Summary
In a previous whole exome sequencing of patients from 41 families with Hodgkin lymphoma, we identified two families with distinct heterozygous rare coding variants in POT1 (D224N and Y36H), both in a highly conserved region of the gene. POT1 D224N mutant did not bind to a single-stranded telomere oligonucleotide in vitro suggesting the mutation perturbs POT1’s ability to bind to the telomeric G-rich overhang. Human HT1080 cells expressing POT1 D224N and lymphoblastoid cells carrying Y36H both showed increased telomere length and fragility in comparison to wild type cells. This strongly suggests that mutant POT1 causes chromosome instability and may play a role in lymphomagenesis in these families.
Keywords: Hodgkin lymphoma, genetic analysis, genomic instability, telomere, POT1
Introduction
Classical Hodgkin lymphoma (HL) is a lymphoproliferative malignancy of B cell origin with an age-adjusted incidence in the United States of 2.6/100,000.(Howlander, et al 2016) Aetiological clues about HL are suggested by 1) the bimodal age distribution, with one peak occurring in young adulthood and a second peak after age 50 years, 2) elevated risk in males, 3) elevated risk in individuals with higher socioeconomic status and from smaller families, 4) the occurrence of Epstein–Barr virus in HL tumour cells and 5) strong familial risk.(Caporaso, et al 2009) We previously analysed data from registries in Scandinavia and found significant familial aggregation of HL.(Goldin, et al 2004) The human leucocyte antigen (HLA) region on chromosome 6 has been associated with HL(McAulay and Jarrett 2015) and other loci have been identified through genome-wide association studies [reviewed in (Kushekhar, et al 2014)] Identification of genes with major susceptibility effects has been more difficult although a few rare variants have been identified in single families.(Ristolainen, et al 2015, Saarinen, et al 2011, Salipante, et al 2009) We recently conducted whole exome sequencing (WES) in HL families and identified a mutation in KDR (kinase insert domain receptor) associated with disease in two families.(Rotunno, et al 2016)
Germline mutations in genes of the shelterin telomere protection complex have been identified in several familial cancers. Notably, germline and somatic mutations have been reported in the key telomere biology gene, POT1 (protection of telomeres 1). POT1 germline mutations have been found in high-risk families with melanoma, colorectal cancer, glioma, Li-Fraumeni like syndrome, and chronic lymphocytic leukaemia [reviewed in (Jones, et al 2016, Speedy, et al 2016)]. Both germline and somatic deleterious mutations tend to be concentrated in the oligonucleotide/oligosaccharide-binding (OB) folds of POT1, regions which are critical to its binding to the single-stranded telomeric DNA and telomere maintenance.(Gu, et al 2016) Here, we demonstrate that two HL families have mutations in POT1 and demonstrate functional consequences of POT1 mutations in vitro and in vivo.
Methods
We conducted WES in 41 families with at least two HL patients (21 with 3–5 HL patients sequenced and 20 with 2 HL patients sequenced) that were participants in an Institutional Review Board approved (NCI-02-C-0210) family study. Clinical, laboratory and informatics methods have been previously described(Rotunno, et al 2016) and details can be found in Appendix S1. Our familial patients had, on average, earlier age of onset and were more likely to be nodular sclerosis subtype (data not shown). Briefly, we sequenced patients and obligate carriers from our families and then filtered high quality variants to keep only non-synonymous variants segregating in patients and obligate carriers and found in less than 1% of European populations using publicly available databases. We used 600 in-house cancer-free control samples from the Prostate, Lung, Colon, and Ovarian (PLCO) and Cancer Prevention Study II (CPS-II) cohorts as another source of allele frequencies.
Results and Discussion
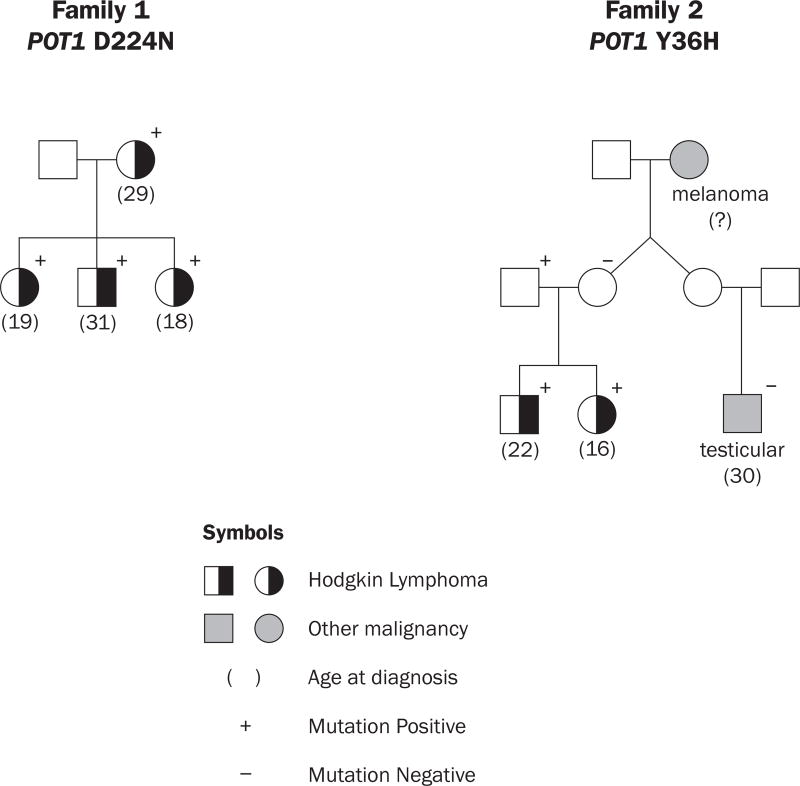
In our original study, we identified one mutation in KDR segregating with HL in two HL families.(Rotunno, et al 2016) Complete results can be found in Rotunno, et al (2016). In the course of our WES study, we identified one HL family (Figure 1a) in which all 4 patients were carriers of the POT1 variant encoding p.Asp224Asn (D224N), which had been previously reported in an American melanoma family.(Shi, et al 2014) Given the importance of POT1 in cancer, we looked for other POT1 mutations in our HL families. Both of the patients in another HL family (Figure 1b) were carriers of the POT1 variant encoding p.Try36His (Y36H). A first cousin with testicular cancer was not a mutation carrier. The patients all had teenage to young adult onset (age 18–31 years, average =22.5; Figure 1). In addition, all 6 patients had the nodular sclerosis subtype. The mutation status of carriers was verified by Sanger sequencing. Both mutations are rare and in highly conserved regions (Table SI). The POT1 D224N variant has a frequency of 7.5×10−5 in the Exac Non-Finnish European population and the POT1 Y36H variant was not seen in any database. Neither variant was present in our 600 laboratory control subjects. The unaffected father of the two patients is a mutation carrier, suggesting that the mutation is associated with incomplete penetrance for HL. Most programs predict the POT1 D224N variant to be deleterious, whereas predictions for the POT1 Y36H variant vary. No other rare coding variants among the shelterin complex genes (including TRF1, TRF2, TERF2IP, TINF2 and ACD) were identified. Because the POT1 D224N variant was also associated with melanoma in one family, and located in the OB2 region of the gene, and predicted to be deleterious, we conducted several functional studies of this mutation. POT1 Y36H and Y36N have both been reported in about 5% of CLL tumours (Landau, et al 2015, Ramsay, et al 2013), and functional studies have been reported for Y36N(Gu, et al 2016, Ramsay, et al 2013). Thus, we conducted a more limited study of POT1 Y36H
Figure 1.
Hodgkin Lymphoma Families with POT1 mutations
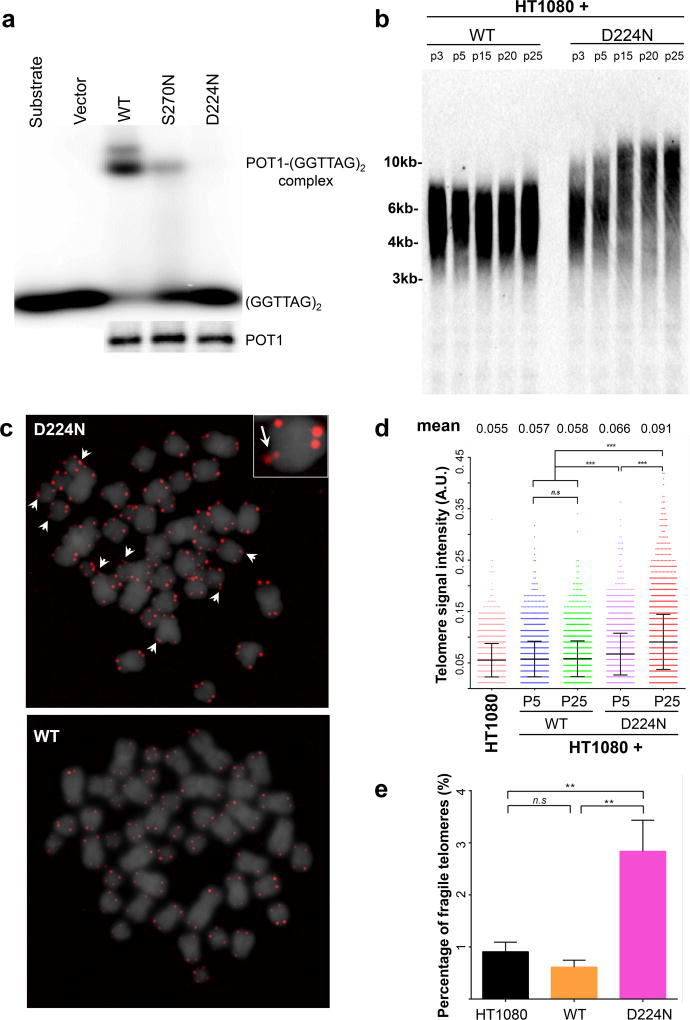
The impact of the POT1 D224N variant on telomere maintenance was determined by various assays, described previously(Shi, et al 2014) and in Appendix S1. We found that both the wild type and mutant POT1 alleles are expressed in the carriers’ peripheral blood mononuclear cells (PBMCs) (Figure S1). POT1 directly binds to single-stranded telomeric G-rich overhang via its N-terminal OB1- and OB2- fold domains. POT1 can also be recruited to double-stranded telomeres via interaction with TPP1 that occurs at its C-terminus. While POT1 p.D224N can be detected at telomeres in human cells (Figure S2), it does not bind to the single-stranded telomere oligonucleotide in vitro (Figure 2a). These data suggest that this N-terminal OB-fold mutation perturbs POT1’s ability to bind to the telomeric single-stranded G-rich overhang, but retains its localization to double-stranded telomeres via POT1-TPP1 interaction. To investigate the impact of the POT1 D224N variant on telomere length in vivo, we examined telomere length in PBMCs from the carriers and non-carriers of the variant by telomere restriction fragment analysis. Our PBMC data are not conclusive which could be due to prior treatment in patients and overall population variability. Nevertheless, the introduction of exogenous POT1 p.D224N into the human fibrosarcoma HT1080 cells leads to telomere lengthening during prolonged culture as shown by telomere restriction fragment analysis (Figure 2b) and telomere-fluorescence in situ hybridisation (FISH) (Figure 2c,d). The expression of exogenous POT1 is confirmed by immunofluorescence (Figure S2) and Western blot (Figure S3). Moreover, both the HT1080 cells and PBMCs expressing the affected POT1 residue show increased telomere fragility, compared to wild-type (WT) POT1 (Figure 2c,d), indicative of telomere replication defects in the mutant. Similarly, telomere-FISH revealed significantly increased telomere signal intensity and fragility in the POT1 Y36H carriers’ lymphoblastoid cells, compared to age-matched WT control (Figure S4). These results demonstrate that POT1 D224N and Y36H variants perturb POT1 function and thus telomere maintenance, including telomere over-lengthening and defective telomere replication in these patients.
Figure 2.
a. Electrophoretic mobility shift assay measures the binding of in vitro translated (i.v.t.) wild-type (WT) and mutant HA-hPOT1 to [p32]-labelled single-stranded telomere oligonucleotides (GGTTAG)2. The amount of i.v.t. hPOT1 for the electrophoretic mobility shift assay was determined by Western blot using the anti-Flag antibody.
b–e. Telomere length and Fragility of POT1 p.D224N and POT1 WT. For comparison purposes, binding to POT1 p.S270N which we reported previously (Shi, et al 2014), is shown.
b, d. Telomere restriction fragment analysis of HT1080 cells using Biotin-labelled telomere probe. Telomere length is demonstrated by telomere signal intensities at higher and lower molecular weights.
c, e. Telomere-FISH analysis of HT1080 cells expressing WT and mutant Flag-POT1 at indicated passages. c. Representative metaphase spread of a lymphoblastoid cell expressing either POT1 WT or p.D224N showing 4′,6-diamidino-2-phenylindole (DAPI) (grey) and telomere (red) fluorescence signals. Arrows indicate fragile telomeres (enlarged view in inset; 2.5× magnification). e. Quantitative measurement of telomere signal intensity in a plot displaying the distribution of telomeres with different signal intensities. Telomere signal intensity is depicted in arbitrary units (A.U.). Mean value denotes the mean telomere signal intensities in the indicated samples. Error bars are shown as mean ± standard deviation. Percentage of fragile telomeres was obtained from at least 30 metaphase spreads per sample. Error bar represents standard error of the mean. P values were obtained from the Wilcoxon rank-sum test. n.s.: not significant.
Of note, telomerase activity was comparable in HT1080 cell lines expressing POT1 WT and p.D224N (Figure S5). Also, the telomere dysfunction-induced damage foci (TIF) were not significantly increased in PBMCs and HT1080 cell lines expressing POT1 p.D224N (Figure S6), which supports the fact that carriers’ PBMCs can be sufficiently activated in culture for the above experiments.
In summary, we have identified two high-risk HL families with germline mutations in POT1. Both mutations are located in the OB folds of POT1, which binds to the telomeric 3’-overhang, and negatively regulate its access by telomerase.(Gu, et al 2016) Here we report the POT1 D224N variant in 3 siblings and a parent with HL. This may be a more general cancer susceptibility mutation since we previously reported 4/5 melanoma patients in one pedigree to be carriers of the variant.(Shi, et al 2014) Among other cancer cases sequenced in our laboratory, the mutation was detected in a patient with bladder cancer who had a family history of bladder cancer. Our functional studies show that the POT1 D224N variant is expressed in the PBMCs of carriers and is probably deleterious since it disrupts POT1 from binding with the telomeric overhang and leads to telomere lengthening and fragility. We showed that POT1 Y36H is also characterized by similar telomere changes. As mentioned above, this variant and another at the same site (Y36N) were reported to be mutated in CLL tumours (Ramsay, et al 2013) with Y36N showing functional consequences.(Gu, et al 2016)
Our study has limitations. To date, sequencing and other genetic studies of HL have identified only a small number of loci that may account for familial HL. Since these account for illness in only a few families, additional loci are likely to be discovered. In fact, other variants could be important in these two families. Tables SII and SIII list all other rare, exonic, high quality variants that were shared among patients within each family. Given that Family 2 only has two patients, there are many other variants shared among them. Our families do not show increased sharing among HLA loci although the experimental platform we used cannot evaluate this region adequately. Since only 2 HL families were identified with POT1 mutations, it is not possible to address questions about disease risk associated with POT1 mutations. Nevertheless, the finding of deleterious germline mutations in POT1 in two HL families shows that disruption of telomere maintenance may operate in the pathway to HL.
Supplementary Material
Acknowledgments
We acknowledge the contribution of Debra Silverman and Nathaniel Rothman on behalf of the New England Bladder Cancer Study and members of the NCI DCEG Cancer Sequencing Working Group: Bari Ballew, Mark H. Greene, Allan Hildesheim, Nan Hu, Jennifer Loud, Phuong Mai, Lisa Mirabello, Lindsay Morton, Philip R. Taylor, Geoffrey S. Tobias, and Guoqin Yu and members of the NCI DCEG Cancer Genomics Research laboratory: Meredith Yeager, Laurie Burdett, Joseph Boland, Sarah Bass, Salma Chowdhury, Michael Cullen, Casey Dagnall, Herbert Higson, Sally Larson, Kerry Lashley, Hyo Jung Lee, Wen Luo, Michael Malasky, Michelle Manning, Jason Mitchell, and David Roberson.
This work was supported by the Intramural Research Program of the U.S National Institutes of Health (NIH), National Cancer Institute (NCI), Division of Cancer Epidemiology and Genetics and National Institute on Aging (NIA).
Footnotes
AUTHORSHIP CONTRIBUTIONS
L.R.G., Y.L., S.A.S., X.R.Y., M.R. A.M.G., and M.T.L performed and interpreted genetic analyses. M.L.M, M.A.T., and N.E.C. directed clinical work for the patients and families. Y.L, C.S. and A.T. performed all POT1 and telomere experiments. K.J., A.V., A. H., B.Z., M.Y. conducted all WES and Sanger sequencing and associated bioinformatics. L.R.G., Y.L., S.A.S. and M.T.L. drafted the manuscript. All authors contributed to the final manuscript.
CONFLICTS OF INTEREST
The authors declare no competing financial interests.
References
- Caporaso NE, Goldin LR, Anderson WF, Landgren O. Current insight on trends, causes, and mechanisms of Hodgkin's lymphoma. Cancer J. 2009;15:117–123. doi: 10.1097/PPO.0b013e3181a39585. [DOI] [PubMed] [Google Scholar]
- Goldin LR, Pfeiffer RM, Gridley G, Gail MH, Li X, Mellemkjaer L, Olsen JH, Hemminki K, Linet MS. Familial aggregation of Hodgkin lymphoma and related tumors. Cancer. 2004;100:1902–1908. doi: 10.1002/cncr.20189. [DOI] [PubMed] [Google Scholar]
- Gu P, Wang Y, Bisht KK, Wu L, Kukova L, Smith EM, Xiao Y, Bailey SM, Lei M, Nandakumar J, Chang S. Pot1 OB-fold mutations unleash telomere instability to initiate tumorigenesis. Oncogene. 2016;36:1939–1951. doi: 10.1038/onc.2016.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlander N, Noone AM, Krapcho M, Miller D, Bishop K, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. Bioinformatics. Vol. 2014. Bethesda, MD: 2016. SEER Cancer Statistics Review, 1975–2013, National Cancer Institute. https://seer.cancer.gov/csr/1975_2013/ [Google Scholar]
- Jones M, Bisht K, Savage SA, Nandakumar J, Keegan CE, Maillard I. The shelterin complex and hematopoiesis. J Clin Invest. 2016;126:1621–1629. doi: 10.1172/JCI84547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushekhar K, van den Berg A, Nolte I, Hepkema B, Visser L, Diepstra A. Genetic associations in classical hodgkin lymphoma: a systematic review and insights into susceptibility mechanisms. Cancer Epidemiol Biomarkers Prev. 2014;23:2737–2747. doi: 10.1158/1055-9965.EPI-14-0683. [DOI] [PubMed] [Google Scholar]
- Landau DA, Tausch E, Taylor-Weiner AN, Stewart C, Reiter JG, Bahlo J, Kluth S, Bozic I, Lawrence M, Bottcher S, Carter SL, Cibulskis K, Mertens D, Sougnez CL, Rosenberg M, Hess JM, Edelmann J, Kless S, Kneba M, Ritgen M, Fink A, Fischer K, Gabriel S, Lander ES, Nowak MA, Dohner H, Hallek M, Neuberg D, Getz G, Stilgenbauer S, Wu CJ. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526:525–530. doi: 10.1038/nature15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAulay KA, Jarrett RF. Human leukocyte antigens and genetic susceptibility to lymphoma. Tissue Antigens. 2015;86:98–113. doi: 10.1111/tan.12604. [DOI] [PubMed] [Google Scholar]
- Ramsay AJ, Quesada V, Foronda M, Conde L, Martinez-Trillos A, Villamor N, Rodriguez D, Kwarciak A, Garabaya C, Gallardo M, Lopez-Guerra M, Lopez-Guillermo A, Puente XS, Blasco MA, Campo E, Lopez-Otin C. POT1 mutations cause telomere dysfunction in chronic lymphocytic leukemia. Nat Genet. 2013;45:526–530. doi: 10.1038/ng.2584. [DOI] [PubMed] [Google Scholar]
- Ristolainen H, Kilpivaara O, Kamper P, Taskinen M, Saarinen S, Leppa S, d'Amore F, Aaltonen LA. Identification of homozygous deletion in ACAN and other candidate variants in familial classical Hodgkin lymphoma by exome sequencing. Br J Haematol. 2015;170:428–431. doi: 10.1111/bjh.13295. [DOI] [PubMed] [Google Scholar]
- Rotunno M, McMaster ML, Boland J, Bass S, Zhang X, Burdett L, Hicks B, Ravichandran S, Luke BT, Yeager M, Fontaine L, Hyland PL, Goldstein AM, Group, N.D.C.S.W., Laboratory, N.D.C.G.R. Chanock SJ, Caporaso NE, Tucker MA, Goldin LR. Whole exome sequencing in families at high risk for Hodgkin lymphoma: identification of a predisposing mutation in the KDR gene. Haematologica. 2016;101:853–860. doi: 10.3324/haematol.2015.135475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarinen S, Aavikko M, Aittomaki K, Launonen V, Lehtonen R, Franssila K, Lehtonen HJ, Kaasinen E, Broderick P, Tarkkanen J, Bain BJ, Bauduer F, Unal A, Swerdlow AJ, Cooke R, Makinen MJ, Houlston R, Vahteristo P, Aaltonen LA. Exome sequencing reveals germline NPAT mutation as a candidate risk factor for Hodgkin lymphoma. Blood. 2011;118:493–498. doi: 10.1182/blood-2011-03-341560. [DOI] [PubMed] [Google Scholar]
- Salipante SJ, Mealiffe ME, Wechsler J, Krem MM, Liu Y, Namkoong S, Bhagat G, Kirchhoff T, Offit K, Lynch H, Wiernik PH, Roshal M, McMaster ML, Tucker M, Fromm JR, Goldin LR, Horwitz MS. Mutations in a gene encoding a midbody kelch protein in familial and sporadic classical Hodgkin lymphoma lead to binucleated cells. Proc Natl Acad Sci U S A. 2009;106:14920–14925. doi: 10.1073/pnas.0904231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Yang XR, Ballew B, Rotunno M, Calista D, Fargnoli MC, Ghiorzo P, Bressac-de Paillerets B, Nagore E, Avril MF, Caporaso NE, McMaster ML, Cullen M, Wang Z, Zhang X, Bruno W, Pastorino L, Queirolo P, Banuls-Roca J, Garcia-Casado Z, Vaysse A, Mohamdi H, Riazalhosseini Y, Foglio M, Jouenne F, Hua X, Hyland PL, Yin J, Vallabhaneni H, Chai W, Minghetti P, Pellegrini C, Ravichandran S, Eggermont A, Lathrop M, Peris K, Scarra GB, Landi G, Savage SA, Sampson JN, He J, Yeager M, Goldin LR, Demenais F, Chanock SJ, Tucker MA, Goldstein AM, Liu Y, Landi MT. Rare missense variants in POT1 predispose to familial cutaneous malignant melanoma. Nat Genet. 2014;46:482–486. doi: 10.1038/ng.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speedy HE, Kinnersley B, Chubb D, Broderick P, Law PJ, Litchfield K, Jayne S, Dyer MJ, Dearden C, Follows GA, Catovsky D, Houlston RS. Germline mutations in shelterin complex genes are associated with familial chronic lymphocytic leukemia. Blood. 2016;128:2319–2326. doi: 10.1182/blood-2016-01-695692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.