Abstract
Enzymes of grasses involved in fructan synthesis are of interest since they play a major role in assimilate partitioning and allocation, for instance in the leaf growth zone. Several fructosyltransferases from tall fescue (Festuca arundinacea) have previously been purified (Lüscher and Nelson, 1995). It is surprising that all of these enzyme preparations appeared to act both as sucrose (Suc):Suc 1-fructosyl transferases (1-SST) and as fructan:fructan 6G-fructosyl transferases. Here we report the cloning of a cDNA corresponding to the predominant protein in one of the fructosyl transferase preparations, its transient expression in tobacco protoplasts, and its functional analysis in the methylotrophic yeast, Pichia pastoris. When the cDNA was transiently expressed in tobacco protoplasts, the corresponding enzyme preparations produced 1-kestose from Suc, showing that the cDNA encodes a 1-SST. When the cDNA was expressed in P. pastoris, the recombinant protein had all the properties of known 1-SSTs, namely 1-kestose production, moderate nystose production, lack of 6-kestose production, and fructan exohydrolase activity with 1-kestose as the substrate. The physical properties were similar to those of the previously purified enzyme, except for its apparent lack of fructan:fructan 6G-fructosyl transferase activity. The expression pattern of the corresponding mRNA was studied in different zones of the growing leaves, and it was shown that transcript levels matched the 1-SST activity and fructan content.
Although starch is the most widespread storage carbohydrate in the plant kingdom, fructans (polymers of Fru) occur as additional storage compounds in a large number of plants from different families, including many members of the Poaceae and Asteraceae (Hendry, 1987). In these plants fructans play an important role in assimilate partitioning and allocation, and their biosynthesis has therefore been studied extensively.
The classic object of study has been Jerusalem artichoke (Helianthus tuberosus), a member of the Asteraceae forming linear β-1,2-linked fructans called inulin. As originally postulated by Edelman and Jefford (1968), inulin is synthesized from Suc via a trisaccharide intermediate by the concerted action of two distinct fructosyltransferases: (a) Suc:Suc 1-fru-ctosyltransferase (1-SST, EC 2.4.1.99), which produces the inulin trisaccharide 1-kestose (isokestose) and Glc from Suc (Koops and Jonker, 1996; Lüscher et al., 1996; Van den Ende et al., 1996; Lüscher et al., 2000; Van den Ende et al., 2000), and (b) fructan:fructan 1-fructosyltransferase (1-FFT, EC 2.4.1.100; Lüscher et al., 1993; Koops and Jonker, 1994), which transfers Fru moieties between fructan molecules.
Typical fructans of Poaceae are graminans, branched polymers containing β-1,2- and β-1,6 linkages. We have previously investigated fructan synthesis in barley (for review, see Wiemken et al., 1995), and have established that in this case, the second key enzyme in addition to 1-SST is Suc:fructan 6-fructosyltransferase (6-SFT). This enzyme transfers fructosyl units from Suc onto acceptor Suc and fructan molecules (Simmen et al., 1993; Duchateau et al., 1995; Sprenger et al., 1995). Some grasses, such as tall fescue (Festuca arundinacea), also form fructans with an oligofructosyl chain attached to the 6-position of the glucosyl residue of Suc (inulin neoseries), a fructan type considered typical of the Liliaceae (Shiomi, 1989). The key enzyme in production of the inulin neoseries in Liliaceae is fructan:fructan 6G-fructosyltransferase (6G-FFT), which transfers a fructosyl moiety from a 1-kestose molecule to the 6-position of the Glc moiety of a Suc molecule, yielding neokestose (Shiomi, 1989; Vijn et al., 1997).
Here we focus on the enzymology of fructan formation in the meristematic growth zone of grass leaves in which water-soluble fructans constitute up to 40% of the dry matter (Schnyder and Nelson, 1989; Roth et al., 1996). This growth zone in which cell proliferation and cell elongation occur is concealed in the sheath of older leaves and lacks photosynthetic activity. It is thought that during the cell proliferation phase, Suc is transported to the growth zone and used as a substrate of fructosyltransferases to form fructans. Although the Fru moiety of Suc is deposited in the form of fructans, the Glc released is available for respiration and cell wall synthesis (Schnyder and Nelson, 1987; Hawker et al., 1991; Roth et al., 1996). Later, the stored fructan is hydrolyzed, and the products serve as osmotica to drive cell elongation and for cell wall synthesis.
In previous biochemical studies the enzymology of fructan synthesis in the leaf growth zone of tall fescue (Lüscher and Nelson, 1995) and in barley (Roth et al., 1996) was investigated. The growth zones were found to have only minor activities of invertase and, in barley, the main activities metabolizing Suc in this tissue were clearly identified as 1-SST and 6-SFT (Roth et al., 1996; Lüscher et al., 2000). In tall fescue three fructosyltransferases (F1, F2, and F3) were purified from the leaf growth zone (Lüscher and Nelson, 1995). Each of these enzyme preparations synthesized, from either Suc or 1-kestose as substrate, a mixture of fructans resembling the low-degree of polymerization (DP) fructan fraction extracted from the same plant tissue.
It is surprising that the products included 1-kestose and neokestose, indicating that the enzyme preparations had 1-SST and 6G-FFT activities. However, the fructosyltransferase preparations did not appear homogeneous on SDS-PAGE gels even after extensive purification; for example, highly purified fructosyl transferase F1 yielded three bands of 80, 58, and 25 kD. This raised the question of whether the preparations consisted of two different, separate enzymes, one with 1-SST and one with 6G-FFT activity, or of one single enzyme with dual function, occurring in an uncleaved (80 kD) and in a cleaved (58 + 25 kD) form. It is well known that many plant invertases (Sturm, 1999) and fructosyl transferases, such as the invertases from barley (Obenland et al., 1993), the 6-SFT from barley (Sprenger et al., 1995), and 1-SSTs from Jerusalem artichoke (Koops and Jonker, 1996; Lüscher et al., 1996) and Cichorium intybus (Van den Ende et al., 1996; Van den Ende et al., 2000) are encoded as approximately 80-kD proteins, which are cleaved during maturation and yield two bands of around 60 and 25 kD on SDS-PAGE gels (Sprenger et al., 1995; Van der Meer et al., 1998).
To resolve this issue we attempted to clone one of the fructosyl transferases from tall fescue, based on peptide sequences derived from the 58-kD band of the purified preparation of fructosyl transferase F1. Here we report the cloning of the cDNA encoding this enzyme, the first grass 1-SST to be described, its functional expression in tobacco protoplasts, and characterization of the recombinant enzyme after overexpression in the methylotrophic yeast Pichia pastoris. Our data show that the enzyme is a typical 1-SST without 6G-FFT activity.
RESULTS
Protein Sequencing of the 58-kD Band of the Fructosyltransferase F1
The fructosyltransferase F1, purified from tall fescue, generated three bands in an SDS-PAGE analysis. In barley it has been shown that the 6-SFT separates into two bands of around 52 and 22 kD, both of which derive from one gene product, with the larger band representing the N-terminal domain and the smaller band the C-terminal domain (Sprenger et al., 1995). Under the assumption that the two transferases were similar, the 58-kD band of the fructosyltransferase F1 preparation, presumed to contain the N-terminal domain of the gene product, was subjected to automated Edman degradation and amino acid identification. A sequence of ten amino acids was obtained: ADGGFPWSNA.
Partial cDNA Fragments
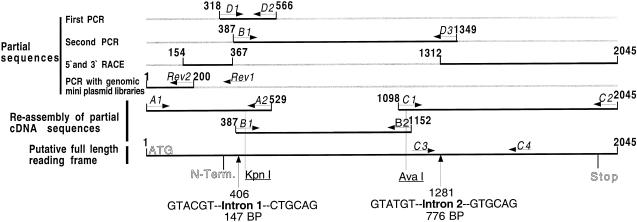
Partially overlapping cDNA sequences were obtained in five steps. The first partial cDNA was obtained by reverse transcriptase (RT)-PCR using a degenerate forward primer D1, coding for the first seven amino acids of the N terminus of the 58-kD band, and a number of different degenerate reverse primers corresponding to highly conserved boxes of plant invertases and fructosyltransferases (Sprenger et al., 1995; Vijn et al., 1997). The combination with the reverse primer D2, coding for the conserved amino acid sequence AMVPDGWY, yielded a DNA fragment of 249 bp. Sequence analysis of the fragment showed high homology with known invertases and transferases, and the three codons 3′ of the D1 primer sequence corresponded exactly to the three amino acids of the N terminus of the 58-kD band. Subsequent 3′- and 5′-RACE to obtain the entire sequence failed at this stage.
A second partial cDNA was obtained using a specific forward primer contained within the 249-bp fragment and a degenerate reverse primer coding for the amino acid sequence of a tryptic fragment of purified 1-SST from barley. Sequencing confirmed that the 179-bp sequence of this fragment between the primers B1 and D3 matched exactly the sequence of the first 249-bp fragment. With the new downstream sequence information of about 900 bp, subsequent 3′-RACE yielded the full 3′ part of the cDNA. About a 150-bp additional upstream sequence information was obtained with 5′-RACE, but it was obvious that when compared with other sequences of fructosyltransferases, the 5′-end had not been reached.
Therefore, another strategy to obtain the translation start was used. Genomic DNA was digested with different restriction enzymes and the fragments were cloned into the Bluescript vector. These “mini-plasmid libraries” served as templates for nested PCR in which two transferase-specific reverse and two vector-specific forward primers were used. With four of these libraries, PCR products were obtained. The longest product of 700 bp was amplified from the library constructed with XbaI and contained the missing 5′-end (Fig. 1). Sequencing confirmed that the 3′ part of this product, consisting of 46 bp, matched exactly the sequence previously determined by 5′-RACE.
Figure 1.
Schematic representation of the 1-SST cloning from tall fescue. Partial cDNA sequences of the tall fescue fructosyltransferase were obtained in five steps: two RT PCRs, 5′- and 3′-RACE, and PCR with genomic mini-plasmid libraries. DNA sequences are in black lines and gray lines represent the missing nucleotide sequence. All attempts to obtain the full-length cDNA with reverse PCR failed. Three short PCR fragments were cloned, cut out, and then re-assembled to a complete cDNA. The positions of the restriction sites KpnI and AvaI are marked. Arrows represent the positions of primers that were used for cloning or to obtain partial cDNAs. Vertical arrows show the positions of the two introns. The translational start and stop codons and the N terminus are marked.
Re-assembly to a Full-Length cDNA and Comparison with Other Fructosyltransferases
To obtain the entire cDNA a 5′-specific forward primer and 3′-specific backward primer were used in RT-PCR. All attempts failed to obtain the full-length cDNA, even with different polymerases. An explanation may be that the GC content of the 5′-end of the 1-SST cDNA is very high and therefore mainly truncated cDNAs might have been produced by reverse transcription. In the subsequent PCR reaction these incomplete cDNAs could not be amplified. The GC content might also be the reason why it was not possible to obtain the 5′-end by RACE. However, the amplification of shorter fragments of the cDNA was successful. These shorter PCR fragments were cloned into the pGEM-T vector. The inserts were cut out and then re-assembled to an entire cDNA by ligation (Fig. 1).
The sequence was compared with the partial cDNA sequences obtained earlier in the first two RT-PCRs that overlap the restriction site junctions KpnI and AvaI. The match with the first RT-PCR fragment was perfect on the amino acid level, but reached 98% at the nucleotide level. For the second fragment the match was 100% in both cases. The full-length clone was 2,045 bp long and the overall GC content was 64%, but went as high as 72% between bp 0 to 200. The sequence (EMBL accession no. AJ297369) contains six putative glycosylation sites. The predicted size of the polypeptide after removal of the sorting sequence amino acids 1 to 106 is 60,861 D. The pI (iso-electric point) calculated for the mature protein is 4.67.
The full-length cDNA was compared with the sequences of other fructosyltransferases. The highest homology on the nucleotide level was found to be with the vacuolar invertase from wheat (accession no. AF069309) most likely because of the GC content, since on the amino acid level the homology was not as striking. On the amino acid level, the highest homology of 67% was with the barley 6-SFT. The tall fescue 1-SST shares the same homologous regions with the other fructosyltransferases and invertases (Fig. 2). The N-terminal sequence from the start Met to the N terminus of the mature protein was relatively long, as in other invertases and fructosyl transferases, and was composed of 106 amino acids.
Figure 2.
Comparison of the deduced amino acid sequence of 1-SST from tall fescue (EMBL accession no. AJ297369) with the 6-SFT from barley (hv6sft: EMBL accession no. X83233), the 1-SST from Jerusalem artichoke (ht1sst: EMBL accession no. AJ009757), Allium cepa (ac1sst: EMBL accession no. AJ006066), and chicory (ci1sst: EMBL accession no. U81520), the 1-FFT from Jerusalem artichoke (htfft: EMBL accession no. AJ009756), and with the 6G-FFT from A. cepa (ac6gfft: EMBL accession no. Y07838). Identical amino acids are highlighted with a black background and similar amino acids with a gray background. The N terminus of the 58-kD protein is marked with an asterisk.
Amplification of Introns
Genomic DNA was amplified by PCR with three primer combinations A1, A2; B1, B2; and C1, C3. With the primer combination A1 and A2, a fragment was amplified that contained 1-SST exon sequence and an intron sequence of 147 bp (EMBL accession no. AJ297370). The consensus sequence at the splice sites, AG↔GTACGT and CTGCAG↔GT, of the intron follows the expected sequence of this highly conserved region (Goodall et al., 1991). It is worth noting that the AT content was relatively low for an intron sequence, reaching only 48%. With the primer combination C1 and C3 another fragment was amplified with 1-SST exon sequence and an intron sequence of 776 bp (EMBL accession no. AJ297371). Again the consensus sequence at the splice sites, AG↔GTATGT and GTGCAG↔G, had the expected conserved sequence. In contrast to the first intron, the AT content was 58% for the second intron. With the primer combination B1 and B2 no product was amplified, indicating that the fragment was probably too large to be amplified by PCR (Fig. 1).
Transient Expression in Tobacco Protoplasts
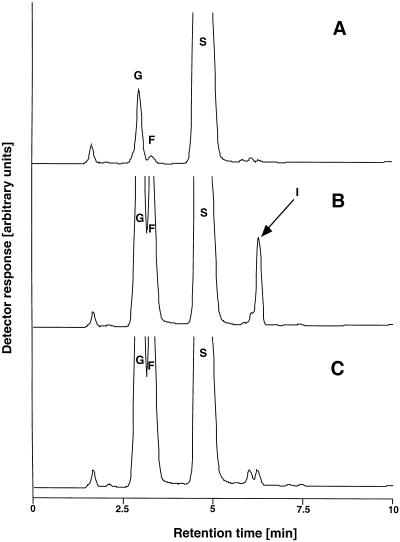
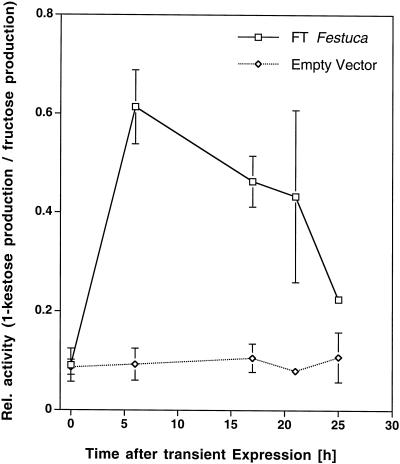
The full-length tall fescue cDNA was transiently expressed in tobacco protoplasts under the control of the cauliflower mosaic virus 35S promoter. At various time points after transfection, protein extracts were tested for transferase activity with Suc as the substrate. Proteins from protoplasts carrying the tall fescue cDNA clearly produced 1-kestose, whereas protein from protoplasts transfected with the empty plasmid was unable to produce trisaccharides (Fig. 3). The highest 1-kestose production as well as the highest 1-kestose:Fru ratio was measured after 6 h (Fig. 4). When proteins from protoplasts were incubated with 1-kestose, or with 1-kestose and Suc in combination, no higher DP fructans were detected in the reaction mixture (data not shown). These results show that the cloned cDNA from tall fescue encodes a functional 1-Suc:sucrose fructosyltransferase.
Figure 3.
HPLC analysis of products formed in enzyme extracts from tobacco protoplasts transiently expressing the 1-SST clone from tall fescue (A and B), or the control with the empty plasmid (C), for 0 h (A), and 12 h (B and C), after incubation with 100 mm Suc for 1 h. G, Glc; F, Fru; S, Suc; I, 1-kestose.
Figure 4.
Ratio of 1-SST and invertase activities in extracts from tobacco protoplasts transiently expressing the 1-SST clone from tall fescue for different time periods. The extracts were incubated with 100 mm Suc for 1 h. The ratio of 1-SST and invertase activity in the extracts is expressed as the ratio of 1-kestose and Fru production.
Expression Pattern in the Growing Leaf
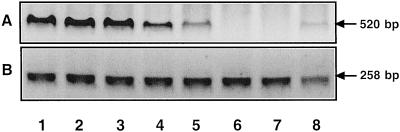
Eight 1-cm long segments were cut, starting from the base of each growing leaf, and from each segment the total RNA was then isolated and the samples were adjusted to contain equal amounts of RNA. A 520-bp fragment of 1-SST cDNA was amplified by RT-PCR from each sample. Primers were chosen that flank an intron to distinguish between genomic and cDNA product. The amplified 520-bp fragment could only be amplified from cDNA, reflecting relative amounts of the 1-SST message. As a control a 258-bp fragment of rRNA was amplified. In extracts from the first 3 cm of the growth zone a large amount of the 1-SST fragment was amplified, then in the next two, the amounts dropped considerably. In the following two segments no product was amplified, whereas in the last sample a trace of the product was detected. The control fragment was amplified to a similar extent in all the samples, indicating that equal amounts of total RNA were present (Fig. 5).
Figure 5.
Transcript level of 1-SST in the base of the growing leaf, estimated by RT-PCR. Eight 1-cm-long segments were cut, starting from the base of each growing leaf, and the total RNA was isolated from each segment. A DNA fragment of 520 bp was amplified by RT-PCR from each sample (lanes 1–8) reflecting relative amounts of the 1-SST message (A), and analyzed by gel electrophoresis. As a control a 258-bp fragment of the rRNA was amplified from each sample (B).
Characterization of Enzyme Produced by P. pastoris
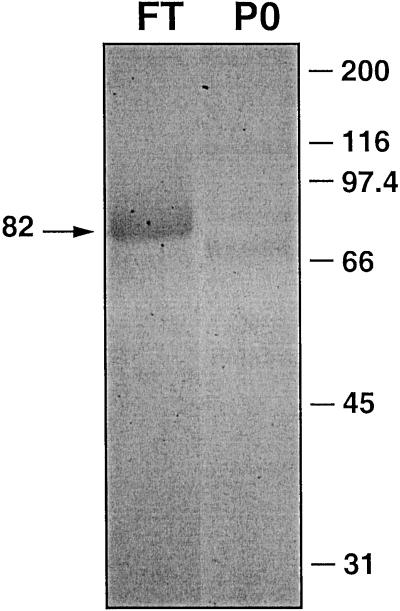
A cDNA encoding the mature 1-SST protein from tall fescue was expressed in P. pastoris for detailed activity characterization. The concentrated supernatant from all transformed colonies after a 48-h methanol induction contained very strong 1-SST activity (data not shown). Concentrated culture medium was analyzed by SDS-PAGE on a 12% (w/v) gel and stained with Coomassie Brilliant Blue (Fig. 6). A single band was found at 82 kD, which did not occur in the controls (concentrated culture media from P. pastoris transformed with empty plasmid).
Figure 6.
SDS-PAGE analysis of concentrated culture medium from transformed P. pastoris expressing tall fescue 1-SST. Concentrated culture medium containing the recombinant 1-SST was separated on a 12% (w/v) gel by SDS-PAGE, and stained with Coomassie Brilliant Blue (lane FT). The control lane P0 contained concentrated medium from P. pastoris transformed with empty plasmid. Mr markers are shown on the right. The arrow at the left indicates the prominent single band of 82 kD.
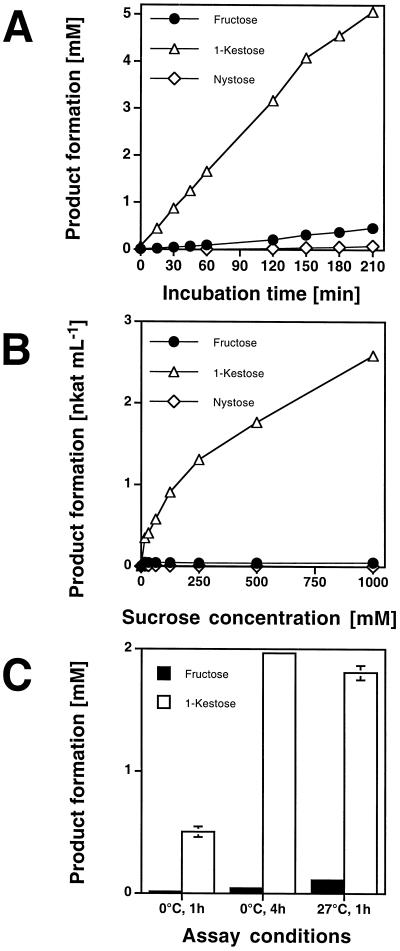
For further characterization the enzyme was desalted, incubated with Suc, and potential products were measured at various time points between 0 and 210 min (Fig. 7A). 1-kestose was produced linearly until 150 min, and was formed at a slightly lower rate from then on. Very little Fru was produced (Glc:Fru ratio of approximately 20:1). Production of nystose was only detectable after 2 h and reached only 0.6% of 1-kestose production.
Figure 7.
Characterization of the recombinant tall fescue 1-SST produced by P. pastoris. A, Time course over a period of 0 to 210 min. 1-SST was incubated with 100 mm Suc as the substrate, and production of Fru, 1-kestose, and nystose was measured. B, Saturation experiment. Recombinant 1-SST was incubated for 60 min with Suc as the substrate with concentrations ranging from 0 to 1,000 mm. Fru, 1-kestose, and nystose production was measured. C, Influence of temperature on enzyme specificity. Enzyme preparation was incubated with 100 mm Suc under the following conditions: 0°C for 60 min, 0°C for 240 min, and 27°C for 60 min. Production of Fru and 1-kestose was determined.
In a saturation experiment the enzyme solution was incubated with increasing Suc concentrations ranging from 0 to 1,000 mm (Fig. 7B). The 1-kestose producing activity could not be saturated, even at a Suc concentration of 1,000 mm. Fru production during this experiment was extremely low (Glc:Fru ratio of approximately 50:1).
The influence of incubation temperature on enzyme characteristics was investigated by incubating at 0°C and 27°C for 60 min. To compensate for the activity decrease due to lower temperatures, another sample was incubated at 0°C for 4 h (Fig. 7C). In both assays the 1-kestose production was similar, but the Fru production was 2-fold higher at the higher temperature, indicating a slight shift from transferase toward hydrolase activity.
Enzyme solution was incubated for 60 min with various substrates: Suc, 1-kestose, nystose, a combination of Suc with 1-kestose, and a 5% (w/v) solution of barley fructan DP 10 to 30 (Table I). With Suc as substrate (100 mm), the main product was 1-kestose (9.065 nkat mL−1). Some Fru (0.490 nkat mL−1) and very little nystose (0.103 nkat mL−1) were formed as well. When incubated with 1-kestose (50 mm), the enzyme acted mainly as a hydrolase, producing Fru (4.277 nkat mL−1) and Suc. A small amount of nystose was also formed, indicating fructosyltransfer onto 1-kestose (1.352 nkat mL−1). When the substrate consisted of a combination of Suc (100 mm) and 1-kestose (50 mm), Fru production doubled, compared with Suc as the sole substrate (0.875 nkat mL−1) and nystose production quadrupled (0.468 nkat mL−1), suggesting a preferred fructosyltransfer from Suc onto 1-kestose to form the tetrasaccharide. Noteworthy in the combination of Suc and 1-kestose as compared with 1-kestose alone is the drastic decrease of Fru production (approximately five times) and of nystose production (approximately three times), indicating that Suc is highly preferred as fructosyl donor as well as fructosyl acceptor substrate. Incubation with nystose yielded mainly hydrolytic products: Fru (0.672 nkat mL−1), Suc (0.090 nkat mL−1), and 1-kestose (0.788 nkat mL−1). However, the activities were quite low compared with the reaction with Suc. No product formation could be determined after incubation with barley fructan DP 10 to 30.
Table I.
Activities of enzyme produced by P. pastoris, incubated with Suc (100 mm), 1-kestose (50 mm), a combination of the two substances (100 and 50 mm, respectively), nystose (50 mm), or barley fructan (5% [w/v], DP 10–30)
| Substrate | Product Formation
|
|||
|---|---|---|---|---|
| Fru | Suc | 1-Kestose | Nystose | |
| nkat mL−1 | ||||
| Suc | 0.490 (±0.002) | a | 9.065 (±0.278) | 0.103 (±0.006) |
| 1-Kestose | 4.277 (±0.088) | 3.399 (±0.050) | a | 1.352 (±0.025) |
| Suc and 1-kestose | 0.875 (±0.023) | a | a | 0.468 (±0.005) |
| Nystose | 0.672 (±0.021) | 0.090 (±0.003) | 0.788 (±0.022) | a |
| Barley fructan (DP 10–30) | b | b | b | b |
Data are given in nkat per milliliter of the enzyme preparation.
Not measured (added as a substrate).
No product formation detectable on HPLC diagram (not analyzed quantitatively).
DISCUSSION
In tall fescue three fructosyltransferases (F1, F2, and F3) have been isolated (Lüscher and Nelson, 1995). Each purified enzyme fraction had 1-SST and 6G-FFT activity and contained three prominent polypeptides when analyzed by SDS-PAGE, e.g. F1 yielded three bands of 80, 58, and 25 kD. In an attempt to determine whether the purified preparations contained two different enzymes with 1-SST and 6G-FFT activities, or one single enzyme with dual function occurring in an uncleaved (80 kD) and in a cleaved form (58 + 25 kD), reverse genetics was used to clone the 58-kD band of F1. The corresponding cDNA was then transiently expressed in tobacco protoplasts, and its functional analysis was carried out in the methylotrophic P. pastoris.
In contrast to the protein preparations purified from tall fescue (F1–F3; Lüscher and Nelson, 1995), the protein produced by transformed tobacco protoplasts (Fig. 3) or P. pastoris (Table I) harboring the tall fescue cDNA was not able to synthesize neokestose from 1-kestose and Suc or higher DP fructan from the inulin neokestose series. In other words, the cloned enzyme had no 6G-FFT activity. Instead, it had all properties of known 1-SSTs (Koops and Jonker, 1996; Lüscher et al., 1996; Van den Ende and Van Laere, 1996; Lüscher et al., 2000): 1-kestose production, lack of 6-kestose production, moderate nystose production, and fructan exohydrolase activity with 1-kestose as the substrate (Table I).
When the 6-SFT from barley was overexpressed in P. pastoris the recombinant protein showed some differences in activity when compared with the plant protein (Hochstrasser et al., 1998). The recombinant tall fescue 1-SST had no abnormal activities that would not fit with a 1-SST, and the investigated properties, such as substrate saturation, linear production, and temperature effects were the same as seen for the plant protein. It could be argued that the recombinant protein might have lost its ability to produce neokestose, but this is unlikely since production of neokestose could also not be detected in the extracts of tobacco protoplasts transiently expressing the cDNA.
With the new results it can be concluded that the purified enzyme preparations previously described probably contained two different enzymes, a 1-SST and 6G-FFT. The combination of the two enzymes would yield neokestose and higher DP fructan when incubated with Suc. There is little doubt that the 58- and 25-kD bands seen on the SDS-PAGE gel represent the 1-SST, but whether the 80-kD band represents the 6G-FFT remains unclear. It cannot be excluded that the two fructosyltransferases cannot be separated by SDS-PAGE, as seen for barley fructosyltransferases (Lüscher et al., 2000). This seems to be unlikely, however, since no double sequence was observed when the amino acid sequence was obtained from the 58-kD band (data not shown). To date, the existence of cofactors for fructosyltransferases has not been reported and it is therefore highly improbable that the purified enzyme contains cofactors that are not present in the expression systems used.
Proteins contained in concentrated culture media of the 1-SST expressing yeast were separated on a 12% (w/v) gel by SDS-PAGE and stained with Coomassie Brilliant Blue (Fig. 6). A single band was found at 82 kD. In control lanes that contained concentrated medium from P. pastoris transformed with empty plasmid, the 82-kD band was absent. Silver staining of an SDS gel prepared the same way revealed several additional bands in the lane containing the recombinant 1-SST (data not shown). However, purification of the 82-kD band from the culture medium showed unequivocally that this protein band represented indeed the recombinant 1-SST enzyme (M. Lüscher and D. Altenbach, unpublished results). Since the protein is not cleaved into two subunits as seen for plant fructosyltransferases (Sprenger et al., 1995; Koops and Jonker, 1996; Lüscher et al., 1996; Van den Ende et al., 1996; Lüscher et al., 2000), it can be concluded that the cleavage occurring in planta is not essential for the transferase activity of the enzyme. The production of the protein in P. pastoris provides a useful tool to learn more about the cleavage process and its function.
The expression pattern in the base of the growing leaf of 1-SST was studied using RT-PCR. In the first 3 cm from the base, a large amount of the 1-SST fragment was amplified. In the next two segments the amounts decreased. In the end, no product at all was amplified in the following two segments (Fig. 5). This result shows clearly that the 1-SST is controlled at the transcriptional level, and the amount of message follows exactly the curve of 1-SST activity (Lüscher and Nelson, 1995) and fructan content (Schnyder and Nelson, 1989). Amplification of a small amount of message could also be found in the sample 8 cm above the base where the tissue is greening.
Genomic DNA was amplified with three different primer pairs. With two of the primer pairs, products were amplified that contained the 1-SST exon sequence and an intron sequence (Fig. 1). With one primer pair, no product could be amplified. In invertases, up to seven introns are found (Sturm, 1999). Therefore, the DNA stretch between this primer pair might contain several introns, preventing the product from being amplified because of its increased size. Most invertases (Sturm, 1999) and the fructosyltransferase 6-SFT (Sprenger, 1997) contain an extremely small exon (exon 2) that codes only for the core tripeptide (DPN) of the conserved F-motif NDPNG. With respect to this small exon the 1-SST gene from tall fescue seems to be an exception, since only one intron was found, namely between NDPN and G. It is not known, however, whether this aberration is of physiological importance.
In conclusion, we have cloned for the first time a cDNA of a grass 1-SST. More work is needed to find out why the purified enzyme preparations F1, F2, and F3 not only act as 1-SST, but also had 6G-FFT activity (Lüscher and Nelson, 1995).
MATERIALS AND METHODS
Plant Material
A clone of tall fescue (Festuca arundinacea) selected for rapid leaf growth was grown as described previously (Lüscher and Nelson, 1995).
Purification of Fructosyltransferases F1, F2, and F3 and Partial Sequencing of the 58-kD Band of the Fructosyltransferase F1
Fructosyl transferases F1, F2, and F3 were purified from leaf growth zones as described previously (Lüscher and Nelson, 1995). Preparations of purified enzymes were subjected to SDS-PAGE, electrotransferred to a polyvinylidene difluoride membrane (Immobilon polyvinylidene difluoride membrane, Millipore, Bedford, MA), and visualized with Ponceau S. Bands of interest were cut out, and their N terminus was sequenced by automated Edman degradation.
RNA Isolation, RT-PCR, and 5′- and 3′-RACE
Six to eight vegetative tall fescue tillers were removed from the pots. Older leaves and sheaths were carefully removed to reveal the base of the growing leaf. A 1-cm-long segment was cut from the base of each growing leaf, frozen in liquid N2, and ground to a fine powder. The total RNA was then isolated using the RNAeasy kit from Qiagen (Basel) according to the manufacturer's manual.
Two micrograms of total RNA was used for RT-PCR (Access kit, Promega, Wallisellen, Switzerland) according to the manufacturer's manual. Products were cloned into the pGEM-T vector (Promega, Madison, WI) and sequenced. Longer cDNA sequences were obtained using the 3′-/5′-RACE kit from Boehringer Mannheim (Mannheim, Germany).
Construction of Genomic Mini-Plasmid Libraries
Genomic DNA was isolated from healthy leaf material with the Nucleon Phytopure kit (Scotlab, Coatbridge, UK). Eight portions were digested with different restriction enzymes: BamHI, EcoRI, XbaI, KpnI, XhoI, SacI, HindIII, and ClaI. The fragments were cloned into the Bluescript vector and these “mini-plasmid libraries” served as templates for PCR.
Partial cDNA Fragments and the Assembly to a Full-Length cDNA
Five partial cDNA sequences of the tall fescue fructosyltransferase were obtained by two RT-PCRs, by 5′- and 3′-RACE, and by PCR with genomic mini-plasmid libraries. A degenerate forward primer D1 corresponding to the N terminus of the 58-kD band of F1 was used in RT-PCR in combination with different reverse primers designed to correspond to boxes highly conserved in plant invertases and fructosyltransferases. One combination of primers, D1 and D2, yielded a PCR fragment of 249 bp. A second partial sequence was obtained with a second RT-PCR using a specific forward primer lying inside the 249-bp fragment, B1, and a degenerate reverse primer, D3, coding for the amino acid sequence of a fragment of 1-SST from barley obtained by tryptic digest (M. Lüscher and V. Galati, unpublished results). The 3′-end of the cDNA was obtained with 3′-RACE and, with 5′-RACE, another partial cDNA sequence was obtained containing an additional 150 bp upstream of the codon corresponding to the N terminus of the 58-kD peptide. The 5′-end of the fructosyltransferase was found on a partial sequence when genomic plasmid mini-libraries were used as templates for nested PCR. The relevant fragment was amplified with two fructosyl-transferase-specific reverse (Rev1 and Rev2) and two vector-specific (Vector1 and Vector2) forward primers. Attempts to obtain the full-length cDNA with reverse PCR failed, but the amplification of shorter fragments was successful. To reconstruct the full-length cDNA, three partial cDNA sequences of the size of 529, 764, and 950 bp were obtained by RT-PCR using the specific primer pairs: A1, A2; B1, B2; and C1, C2. Each of these PCR fragments was cloned into the pGEM-T vector. The inserts were cut out and then re-assembled to an entire cDNA by ligation at the two restriction sites KpnI (437) and AvaI (1,134). The sequences of all primers are listed in Table II.
Table II.
PCR primers
| Name | Sequence |
|---|---|
| Cloning of first sequencesa | |
| D1 | GGN TTY CCN TGG AGY GAY GC |
| D2 | TAC CAY TGR TCN GCN ACC AT |
| D3 | TCY TCN AYN GGC CAY TGD AT |
| Search of the 5′-end using genomic DNA | |
| Vector 1 | CAC ACA GGA AAC AGC TAT GAC CAT G |
| Vector 2 | CGC CAA GCT CGA AAT TAA CCC TCA C |
| Rev 1 | GCC GAC GCC ACG TCT CCT C |
| Rev 2 | CGG ACC ACG ATC CAC CCT GC |
| Assembly of the fa1sst cDNA | |
| A1 | GAT GGA GTC CAG CGC CGT CG |
| A2 | GCC AGT TGA CCA TGT CCT TGG |
| B1 | GAG AAG CAC TAC ATG AAC GA |
| B2 | GTA CCT GAG CCC GAT CCC GAG |
| C1 | CAA CAT CTG GAC ACC CAT CGA |
| C2 | AAA AAA AAG CTT GCC AAT GAA TGT GTA G |
| Amplification of introns and expression pattern | |
| C3 | CTG CCA ATC ATC CTA TTC ACA |
| C4 | CAG AAG AAG AAC CGC CGC ATC |
| R1 | ATC TCG GCT CTC GCA TCG ATG |
| R2 | ACG CGC TGT GTC CGG CTC |
| Cloning into P. pastoris plasmidb | |
| P1 | GAA GGC GTC GGG GGC GAA TTC CGC CGA CGG CGG GTT CCC GTG G |
| P2 | GAC AGG GGC GAT CAG CGG CCG CCA TGA ATG CAA CAA GAG CTC G |
Degenerate primers: N = (A, C, G, T); Y = (C, T); R = (A, G); D = (A, G, T).
Restriction sites indicated in boldface (EcoRI on P1, Not1 on P2).
Transient Expression in Tobacco Protoplasts
The fructosyltransferase cDNA was cloned into the transient expression vector pDH51 (Pietrzak et al., 1986) and wild tobacco protoplasts were transfected essentially as before (Sprenger et al., 1995). Control protoplasts were transfected with the empty plasmid. The protoplasts were harvested after various time points by centrifugation and immediately frozen in liquid nitrogen and kept at −70°C until further analysis. Fifty microliters of MES [2-(N-morpholino)-ethanesulfonic acid] (NaOH) buffer (100 mm, pH 5.75) was added to each sample and the protoplast proteins were extracted with a small pestle. The slurry was centrifuged and the supernatant was desalted over Biogel P6 (Bio-Rad, Glattbrugg, Switzerland). Proteins were incubated with Suc (100 mm), 1-kestose (50 mm), or both sugars overnight at 25°C. The reaction was quenched with 0.5 m NaOH and the products formed were analyzed by HPLC (Lüscher et al., 2000).
Amplification of Introns
Genomic DNA was amplified by PCR with the three primer combinations: A1, A2; B1, B2; and C1, C3. Products were cloned and sequenced.
Transcript Levels in the Growing Leaf
Six growing tall fescue leaves were prepared as described above. Eight consecutive 1-cm-long segments were cut, starting from the base of each growing leaf, frozen in liquid N2, and ground to a fine powder. Total RNA was isolated from each segment and the samples were adjusted to contain equal amounts of RNA. A cDNA fragment of 520 bp was amplified with 35 cycles from each sample by RT-PCR with 900 ng of RNA using the primers C3 and C4. As a control, a fragment of the 5.8S rRNA from tall fescue (accession no. AJ240154; 258 bp) was amplified with 90 ng of RNA and 28 cycles using the primers R1 and R2.
Expression of the 1-SST cDNA in Pichia pastoris
A cDNA encoding the mature 1-SST from tall fescue was expressed in P. pastoris for detailed activity characterization. The sequence coding for the mature protein stretches from codon 106 to 655 (nucleotides 318–1,965), counting from the start of the open reading frame. A 1-SST cDNA for this mature protein was created by PCR and ligated into the expression vector pPICZαC. The cDNA was ligated in frame behind the N-terminal signal sequence of the Saccharomyces cerevisiae α-factor, which is contained in vector pPICZαC, to allow entry into the secretory pathway. During secretion the α-factor sequence is cleaved off the new protein (Clare et al., 1991).
The insertion of the cDNA into the shuttle vector and the transformation of P. pastoris were performed as described (Hochstrasser et al., 1998). In brief, the 1-SST cDNA was amplified by PCR with a forward primer (P1) coding for the N terminus of the mature protein and a reverse primer (P2) coding for the 3′-end. The primers were designed to introduce an EcoRI site at the 5′-end and a NotI site at the 3′-end. The resulting product could be ligated in frame behind the α-factor signal sequence of the expression vector (EasySelect Pichia expression kit from Invitrogen bv, Leek, NL) to obtain the plasmid pF1. P. pastoris strain X-33 was transformed with 3 μg of Pme I-linearized pF1 and plated on selective YPDS/Zeocin plates.
For activity screening, single colonies of transformants were inoculated on fresh YPDS/Zeocin plates. Some of the newly grown colonies were inoculated in liquid culture and induced with methanol. After 48 h the supernatant of the culture medium was concentrated from 50 mL to 800 μL by ultrafiltration and desalted into 100 mm MES (NaOH) buffer (pH 5.75). The concentrated media of the different transformants were tested for 1-SST activity. Transformants producing highest amounts of 1-SST were kept for further analysis.
Characterization of the Recombinant 1-SST
Recombinant 1-SST was produced as described above from the best producing transformant. Enzyme assays were performed in triplicate for 60 min at 27°C, with 100 mm Suc or with 50 mm of all other substrates, unless stated otherwise. The products formed were analyzed by HPLC as described (Lüscher et al., 2000).
Substrate specificity was tested with Suc, 1-kestose, nystose, a combination of Suc with 1-kestose, and with a 5% (w/v) solution of barley fructan containing oligosaccharides of DP 10 to 30. Time-dependent product formation was determined with Suc as the substrate for various periods of time, whereas the influence of temperature on product formation was tested with Suc under the following conditions: 0°C for 60 min, 0°C for 240 min, and 27°C for 60 min.
ACKNOWLEDGMENTS
We thank Mr. J.H. Coutts (University of Missouri, Columbia) for technical assistance, Mr. M. Müller (Friedrich Miescher-Institute, Basel) for the preparation of the tobacco protoplasts, and Dr. M.A. Collinge (Friedrich Miescher-Institute, Basel) for critical reading of the manuscript.
Footnotes
This work was supported by the Swiss Federal Office of Education and Science in the context of the European Union project FAIR–CT96–1896 and by the Swiss National Science Foundation.
LITERATURE CITED
- Clare JJ, Romanos MA, Rayment FB, Rowedder JE, Smith MA, Payne MM, Sreekrishna K, Henwood CA. Production of mouse epidermal growth factor in yeast: high-level secretion using Pichia pastoris strains containing multiple gene copies. Gene. 1991;105:205–212. doi: 10.1016/0378-1119(91)90152-2. [DOI] [PubMed] [Google Scholar]
- Duchateau N, Bortlik K, Simmen U, Wiemken A, Bancal P. Sucrose:fructan 6-fructosyltransferase, a key enzyme for diverting carbon from sucrose to fructan in barley leaves. Plant Physiol. 1995;107:1249–1255. doi: 10.1104/pp.107.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman J, Jefford TG. The mechanism of fructosan metabolism in higher plants as exemplified in Helianthus tuberosus L. New Phytol. 1968;67:517–531. [Google Scholar]
- Goodall G, Kiss T, W. F. Nuclear RNA splicing and small nuclear RNAs and their genes in higher plants. Oxf Surv Plant Mol Cell Biol. 1991;7:255–296. [Google Scholar]
- Hawker JS, Jenner CF, Niemietz CM. Sugar metabolism and compartmentation. Aust J Plant Physiol. 1991;18:227–237. [Google Scholar]
- Hendry G. The ecological significance of fructan in contemporary flora. New Phytol. 1987;106:201–216. [Google Scholar]
- Hochstrasser U, Lüscher M, De Virgilio C, Boller T, Wiem-ken A. Expression of a functional barley sucrose-fructan 6-fructosyltransferase in the methylotrophic yeast Pichia pastoris. FEBS Lett. 1998;440:356–360. doi: 10.1016/s0014-5793(98)01487-2. [DOI] [PubMed] [Google Scholar]
- Koops AJ, Jonker HH. Purification and characterization of the enzymes of fructan biosynthesis in tubers of Helianthus tuberosus ‘Colombia’: I. Fructan:fructan fructosyltransferase. J Exp Bot. 1994;45:1623–1631. doi: 10.1104/pp.110.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koops AJ, Jonker HH. Purification and characterization of the enzymes of fructan biosynthesis in tubers of Helianthus tuberosus ‘Colombia’: II. Purification of sucrose: sucrose 1-fructosyl transferase and reconstitution of fructan synthesis in vitro with purified sucrose:sucrose 1-fructosyl transferase and fructan:fructan fructosyl transferase. Plant Physiol. 1996;110:1167–1175. doi: 10.1104/pp.110.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher M, Erdin C, Sprenger N, Hochstrasser U, Boller T, Wiemken A. Inulin synthesis by a combination of purified fructosyltransferases from tubers of Helianthus tuberosus. FEBS Lett. 1996;385:39–42. doi: 10.1016/0014-5793(96)00343-2. [DOI] [PubMed] [Google Scholar]
- Lüscher M, Frehner M, Nösberger J. Purification and characterization of fructan:fructan fructosyltransferase from Jerusalem artichoke (Helianthus tuberosus L.) New Phytol. 1993;123:717–724. [Google Scholar]
- Lüscher M, Hochstrasser U, Boller T, Wiemken A. Isolation of sucrose:sucrose 1-fructosyltransferase (1-SST) from barley (Hordeum vulgare) New Phytol. 2000;145:225–232. [Google Scholar]
- Lüscher M, Nelson CJ. Fructosyltransferase activities in the leaf growth zone of tall fescue. Plant Physiol. 1995;107:1419–1425. doi: 10.1104/pp.107.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenland DM, Simmen U, Boller T, Wiemken A. Purification and characterization of three soluble invertases from barley (Hordeum vulgare L.) leaves. Plant Physiol. 1993;101:1331–1339. doi: 10.1104/pp.101.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrzak M, Shilito RD, Hohn T, Potrykus I. Expression in plants of two bacterial antibiotic resistance genes after protoplast transformation with a new plant expression vector. Nucleic Acids Res. 1986;14:5857–5868. doi: 10.1093/nar/14.14.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Lüscher M, Sprenger N, Boller T, Wiemken A. Fructan and fructan-metabolizing enzymes in the growth zone of barley leaves. New Phytol. 1996;136:73–79. [Google Scholar]
- Schnyder H, Nelson CJ. Growth rates and carbohydrate fluxes within the elongation zone of tall fescue leaf blades. Plant Physiol. 1987;85:548–553. doi: 10.1104/pp.85.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnyder H, Nelson CJ. Growth rates and assimilate partitioning in the elongation zone of tall fescue leaf blades at high and low irradiance. Plant Physiol. 1989;90:1201–1206. doi: 10.1104/pp.90.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi N. Properties of fructosyltransferases involved in the synthesis of fructan in Liliaceous plants. J Plant Physiol. 1989;134:151–155. [Google Scholar]
- Simmen U, Obenland D, Boller T, Wiemken A. Fructan synthesis in excised barley leaves: identification of two sucrose-sucrose fructosyltransferases induced by light and their separation from constitutive invertases. Plant Physiol. 1993;101:459–468. doi: 10.1104/pp.101.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger N. Fructan biosynthesis in grasses: the key role of sucrose:fructan 6-fructosyltransferase. PhD thesis. Basel: University of Basel; 1997. [Google Scholar]
- Sprenger N, Bortlik K, Brandt A, Boller T, Wiemken A. Purification, cloning, and functional expression of sucrose:fructan 6-fructosyltransferase, a key enzyme of fructan synthesis in barley. Proc Natl Acad Sci USA. 1995;92:11652–11656. doi: 10.1073/pnas.92.25.11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A. Invertases: primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol. 1999;121:1–8. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, Van Wonterghem D, Vergauwen R, Van Laere A. Cloning, developmental, and tissue-specific expression of sucrose:sucrose 1-fructosyl transferase from Taraxacum officinale: fructan localization in roots. Plant Physiol. 2000;123:71–80. doi: 10.1104/pp.123.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, Van Laere A. Fructan synthesizing and degrading activities in chicory roots (Cichorium intybus L.) during field-growth, storage and forcing. J Plant Physiol. 1996;149:43–50. [Google Scholar]
- Van den Ende W, Van Wonterghem D, Dewil E, Verhaert P, Deloof A, Van Laere A. Purification and characterization of 1-SST, the key enzyme initiating fructan biosynthesis in young chicory roots (Cichorium intybus L.) Physiol Plant. 1996;98:455–466. [Google Scholar]
- Van der Meer IM, Koops AJ, Hakkert JC, Van Tunen AJ. Cloning of the fructan biosynthesis pathway of Jerusalem artichoke. Plant J. 1998;15:489–500. doi: 10.1046/j.1365-313x.1998.00230.x. [DOI] [PubMed] [Google Scholar]
- Vijn I, van Dijken A, Sprenger N, van Dun K, Weisbeek P, Wiemken A, Smeekens S. Fructan of the inulin neoseries is synthesized in transgenic chicory plants (Cichorium intybus L.) harbouring onion (Allium cepa L.) fructan:fructan 6G-fructosyltransferase. Plant J. 1997;11:387–98. doi: 10.1046/j.1365-313x.1997.11030387.x. [DOI] [PubMed] [Google Scholar]
- Wiemken A, Sprenger N, Boller T. Fructan: an extension of sucrose by sucrose. In: Pontis HG, Salerno GL, Echeverria EJ, editors. International Symposium on Sucrose Metabolism. Rockville, MD: American Society of Plant Physiologists; 1995. pp. 179–188. [Google Scholar]