P, Barry JD, et al (2012). NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol , e1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstler M, Boshart M. (2004). Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev , 2798–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res , W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell , 63–73. [DOI] [PubMed] [Google Scholar]
- Galy V, Olivo-Marin JC, Scherthan H, Doye V, Rascalou N, Nehrbass U. (2000). Nuclear pore complexes in the organization of silent telomeric chromatin. Nature , 108–112. [DOI] [PubMed] [Google Scholar]
- Gravely BR. (2000). Sorting out the complexity of SR protein functions. RNA , 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. (2002). Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell , 1282–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol , 307–321. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Chikne V, Eliaz D, Tkacz ID, Naboishchikov I, Carmi S, Waldman H, Ben-Asher, Michaeli S. (2014). Two splicing factors carrying serine-arginine motifs, TSR1 and TSR1IP, regulate splicing, mRNA stability, and rRNA processing in Trypanosoma brucei. RNA Biol , 715–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. (1994). Axenic culture of African trypanosome bloodstream forms. Parasitol Today , 80–84. [DOI] [PubMed] [Google Scholar]
- Holden JM, Koreny L, Obado S, Ratushny AV, Chen WM, Chiang JH, Kelly S, Chait BT, Aitchison JD, Rout MP, Field MC. (2014). Nuclear pore complex evolution: a trypanosome Mlp analogue functions in chromosomal segregation but lacks transcriptional barrier activity. Mol Biol Cell , 1421–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra A, Benner C, Tyagi S, Cool J, Hetzer MW. (2016). Nucleoporin-mediated regulation of cell identity genes. Genes Dev , 2253–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW. (2002). The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol , 807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. (2002). Chromatin boundaries in budding yeast: the nuclear pore connection. Cell , 551–562. [DOI] [PubMed] [Google Scholar]
- Ismaïli N, Pérez-Morga D, Walsh P, Cadogan M, Pays A, Tebabi P, Pays E. (2000). Characterization of a Trypanosoma brucei SR domain-containing protein bearing homology to cis-spliceosomal U1 70 kDa proteins. Mol Biochem Parasitol , 109–120. [DOI] [PubMed] [Google Scholar]
- Ismaïli N, Pérez-Morga D, Walsh P, Mayeda A, Pays A, Tebabi P, Krainer AR, Pays E. (1999). Characterization of a SR protein from Trypanosoma brucei with homology to RNA-binding cis-splicing proteins. Mol Biochem Parasitol , 103–115. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. (2010). Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell , 360–371. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. (1999). CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol , 764–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res , 511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly S, Reed J, Kramer S, Ellis L, Webb H, Sunter J, Salje J, Marinsek N, Gull K, Wickstead B, Carrington M. (2007). Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol , 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev NG, Ramey-Butler K, Cross GAM, Ullu E, Tschudi C. (2012). Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science , 1352–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Bannerman-Chukualim B, Ellis L, Boulde NE, Kelly S, Field MC, Carrington M. (2013). Differential localization of the two T. brucei poly(A) binding proteins to the nucleus and RNP granules suggests binding to distinct mRNA pools. PLoS ONE , e54004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S, Dorries J, Boysen B, Reidenbach S, Magnius L, Norder H, Thyberg J, Cordes VC. (2010). Protein Tpr is required for establishing nuclear pore- associated zones of heterochromatin exclusion. EMBO J , 1659–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. (2007). SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J , 4956–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Navarro M. (2007). Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei. J Cell Biol , 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP. (2009). The sequence alignment/map format and SAMtools. Bioinformatics , 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Liu L, Tschudi C, Michaeli S. (2006). Analysis of spliceosomal complexes in Trypanosoma brucei and silencing of two splicing factors Prp31 and Prp43. Mol Biochem Parasitol , 29–39. [DOI] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. (2013). Dynamic association of NUP98 with the human genome. PLoS Genet , e1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH. (2013). A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol , e1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manger ID, Boothroyd JC. (1998). Identification of a nuclear protein in Trypanosoma brucei with homology to RNA-binding proteins from cis-splicing systems. Mol Biochem Parasitol , 1–11. [DOI] [PubMed] [Google Scholar]
- Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, Andrews B, Santangelo GM. (2005). Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci USA , 5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morchoisne-Bolhy S, Geoffroy MC, Bouhlel IB, Alves A, Audugé N, Baudin X, Van Bortle K, Powers MA, Doye V. (2015). Intranuclear dynamics of the Nup107-160 complex. Mol Biol Cell , 2343–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro M, Gull K. (2001). A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature , 759–763. [DOI] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM. (2010). Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One , e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepel M, Molloy K, Williams RR, Farr JC, Meinema AC, Vecchietti N, Cristea IM, Chait BT, Rout MP, Strambio-De-Castillia C. (2013). The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell , 3920–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obado S, Brillantes B, Uryu K, Zhang W-Z, Ketaren NE, Chait BT, Field MC, Rout MP. (2016). Interactome mapping reveals the evolutionary history of the nuclear pore complex. PLoS Biol , e1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer M, Morand S, Kunz S, Seebeck T. (2006). A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol , 117–120. [DOI] [PubMed] [Google Scholar]
- Ouellette M, Papadopoulou B. (2009). Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol , 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palenchar JB, Bellofatto V. (2006). Gene transcription in trypanosomes. Mol Biochem Parasitol , 135–141. [DOI] [PubMed] [Google Scholar]
- Park JH, Jensen BC, Kifer CT, Parsons M. (2001). A novel nucleolar G-protein conserved in eukaryotes. J Cell Sci (Pt 1), 173–185. [DOI] [PubMed] [Google Scholar]
- Rasband WS, Bright DS. (1995). NIH Image—a public domain image processing program for the Macintosh. Microbeam Analysis , 137–149. [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol , 635–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods , 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A, Ellis L, Luther J, Carrington M, Stoecklin G, Clayton C. (2008). A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucleic Acids Res , 3374–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede A, Manful T, Jha BA, Helbig C, Bercovich N, Stewart M, Clayton C. (2009). The role of deadenylation in the degradation of unstable mRNAs in trypanosomes. Nucleic Acids Res , 5511–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanne TM, Kushwaha M, Wand M, Taylor JE, Rudenko G. (2011). TbISWI regulates multiple polymerase I (Pol I)-transcribed loci and is present at Pol II transcription. Eukaryotic Cell , 964–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tous C, Rondón AG, García-Rubio M, González-Aguilera C, Luna R, Aguilera A. (2011). A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J , 1953–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol , 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utter CJ, Garcia SA, Milone J, Bellofatto V. (2011). PolyA-specific ribonuclease (PARN-1) function in stage-specific mRNA turnover in Trypanosoma brucei. Eukaryot Cell , 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. (2010). Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet , e1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassella E, Den Abbeele JV, Bütikofer P, Renggli CK, Furger A, Brun R, Roditi I. (2000). A major surface glycoprotein of trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev , 615–626. [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GA. (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol , 89–101. [DOI] [PubMed] [Google Scholar]
- Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Chen CW, Chu SH, Brien GL, Park CY, et al (2016). NUP98 fusion proteins interact with the NSL and MLL1 complexes to drive leukemogenesis. Cancer Cell , 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. (2009). RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell , 99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.