Abstract
Cancer aggressiveness may result from the selective pressure of a harsh nutrient-deprived microenvironment. Here we illustrate how such conditions promote chemotherapy resistance in pancreatic ductal adenocarcinoma (PDAC). Glucose or glutamine withdrawal resulted in a 5- to 10-fold protective effect with chemotherapy treatment. PDAC xenografts were less sensitive to gemcitabine in hypoglycemic mice compared with hyperglycemic mice. Consistent with this observation, patients receiving adjuvant gemcitabine (n = 107) with elevated serum glucose levels (HgbA1C > 6.5%) exhibited improved survival. We identified enhanced antioxidant defense as a driver of chemoresistance in this setting. ROS levels were doubled in vitro by either nutrient withdrawal or gemcitabine treatment, but depriving PDAC cells of nutrients before gemcitabine treatment attenuated this effect. Mechanistic investigations based on RNAi or CRISPR approaches implicated the RNA binding protein HuR in preserving survival under nutrient withdrawal, with or without gemcitabine. Notably, RNA deep sequencing and functional analyses in HuR-deficient PDAC cell lines identified isocitrate dehydrogenase 1 (IDH1) as the sole antioxidant enzyme under HuR regulation. HuR-deficient PDAC cells lacked the ability to engraft successfully in immunocompromised mice, but IDH1 overexpression in these cells was sufficient to fully restore chemoresistance under low nutrient conditions. Overall, our findings highlight the HuR–IDH1 regulatory axis as a critical, actionable therapeutic target in pancreatic cancer.
Introduction
Low nutrient availability is a hallmark feature of pancreatic ductal adenocarcinoma (PDAC) cells (1–3), and PDAC cells are particularly well adapted to survive austere conditions as compared to other aggressive cancers (4, 5). In addition, PDAC cells are relatively resistant to conventional chemotherapy (5, 6). Because mathematical models implicate the harsh microenvironment as a principal driver of aggressive cancer biology (7), we reasoned that insights into PDAC molecular reprograming in response to metabolic stress could shed light on chemotherapy resistance mechanisms. For example, both low nutrient conditions and chemotherapy are potent inducers of reactive oxygen species (ROS; refs. 8, 9), suggesting that adaptive prosurvival reprogramming in response to oxidative stress caused by nutrient withdrawal, may also prime or prepare PDAC cells against additional oxidative stressors like chemotherapy.
HuR (ELAVL1) is an acute stress response and RNA binding protein that has been reported to have important regulatory functions under oxidative stress (10, 11). In fact, detailed x-ray crystallography reveals that activation of the protein, reflected in nuclear to cytoplasmic translocation, occurs only after homodimerization mediated by an oxidized disulfide bridge (12). These findings invite speculation that the protein may act as a redox sensor. We previously established a role for HuR as a regulator of chemotherapy resistance in PDAC cells (13), and more recently as a prosurvival protein in response to hypoxia (14) and glucose withdrawal (15). On the basis of this work, we hypothesized that PDAC cells enhance antioxidant defense under diverse metabolic stressors (like nutrient withdrawal and chemotherapy) by recruiting HuR.
Herein, we observe for the first time that nutrient deprivation alone (which is a ubiquitous physiologic stress in PDAC; refs. 1–3) results in chemotherapy resistance in pancreatic cancer, through the activation of an HuR-mediated prosurvival network. This finding has important implications for current standard of care, and could shed light on why PDAC cells are so refractory to conventional cytotoxic agents (16) compared to other cancer types (4). We present data that PDAC survival under low nutrient conditions, as well as chemotherapy, is attributable to rapid mobilization of an antioxidant defense program, and that the NADPH generating enzyme, isocitrate dehydrogenase 1 (IDH1), is the critical regulatory target under HuR control for this purpose.
Materials and Methods
Cell lines and cell culture
MiaPaCa2, Panc-1, BxPC3, and HS-766T PDAC cell lines were obtained from the ATCC (2013). All cell lines were tested routinely and prior to all metabolomic analyses, for mycoplasma contamination and grown at 37°C and 5% CO2. Parental lines and their genetic modifications are summarized in Supplementary Table S1. Early passages (3–5) of these cultured cells were used for all the experiments. Under standard culture conditions, DMEM (containing 25 mmol/L glucose or 450 mg/dl, and 4 mmol/L glutamine or 36 mg/dl) was used. Media was supplemented with 10% FBS, 1% l-glutamine (200 mmol/L), and 1% penicillin/streptomycin (Invitrogen). In this study, 5 mmol/L glucose and 500 µmol/L glutamine were typically used to simulate nutrient withdrawal. Although the literature quotes these as normal levels in patient serum, they more accurately produce a relative nutrient withdrawal in cell line cultures chronically adapted to nutrient abundance. Lower doses may result in excessive toxicity and therefore can confound interpretation of biologic assays (15). Moreover, nutrient levels decline steadily over multiday experiments well into the subnormal range (15). For all in vitro assays, experiments were performed in triplicate.
MiaPaca2 cells with stable doxycycline inducible suppression of HuR were generated using lentiviral transduction of shRNAs using a Tet-pLKO-puro backbone plasmid (Addgene; 21915), as described (17). CRISPR/Cas9-mediated knockout of HuR in MiaPaCa2 and HS-766T cells was accomplished using a guide RNA targeting HuR, fused with CRISPR/Cas9 and GFP protein (18). Plasmids were designed and purchased from (Sigma-Aldrich) along with the CRISPR Universal Negative Control plasmid (CRISPR06-1EA). Stable cell line cultures with IDH1 overexpression were generated using MiaPaCa2 cells previously modified as CRISPR/Cas9 HuR knockouts. IDH1 transduction was performed in these cells using retroviral transduction of a pBABE-puro-WT.IDH1 plasmid, generously provided by Kun-Liang Guan (Moores Cancer Center, University of California, San Diego, CA). Scrambled pBABE-puro was used as a negative control plasmid (Addgene; 1764).
Drug sensitivity assays
Gemcitabine, oxaliplatin, and N-acetyl cysteine were purchased from Sigma-Aldrich. Drugs were dissolved in DMSO, PBS, and cell culture media, respectively. Cells were plated in 96-well plates at 103 cells per well and assayed using Quant-it PicoGreen dsDNA Assay Kit (Invitrogen) at 5 days (13). To estimate cell death, cells were trypsinized at 24 hours and counted after Trypan blue staining (Invitrogen) with a Hausser bright-line hemocytometer (Fisher Scientific).
Small RNA interference, cDNA transfections
HuR overexpression and siRNA transfections were performed as previously described (13, 15). Overexpression (OE) and empty vector (EV) plasmids were purchased from OriGene Technologies (pCMV6-XL5; SC116430). siRNA oligos were purchased from Life Technologies (siIDH1, 7121; siCTRL, AM4635). The siRNA against HuR (referenced herein as si.HuR) was a customized oligo designed to minimize off-target effects (sequence CCAUUAAGGUGUCGUAUGCUCUU).
Western blot analysis, immunofluorescence, ribonucleoprotein immunoprecipitation, half-life assays, and RNA quantitation
Protein detection and RNA quantitation (RT-qPCR) were performed using standard methodology, as previously described (13). When subcellular fractionation was performed for protein extraction, samples were prepared using the NE-PER Kit (Thermo Fisher Scientific). Membranes for immunoblotting were probed with antibodies against HuR (Santa Cruz Biotechnologies; 5261 clone 3A2), IDH1 (Abcam; ab 184615), GAPDH (Cell Signaling Technology; 5014), and α-Tubulin (Invitrogen; 32-2500). Actinomycin D (a transcription inhibitor) was used for half-life assays (ActD; 5 µg/mL; Fisher Scientific).
For immunofluorescence, cells were cultured at 5,000 cells per well on coverslips in 24-well plates. After appropriate treatments, cell were fixed with 3.7% paraformaldehyde for 10 minutes, permeabilized with 0.1% Triton-X 100 for 30 minutes, blocked with 5% goat serum for 1 hour at room temperature, and incubated with primary antibody (γH2AX; Millipore; JBW301; 1:500, HuR; Santa Cruz Biotechnologies; 5261 clone 3A2; 1:200, IDH1; Abcam; ab 184615; 1:300) overnight at 4°C Alexa Fluor 488 F anti-mouse secondary antibody was applied to coverslips for 1 hour the following day, nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) and mounted (ProLong Gold, Life Technologies) for analysis with a Zeiss LSM-510 Confocal Laser Microscope. After confocal acquisition, foci were quantified blindly using single plane images and plotted ± SEM (n = 30 foci counted).
Metabolomic profiling
For steady-state metabolomic analyses, cells were grown to ∼50% confluence in complete growth media on six-well plates in biological triplicates. The following day, complete media was exchanged with 25 or 5 mmol/L glucose media containing the [1,2-13C2]glucose tracer (Cambridge Isotope Labs), and cells were incubated for 24 hours prior to metabolite extraction. Metabolite fractions were extracted and analyzed by targeted gas chromatography-mass spectroscopy (GC-MS) via selected reaction monitoring (SRM), as previously described (19). Resulting mass isotopomer distributions were integrated and corrected for natural abundance using in-house algorithms.
Bioenergetics other metabolic assays
Measurements of mitochondrial function were undertaken using the Seahorse XF24 Extracellular Flux Analyzer (Seahorse Bioscience) as detailed in the supplemental section. The NADP+/NADPH ratio (Abcam; ab65349), intracellular GSSG/GSH ratios (Abcam; ab138881), α-ketoglutarate (BioVision; K677-100), and ATP (Abcam; 83355) were performed per the manufacturer’s instructions. For ROS assays, cells were incubated with 5 µmol/L 2′,7′-dichlorodihydrofluorescein diacetate (DCFDA; Invitrogen) and fluorescent DCF was measured per the manufacturer’s instructions. For the mitochondrial ROS levels, cells were incubated with 5 µmol/L MitoSOX Red mitochondrial superoxide indicator (Invitrogen; M36008) and the fluorescence measured per the manufacturer’s instructions using the GloMax Explorer system (Promega). Values were normalized to total protein.
Xenograft studies
All experiments involving mice were approved by the Thomas Jefferson University Institutional Animal Care Regulations and Use Committee. Six-week-old, male, athymic nude mice (Nude-Foxn1nu) were purchased from Harlan Laboratories (6903M). MiaPaca2 cells, or genetically modified variants of this cell line, were prepared in 100 µL solution comprised of 70% DPBS and 30% Matrigel. Suspensions of 5 × 106 cells were then injected subcutaneously into the left and right flanks of mice. Tumor volumes were measured three times per week using a caliper (volume = length × width2/2), along with body weights. Additional details are provided in supplemental experimental procedures.
Luciferase reporter assays
The full-length IDH1 3′UTR was subcloned into the psiCheck2 vector (Promega) at the XhoI and NotI restriction enzyme sites. A deletion series of the IDH1 3′UTR based on HuR binding predictions was generated as previously described (13). Renilla luciferase activity was normalized to firefly luciferase activity.
Immunohistochemistry
A tissue microarray (TMA) of resected PDAC and other periampullary cancer samples was prepared with formalin and embedded in paraffin, using tissues from previously consented patients undergoing treatment at Thomas Jefferson University Hospital. Standard immunolabeling techniques and IHC scoring were performed as described (20, 21) and detailed in the Supplemental section.
RNA sequencing
RNA seq was performed on an Illumina NextSeq 500 machine, and RNAs were interrogated from two different PDAC cell lines (MiaPaCa2 and HS-766T), both subjected to HuR deletion by CRISPR/Cas9 gene editing. Details of the techniques and bioinformatics are provided in the Supplementary section.
Clinical outcome data
All human subjects gave their written informed consent. For the clinical correlation analysis, we analyzed records from 724 consecutive patients with PDAC resected at Thomas Jefferson University Hospital between 2002 and 2014, after obtaining IRB approval. Clinical outcome analyses were separated into two related clinical cohorts, with details provided in the Supplementary section. In a cohort of 345 patients, HbA1C levels were available, along with pathologic descriptors (e.g., tumor size, lymph node status) of resected PDAC specimens. Patients were stratified by glycemic index (HbA1C or hemoglobin A1C) and correlation studies were performed to test for associations between HbA1C and pathologic features. For the survival analysis, 107 patients with resected PDAC who also underwent adjuvant chemotherapy were categorized into high glucose (HG, n = 86) and normal glucose (NG, n = 21) groups using HbA1C when values were available or a prior clinical diagnosis of diabetes as additional criteria of glycemic status. Recurrence data were available for these patients, and Cox multivariate hazard models were computed to estimate disease-free survival. Demographic details of the cohort are summarized (Supplementary Table S2).
Additional methodologic details for mouse studies, bioenergetics analyses, IDH1 mRNA analyses in patient specimens (from published datasets), IHC, RNA sequencing, and clinical outcome data are provided in the Supplementary Information.
Results
Low nutrient levels promote chemotherapy resistance in vitro
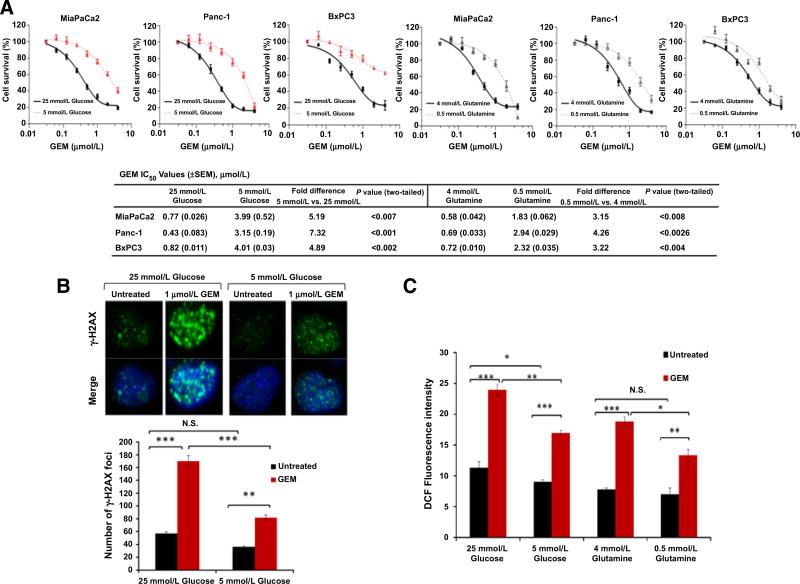
PDAC cells exhibited a 5- to 10-fold increased resistance to either gemcitabine (Fig. 1A) or oxaliplatin (Supplementary Fig. S1A) in vitro when exposed to low levels of glucose and glutamine over the course of the experiment. In the absence of chemotherapy however, PDAC cells had decreased cell survival under low glucose or glutamine conditions, compared to nutrient abundance (Supplementary Fig. S1B). As further evidence of nutrient withdrawal-associated chemoresistance, PDAC cells that were first precultured under low glucose conditions for 30 hours experienced less DNA damage with subsequent chemotherapy, as detected by a reduction in gamma H2AX foci formation, compared to cells cultured for the entire time under normal (supra-physiologic) culture conditions (Fig. 1B). As observed in prior studies (8, 9), nutrient withdrawal and chemotherapy proved to be potent inducers of ROS in PDAC cells (as measured by DCF fluorescence), with the greatest increase observed after simultaneous exposure of PDAC cells to both stressors (Supplementary Fig. S1C). However, when cells were again precultured initially for 30 hours with low glucose or glutamine, subsequent gemcitabine treatment had an attenuated effect on ROS levels (Fig. 1C). Together, these data suggest that nutrient deprivation (a feature of the normal PDAC microenvironment) slows PDAC cell growth in the absence of treatment, but may initiate an adaptive pro-survival and antioxidant program that renders PDAC cells resistant to additional oxidative insults, such as chemotherapy.
Figure 1. Low nutrient levels promote chemotherapy resistance in PDAC (in vitro model).
A, Survival of PDAC cell lines treated with the indicated doses of gemcitabine (GEM). IC50 values are provided. Cell survival was calculated by measurement of dsDNA content using PicoGreen. B, γ-H2AX foci. MiaPaCa2 cells cultured for 48 hours and treated with gemcitabine for the final 18 hours. C, ROS levels in MiaPaCa2 cells by DCF fluorescence and under the indicated conditions for 48 hours. Cells were treated with gemcitabine (1 µmol/L) for the final 18 hours. Error bars represent ± SEM of triplicate wells from a representative experiment. (N.S., nonsignificant; *, P < 0.05; **, P < 0.01; ***, P < 0.001). See also Supplementary Fig. S1.
Low nutrient levels promote chemotherapy resistance in vivo
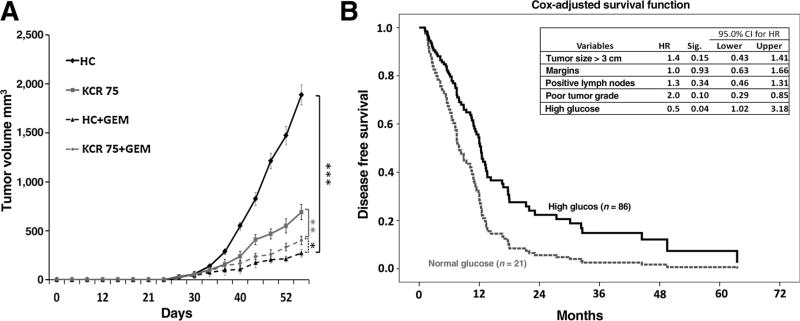
We further evaluated whether nutrient deprivation contributes to chemotherapy resistance in vivo. Isolated manipulation of nutrient levels in the tumor alone (vs. the whole mouse) is a technical challenge. Therefore, we examined the impact of modulating peripheral glucose levels in mice on tumor growth, as prior studies have demonstrated that peripheral glucose levels vary directly with intratumoral glucose levels (22, 23). These studies reveal that small changes in serum glucose levels result in even larger changes in xenograft glucose levels (and in the same direction). Mice fed a high carbohydrate (HC) diet experienced more than a two-fold increase in peripheral glucose levels (median 173 mg/dL vs. 71 mg/dL by 4 weeks, P < 0.0001) compared to mice fed a ketogenic and calorie restricted diet (fed 75% of baseline calories, referred to herein as KCR75; Supplementary Fig. S2A). In the absence of chemotherapy treatment, hyperglycemic mice experienced a higher xenograft growth rate than hypoglycemic mice (top two curves, Fig. 2A). However, these proliferative tumors exposed to higher ambient glucose levels were much more responsive to gemcitabine (1,887.04 mm vs. 289.92 mm, 84% reduction, P = 0.0008) than slower growing tumors in hypoglycemic mice (690.24 vs. 400.81, 41% reduction, P = 0.003; Fig. 2A and Supplementary Fig. S2B). These data suggest that in this animal model, like the in vitro model, nutrient withdrawal slows growth, yet induces a chemoresistant phenotype.
Figure 2. Low nutrient conditions promote chemotherapy resistance in PDAC (in vivo mouse and in patients).
A, Tumor growth curves of MiaPaCa2 PDAC xenografts in mice fed an HC or a ketogenic and calorie restricted diet (75% of the average caloric intake, KCR75). Each data point represents the mean ± SEM (n= 5 per group). *, P < 0.05; **, P < 0.01; ***, P < 0.001. B, Cox multivariate disease-free survival in patients receiving gemcitabine after resection for PDAC. See also Supplementary Fig. S2. GEM, gemcitabine.
To further investigate the clinical relevance of this observation, we examined pathology and outcome data in a large cohort of patients who underwent surgery for pancreatic cancer. On the basis of the above preclinical studies, we hypothesized that high ambient glucose levels in patients may promote tumor growth in the absence of chemotherapy, but also render tumors more vulnerable to chemotherapy, compared to nutrient-deprived PDACs in normoglycemic patients. To test this hypothesis, patients were stratified by HbA1C levels, which is a chronic biomarker of glycemic status (24). As expected, tumors in hyperglycemic patients had worse pathologic features in their resected specimens. Specifically, patients with elevated HbA1C levels had larger tumors ssand increased regional lymph node spread observed at resection, prior to any exposure to chemotherapy (Supplementary Figs. S2C and S2D). Both of these pathologic features are validated prognostic predictors of poor outcome (25, 26), demonstrating that PDACs exposed to HG environments behave more aggressively, have increased growth, and even are more likely to metastasize (keeping with the trend observed in the mouse study). Despite this finding, in a subgroup of patients who later received gemcitabine adjuvant therapy, patients with HG levels (i.e., the more aggressive cancers) surprisingly experienced superior disease-free survival compared to patients with NG levels, even in an adjusted multivariate model that included important prognostic features (Fig. 2B and refs. 25, 26). The model in Supplementary Figure S2E summarizes the observations across these diverse PDAC systems (in vitro, in vivo, patients); whereas PDAC cells grow more slowly under nutrient withdrawal, harsh conditions (i.e., a low glucose environment) induce chemotherapy resistance.
Silencing HuR combined with nutrient withdrawal inhibits tumor growth in vivo
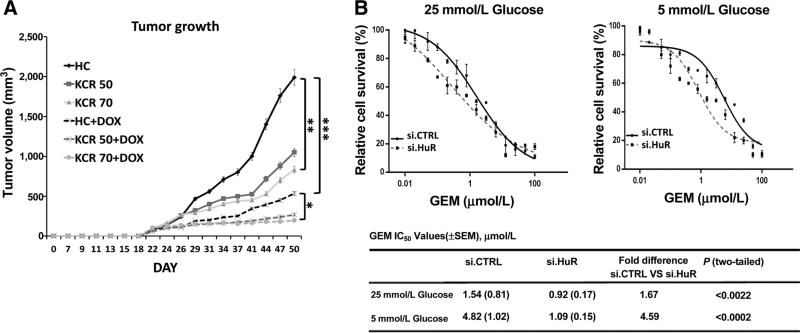
We pursued the regulatory mechanism behind PDAC adaptation to low nutrient conditions, both in the presence and absence of chemotherapy. Recent work by our group demonstrated a protective role for the RNA binding and regulatory protein, HuR, for PDAC cell survival under low glucose conditions (15). This prior work included in vitro experiments performed under glucose withdrawal, but did not further investigate the impact of HuR on PDAC cell viability after withdrawal of other nutrients, with chemotherapy treatment, or in an animal model (all addressed in this study). As a first step towards answering these questions, we used a previously generated doxycycline inducible shRNA PDAC xenograft system (MiaPaCa2.sh.HuR) to reversibly silence HuR with oral doxycycline (17). Cell culture validation of the system is provided in Supplementary Figure S3A. Again, mice fed a ketogenic and calorie-restricted diet had markedly lower peripheral glucose levels (Supplementary Fig. S3B). Mice fed the KCR diet had a median glucose level of 69 mg/dL at 4 weeks, as compared to 171 mg/dL in mice with HG levels (P < 0.0001). Decreased tumor growth was observed in mice with low peripheral glucose levels in the absence of doxycycline (HuR-proficient xenografts; Fig. 3A), but the combination of low glucose levels and HuR silencing (with doxycycline) had the greatest impact, essentially thwarting any additional xenograft growth beyond 4 weeks (Fig. 3A and Supplementary Fig. 3C). HuR mRNA and protein silencing in the xenografts was confirmed by both qPCR and immunoblot, from xenograft lysates obtained after 50 days (Supplementary Fig. S3D).
Figure 3. HuR protects PDAC cells against nutrient withdrawal and chemotherapy.
A, Tumor growth curves of MiaPaCa2 PDAC xenografts containing a doxycycline (DOX)-inducible shRNA construct against HuR (MiaPaCa2.sh.HuR). Mice were fed an HC diet, a ketogenic diet with 75% caloric intake (KCR75), or a ketogenic diet with 50% caloric intake (KCR50). Each data point represents the mean ± SEM (n= 8 per group). B, PicoGreen cell survival and drug sensitivity assays in MiaPaCa2 cells treated with gemcitabine (GEM). See also Supplementary Figs. S3 and S4.
Like glucose withdrawal, glutamine withdrawal activates HuR
The impact of glutamine withdrawal (as an alternative to glucose withdrawal) on HuR biology was assessed to determine if adaptive reprogramming by HuR was applicable to other nutrient sources, beyond glucose deprivation. Glutamine withdrawal indeed induced HuR translocation from the nucleus to the cytoplasm in MiaPaCa2 parental cells (similar to glucose withdrawal; ref. 15), as detected by both immunofluorescence (Supplementary Fig. S4A) and immunoblot of cytoplasmic lysates (Supplementary Fig. S4B). In both MiaPaCa2.si.HuR and Panc1. si.HuR cells, HuR silencing impaired cell viability with decreasing levels of glutamine in the absence of chemotherapy (PicoGreen assay, seven-fold decrease in the IC50; Supplementary Fig. S4C). In contrast, HuR overexpression (MiaPaCa2.HOE and Panc1.HOE) was protective under these conditions (8.65-fold increase in IC50; Supplementary Fig. S4C). These findings were further validated by a Trypan Blue assay, with a 15% to 30% increase in cell death by 72 hours (Supplementary Fig. S4D). Importantly, HuR also affected chemo-response under low nutrient conditions. HuR silencing not only prevented nutrient withdrawal-associated chemoresistance observed above in parental cells (a five-fold affect, see Fig. 1A), but rather sensitized MiaPaCa2.si.HuR cells to gemcitabine by an additional five-fold compared to MiaPaCa2.si.CTRL (Fig.3B and Supplementary Fig. S4E).
HuR minimizes the levels of ROS in PDAC cells
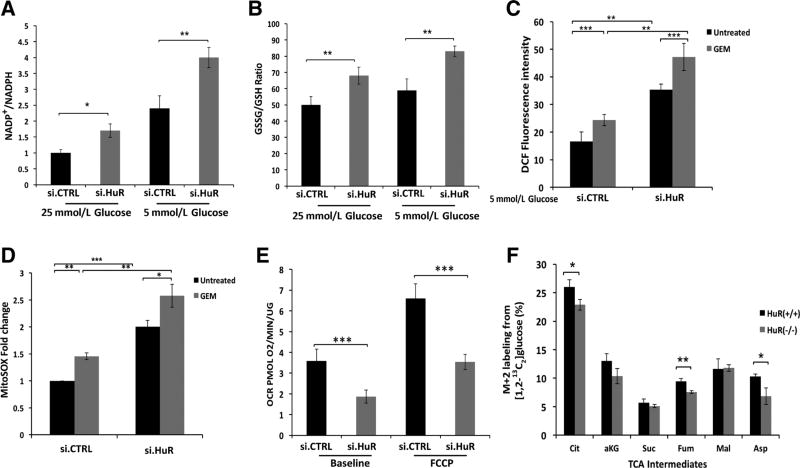
Through multiple independent assays, HuR expression reduced ROS levels under low glucose, whereas HuR silencing augmented ROS levels. For instance, compared to MiaPaCa2.si.CTRL cells, MiaPaCa2.si.HuR cells had a rise in the NADP+/NADPH ratio, GSSG/GSH ratio, DCF fluorescence, and mitochondrial ROS (Fig. 4A – D). Oxidative stress trended the highest when PDAC cells were simultaneously exposed to both glucose withdrawal and gemcitabine (Fig. 4C).
Figure 4. HuR minimizes ROS levels, while enhancing mitochondrial function.
A, NADP+/NADPH ratio in MiaPaCa2 cells cultured under the indicated conditions for 24 hours. B, GSSG/GSH ratio in MiaPaCa2 cells under the same conditions. C, ROS levels, detected by DCF fluorescence in MiaPaCa2 cells cultured in 5 mmol/L glucose media for 24 hours, and gemcitabine (GEM; 1 µmol/L) as indicated. D, Mitochondrial-specific ROS were analyzed using MitoSOX in MiaPaCa2 cells cultured in 5 mmol/L glucose media for 24 hours, and gemcitabine (1 µmol/L) as indicated. E, Oxygen consumption rates (OCR) in MiaPaCa2 cells, cultured in 5 mmol/L glucose for 24 hours. F, Quantitative 13C-isotope tracer labeling of TCA cycle intermediates (or surrogate metabolites, e.g., Asp) in MiaPaca2 cells that were modified by CRISPR/Cas9 against HuR(−/−), or isogenic controls (+/+). Cells were cultured in 5 mmol/L glucose for 24 hours in the presence of [1,2-13C6]glucose. Each data point represents the mean ± SEM of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Supplementary Fig. S5.
Conceptually, HuR could possibly mitigate ROS levels by either reducing ROS production or by enhancing ROS clearance. Because mitochondria are the dominant producers of ROS in cancer cells (27), we tested the former possibility first by examining the effect of HuR on diverse mitochondrial functions. Diminution of mitochondrial function would be one strategy for cancer cells to lower ROS production. Although glucose withdrawal increased oxygen consumption in parental MiaPaCa2 cells (Seahorse XF analysis; Supplementary Fig. S5A), the increase was effectively blocked by transient HuR silencing (Fig. 4E). In addition, HuR silencing minimized cellular ATP levels under low glucose conditions (Supplementary Fig. S5B). We traced the fate of 13C from [1, 2-13C2]glucose by GC-MS in HuR-knockout cells [Mia.HuR(−/−) vs. Mia.HuR(+/+)], and observed a mild attenuation of carbon flux in the TCA cycle under low glucose conditions in Mia.HuR (−/−), as evidenced by lower M2 fractions of multiple relevant metabolites (Fig. 4F and Supplementary Figs. S5C). Because HuR generally augments mitochondrial activity according to these findings, increased HuR expression or activity is unlikely to attenuate mitochondrial ROS production. Instead, the data collectively point to enhanced ROS clearance (as opposed to diminished ROS production) as the principal mechanism of HuR-mediated ROS control in glucose-deprived PDAC (as summarized in Supplementary Fig. S5D).
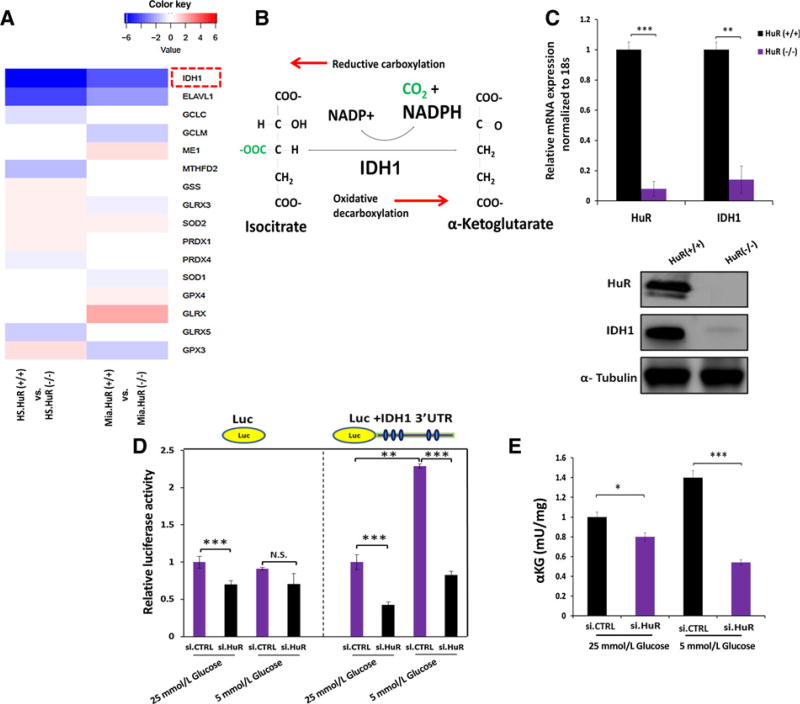
HuR impacts antioxidant defense by regulating IDH1
We performed unsupervised deep RNA sequencing in two different PDAC cell lines modified by CRISPR/Cas9 editing to delete HuR expression (MiaPaCa2 and HS-766T). RNA expression was compared in HuR(−/−) and HuR(+/+) cells (Supplementary Table S3 lists the results for coding transcripts), with a particular focus on 40 established antioxidant enzymes (Supplementary Tables S4 and S5). A total of 15 enzymes were significantly dysregulated in at least one of the two HuR(−/−) cell lines compared to the HuR(+/+) lines, but only IDH1 (isocitrate dehydrogenase 1) was suppressed in both Mia.HuR(−/−) and HS.HuR(−/−) cells (Fig. 5A). Moreover, the magnitude of IDH1 suppression in HuR(−/−) cells compared to HuR(+/+) cells was even greater than the decrease in HuR expression (or ELAVL1, included in Fig. 5A as a reference). IDH1 is a cytosolic enzyme that catalyzes the interconversion of isocitrate and α-ketoglutarate, and couples this redox reaction to NADP+/NADPH (Fig. 5B). Oxidative decarboxylation (rightward direction in Fig. 5B) produces NADPH. We validated the RNA seq findings through a separate HuR silencing experiment in MiaPaCa2 cells under low glucose conditions. Out of the eight different NADPH generating enzymes listed at the top of Supplementary Table S4, only IDH1 was markedly downregulated by HuR silencing (Supplementary Fig. S6A). The importance of IDH1 under acute glucose withdrawal, compared to these other NADPH generating enzymes, was further suggested by the observation that it was one of just two enzymes to increase in expression with this metabolic stress (Supplementary Fig. S6B). Downregulation of IDH1 was further confirmed in a separate in vivo experiment using the doxycycline-inducible sh.HuR system; IDH1 mRNA levels were indeed reduced in MiaPaCa2.sh.HuR cells when mice were fed DOX-supplemented chow for 43 days (Supplementary Fig. S6C). Next, an HuR-specific ribonucleoprotein immunoprecipitation (RNP-IP) assay was performed in MiaPaCa2 cells, providing evidence that HuR binds directly to the IDH1 mRNA transcript, similar to DCK, an established HuR target (ref. 20; Supplementary Fig. S6D, GLUT1 serves as a negative control; ref. 15). IDH1 mRNA and protein expression were virtually undetectable in both Mia. HuR(−/−) and HS.HuR(−/−) compared to the control cells (Fig. 5C and Supplementary Fig. S6E), proving that HuR is required for IDH1 expression in these models. As confirmation that HuR regulates IDH1, reintroduction of HuR into Mia.HuR(−/−) and HS.HuR(−/−) cells restored IDH1 expression (Supplementary Fig. S6F).
Figure 5. HuR regulates IDH1 expression in PDAC.
A, RNA sequencing was performed in two PDAC cell lines (HS-766T and MiaPaCa2) modulated by CRISPR/Cas9 against HuR. The heatmap displays transcripts encoding antioxidant enzymes (out of 40 directly involved in ROS detoxification), with significant changes in HuR(−/−) cells as compared with the HuR(+/+) control cells. HuR (ELAVL1, the CRISPR deletion target) is included in the table as reference. B, Schematic of the reversible IDH1 catalytic reaction. NADPH is produced by oxidative decarboxylation. C, HuR and IDH1 mRNA levels in MiaPaca2 CRISPR HuR(+/+) or HuR(−/−) cells; immunoblot analysis of IDH1 and HuR protein in the same cells. D, MiaPaCa2 cells were cotransfected with siRNAs and luciferase reporter constructs (luciferase control or fused with IDH1 3′ UTR). Cells were cultured as indicated for 24 hours. E, αKG levels in MiaPaCa2 cells cultured under the indicated conditions for 24 hours. Error bars, ± SEM of triplicate wells from a representative experiment. *, P < 0.05; **, P < 0.01; ***, P < 0.001. See also Supplementary Figs. S6 and S7.
A computational analysis of a publically available HuR-crosslinking immunoprecipitation (CLiP) Seq dataset (28) identified five AREs in the 3′UTR of the IDH1 transcript as potential HuR binding sites (Supplementary Fig. S7A). We therefore subcloned the entire 3′UTR containing these sequences into a luciferase reporter plasmid and measured the impact of HuR expression on reporter activity in MiaPaCa2 cells, as definitive proof of a regulatory interaction. Transient HuR silencing and overexpression impacted luciferase activity in an HuR-dependent manner, especially under low glucose conditions (Figs. 5D; Supplementary Fig. S7B). Subcloning of multiple deletion constructs derived from the full length IDH1 3′UTR further honed the critical regulatory sequence to the upstream 192 bp region, with functional contributions from each of the three predicted AREs within this sequence (Supplementary Fig. S7C). Treatment of MiaPaCa2. si.HuR cells with actinomycin D (transcriptional inhibition) revealed a decrease in IDH1 mRNA stability, confirming that control of IDH1 transcript levels by HuR was post-transcriptional (Supplementary Fig. S7D).
HuR’s regulation of IDH1 enhances PDAC reductive power
We observed an increase in the α-ketoglutarate/citrate ratio in HuR-proficient Mia.HuR(+/+) cells upon glucose withdrawal (detected by GC-MS). However, the same increase was not observed in the Mia.HuR(−/−) cells (Supplementary Fig. S7E), providing biochemical evidence that the IDH reaction predominantly proceeds in the oxidative direction under low glucose to generate NADPH (rightward direction in Fig. 5B). Similarly, we observed a reduction in α-ketoglutarate levels in MiaPaCa2.si. HuR cells under glucose withdrawal (vs. MiaPaCa2.si.CTRL), using a separate redox-coupled colorimetric assay (Fig. 5E). These findings suggest that HuR’s regulation of IDH1 supports the generation of NADPH and reductive power under low glucose conditions.
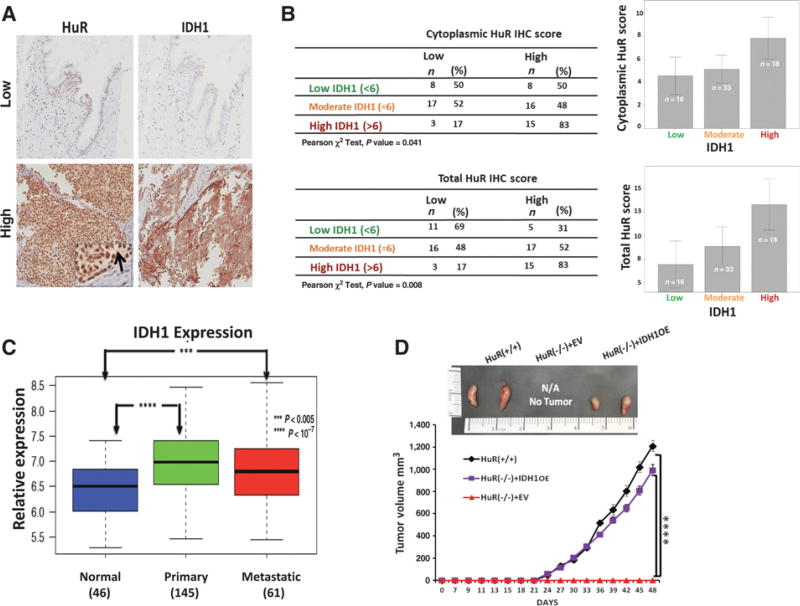
HuR and IDH1 expression are associated in clinical specimens
The interaction between HuR and IDH1 was next assessed in patient clinical samples using a TMA derived from resected specimens of PDAC and other periampullary cancers. Both total and cytoplasmic HuR (the latter being the best marker of the activated protein; ref. 21) correlated with IDH1 protein expression by immunohistochemistry (Fig. 6A and B). In addition, we interrogated a publically reported gene expression microarray (29) for IDH1 mRNA expression, and observed increased levels of the transcript in both primary and metastatic PDAC compared to normal tissue (Fig. 6C). These findings parallel previous observations by our group (20), and by others (30), which revealed that cytoplasmic HuR protein expression (i.e., the sub-cellular location where the protein most affects RNA stability of its bound mRNA targets) is increased in cancer tissues.
Figure 6. HuR is associated with IDH1 expression in clinical PDAC samples.
A, Immunohistochemistry of HuR and IDH1 in formalin-fixed, paraffin-embedded PDAC tumors. The inset and arrow indicate cytoplasmic HuR expression. B, Tabulation and graphing of IHC scores from the TMA dataset shows cytoplasmic and total HuR IHC scores to be associated with IDH1 scores. Spearman correlation test yielded a significant correlation between the scores (P = 0.33, P < 0.01). Error bars display 95% confidence interval. C, IDH1 mRNA expression levels in normal pancreatic tissue, primary PDAC, and metastatic PDAC. For each boxplot, median expression value, and first and third quartiles are indicated. ***, P < 0.005; ****, P < 0.0000001. D, Representative images of subcutaneous MiaPaca2 xenografts (day 48) along with tumor growth curves. Each data point represents the mean ± SEM (n= 8 per group; ****, P < 0.0001). See also Supplementary Fig. S8.
IDH1 can phenocopy HuR both in vitro and in vivo
We observed in vivo that CRISPR deletion of the HuR gene from PDAC cells [Mia.HuR(−/−)] abrogated xenograft transplantation and uptake in the flanks of nude mice (18), underscoring the profound phenotypic impact of HuR expression on tumor growth in an animal model. In this study, stable reconstitution of IDH1 alone by retroviral transduction in these cells [Mia.HuR(−/−). IDH1OE] completely rescued engraftment potential (Fig. 6D).
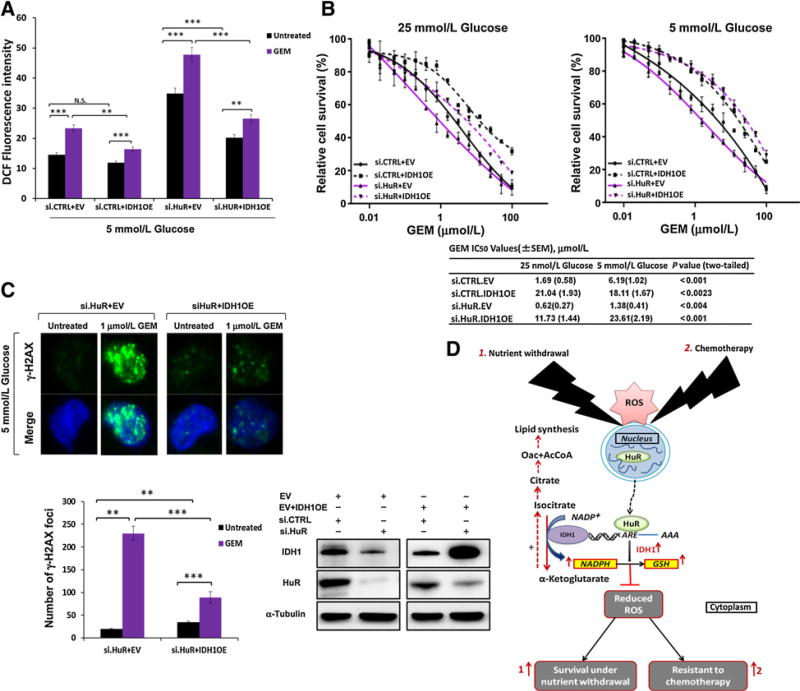
Similar to MiaPaCa2.si.HuR in vitro (Fig. 4C), MiaPaCa2.si. IDH1 had increased ROS levels (DCF fluorescence) compared to MiaPaCa2.si.CTRL after nutrient withdrawal (either glucose or glutamine) and chemotherapy treatment (Supplementary Fig. S8A). Accordingly, IDH1 overexpression reduced ROS levels under these conditions (Supplementary Fig. S8B). IDH1 over-expression also enhanced NADPH levels under low glucose conditions in two PDAC cell lines (Supplementary Fig. S8C). Similar to MiaPaCa2.si.HuR above (Figs. 3B and Supplementary Fig. S4E), MiaPaCa2.si.IDH1 cells showed decreased cell survival with gemcitabine treatment (Supplementary Fig. S8D). The effect was particularly pronounced under glucose withdrawal. Moreover, increased DNA damage with gemcitabine was observed with MiaPaCa2.si.IDH1, whereas IDH1 overexpression (MiaPaCa2. IDH1OE) was protective against DNA damage by gemcitabine (Supplementary Fig. S8E). The antioxidant, n-acetyl cysteine, was protective to both MiaPaCa2.si.IDH1 and MiaPaCa2.si.HuR cells after treatment with gemcitabine, under low glucose conditions (Supplementary Fig. S8F), further linking the HuR-IDH1 regulatory axis to antioxidant defense.
We performed IDH1 rescue experiments in an HuR-deficient background, by cotransfecting IDH1 cDNA plasmids with siHuR oligos, or by transfecting the plasmids into HuR-knockout cells [Mia.HuR(−/−)]. In the presence of gemcitabine treatment, IDH1 overexpression returned ROS levels, cell death, and DNA damage close to levels observed in untreated HuR-proficient cells (Fig. 7A–C and Supplementary Fig. S8G), to effectively restore chemo-resistance in these models.
Figure 7. IDH1 rescues HuR-deficient PDAC cells from metabolic stress.
A, ROS levels in MiaPaCa2 as measured by DCF fluorescence. The indicated culture conditions lasted 48 hours, with gemcitabine (GEM; 1 µmol/L) treatment for the final 18 hours, as indicated. Cotransfections with siRNAs and overexpression plasmids were performed as indicated and repeated for B and C. B, PicoGreen cell survival and drug sensitivity assays in MiaPaCa2 cells. C, γ-H2AX foci in MiaPaCa2 cells. Culture and treatments paralleled the experiment in A. D, A schematic model of how HuR’s regulation of IDH1 promotes survival under nutrient withdrawal and chemotherapy treatment by enhancing reductive power and antioxidant defense. Error bars, ± SEM of triplicate wells from a representative experiment (N.S., nonsignificant; **, P < 0.01; ***, P < 0.001). See also Supplementary Fig. S8.
Discussion
Pancreatic cancer lethality cannot be blamed solely on ineffective cancer screening and late cancer diagnosis. Rather, the evidence indicates that the disease is more aggressive than most other common cancers. For instance, survival with advanced disease is significantly worse than other cancers (31), and early lesions with microinvasion are more likely to spread and recur (32). Similarly, cytotoxic chemotherapy is less effective in PDAC (4, 5). The molecular changes that govern PDAC fitness in the face of severe stress like harsh conditions (1) or chemotherapy have not been fully elucidated. In recent years, there have been a growing number of studies addressing the question of how PDAC cells overcome the “desert”-like conditions present in the micro-environment. These studies were motivated by a presumption that metabolic dependencies in nutrient-deprived tumor tissue (1) may be less important or deprioritized in well perfused normal tissue. Thus, molecular insights into the metabolic dependencies of PDAC could uncover a natural therapeutic window. For example, Birsoy and colleagues observed in cancer cells that are well adapted to low glucose conditions, that mitochondrial function is enhanced in order to optimize ATP yield when nutrients are scarce (33). Commisso and colleagues, described macropinocytosis as an important mechanism for amino acid scavenging under nutrient deprived conditions in PDAC (34). Sousa and colleagues recently demonstrated that pancreatic stellate cells in the microenvironment provide alanine as an alternative carbon source under glucose-limiting conditions (35). In this study, we highlight two promising therapeutic targets (HuR and IDH1) with increased biologic importance for PDAC under physiologic and nutrient-deprived conditions. Moreover, we propose that the HuR-IDH1 regulatory axis recruited in this context further protects at risk PDAC cells from additional oxidative insults, like chemotherapy, leading to a chemoresistant phenotype.
From a more broad perspective, we demonstrate using multiple different model systems (in vitro, in vivo, patients) that low nutrient conditions drives PDAC chemoresistance. For instance, we show in three different PDAC cell lines, that regardless of the reduced nutrient (glucose or glutamine), PDAC cells are substantially more refractory to cytotoxic therapy. Moreover, this pattern was not dependent on the mechanism of chemotherapy drug action (gemcitabine is an anti-metabolite and oxaliplatin is a DNA cross-linker). Further support for these findings was observed in both an animal model and actual patient data. Although the in vivo studies do not exactly parallel the cell culture experiments, in each model, more austere conditions (withdrawal of glucose from the culture media, a low glucose mouse diet, or normoglycemia in patients) resulted in a diminished treatment response. Therefore, these findings may collectively offer insight into why current pharmacologic agents are largely ineffective under physiologic conditions. The data suggest that PDAC cells recruit a promiscuous prosurvival program under metabolic stress, which also confers resistance to conventional therapeutics.
In this work, we highlight antioxidant defense as one potential adaptive strategy used by PDAC cells to survive both harsh conditions and chemotherapeutic stress. More specifically, we demonstrate that HuR’s regulation of IDH1 is central to this task. IDH1 has been the subject of numerous studies in the cancer research field because of its high mutation frequency in many cancers (36). Of note, this genetic abnormality is exceptionally rare in PDAC, and has only recently been reported in a single case by our group (37). We theorize that the rarity of this mutation in PDAC is related to the deleterious effect that has been ascribed to haploinsufficiency of the wild type allele under very austere conditions (38). IDH1 mutations divert α-ketogluatrate to 2-hydroxyglutarate, and consume NADPH in the process. Although this chemical reaction is oncogenic in affected tumors, it can have devastating consequences on the reductive capacity of PDAC cells living in a highly oxidative environment. In fact, IDH1 mutant tumors tend to be less necrotic, less aggressive, and less chemoresistant (i.e., less “PDAC-like”) than their wild-type counterparts (39). It stands to reason that a lack of IDH1 mutations in PDAC provides indirect support for the importance of wild type IDH1 biology in PDAC.
Prior work has already demonstrated a prosurvival role for wild-type IDH1 in stressed cancer cells. For example, IDH1 activity promoted reductive carboxylation of α-ketogluatrate under hypoxia as a compensatory lipogenic pathway in the context of impaired mitochondria (19). In addition, lower ROS levels attributed to wild type IDH expression enabled in vitro spheroid growth (a cell culture model where nutrient gradients exist; ref. 40). In a separate study, IDH1-deficient mice were especially susceptible to oxidative liver damage with LPS administration (41). Because IDH1 is just one of eight NADPH generating enzymes, from four separate metabolic pathways (Supplementary Fig. S6A), its function on the surface may appear expendable. However, only IDH1 is regulated by the acute stress response and prosurvival protein, HuR. In fact, stable IDH1 overexpression restored xenograft uptake and growth in HuR-null PDAC cells, and IDH1 overexpression restored chemotherapy resistance in HuR-deficient PDAC cells exposed to nutrient withdrawal. Thus, PDAC cells may be particularly dependent on the HuR–IDH1 regulatory axis for adaptive survival under acute stress (as summarized in Fig. 7D), highlighting a potential metabolic vulnerability and therapeutic opportunity.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by American Cancer Society Mentored Research ScholarGrant-14-019-01-CDD (J.M. Winter) and Gail V. Coleman-Kenneth M. Bruntel Pancreatic Cancer Research Fund (J.M. Winter).
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
C.M. Metallo is a consultant/advisory board member of Agios Pharmaceuticals. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: M. Zarei, C.J. Yeo, J.R. Brody, J.M. Winter
Development of methodology: M. Zarei, S. Lal, N.C. Mambelli-Lisboa, M. Jimbo, J.M. Winter
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M. Zarei, S. Lal, S.J. Parker, A. Nevler, K. Dukleska, C. Moffat, F.F. Blanco, S.N. Chand, M. Jimbo, W. Jiang, E.L. Seifert, C.M. Metallo, J.M. Winter
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M. Zarei, S. Lal, S.J. Parker, A. Nevler, A. Vaziri-Gohar, K. Dukleska, C. Moffat, F.F. Blanco, S.N. Chand, W. Jiang, E.R. Londin, E.L. Seifert, C.M. Metallo, J.R. Brody, J.M. Winter
Writing, review, and/or revision of the manuscript: M. Zarei, M. Jimbo, W. Jiang, C.J. Yeo, C.M. Metallo, J.R. Brody, J.M. Winter
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. Zarei, C. Moffat, J.A. Cozzitorto, J.M. Winter
Study supervision: M. Zarei, C.J. Yeo, J.R. Brody, J.M. Winter
Other (helped with experiments): A. Vaziri-Gohar
References
- 1.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 2015;75:544–53. doi: 10.1158/0008-5472.CAN-14-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Provenzano PP, Cuevas C, Chang AE, Goel VK, Von Hoff DD, Hingorani SR. Enzymatic targeting of the stroma ablates physical barriers to treatment of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:418–29. doi: 10.1016/j.ccr.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ros PR, Mortele KJ. Imaging features of pancreatic neoplasms. JBR-BTR. 2001;84:239–49. [PubMed] [Google Scholar]
- 4.Morgan G, Ward R, Barton M. The contribution of cytotoxic chemotherapy to 5-year survival in adult malignancies. Clin Oncol. 2004;16:549–60. doi: 10.1016/j.clon.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25. doi: 10.1056/NEJMoa1011923. [DOI] [PubMed] [Google Scholar]
- 6.Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691–703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson AR, Weaver AM, Cummings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective pressure from the microenvironment. Cell. 2006;127:905–15. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 8.Ahmad IM, Aykin-Burns N, Sim JE, Walsh SA, Higashikubo R, Buettner GR, et al. Mitochondrial O2*- and H2O2 mediate glucose deprivation-induced stress in human cancer cells. J Biol Chem. 2005;280:4254–63. doi: 10.1074/jbc.M411662200. [DOI] [PubMed] [Google Scholar]
- 9.Sangeetha P, Das UN, Koratkar R, Suryaprabha P. Increase in free radical generation and lipid peroxidation following chemotherapy in patients with cancer. Free Radical Biol Med. 1990;8:15–9. doi: 10.1016/0891-5849(90)90139-a. [DOI] [PubMed] [Google Scholar]
- 10.Dong R, Lu JG, Wang Q, He XL, Chu YK, Ma QJ. Stabilization of Snail by HuR in the process of hydrogen peroxide induced cell migration. Biochem Biophys Res Commun. 2007;356:318–21. doi: 10.1016/j.bbrc.2007.02.145. [DOI] [PubMed] [Google Scholar]
- 11.Mehta M, Basalingappa K, Griffith JN, Andrade D, Babu A, Amreddy N, et al. HuR silencing elicits oxidative stress and DNA damage and sensitizes human triple-negative breast cancer cells to radiotherapy. Oncotarget. 2016;7(40):64820. doi: 10.18632/oncotarget.11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benoit RM, Meisner NC, Kallen J, Graff P, Hemmig R, Cebe R, et al. The x-ray crystal structure of the first RNA recognition motif and site-directed mutagenesis suggest a possible HuR redox sensing mechanism. J Mol Biol. 2010;397:1231–44. doi: 10.1016/j.jmb.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 13.Lal S, Burkhart RA, Beeharry N, Bhattacharjee V, Londin ER, Cozzitorto JA, et al. HuR posttranscriptionally regulates WEE1: implications for the DNA damage response in pancreatic cancer cells. Cancer Res. 2014;74:1128–40. doi: 10.1158/0008-5472.CAN-13-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blanco FF, Jimbo M, Wulfkuhle J, Gallagher I, Deng J, Enyenihi L, et al. The mRNA-binding protein HuR promotes hypoxia-induced chemoresistance through posttranscriptional regulation of the proto-oncogene PIM1 in pancreatic cancer cells. Oncogene. 2015;35:2529–41. doi: 10.1038/onc.2015.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burkhart RA, Pineda DM, Chand SN, Romeo C, Londin ER, Karoly ED, et al. HuR is a post-transcriptional regulator of core metabolic enzymes in pancreatic cancer. RNA Biol. 2013;10:1312–23. doi: 10.4161/rna.25274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oettle H, Neuhaus P, Hochhaus A, Hartmann JT, Gellert K, Ridwelski K, et al. Adjuvant chemotherapy with gemcitabine and long-term outcomes among patients with resected pancreatic cancer: the CONKO-001 randomized trial. JAMA. 2013;310:1473–81. doi: 10.1001/jama.2013.279201. [DOI] [PubMed] [Google Scholar]
- 17.Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL, et al. Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells. Oncotarget. 2015;6:27312–31. doi: 10.18632/oncotarget.4743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, et al. CRISPR knockout of the HuR gene causes a xenograft lethal phenotype. Mol Cancer Res. 2017 doi: 10.1158/1541-7786.MCR-16-0361. molcanres–0361.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–4. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, et al. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009;69:4567–72. doi: 10.1158/0008-5472.CAN-09-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards NG, Rittenhouse DW, Freydin B, Cozzitorto JA, Grenda D, Rui H, et al. HuR status is a powerful marker for prognosis and response to gemcitabine-based chemotherapy for resected pancreatic ductal adenocarcinoma patients. Ann Surg. 2010;252:499–505. doi: 10.1097/SLA.0b013e3181f1fd44. discussion −6. [DOI] [PubMed] [Google Scholar]
- 22.Peltz C, Schroeder T, Dewhirst MW. Monitoring metabolite gradients in the blood, liver, and tumor after induced hyperglycemia in rats with R3230 flank tumors using microdialysis and bioluminescence imaging. Adv Exp Med Biol. 2005;566:343–8. doi: 10.1007/0-387-26206-7_45. [DOI] [PubMed] [Google Scholar]
- 23.Schroeder T, Yuan H, Viglianti BL, Peltz C, Asopa S, Vujaskovic Z, et al. Spatial heterogeneity and oxygen dependence of glucose consumption in R3230Ac and fibrosarcomas of the Fischer 344 rat. Cancer Res. 2005;65:5163–71. doi: 10.1158/0008-5472.CAN-04-3900. [DOI] [PubMed] [Google Scholar]
- 24.Florkowski C. HbA1c as a diagnostic test for diabetes mellitus - reviewing the evidence. Clin Biochem Rev. 2013;34:75–83. [PMC free article] [PubMed] [Google Scholar]
- 25.Winter JM, Cameron JL, Campbell KA, Arnold MA, Chang DC, Coleman J, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: a single-institution experience. J Gastrointest Surg. 2006;10:1199–210. doi: 10.1016/j.gassur.2006.08.018. discussion 210-1. [DOI] [PubMed] [Google Scholar]
- 26.Pawlik TM, Gleisner AL, Cameron JL, Winter JM, Assumpcao L, Lillemoe KD, et al. Prognostic relevance of lymph node ratio following pancreati-coduodenectomy for pancreatic cancer. Surgery. 2007;141:610–8. doi: 10.1016/j.surg.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Holmstrom KM, Finkel T. Cellular mechanisms and physiological consequences of redox-dependent signalling. Nat Rev Mol Cell Biol. 2014;15:411–21. doi: 10.1038/nrm3801. [DOI] [PubMed] [Google Scholar]
- 28.Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell. 2011;43:327–39. doi: 10.1016/j.molcel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdisciplinary Rev RNA. 2010;1:214–29. doi: 10.1002/wrna.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: A Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 32.Winter JM, Jiang W, Basturk O, Mino-Kenuduson M, Fong ZV, Tan WP, et al. Recurrence and survival after resection of small intraductal papillary mucinous neoplasm-associated carcinomas (</=20-mm invasive component): a multi-institutional analysis. Ann Surg. 2015;263:793–801. doi: 10.1097/SLA.0000000000001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–12. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. 2013;497:633–7. doi: 10.1038/nature12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sousa CM, Biancur DE, Wang X, Halbrook CJ, Sherman MH, Zhang L, et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 2016;536:479–83. doi: 10.1038/nature19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19:764–72. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 37.Brody JR, Yabar CS, Zarei M, Bender J, Matrisian LM, Rahib L, et al. Identification of a novel metabolic-related mutation (IDH1) in metastatic pancreatic cancer. Cancer Biol Thera. 2016 doi: 10.1080/15384047.2016.1210743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grassian AR, Parker SJ, Davidson SM, Divakaruni AS, Green CR, Zhang X, et al. IDH1 mutations alter citric acid cycle metabolism and increase dependence on oxidative mitochondrial metabolism. Cancer Res. 2014;74:3317–31. doi: 10.1158/0008-5472.CAN-14-0772-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L, Shestov AA, Swain P, Yang C, Parker SJ, Wang QA, et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature. 2016;532:255–8. doi: 10.1038/nature17393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itsumi M, Inoue S, Elia AJ, Murakami K, Sasaki M, Lind EF, et al. Idh1 protects murine hepatocytes from endotoxin-induced oxidative stress by regulating the intracellular NADP(+)/NADPH ratio. Cell Death Differ. 2015;22:1837–45. doi: 10.1038/cdd.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.