Abstract
Antibiotic resistant infections lead to 700,000 deaths per year worldwide1. The roles of phenotypically diverse subpopulations of clonal bacteria in the progression of diseases are unclear. We found that the increasingly pathogenic and antibiotic resistant pathogen, Acinetobacter baumannii, harbors a highly virulent subpopulation of cells responsible for disease. This virulent subpopulation possesses a thicker capsule and is resistant to host antimicrobials, which drive its enrichment during infection. Importantly, bacteria harvested from the bloodstream of human patients belong exclusively to this virulent subpopulation. Furthermore, the virulent form exhibits increased resistance to hospital disinfectants and desiccation, indicating a role in environmental persistence and the epidemic spread of disease. We identified a transcriptional “master regulator” of the switch between avirulent and virulent cells, and whose overexpression abrogated virulence. Further, the overexpression strain vaccinated mice against lethal challenge. This work highlights a phenotypic subpopulation of bacteria that drastically alters the outcome of infection, and illustrates how knowledge of the regulatory mechanisms controlling such phenotypic switches can be harnessed to attenuate bacteria and develop translational interventions.
Acinetobacter baumannii has become a major healthcare threat worldwide, responsible for both hospital and community acquired infections 2–6. These infections have become increasingly virulent 7–10 and exceedingly difficult to treat due to high levels of antibiotic resistance (63% of isolates in the U.S. are multidrug-resistant, and some isolates are even pan-resistant) 11–15. A. baumannii is also notoriously difficult to eradicate in hospital settings 2,14 and contamination of intensive care wards is a frequent problem 6,16,17. The remarkable ability of A. baumannii to persist in this environment is due to both its intrinsic resistance to commonly used disinfectants and its ability to survive long periods of desiccation 18–21. However, the molecular mechanisms controlling virulence, resistance to disinfectants, and desiccation tolerance remain poorly understood.
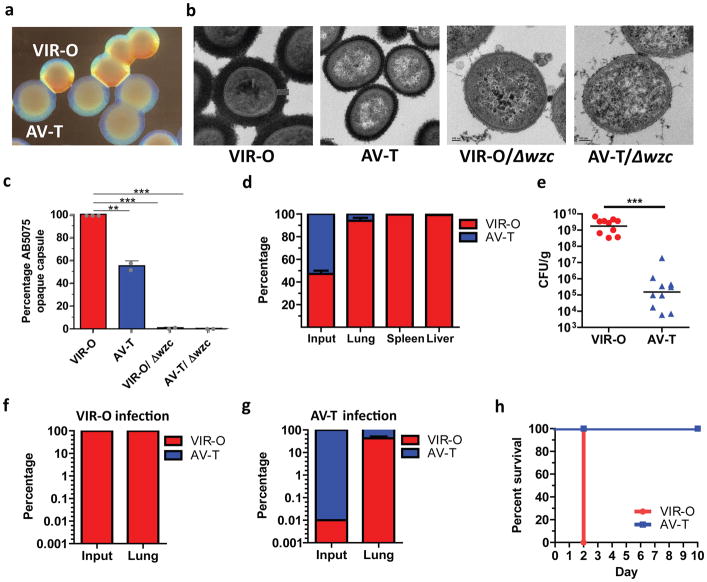
Bacteria can exhibit genotypic and/or phenotypic heterogeneity, the latter being traits expressed by some cells within a genetically homogenous population. Characterization of the highly virulent A. baumannii isolate AB507522 revealed that it exhibits phenotypic heterogeneity by rapidly interconverting between cells capable of forming opaque or translucent colonies23 (Fig. 1a). A single colony can be sequentially propagated between opaque and translucent states with switching frequencies of ~4–13% in 24 hr colonies (Supplementary Fig. 1) and 20–40% in 48 hr colonies (Supplementary Fig. 2). We hypothesized that differences in capsule contributed to the opacity differences between VIR-O and AV-T cells. Examination of cells by electron microscopy after staining for capsule with ruthenium red revealed the VIR-O cells produced a capsule with a 2-fold increased thickness compared to that of AV-T cells (Figs 1b and 1c). By comparison, VIR-O and AV-T cells missing the Wzc tyrosine kinase required for capsule synthesis (Δwzc) failed to produce any capsular material and exhibited a translucent phenotype that was more pronounced than the AV-T cells 24.
Figure 1. A highly virulent opaque (VIR-O) population is responsible for causing disease during in vivo pulmonary infection of mice.
(a), Representative A. baumannii strain AB5075 wild-type virulent opaque (VIR-O) and avirulent translucent (AV-T) colonies. (b) Strains were stained for capsule with ruthenium red and imaged by transmission electron microscopy. Representative images are shown for each strain. Scale bars in each image represent 100 nanometers. (c) Capsule abundance of the indicated strains was determined by capsule extraction and quantitation on SDS-PAGE gels stained with Alcian blue. Values were obtained from three biological replicates and error bars represent standard deviation of the mean. p-values (**p < 0.005; ***p < 0.0005) were determined using one-way ANOVA. (d), Mice were infected with a 1:1 mixture of VIR-O (red) and AV-T (blue) strains (n=5/group). At 24 hours post-infection, organs were harvested and plated to assess the percentage of VIR-O and AV-T cells present. (e), Mice were infected with VIR-O and AV-T (n=5/group). Presented data were pooled from two separate experiments and repeated at least 10 times. At 24 hours post-infection, lungs were harvested and plated for colony forming units (e). Bacteria recovered from the (f) VIR-O and (g) AV-T-infected lungs were assessed for the percentage of VIR-O and AV-T cells present, respectively. (h), Survival of mice infected with VIR-O and AV-T (n=5/group). This experiment was repeated 3 times. Error bars represent standard deviation of the mean in (d, f and g); Error bars represent geometric mean and significance was determined using a two-tailed Mann-Whitney test (***p < 0.0005) in (e).
The role of these two phenotypic subpopulations in the progression of disease was unclear. After intranasal inoculation of mice with a 1:1 mixture of the two types of cells, the cells that form opaque colonies (VIR-O; virulent, opaque) vastly outcompeted those that form translucent colonies (AV-T cells; avirulent, translucent) in all organs tested (lungs, spleen, liver) at 24 hours post-infection (h.p.i.) (Fig. 1d). Similar results were obtained in single infection experiments where mice infected with VIR-O cells harbored over 10,000-fold more bacteria in the lungs than mice infected with AV-T cells (Fig. 1e). In addition, 1,000-fold more bacteria were recovered in the spleens and livers of the VIR-O-infected mice than those infected with AV-T cells (Supplementary Fig. 3). Surprisingly, while bacteria recovered from VIR-O-infected mice remained in the VIR-O form (Fig. 1f), there was a greater than 3,000-fold increase in the frequency of VIR-O cells recovered from the AV-T-infected mice, as compared to the inoculum (0.01% VIR-O cells in the inoculum of AV-T-infected mice) (Fig. 1g). Furthermore, VIR-O cells derived from AV-T colonies regained virulence in mice (Supplementary Fig. 4), confirming that the attenuation of AV-T cells was not due to random mutations within the genome. These results revealed a strong selective pressure for the VIR-O population in vivo. In survival experiments, VIR-O-infected mice rapidly succumbed to disease by day 2, whereas AV-T infected-mice survived (Fig. 1h). VIR-O and AV-T variants exhibited similar growth rates in both rich and defined media, indicating that intrinsic growth rate differences do not account for the virulence defect of AV-T variants (Supplementary Fig. 5). Taken together, these data indicate that the VIR-O population is virulent and predominates during in vivo infection, whereas the AV-T population is unable to cause acute disease.
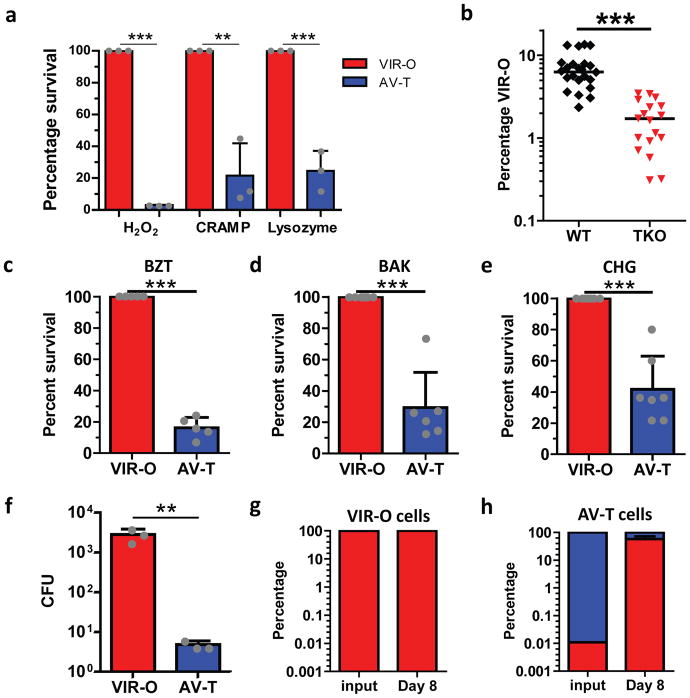
It was unclear which factors were responsible for the enrichment of the VIR-O population following in vivo lung infection (Fig. 1e). The lung is replete with innate immune antimicrobials including lysozyme, antimicrobial peptides such as CRAMP, and reactive oxygen species 25,26. We hypothesized that the VIR-O cells might be more resistant than AV-T to such antimicrobials. To test this, we conducted time-kill assays with hydrogen peroxide (which leads to the generation of reactive oxygen species), lysozyme, and CRAMP. After 1 hour of treatment with each of the three antimicrobials, VIR-O cells vastly outnumbered AV-T cells (Figure 2a). Moreover, VIR-O cells also outnumbered AV-T after treatment with LL-37, the human orthologue of CRAMP (Supplementary Fig. 6). These data highlight that VIR-O cells are more resistant than AV-T cells to diverse innate immune antimicrobials.
Figure 2. The host antimicrobial, hospital disinfectant and desiccation-resistant VIR-O cells are selected during in vivo infection.
(a), VIR-O (red) or AV-T (blue) were treated with H2O2, CRAMP or lysozyme, and percent survival relative to VIR-O was calculated. The reported values represent the mean of three replicates with standard deviations. Repeated experiments gave similar results. (b), Wild-type (WT, black) or triple knockout (TKO; red) mice lacking the gp91 subunit of the NADPH oxidase, lysozyme and CRAMP were infected with AV-T (n= 4 to 8/group). At 8 hours post-infection, lungs were harvested and plated to assess the percentage of VIR-O cells present. Presented data were pooled from three separate experiments and repeated at least 5 times for a total of 18–23 mice/per group. (c–e), VIR-O or AV-T was treated with the indicated amounts of disinfectants: (c) benzethonium chloride (BZT) 0.01%, (d) benzalkonium chloride (BAK) 0.004% and (e) chlorhexidine gluconate (CHG) 0.008%, and percent survival relative to VIR-O was calculated. Presented data were pooled from three separate experiments and a total of 5 replicates were used for (c), 6 replicates for (d) and 7 replicates for (e). (f–h), VIR-O and AV-T survival after desiccation. (f), Bacteria were rehydrated and plated on day 8 of desiccation to determine viability. Values represent the mean of three replicates. Recovered bacteria from the (g) VIR-O cells and (h) AV-T cells were assessed for the percentage of VIR-O and AV-T cells present. Error bars represent standard deviation of the mean and Student’s two-tailed t-test (**p < 0.005; ***p < 0.0005) in (a) and (c–h); Error bars represent geometric mean and a two-tailed Mann-Whitney test was to determine significance (***p < 0.0005) in (b).
To test whether these host defenses contributed to the enrichment of the VIR-O subpopulation during in vivo infection, wild-type (WT) mice and triple knockout (TKO) mice lacking a functional NADPH oxidase (cybb−/−; required for the production of reactive oxygen species), CRAMP (cramp−/−), and lysozyme (lysM−/−) were infected with an inoculum of AV-T cells. At 8hr post-infection, lungs from WT mice harbored a higher percentage of VIR-O cells than those from TKO mice (Fig. 2b). This was despite the overall levels of bacteria being higher in TKO mice than WT mice (Supplementary Fig. 7). These data indicate that reactive oxygen species, CRAMP, and lysozyme specifically contribute to the enrichment of the virulent VIR-O subpopulation during early infection. However, since the VIR-O population is still enriched in the TKO mice, this indicates that other host factors are involved as well.
Since the VIR-O subpopulation displayed increased resistance to host-derived antimicrobials, we hypothesized that the cells might also exhibit resistance to hospital disinfectants. The VIR-O and AV-T subpopulations were tested for sensitivity to three of the most commonly used disinfectants; benzethonium chloride (BZT), benzalkonium chloride (BAK) and chlorhexidine gluconate (CHG) and the VIR-O cells displayed increased resistance to all three agents compared to the AV-T cells (Fig. 2c–e). In addition to resistance to disinfectants, desiccation resistance is a major contributor to the persistence of A. baumannii in the hospital environment. Following eight days of desiccation, VIR-O cells survived better on dry surfaces compared to AV-T cells (Fig. 2f). Moreover, while viable bacteria recovered from VIR-O cells remained in the VIR-O form (Fig. 2g), there was a greater than 5,000-fold increase in the frequency of VIR-O cells recovered from the desiccated AV-T cells (Fig. 2h). Taken together, these data link increased virulence and environmental persistence (resistance to hospital disinfectants and desiccation) to a single phenotypic subpopulation of bacterial cells.
To elucidate factors controlling the virulent VIR-O and avirulent AV-T subpopulations, we performed genome-wide transcriptional profiling using RNA-seq. This revealed that a predicted TetR-type transcriptional regulator, ABUW_1645 (or “1645”), was among the most differentially expressed regulatory genes between the VIR-O and AV-T subpopulations (Supplementary Table 1). qRT-PCR confirmed that the AV-T population expressed 60-fold higher levels of 1645 as compared to VIR-O cells (Supplementary Fig. 8). Intriguingly, ABUW_1645 expression was reduced significantly as AV-T cells gradually switched to VIR-O (Supplementary Fig. 9a), whereas its expression level in the opaque cells remained low (Supplementary Fig. 9b). An in-frame deletion of ABUW_1645 did not alter the rate of VIR-O to AV-T switching, but the rate of AV-T to VIR-O switching was increased 18-fold relative to wild-type, indicating a role for ABUW_1645 in maintaining the AV-T state (Supplementary Fig. 10).
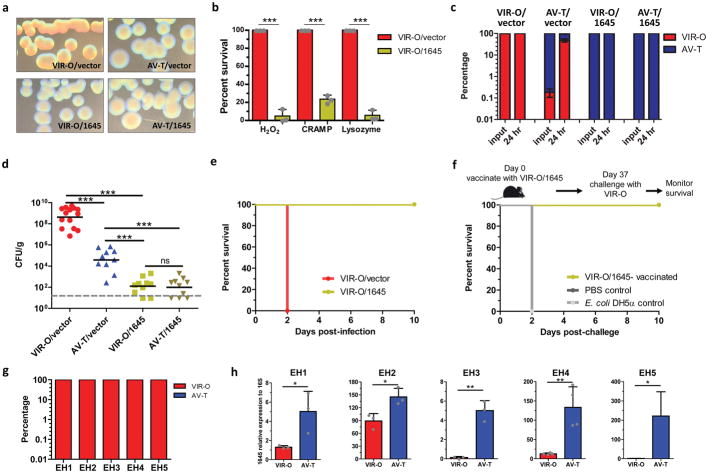
Strikingly, overexpression of 1645 in VIR-O cells (VIR-O/1645) led to a translucent colony morphology, and a complete inability to switch back to VIR-O (Fig. 3a). AV-T cells overexpressing 1645 (AV-T/1645) were similarly “locked”, and unable to switch to VIR-O cells (Fig. 3a). Transcriptional profiling of VIR-O/vector and VIR-O/1645 cells revealed that 1645 controlled ~70% of the genes differentially expressed between the VIR-O and AV-T cells (Supplementary Table 1). Taken together, these data identify 1645 as a key regulator of the VIR-O/AV-T high frequency phenotypic switch.
Figure 3. ABUW_1645 is a global regulator in mediating phenotypic switching, virulence and resistance to host defenses.
(a), Representative colonies of VIR-O or AV-T cells overexpressing ABUW_1645 or with empty vector. (b), VIR-O/vector (red) and VIR-O/1645 (green) were treated with lysozyme, H2O2 or CRAMP, and percent survival relative to VIR-O/vector was calculated. The reported values represent the mean of three independent replicates with standard deviations. Repeated experiments gave similar results. (c, d), Mice were infected with VIR-O/vector (red), AV-T/vector (blue), VIR-O/1645 (green) or AV-T/1645 (gold) strains (10 mice/group). At 24 hours post-infection, lungs were harvested and plated for colony forming units (d) and assessed for the percentage of VIR-O and AV-T cells present (c). (e), Survival of mice infected with VIR-O/vector and VIR-O/1645 (n=5/group). This experiment was repeated two times with identical results. (f), Survival of VIR-O/1645 and E. coli-vaccinated mice after lethal challenge (n=5/group). This experiment was repeated two times with identical results. (g) Blood cultures from A. baumannii-infected patients were plated directly on 0.5× LB agar plates to assess the percentage of VIR-O and AV-T cells present. (h) For each isolate, AV-T variants were isolated from the VIR-O colonies and the expression of ABUW_1645 was determined in each variant by quantitative real-time PCR. Data represents the mean from three replicates and error bars represent standard deviations of the mean. Student’s two-tailed t-test (*p < 0.05; **p <0.005; ***p < 0.0005) was used in (b and h). A two-tailed Mann-Whitney test was used in (d)(***p < 0.0005). ns, not significant.
Previously identified mutations that either increased (ompR)27 or decreased (arpB)28 the rate of VIR-O to AV-T switching did not alter ABUW_1645 expression (Supplementary Fig. 11a). Furthermore, the 1645 deletion did not alter the hyper-switching phenotype of an ompR::Tc mutant (Supplementary Fig. 11b). In addition, RNA-Seq data indicated that the expression of ompR or arpB was not altered by 1645 overexpression (Supplementary Table 1). Therefore, ABUW_1645 regulates phenotypic switching by a pathway separate from that regulated by OmpR or ArpB.
Overexpression of 1645 in VIR-O cells completely reversed resistance to host antimicrobials (Fig. 3b), disinfectants (Supplementary Fig. 12a–b) and desiccation (Supplementary Fig. 12c–d). In single infection experiments, VIR-O cells were recovered from mice infected with either VIR-O/vector or AV-T/vector cells (Fig. 3c). In contrast, only AV-T cells were recovered from mice infected with 1645 overexpression strains (Fig. 3c). This was associated with drastic attenuation of both VIR-O/1645 and AV-T/1645 strains, which exhibited a 7-log reduction in bacterial levels in the lungs relative to VIR-O/vector-infected mice (Fig. 3d). Furthermore, since the 1645 overexpression strains are even less virulent than AV-T cells, these data indicate that the residual colonization of mice during AV-T infection is in fact due to VIR-O cells, which become enriched during infection. This strongly indicates that AV-T cells do not have the ability to replicate and/or survive during in vivo infection, whereas VIR-O cells are responsible for causing disease. In agreement with these findings, infection of mice with the 1645 deletion mutant led to the recovery of VIR-O cells and this strain was not attenuated compared to wild-type (Supplementary Fig. 13), as was the 1645 overexpression strain. VIR-O/1645 bacteria were unable to cause a lethal infection, as compared to VIR-O/vector cells, which caused a rapidly lethal infection (Fig. 3e). These data highlight 1645 as a critical regulator of virulence as well as traits associated with persistence in the hospital environment.
Since the VIR-O/1645 strain was completely attenuated in vivo, we tested whether it might protect mice against subsequent lethal challenge with the virulent VIR-O/vector cells. While control mice vaccinated with PBS or Escherichia coli K-12 rapidly succumbed to challenge on day 2, mice vaccinated with VIR-O/1645 were completely protected (Fig. 3f). These data demonstrate that a strain engineered to be locked to produce only avirulent cells can effectively provide protection against otherwise lethal infection, serving as a live attenuated vaccine against A. baumannii.
Because VIR-O cells clearly predominate during experimental in vivo murine infection, as well as in conditions that simulate stresses encountered in the hospital environment, we set out to test their abundance in samples from hospitalized human patients. Blood cultures from 5 patients with systemic A. baumannii infections were directly plated on agar and only VIR-O cells were detected from these clinical isolates (Fig. 3g). Therefore, similarly to experimental infections in mice, VIR-O cells vastly predominate in human infections. In addition, AV-T variants could be selected from the VIR-O cells of each clinical isolate, indicating that this phenotypic switch is present in A. baumannii isolates other than AB5075. Moreover, the expression of ABUW_1645 was significantly higher in the AV-T subpopulation from each isolate, relative to the corresponding VIR-O subpopulation, further supporting the key role of this regulator in the VIR-O to AV-T switch (Fig. 3h). These results provide clinical evidence from diverse isolates that highlights the critical role of the phenotypic subpopulation of VIR-O cells in human disease.
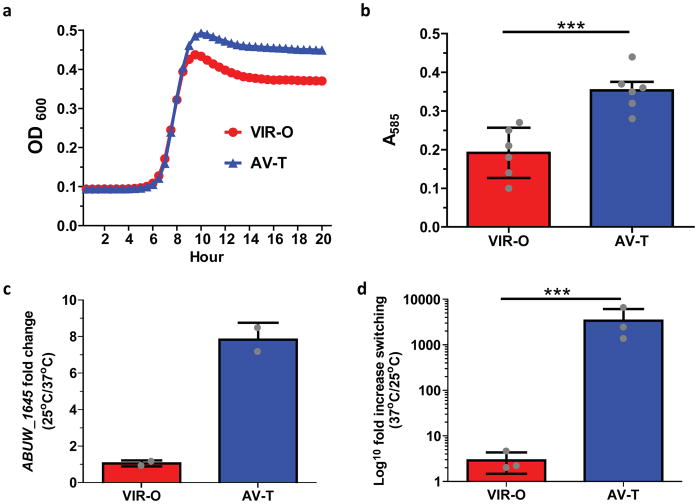
A remaining question is what advantages does the AV-T subpopulation confer to A. baumannii? A subset of genes upregulated in the AV-T cells are predicted to be involved in the catabolism of aromatic compounds (ABUW_0066, ABUW_0068, ABUW_0070), generation of free phosphate and sulfur from organic sources (ABUW_0904, ABUW_2921), iron storage (ABUW_3125) and nutrient transport (ABUW_0143, ABUW_3403, ABUW_1660). This raises the possibility that the AV-T subpopulation is better suited for natural environments outside the host, where nutrients are limited and bacteria may rely on the uptake and catabolism of atypical compounds for use as nutrients. Indeed, AV-T cells grew to higher yields in a nutrient poor media (Chamberlain’s) relative to VIR-O cells (Fig. 4a). Biofilm formation may facilitate colonization of natural environments, such as in soil and water, and AV-T cells were shown to be more proficient than VIR-O cells at biofilm formation at low temperature (25°C) (Fig. 4b). Furthermore, at low temperature outside the host, ABUW_1645 expression in AV-T cells was increased, and was subsequently reduced in cells grown at 37°C (Fig. 4c). Correspondingly, when AV-T cells were grown at host temperature (37°C) as would occur during infection, their rate of switching to VIR-O cells increased more than 3,400-fold (Fig. 4d). These data are in agreement with those from mouse infections (Fig. 1) and human samples (Fig. 3), in which VIR-O cells vastly outnumber AV-T cells. Taken together, these results suggest that AV-T cells are better suited to survive in certain environmental conditions, and their reduced levels of capsule may even render these cells resistant to bacteriophages29.
Figure 4. AV-T specific phenotypes.
(a) Growth curves of VIR-O and AV-T in Chamberlain’s minimal media. Values represent (b) Biofilm formation of VIR-O and AV-T cells grown for 24 hours at 25°C. Values represent the average of 6 replicates for each strain. Student’s two-tailed t-test was used to determine significance ***p < 0.0005) (c) Ratio of ABUW_1645 expression in VIR-O and AV-T cells at low (25°C) and high temperature (37°C). Values represent the averages of two independent biological replicates. (d) Phenotypic switching of VIR-O and AV-T cells at low (25°C) and high temperature (37°C) in LB media harvested at an optical density A600 of 1.6. Values represent three biological replicates. A two-tailed paired t-test (***p < 0.001) was used in (d). All error bars in panels (b–d) represent standard deviations.
This study reveals a phenotypic switch linking resistance to host antimicrobials, virulence and environmental persistence. The data highlight an unexpected role for specific host innate immune antimicrobials (CRAMP, lysozyme and H2O2) in enriching the virulent VIR-O subpopulation during murine infection. Our data clearly demonstrate that the VIR-O cells are responsible for causing disease in vivo. In addition to being the predominant cells recovered from infected mice, only VIR-O cells were found in the bloodstream of infected human patients, highlighting their critical role in virulence. It is therefore likely that bacterial cells shed into the environment by infected patients are in the VIR-O form. Interestingly, VIR-O cells also have an increased ability to resist disinfectants and desiccation, two stresses encountered in the hospital environment. This suggests that alarmingly, human infection selects for cells that are best-suited for environmental persistence, and that likewise, the bacterial cells persisting in the environment are those that are most virulent when transmitted to future human hosts. Therefore, the VIR-O subpopulation likely underlies a worrisome cycle present in hospital wards, accounting for the difficulty in eradicating A. baumannii as well as the high virulence of many of these infections.
Phenotypic switches that control virulence have been identified in both bacterial and fungal pathogens and this may also represent a bet-hedging strategy where the avirulent subpopulation has a selective advantage outside the host 30–35. Interestingly, a phenotypic switch in Photorhabdus luminescens has similarities to A. baumannii, where a virulent subpopulation is more resistant to host antimicrobials34. In addition, in the eukaryote Candida albicans, a phenotypic switch that controls the white-opaque colony transition also has parallels with the A. baumannii system, including subpopulation differences in virulence, cell shape and biofilm formation35.
Overexpression of 1645 completely converted VIR-O cells to AV-T cells, and reversed the virulence and environmental persistence attributes of VIR-O cells (Fig. 3). Thus, while the VIR-O/AV-T phenotypic switch is critical for facilitating A. baumannii virulence, our data suggest that it could be subverted and turned into an “Achilles’ heel”. Small molecules that drive cells into the AV-T form would render cells avirulent and could represent a potential therapeutic for the treatment of infections. Furthermore, we demonstrate that the highly attenuated 1645 overexpression strain is a promising live-attenuated vaccine candidate against A. baumannii, exerting striking protection against lethal challenge. These findings highlight how knowledge of phenotypic traits endowed by subpopulations of cells can be harnessed to facilitate translational interventions.
Materials and Methods
Bacterial strains
A. baumannii strain AB5075 was used in this study. All experiments reported in study were done using the same glycerol stock of VIR-O and AV-T cells that were at least 99.9% pure. For each mouse infection, the bacterial inoculum was checked to verify that each culture maintained the VIR-O or AV-T phenotype. Escherichia coli strain EC100D (Epicentre) was used for all cloning experiments.
Methods to distinguish VIR-O and AV-T variants
The ability to distinguish between VIR-O and AV-T variants requires the use of stereo (dissecting) microscope that illuminates the plates from below with a light source with an adjustable angle (oblique lighting). A correct light angle is required to observe the opacity differences and an improper light angle can make O colonies look like T and vice-versa. Each microscope should be standardized with VIR-O and AV-T variants to set the proper angle. VIR-O and AV-T stocks will be made available upon request. Colonies should be grown on agar plates composed of 5 g tryptone, 2.5 g yeast extract, 2.5 g sodium chloride and 8 g agar per liter (0.5× LB/0.8% agar). The VIR-O and AV-T phenotypes can be observed on regular LB plates, but it is much harder to distinguish the variants. Importantly, the VIR-O and AV-T opacity phenotypes can only be accurately distinguished at high colony density (~100/plate). At low colony density, both variants will look like VIR-O. In addition, VIR-O colonies typically give rise to translucent sectors after 24 hours. AV-T colonies rarely sector to VIR-O, yet AV-T colonies can have up to 50% VIR-O cells at 48 hours.
Electron Microscopy
Colonies of bacteria were collected from culture plates and placed in 0.1 M sodium cacodylate (pH 7.4) buffered fixative that contained 2% paraformaldehyde, 2.5% glutaraldehyde, 0.075 grams of ruthenium red, and 1.55 grams of L-lysine acetate. After 20 minutes of fixation on ice, samples were centrifuged and washed twice in sodium cacodylate/ruthenium red buffer. Samples were then fixed for a second time for 2 hours with fixative that did not include the L-lysine acetate. Following two additional sodium cacodylate/ruthenium red buffer washes, samples were placed in 1% osmium tetroxide in sodium cacodylate/ruthenium red buffer for 1 hour at room temperature. The fixed bacterial samples were then washed, dehydrated through a graded ethanol series and placed in 100% ethanol. The samples were infiltrated with a mixture of Eponate 12 resin (Ted Pella, Inc.; Redding, CA) and propylene oxide before being placed in pure Eponate 12 resin overnight. All resin-infiltrated bacterial samples were polymerized for 48 hours in a 60°C oven. The resin-embedded bacterial samples were sectioned to 70–80 nm thick sections using a Leica UltraCut microtome (Leica Biosystems; Buffalo Grove, IL) and subsequently stained with 5% uranyl acetate and 2% lead citrate. The ultrathin sections were imaged with a JEOL JEM-1400 transmission electron microscope (JEOL Ltd; Tokyo, Japan) operated at 80 kV and equipped with a Gatan US1000 CCD camera (Gatan; Pleasanton, CA).
Mice
WT C57BL/6J mice were purchased from Jackson Laboratories and used at age 8–10 weeks; all experiments used age- and sex-matched mice. TKO deficient in the gp91 component of the NADPH oxidase, lysozyme and CRAMP, were derived by crossing cybb−/− (gp91, Jackson Laboratories), lysM−/− (lysozyme, generously provided by D. Portnoy, UC Berkeley) and cramp−/− (CRAMP; Jackson Laboratories) mice. Mice were housed under specific pathogen-free conditions at Yerkes National Primate Center, Emory University. All experimental procedures were approved by the Emory University Institutional Animal Care and Use Committee. Mouse sample size was determined based on previous studies that generated highly statistically significant results, while also minimizing the number of animals used (5 mice per group for the majority of experiments). Females were used for the majority of the animal experiments. Male mice were used in some experiments to match available knockout mice. Groups of mice with different genetic backgrounds had to be housed separately, precluding randomization and blinding.
Construction of an ABUW_1645 (TetR) expression vector
To generate an expression plasmid for the ABUW_1645 open reading frame, a 697-bp DNA fragment, which began 60 bp upstream from the predicted ABUW_1645 start codon and ended 82 bp downstream from the predicted ABUW_1645 stop codon, was amplified by PCR using chromosomal DNA from A. baumannii strain AB5075 as the template (Phusion Hot Start Polymerase; Thermo Scientific, Waltham, MA). Oligonucleotide primers (1645 Exp. 1.1; 5′-GAGTGACGGCATGTCTATCT-3′ and 1645 Exp. 2.2; 5′-CTTATAGCCATAAGTGGTAATTGAG-3′) were treated with T4 Polynucleotide Kinase (New England Biolabs, Ipswich, MA) to add 5′-phosphates prior to PCR amplification. The fragment was purified from an agarose gel slice and ligated (Fast-Link Ligase; Epicentre, Madison, WI) into pWH126636 that had been digested with ScaI (New England Biolabs) and subsequently treated with shrimp alkaline phosphatase (New England Biolabs) to dephosphorylate linearized vector. The ligation was transformed into E. coli Transformax EC100D competent cells (Epicentre) and plated on LB+Tet (10 μg/mL) plates, resulting in the expression vector pKT1645.
RNA isolation and RNAseq analysis
Cultures of A. baumannii strain AB5075 VIR-O cells harboring empty pWH1266 or pKT1645 and strain AB5075 AV-T cells harboring empty pWH1266 or pKT1645 were grown in LB+Tet (5 μg/mL) at 37°C with shaking to an OD600 ~0.75. The cells were harvested from cultures by centrifugation and RNA was isolated using a MasterPure RNA purification kit according to the manufacturer’s protocol (Epicentre). Contaminating DNA was removed by treatment with Turbo DNA-free according to the manufacturer’s protocol (Ambion, Waltham, MA). RNA concentration was quantified with a NanoDrop ND-1000 spectrophotometer. RNA purity was assessed with qRT-PCR analysis of clpX expression in samples with/out reverse transcriptase.
RNA sequencing and analysis
RNA samples were first depleted of ribosomal RNAs using the RiboZero rRNA Removal kit (Illumina). RNA libraries were prepared using the NEBNext® Ultra™ RNA library prep kit for Illumina (NEB) and run on a single multiplexed HiSeq4000 150PE lane (University of Maryland Genomics Resource Center). Paired-end Illumina libraries were mapped against the A. baumannii AB5075-UW genome using Bowtie aligner (v0.12.9) and differential gene expression was quantified by DESeq (v1.5.25) (University of Maryland Genomics Resource Center). The Fisher’s Exact Test (modified by DESeq) was used to calculate the p-values, which were adjusted for multiple-testing with the Benjamini-Hochberg method. Differentially expressed transcripts with a p-value of ≤ 0.05, FDR ≤ 0.05 and log2 fold change ≥ 1.7 were used in this study. Sequence reads were deposited at NCBI under Bioproject no. PRJNA400082 as BioSamples SAMN07562376, SAMN07562377, SAMN07562378, SAMN07562379, SAMN07562380, SAMN07562381, SAMN07562382, SAMN07562383 and SAMN07562384.
Mouse pulmonary infection models
Approximately 5 × 107 CFU (24 hour time point) and 5 × 108 CFU (8 hour time point) were administered per mouse for infections to quantify the bacterial load, and approximately 3 × 108 CFU were administered for survival experiments. For mouse infections, overnight standing bacterial cultures at room temperature were sub-cultured in LB broth and grown at 37°C with shaking to an OD600 ~0.15, washed and re-suspended in PBS. Fifty μL of bacterial inocula were inoculated intranasally (i.n.) to each mouse. Mice were anesthetized with isoflurane immediately prior to intranasal inoculation. At each time point, the mice were sacrificed and the lungs, spleen and liver were harvested, homogenized, plated for CFU on 0.5× LB plates.
Antimicrobial killing assays
An equal mixture of VIR-O and AV-T variants were grown to early-log phase in 2 mL LB broth, or LB with tetracycline 5 μg/mL for strains with plasmids. A 5 μL aliquot of each culture was mixed together in 250 μL of 0.1× tryptone/yeast extract (1 g tryptone, 0.5 g yeast extract/liter). Cells were mixed by vortexing and serial dilutions were plated on 0.5× LB agar plates to assess the initial CFU’s for both VIR-O and AV-T variants. Various antimicrobials were then added at the following final concentrations: CRAMP (10 μg/mL), lysozyme (10 mg/mL), LL-37 (15 μg/mL) and H2O2. (0.01%). Cells were treated for 1 hour and dilutions were plated on 0.5× LB agar plates to enumerate the surviving VIR-O and AV-T variants.
Desiccation assay
Overnight standing bacterial cultures at room temperature were sub-cultured in LB broth and grown at 37°C with shaking to an OD600 ~0.15, washed and re-suspended in PBS. Twenty-five μL bacterial inocula were desiccated on a 96-well flat bottom polystyrene plate at room temperature. At each time point following desiccation, wells were rehydrated in 100 μL sterile PBS for 30 min, then serially diluted and plated for CFU on 0.5× LB plates to enumerate the surviving VIR-O and AV-T variants.
Sensitivity to disinfectants
Overnight standing bacterial cultures at room temperature were sub-cultured in LB broth and grown at 37°C with shaking to an OD600 ~0.15. Nine hundred ninety microliters of culture were then added to tubes containing 10 μL of the appropriated disinfectants, BZT (Sigma-Aldrich), BAK (Sigma-Aldrich) and CHG (Sigma-Aldrich), respectively. Tubes were incubated at room temperature for 30 min. Cultures were then serially diluted and plated for CFU enumerations.
Construction of an ABUW_1645 deletion
Mutant A. baumannii strains were generated as previously described by Hoang et al. 37. The ABUW_1645 deletion was generated by PCR (Phusion polymerase, Thermo-Fisher Scientific) amplification of approximately 2 kbp up- and downstream fragments of the ABUW_1645 gene using A. baumannii strain AB5075 genomic DNA as template. Oligonucleotide primers 1645 Up-1.1 (AAAAAGGATCCCTACAGACCTTAAATAACGGTG) and 1645 Down-2.1 (AAAAAGGATCCTGGTCAAACTTTACGTGGT) were designed to contain BamHI restriction sites near the 5′ end and were paired with 1645 Up-2 (TGCTCTAAATGAAGCTTCTAA) and 1645 Down-1 (ATAATACTGTCCTAGATTAAAAATAAAAGC) primers, respectively. The up- and downstream fragments were gel purified (UltraClean 15 DNA Purification Kit, MoBio Laboratories) and then ligated (Fast-Link DNA Ligation Kit, Epicentre Biotechnologies) to produce an approximately 4 kbp deletion allele. This allele contains a deletion corresponding to amino acids 10 to 180 (92% of the protein sequence) of ABUW_1645. The Δ1645 deletion allele was gel purified after ligation and re-amplified via PCR with 1645 Up-1.1 and Down-2.1 primers. Gel-purified Δ1645 deletion allele and pEX18Tc were digested with BamHI and subsequently gel purified. Digested fragments were ligated and transformed into competent E. coli Transformax EC100D cells (Epicentre Biotechnologies). This ligation produced the suicide vector pΔ1645/EX18Tc.
To transfer the deletion allele to the chromosome of A. baumannii strain AB5075, suicide vector was electroporated into competent AB5075 cells which had been grown in LB medium and washed with 300 mM sucrose, as described by Choi and Schweizer 38. Integrants were selected on LB+tetracycline (5 μg/ml). Counterselection was carried out at room temperature on LB medium without sodium chloride supplemented with 10% sucrose. Potential mutants were screened by PCR amplification and confirmed by DNA sequencing.
An ompR::Tc/Δ1645 double mutant was generated by transformation of the Δ1645 mutant with chromosomal DNA from an ompR::Tc mutant obtained from the University of Washington strain collection.
Biofilm analysis
VIR-O and AV-T cells were taken directly from freezer stocks and grown in 2 ml 0.5× LB without shaking at room temperature to an optical density A600 of 0.1. Each tube was then used to inoculate 6 wells of a 96 well microtiter plate with 150 μl of culture. Plates were incubated stationary at 25°C for 24 hours. Cells were removed for the well and the optical density of each well was read at A600 for cell growth. To stain biofilms, 250 μl of 10% crystal violet was added to each well for 30 minutes. The crystal violet was gently decanted and each well was gently washed 3 times with distilled water. 300 μl of 33% acetic acid was added to each well to solubilize the crystal violet and this was then added to 600 μl 33% acetic acid. The absorbance of sample was read at A585.
Growth of A. baumannii
The A. baumannii VIR-O and AV-T strains were sub-cultured to an OD600 of 0.03 in Chamberlain’s Defined Media (Teknova), lysogeny broth (LB) (BD Biosciences), or minimal media (M9) supplemented with 0.2% casamino acids. The iron chelator, 2,2′-dipyridyl disulfide (156 μM) was added as indicated. Subcultures were placed at 37°C with aeration in a Biotek Synergy MX plate reader (Applied Biosystems) and OD600 was measure every 30 min for 20 hours.
Statistics
Statistical analyses were performed using Prism 5 (GraphPad Software). The significance of the mouse experiments was determined with the Mann-Whitney test, as not all data were normally distributed, and all in vitro experiments were analyzed using the two-tailed unpaired student’s t-test (for data with a normal distribution). All experiments were repeated at least two to three times. All replicates shown are biological replicates.
Data availability
All data supporting this study will be available upon request. Compiled RNA-Seq data is available as a supplementary Excel file (Supplementary Table 1.xlsx) and sequence reads were deposited at NCBI under Bioproject no. PRJNA400082 as BioSamples SAMN07562376, SAMN07562377, SAMN07562378, SAMN07562379, SAMN07562380, SAMN07562381, SAMN07562382, SAMN07562383 and SAMN07562384.
Supplementary Material
Acknowledgments
The authors thank the Genomics Resource Center at University of Maryland for help with RNA sequencing and analysis, Hannah Ratner for mice experiments, D. Bonenberger for breeding knockout mice and Dr. William Shafer for comments on the manuscript. This study was supported in part by the Robert P. Apkarian Integrated Electron Microscopy Core (RPAIEMC), which is subsidized by the Emory College of Arts and Sciences and the Emory University School of Medicine and is one of the Emory Integrated Core Facilities. Additional support was provided by the Georgia Clinical & Translational Science Alliance of the National Institutes of Health under award number UL1TR000454. P.N.R is supported by NIH grants R21AI115183 and R01072219, VA Merit award I01 BX001725 and a Research Career Scientist Award from the Department of Veterans Affairs. D.S.W. is supported by a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Disease award, VA Merit award I01 BX002788 and National Institutes of Health (NIH) grant AI098800. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH and the Department of Veterans Affairs.
Footnotes
Author contributions
Experiments were conducted by C.Y.C., K.A.T., and M.F. The manuscript was prepared by C.Y.C., K.A.T, D.S.W., and P.N.R. Samples from human patients were provided by E.M.B. The study was planned and directed by D.S.W. and P.N.R.
Competing Interests. The authors declare no competing interests.
References
- 1.O’Neill J. Review on Antimicrobial Resistance: Tackling drug resistant infections globally. London: 2014. https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf. [Google Scholar]
- 2.Bergogne-Berezin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clinical microbiology reviews. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathogens and disease. 2014;71:292–301. doi: 10.1111/2049-632x.12125. [DOI] [PubMed] [Google Scholar]
- 4.Dijkshoorn L, Nemec A, Seifert H. An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nature reviews. Microbiology. 2007;5:939–951. doi: 10.1038/nrmicro1789. [DOI] [PubMed] [Google Scholar]
- 5.Joly-Guillou ML. Clinical impact and pathogenicity of Acinetobacter. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2005;11:868–873. doi: 10.1111/j.1469-0691.2005.01227.x. [DOI] [PubMed] [Google Scholar]
- 6.Murray CK, Hospenthal DR. Acinetobacter infection in the ICU. Critical care clinics. 2008;24:237–248. vii. doi: 10.1016/j.ccc.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Charnot-Katsikas A, et al. Two cases of necrotizing fasciitis due to Acinetobacter baumannii. Journal of clinical microbiology. 2009;47:258–263. doi: 10.1128/jcm.01250-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerrero DM, et al. Acinetobacter baumannii-associated skin and soft tissue infections: recognizing a broadening spectrum of disease. Surgical infections. 2010;11:49–57. doi: 10.1089/sur.2009.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowman W, Kalk T, Menezes CN, John MA, Grobusch MP. A case of community-acquired Acinetobacter baumannii meningitis - has the threat moved beyond the hospital? Journal of medical microbiology. 2008;57:676–678. doi: 10.1099/jmm.0.47781-0. [DOI] [PubMed] [Google Scholar]
- 10.Telang NV, Satpute MG, Dhakephalkar PK, Niphadkar KB, Joshi SG. Fulminating septicemia due to persistent pan-resistant community-acquired metallo-beta-lactamase (IMP-1)-positive Acinetobacter baumannii. Indian journal of pathology & microbiology. 2011;54:180–182. doi: 10.4103/0377-4929.77397. [DOI] [PubMed] [Google Scholar]
- 11.Boucher HW, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 12.Doi Y, Husain S, Potoski BA, McCurry KR, Paterson DL. Extensively drug-resistant Acinetobacter baumannii. Emerging infectious diseases. 2009;15:980–982. doi: 10.3201/eid1506.081006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottig S, et al. Detection of pan drug-resistant Acinetobacter baumannii in Germany. The Journal of antimicrobial chemotherapy. 2014;69:2578–2579. doi: 10.1093/jac/dku170. [DOI] [PubMed] [Google Scholar]
- 14.Lei J, et al. Extensively drug-resistant Acinetobacter baumannii outbreak cross-transmitted in an intensive care unit and respiratory intensive care unit. American journal of infection control. 2016 doi: 10.1016/j.ajic.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 15.Park YK, et al. Extreme drug resistance in Acinetobacter baumannii infections in intensive care units, South Korea. Emerging infectious diseases. 2009;15:1325–1327. doi: 10.3201/eid1508.080772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maragakis LL, Perl TM. Acinetobacter baumannii: epidemiology, antimicrobial resistance, and treatment options. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2008;46:1254–1263. doi: 10.1086/529198. [DOI] [PubMed] [Google Scholar]
- 17.Villegas MV, Hartstein AI. Acinetobacter outbreaks, 1977–2000. Infection control and hospital epidemiology. 2003;24:284–295. doi: 10.1086/502205. [DOI] [PubMed] [Google Scholar]
- 18.Fernandez-Cuenca F, et al. Reduced susceptibility to biocides in Acinetobacter baumannii: association with resistance to antimicrobials, epidemiological behaviour, biological cost and effect on the expression of genes encoding porins and efflux pumps. The Journal of antimicrobial chemotherapy. 2015;70:3222–3229. doi: 10.1093/jac/dkv262. [DOI] [PubMed] [Google Scholar]
- 19.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. Journal of clinical microbiology. 1998;36:1938–1941. doi: 10.1128/jcm.36.7.1938-1941.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan KA, et al. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:20254–20259. doi: 10.1073/pnas.1317052110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks SE, Walczak MA, Hameed R, Coonan P. Chlorhexidine resistance in antibiotic-resistant bacteria isolated from the surfaces of dispensers of soap containing chlorhexidine. Infection control and hospital epidemiology. 2002;23:692–695. doi: 10.1086/501996. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs AC, et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio. 2014;5:e01076–01014. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tipton KA, Dimitrova D, Rather PN. Phase-Variable Control of Multiple Phenotypes in Acinetobacter baumannii Strain AB5075. Journal of bacteriology. 2015;197:2593–2599. doi: 10.1128/jb.00188-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geisinger E, Isberg RR. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS pathogens. 2015;11:e1004691. doi: 10.1371/journal.ppat.1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin TR, Frevert CW. Innate immunity in the lungs. Proceedings of the American Thoracic Society. 2005;2:403–411. doi: 10.1513/pats.200508-090JS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiemstra PS, McCray PB, Jr, Bals R. The innate immune function of airway epithelial cells in inflammatory lung disease. The European respiratory journal. 2015;45:1150–1162. doi: 10.1183/09031936.00141514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tipton KA, Rather PN. An ompR/envZ Two-Component System Ortholog Regulates Phase Variation, Osmotic Tolerance, Motility, and Virulence in Acinetobacter baumannii strain AB5075. Journal of bacteriology. 2016 doi: 10.1128/jb.00705-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tipton KA, Farokhyfar M, Rather PN. Multiple roles for a novel RND-type efflux system in Acinetobacter baumannii AB5075. Microbiology Open. 2016 doi: 10.1002/mbo3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Regeimbal JM, et al. Personalized Therapeutic Cocktail of Wild Environmental Phages Rescues Mice from Acinetobacter baumannii Wound Infections. Antimicrobial agents and chemotherapy. 2016;60:5806–5816. doi: 10.1128/aac.02877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slutsky B, et al. “White-opaque transition”: a second high-frequency switching system in Candida albicans. Journal of bacteriology. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner KH, Vallet-Gely I, Dove SL. Epigenetic control of virulence gene expression in Pseudomonas aeruginosa by a LysR-type transcription regulator. PLoS genetics. 2009;5:e1000779. doi: 10.1371/journal.pgen.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diard M, et al. Stabilization of cooperative virulence by the expression of an avirulent phenotype. Nature. 2013;494:353–356. doi: 10.1038/nature11913. [DOI] [PubMed] [Google Scholar]
- 33.Ronin I, Katsowich N, Rosenshine I, Balaban NQ. A long-term epigenetic memory switch controls bacterial virulence bimodality. eLife. 2017;6 doi: 10.7554/eLife.19599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouammine A, et al. An antimicrobial peptide-resistant minor subpopulation of Photorhabdus luminescens is responsible for virulence. Scientific reports. 2017;7:43670. doi: 10.1038/srep43670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bommanavar SB, Gugwad S, Malik N. Phenotypic switch: The enigmatic white-gray-opaque transition system of Candida albicans. Journal of oral and maxillofacial pathology : JOMFP. 2017;21:82–86. doi: 10.4103/0973-029x.203781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hunger M, Schmucker R, Kishan V, Hillen W. Analysis and nucleotide sequence of an origin of DNA replication in Acinetobacter calcoaceticus and its use for Escherichia coli shuttle plasmids. Gene. 1990;87:45–51. doi: 10.1016/0378-1119(90)90494-c. [DOI] [PubMed] [Google Scholar]
- 37.Hoang T, Karkhoff-Schweizer R, Kutchma A, Schweizer H. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene. 1998;212:77–86. doi: 10.1016/s0378-1119(98)00130-9. [DOI] [PubMed] [Google Scholar]
- 38.Choi K, Schweizer H. mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nature Protocols. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting this study will be available upon request. Compiled RNA-Seq data is available as a supplementary Excel file (Supplementary Table 1.xlsx) and sequence reads were deposited at NCBI under Bioproject no. PRJNA400082 as BioSamples SAMN07562376, SAMN07562377, SAMN07562378, SAMN07562379, SAMN07562380, SAMN07562381, SAMN07562382, SAMN07562383 and SAMN07562384.