Abstract
Excess lipid accumulation is an early signature of nonalcoholic fatty liver disease (NAFLD). Although liver receptor homolog 1 (LRH-1) (encoded by NR5A2) is suppressed in human NAFLD, evidence linking this phospholipid-bound nuclear receptor to hepatic lipid metabolism is lacking. Here, we report an essential role for LRH-1 in hepatic lipid storage and phospholipid composition based on an acute hepatic KO of LRH-1 in adult mice (LRH-1AAV8-Cre mice). Indeed, LRH-1–deficient hepatocytes exhibited large cytosolic lipid droplets and increased triglycerides (TGs). LRH-1–deficient mice fed high-fat diet displayed macrovesicular steatosis, liver injury, and glucose intolerance, all of which were reversed or improved by expressing wild-type human LRH-1. While hepatic lipid synthesis decreased and lipid export remained unchanged in mutants, elevated circulating free fatty acid helped explain the lipid imbalance in LRH-1AAV8-Cre mice. Lipidomic and genomic analyses revealed that loss of LRH-1 disrupts hepatic phospholipid composition, leading to lowered arachidonoyl (AA) phospholipids due to repression of Elovl5 and Fads2, two critical genes in AA biosynthesis. Our findings reveal a role for the phospholipid sensor LRH-1 in maintaining adequate pools of hepatic AA phospholipids, further supporting the idea that phospholipid diversity is an important contributor to healthy hepatic lipid storage.
Keywords: Hepatology, Metabolism
Keywords: Diabetes
LRH-1 plays a role in preserving normal hepatic lipid storage and phospholipid composition, implicating LRH-1 as a potential therapeutic target for NAFLD.
Introduction
Nonalcoholic fatty liver disease (NAFLD) has emerged as a major chronic problem in Western societies, and is tightly coupled with central obesity, metabolic syndrome, and polycystic ovarian disease, as reviewed in refs. 1, 2. Except for sustained lifestyle changes that are difficult to achieve, effective therapies to eliminate NAFLD remain limited. An elevation in hepatic triglycerides (TGs), known as nonalcoholic fatty liver, can progress to more serious nonalcoholic steatohepatitis (NASH) disease that involves hepatic inflammation and injury (3, 4), a transition from microvesicular to macrovesicular steatosis, and/or changes in hepatic phospholipid composition (5, 6).
Excess hepatic lipid accumulation results from disruption in import, export, or oxidation of lipids as well as changes in de novo lipid synthesis. Evidence in mice also suggests that maintenance of phospholipid diversity is an important factor in hepatic fat deposition and, specifically, that adequate levels of arachidonoyl (AA) phospholipid species ensure proper lipid trafficking and storage in both the liver (7–9) and intestine (10). Conditional KO of lysophospholipid acyltransferase LPCAT3 that generates hepatic AA phospholipids, results in fatty liver due to impaired TG packaging into VLDL particles. LPCAT3 is an LXR target that exhibits strong specificity for arachidonic acid (20:4) as well as linoleic acid (LA; 18:2) (11–13). Hepatic AA phospholipids are also diminished in the global KO of ELOVL5, an elongase required for de novo arachidonic acid synthesis from dietary LA (14), and in a mouse model that diminishes desaturase activity encoded by Fads2 (15); ElOVL5-null mutant mice develop hepatic steatosis due to increased de novo lipogenesis (9). These studies, together with the stepwise decrease in AA phospholipids in human NAFLD and NASH (5), support the concept that maintenance of hepatic AA phospholipid composition and phospholipid diversity are critical for hepatic lipid homeostasis.
Expression profiling reported that the liver receptor homolog 1 (LRH-1, encoded by NR5A2) is significantly downregulated in human patients with either simple bland hepatic steatosis or NASH (16). Despite this clinical correlation, the role of LRH-1 in hepatic lipid metabolism is not well delineated. LRH-1 was originally linked to bile acid synthesis based on cellular studies showing LRH-1 involvement in the classical or neutral bile acid synthesis via Cyp7a1 and in the alternative or acidic bile acid synthetic pathway via Cyp8b1 (17, 18). Unexpectedly, multiple liver-specific mouse KOs of LRH-1 using the albumin-Cre driver (Lrh-1Alb-Cre) report only modest changes in acidic bile acids (19–21). Moreover, glucose homeostasis and hepatic TGs are unchanged in Lrh-1Alb-Cre mice. Collectively, these studies imply that LRH-1 plays only a minor role in hepatic lipid metabolism when compared with other classes of nuclear receptors, such as FXR, LXR, and PPARα, that have been targeted for NAFLD therapy (as reviewed in ref. 1).
Given that LRH-1 as well as its close homolog steroidogenic factor 1 (SF-1, also known as NR5A1) possess large ligand-binding pockets able to bind phospholipids (22–26), we asked if LRH-1 helps preserve normal hepatic phospholipid composition. To test this hypothesis, we chose to acutely KO LRH-1 in the adult hepatocyte using viral-mediated AAV8-TBG-Cre deletion. Here, we report that, in contrast to Lrh-1Alb-Cre mice, eliminating LRH-1 in the adult hepatocyte (Lrh-1AAV8-Cre) profoundly affects hepatic lipid metabolism, leading to stark macrovesicular steatosis, mild liver injury, and poor glucose tolerance. Based on these phenotypic data coupled with genomic and lipidomic analyses, we propose that LRH-1 maintains the pool of AA phospholipids, thus ensuring phospholipid diversity and normal lipid homeostasis in the adult liver.
Results
Acute loss of LRH-1 results in hepatic steatosis in mice, independent of diet.
While hepatic TGs are unchanged in the conventional liver-specific KO of LRH-1 (Lrh-1Alb-Cre), increasing LRH-1 activity in adult mice appears to lower hepatic TGs after prolonged periods of overnutrition (19). To assess how loss of LRH-1 affects adult liver, and to avoid compensatory effects from the developmental Lrh-1Alb-Cre KO (LKO) model (27) or off-target effects of tamoxifen (28), we generated an adult liver-specific LRH-1 model (Lrh-1AAV8-Cre) by infecting 6-week-old Lrh-1fl/fl male mice with an AAV8 vector expressing Cre recombinase (AAV8-Cre) under the control of the thyroxine-binding globulin (TBG) promoter that restricts Cre activity to hepatocytes (29). Lrh-1fl/fl male mice infected with AAV8-GFP served as controls (Lrh-1AAV8-GFP). Loss of Lrh-1 transcripts in liver and in primary hepatocytes obtained from Lrh-1AAV8-Cre mice was detected as early as 2 weeks after infection, with knockdown efficiency at 8 weeks after infection comparable to that observed for the developmental LKO model (Figure 1A).
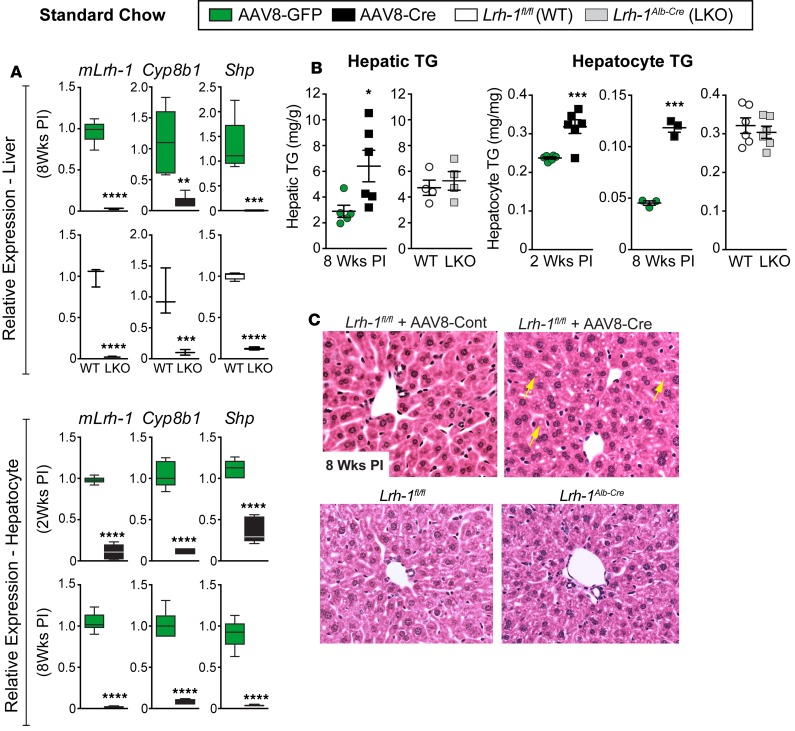
Figure 1. Deleting LRH-1 in adult liver promotes hepatic lipid accumulation.
(A) Relative expression by qPCR analysis of Lrh-1, Cyp8b1, and Shp in livers or primary hepatocytes isolated from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre male mice or from age-matched Lrh-1fl/fl or Lrh-1Alb-Cre mice (n = 5 and n = 6 for Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice, respectively, and n = 3 for both Lrh-1fl/fl and Lrh-1Alb-cre mice). (B) TG content in livers or primary hepatocytes at two time points after infection (PI) from male mice with groups indicated in the figure. (C) Representative images (original magnification, ×20) of H&E-stained livers from Lrh-1AAV8-Cre and Lrh-1Alb-Cre mice; yellow arrows highlight lipid droplets in Lrh-1AAV8-Cre liver. For primary hepatocytes n = 2 for all genotypes or groups done in triplicate. Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired Student’s t tests.
As expected, viral-induced loss of LRH-1 severely attenuated two well-characterized downstream targets, Shp and Cyp8b1, in both Lrh-1AAV8-Cre livers and primary hepatocytes (Figure 1A). Unexpectedly, and in contrast to the developmental LKO model, TGs were elevated in livers from Lrh-1AAV8-Cre mice fed standard diet (SD) (Figure 1B). This increase in hepatic TGs was also observed in primary hepatocytes isolated from Lrh-1AAV8-Cre livers just 2 weeks or 8 weeks after infection, with a greater difference observed at the later time point. Lipid levels were unaffected in Lrh-1Alb-Cre livers and hepatocytes (Figure 1B). Consistent with elevated TG levels, histological analyses revealed small lipid droplets in H&E-stained liver sections from Lrh-1AAV8-Cre mice but not from Lrh-1Alb-Cre LKO mice (Figure 1C). Collectively, these data obtained in both primary hepatocytes and adult male mice suggest that loss of LRH-1 promotes abnormal hepatic lipid accumulation.
Macrovesicular steatosis and liver injury after HFD in Lrh-1AAV8-Cre mice.
Based on the elevated hepatic TGs observed in Lrh-1AAV8-Cre mice, we challenged these mice to high-fat diet (HFD) (Research Diet, 60% Kcal from fat) for a 6-week period after first infecting them with AAV8-Cre for 2 weeks. Lrh-1AAV8-Cre mice exhibited increased liver weight, increased hepatic TGs, and impaired glucose tolerance (Figure 2, A and B); other parameters, such as body weight, hepatic bile acids, and insulin tolerance, were unchanged from control mice (Supplemental Figure 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.96151DS1). Fasting plasma FFA was significantly elevated in mutant mice fed SD, with a similar trend observed on HFD (Figure 2A). Lrh-1AAV8-Cre livers were enlarged and fatty, with severe periportal macrovesicular steatosis and an increase in periportal Sirius red staining (Figure 2C). In addition, Lrh-1AAV8-Cre livers showed signs of mild liver injury, with a near-significant rise in plasma alanine transaminase (ALT) and an elevation of two markers of hepatic stellate cell activation, αSma and ColA1 (Figure 2D). We also confirmed that hepatic lipid accumulation and liver histology were normal in age-matched Lrh-1Alb-Cre male mice placed on HFD for 6 weeks (Figure 2C and Supplemental Figure 3). The hepatic steatosis observed in Lrh-1AAV8-Cre mice did not result from impaired VLDL secretion, as fasting plasma TG levels and secretion were unchanged from Lrh-1AAV8-GFP mice (Figure 2A and Supplemental Figure 2). Thus, our collective data from Lrh-1AAV8-Cre mice maintained on either SD or HFD imply that loss of LRH-1 increases hepatic lipid storage.
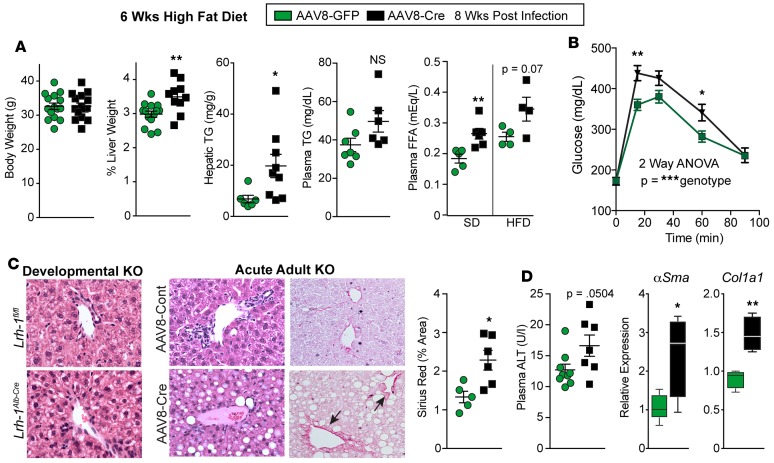
Figure 2. Lrh-1AAV8-Cre mice exhibit hepatic steatosis and liver injury after dietary challenge.
(A) Body weight, percentage liver weight, hepatic/plasma TG levels, and plasma free fatty acids (FFA) obtained for Lrh-1AAV8-GFP and Lrh-1AAV8-Cre male mice after 6 weeks of HFD. For body and liver weight, n = 15 and 13 per group; for plasma/hepatic TG, n = 7 and 9 per group; and for plasma FFA, n = 4 per group. Plasma FFA levels are also shown for mice fed SD (Lrh-1AAV8-GFP, n = 5, and Lrh-1AAV8-Cre, n = 6). (B) GTT (i.p.) for Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice after 6 weeks HFD challenge (Lrh-1AAV8-GFP, n = 12, and Lrh-1AAV8-Cre, n = 6 per group). (C) Representative images (original magnification, ×20) of H&E-stained liver sections from the developmental KO mice and controls (Lrh-1fl/fl and Lrh-1Alb-Cre) and adult acute KO mice and controls (Lrh-1AAV8-Cre and Lrh-1AAV8-GFP). All groups of mice were fed HFD. Sirius red staining and quantification are shown for adult KO mice, with arrows highlighting periportal collagen. (D) Plasma ALT and expression of αSma and Col1a1 in livers from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice. For plasma ALT, n = 9 and 7 per group; for qPCR, n = 5 and 4 per group. Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test (A, B, and D) and 2-way ANOVA with Sidak (C).
We also noted that Lrh-1AAV8-Cre mice exhibit lower fasting plasma cholesterol on both SD and HFD, with hepatic cholesterol increased only after HFD (Supplemental Figures 1 and 2). Fast protein liquid chromatography analyses of plasma cholesterol verified that LDLs are reduced, with a corresponding increase in HDLs in Lrh-1AAV8-Cre mutant cohorts (Supplemental Figure 2). This decline in plasma cholesterol can be partially attributed to suppression of Pcsk9, a negative regulator of LDL receptor and new target for cholesterol-lowering biologics (30–32) (Supplemental Figure 2), suggesting that LRH-1 might participate in regulating this branch of cholesterol flux.
Increased lipid accumulation and storage and decreased fatty acid oxidation in Lrh-1AAV8-Cre hepatocytes.
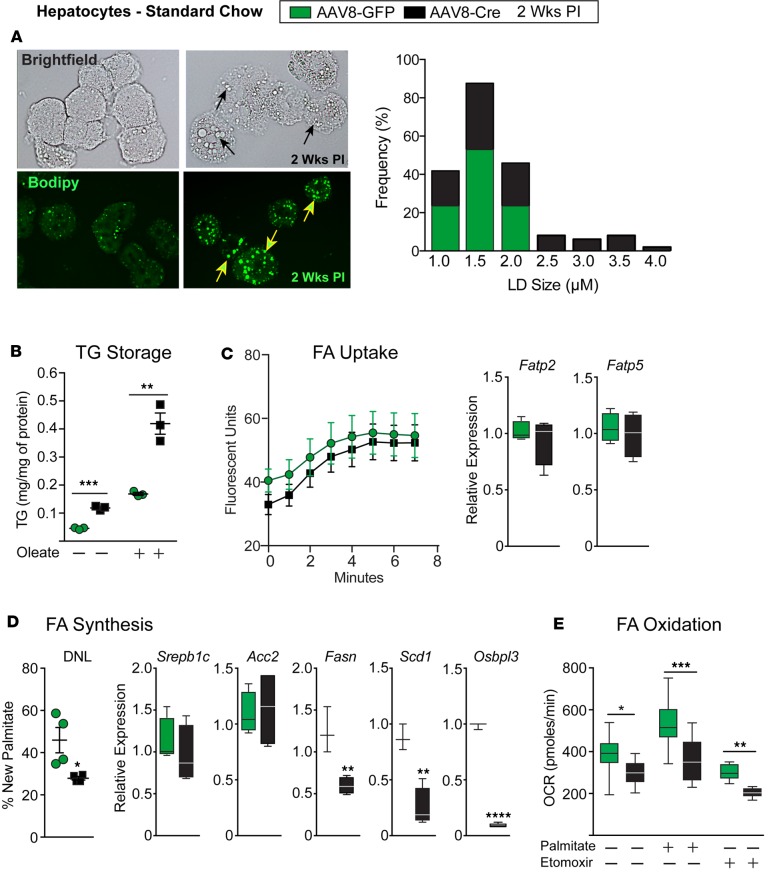
To better understand the NAFLD phenotype observed in Lrh-1AAV8-Cre mice, primary hepatocytes were used to assess whether lipid uptake, synthesis, oxidation, and/or storage were affected. Increased lipid accumulation in Lrh-1AAV8-Cre hepatocytes was associated with large intracellular lipid droplets. Quantification of BODIPY-stained hepatocytes confirmed a higher frequency of larger lipid droplets in Lrh-1AAV8-Cre hepatocytes compared with control Lrh-1AAV8-GFP hepatocytes (Figure 3A). Consistent with the elevated hepatic TGs in Lrh-1AAV8-Cre mice fed HFD, supplementing Lrh-1AAV8-Cre hepatocytes with oleic acid greatly increased their storage of lipids (Figure 3B). FFA uptake, as assayed by incorporation of fluorescent-labeled substrate (33), was unchanged in Lrh-1AAV8-Cre hepatocytes, as were Fatp2 and Fatp5, which encode two major hepatic fatty acid (FA) transport proteins (Figure 3C). De novo lipogenesis quantified by new palmitate synthesis after heavy water labeling was decreased in Lrh-1AAV8-Cre livers 2 weeks after infection. In addition, two SREBP-1c targets, Fasn and Scd1, were also downregulated (Figure 3D). We noted that Osbpl3, an LRH-1 target gene proposed to increase fatty liver via SREBP-1c processing (34), was significantly reduced in Lrh-1AAV8-Cre livers. FA oxidation was impaired in Lrh-1AAV8-Cre hepatocytes, basally and after supplementing with fatty exogenous palmitic acid (Figure 3E), likely reflecting impair turnover of stored neutral lipids (35), due to large lipid droplets present in mutant hepatocytes. Etomoxir was able to suppress basal FA oxidation in Lrh-1AAV8-Cre hepatocytes, indicating that mitochondrial oxidation persists in mutant hepatocytes. These collective data show that hepatocytes lacking LRH-1 preferentially store FAs in large cytosolic lipid droplets independent of increased FA uptake or new FA synthesis.
Figure 3. Lrh-1AAV8-Cre hepatocytes preferentially store fatty acids in large lipid droplets.
(A) Representative bright-field images and images of BODIPY-stained primary hepatocytes from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre male mice 2 weeks after infection; black or yellow arrows highlight lipid droplets in Lrh-1AAV8-Cre hepatocytes. Quantification of lipid droplet size and frequency is shown, as measured by ImageJ. (B) TG content obtained from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mouse hepatocytes with or without FA loading with oleic acid for 16 hours. (C) Measurement of FA uptake and expression of hepatic fatty acid transporters (Fatp2 and Fatp5) in Lrh-1AAV8-GFP and Lrh-1AAV8-Cre hepatocytes. (D) De novo lipogenesis (DNL) in hepatic tissue 2 weeks after infection, as quantified by the percentage of new hepatic palmitate (n = 4 per group), and expression of key lipogenic genes in hepatocytes isolated from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice. (E) Fatty acid oxidation in hepatocytes from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice. Primary hepatocytes were analyzed from at least 2–4 males per group, with each assay done in triplicate for B–E. Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, **P < 0.01, ***P < 0.001, unpaired Student’s t test for B, D, and E.
Loss of LRH-1 alters gene pathways in lipid storage and phospholipid metabolism.
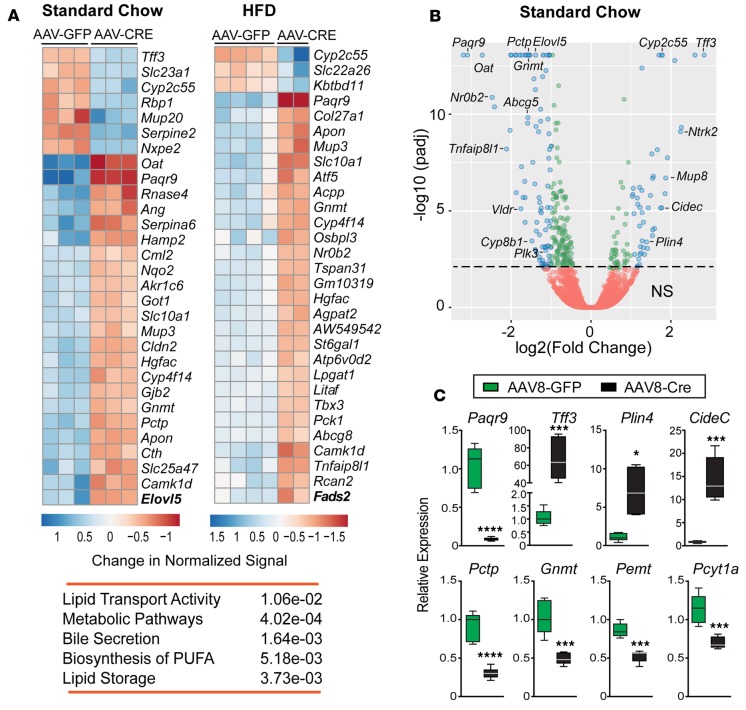
To gain a global perspective of gene changes associated with increased hepatic lipid accumulation in Lrh-1AAV8-Cre mice, livers from mutant and control groups fed either SD or HFD for 6 weeks were profiled by RNA sequencing (RNA-Seq). The number of differentially expressed genes in mutant livers (DEGs, adjusted P ≤ 0.05) was 483 and 365 transcripts on SD or HFD, respectively, with 26% genes overlapping among all DEGs (P < 0.05) (Supplemental Data Set 1). Heatmaps of the top 30 DEGs significantly changed in mutant mice fed SD and HFD are shown for comparison (Figure 4A), with the corresponding volcano plots for SD (Figure 4B) and HFD (Supplemental Figure 4). Aside from well-known targets, such as Nr0b2, Slc10a1, Atf5, Abcg8, and Gnmt, other robust targets emerged, including Paqr9 and Tff3, which were later confirmed by qPCR (Figure 4C and Supplemental Figure 5). Functional and pathway analyses of DEGs indicate an association of LRH-1 with lipid storage and synthesis of phospholipids and polyunsaturated FAs (Figure 4A). Indeed, CideC, which influences lipid droplet size via lipid exchange, and Plin4 (36, 37), which coats nascent lipid droplets in adipocytes, are markedly upregulated in Lrh-1AAV8-Cre livers (Figure 4C). Genes significantly downregulated included those affecting phosphatidylcholine metabolism (Pcyt1a, Gnmt, Pemt, and Pctp; Figure 4C) and two genes associated with human NAFLD (Tm6sf2 and Gckr; Supplemental Figure 5). Interestingly, despite the large lipid droplets, major transcripts in lipid droplet biogenesis, such as Bscl2, Fitm1, Fitm2, Gpat4, and Dgat2 (38–41), are unchanged in Lrh-1AAV8-Cre livers. Additionally, autophagy genes, such as Atg7, which when deleted can lead to hepatic steatosis (42), were unchanged after acutely deleting LRH-1 in adult livers.
Figure 4. Lipid storage and phospholipid metabolic pathways are affected in Lrh-1AAV8-Cre mice.
(A) Top 30 differentially expressed genes (DEGs) and significant KEGG pathways in livers of Lrh-1AAV8-Cre male mice fed standard diet (SD) or high-fat diet (HFD). (B) Volcano plot of DEGs between Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice fed SD, with DEGs of adjusted P < 0.01 (green circles) and those with adjusted P < 0.01 and absolute log2 fold change >1.0 (blue circles). Annotated genes include established LRH-1 targets (Cyp8b1, Nr0b2, and Plk3), top DEGs (Paqr9 and Tff3), and DEGs associated with lipid droplet size and formation (Plin4 and Cidec) and phospholipid biosynthesis (Pctp and Gnmt). (C) Validation of select DEGs by qPCR analysis in livers from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice fed SD (Lrh-1AAV8-GFP, n = 5, and Lrh-1AAV8-Cre, n = 6). Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, ***P < 0.001, ****P < 0.0001, unpaired Student’s t test.
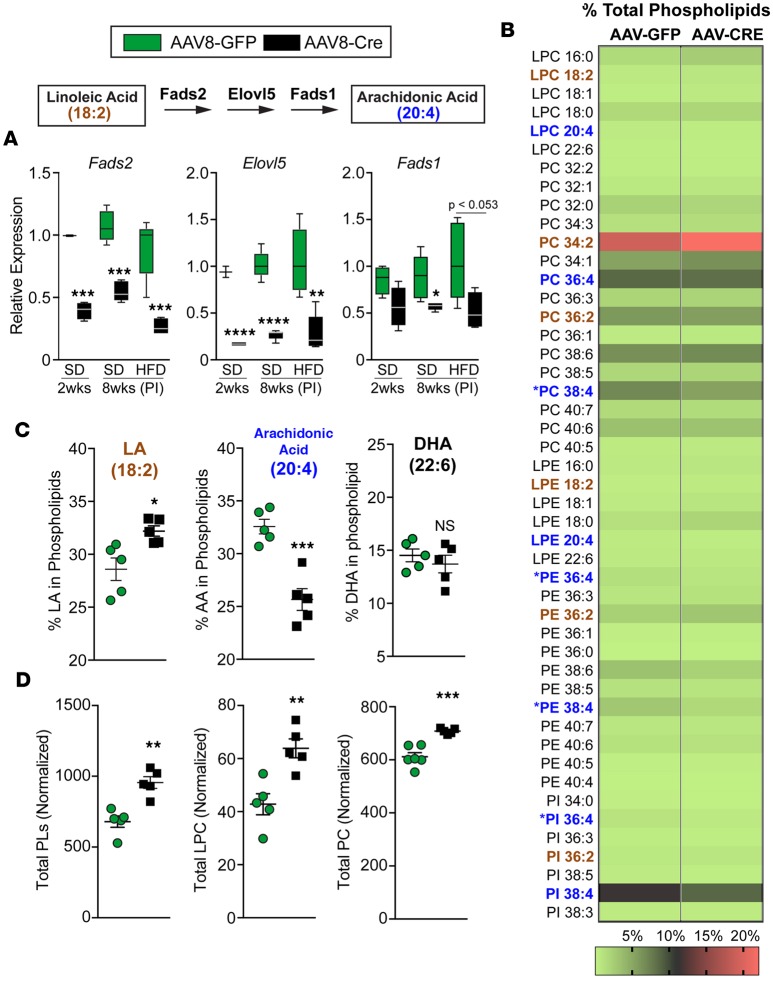
Two new potential LRH-1 target genes identified here include long-chain FA desaturase (Fads2) and elongase (Elovl5) (Figure 4A and Figure 5A), used to synthesize arachidonic acid (20:4) from the dietary FA LA (18:2). These two polyunsaturated FAs are used by LPCATs to generate AA phospholipids, as described above. Horton and colleagues showed previously that the global KO of Elovl5 and reduction of hepatic AA phospholipids promotes fatty liver (9). We found that expression of Fads2 and Elovl5 as well as Fads1 was significantly downregulated in Lrh-1AAV8-Cre livers, independent of diet (Figure 5A). Targeted lipidomics analyses of total hepatic phospholipids in Lrh-1AAV8-Cre mice fed HFD confirmed a marked attenuation of AA phospholipid species (phosphatidylcholines [PCs], phosphatidylethanolamine [PE], phosphatidylinositol), with a concomitant increase in LA phospholipid species containing 18:2 (Figure 5, B and C). FADS2 and ELOVL5 also convert α-linolenic acid (18:3) to decosahexaenoic acid (DHA), another key component of hepatic phospholipids. Despite changes in AA phospholipids, incorporation of DHA into phospholipid species was unchanged in Lrh-1AAV8-Cre mice (Figure 5C). A survey of all major phospholipid species revealed that the total pool of PCs was significantly higher in Lrh-1AAV8-Cre livers with an increase in monounsaturated lysophosphatidylethanolamine and lysophosphatidylcholine (LPC) lysophospholipid species (Figure 5D and Supplemental Figure 6 ).
Figure 5. Loss of LRH-1 alters the phospholipid remodeling program and reduces hepatic arachidonic acid phospholipids.
(A) Schematic of the arachidonic acid biosynthetic pathway. Hepatic expression of Fads2, Elovl5, and Fads1 from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre mice fed SD 2 weeks or 8 weeks after infection (PI) or fed HFD 8 weeks after infection. (B) Heatmap of lipidomics analysis of liver tissue from Lrh-1AAV8-GFP and Lrh-1AAV8-Cre male mice fed HFD for 6 weeks shown, with each lipid species depicted as a percentage of total phospholipids; the scale is shown at the bottom. Lipid species highlighted in brown represent linoleic acid phospholipids, and those highlighted in blue represent arachidonic acid phospholipid species. Substantially changed arachidonic acid species are indicated. (C) Percentages of phospholipid species containing arachidonic acid, linoleic acid (LA), or docosahexaenoic acid (DHA) in Lrh-1AAV8-GFP and Lrh-1AAV8-Cre livers. (D) Sum of total phospholipids (PL), lysophosphatidylcholine (LPC), and phosphatidylcholine (PC) (n = 5 per group). Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired Student’s t test for A, C, and D.
Expression of hLRH-1 normalizes hepatic lipids and improves glucose homeostasis.
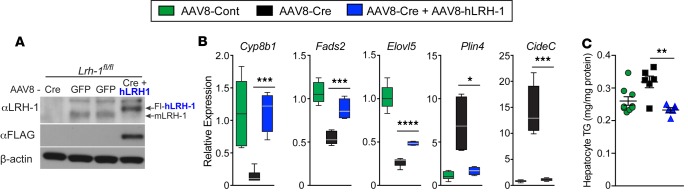
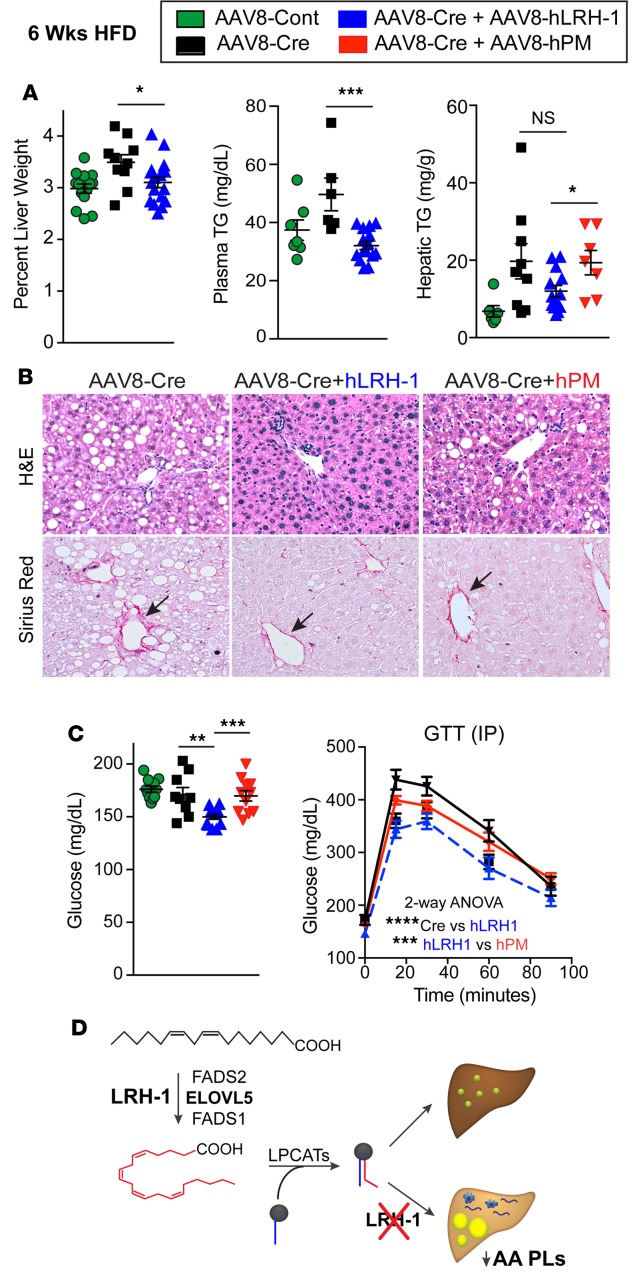
Distinct structural features at the mouth of the ligand-binding pocket allow human LRH-1 (hLRH-1) to efficiently bind phospholipid ligands compared with mouse LRH-1 (mLRH-1) (23, 43). To test whether hLRH-1 rescues hepatic phenotypes observed in Lrh-1AAV8-Cre mice, we humanized mouse livers by simultaneously infecting mice with AAV8-Cre– and AAV8-FLAG–tagged hLRH-1 (Lrh-1AAV8-Cre+hLrh1), which eliminates mLRH-1, while introducing epitope-tagged hLRH-1 in approximately 80% of hepatocytes (44), as shown in Figure 6A. Known targets, such as Cyp8b1 and Shp, as well as targets identified in this study respond appropriately after expression of hLRH-1 (Figure 6B and Supplemental Figure 7). Expression of hLRH-1 lowered Cidec and Plin4 expression and reversed elevated TGs in Lrh-1AAV8-Cre hepatocytes (Figure 6, B and C). Further examination of liver parameters revealed that expressing hLRH-1 resulted in reduced collagen deposition and importantly resolved macrovesicular steatosis, with a trend toward lower hepatic TGs on HFD (Figure 7, A and B, and Supplemental Figure 7). Further, while Lrh-1AAV8-Cre+hLrh1 mice exhibited lowered fasting glucose after a 6-hours fast and performed better in glucose tolerance testing relative to controls, expressing a ligand-deficient mutant of hLRH-1 (hPM) unable to bind lipid ligands (23) failed to reverse fatty liver or glucose intolerance (Figure 7, A–C). These data establish that hLRH-1 is able to complement mLRH-1 and mitigate hepatic steatosis in a ligand-dependent manner.
Figure 6. Human LRH-1 complements mouse LRH-1 and prevents hepatocyte TG accumulation.
(A) Expression of mouse LRH-1 or flag-tagged human LRH-1 in liver 8 weeks after infection with AAV8-Cre (CRE), AAV8-GFP (GFP), or AAV8-CRE+Fl-hLRH-1 (CRE+hLRH-1). (B) Relative expression of known or new LRH-1 gene targets and expression of lipid droplet–associated genes in livers in 3 groups: Lrh-1AAV8-GFP, Lrh-1AAV8-Cre, and Lrh-1AAV8-Cre+hLrh1. The number per group for all measurements is n = 5, 6, and 4. (C) TG levels in hepatocytes isolated 2 weeks after infection from at least 2 mice, performed in triplicate. Error bars represent ± SEM. For box-and-whisker plots, maximum and minimum values are shown with median. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired Student’s t test for B and C.
Figure 7. Replacing mouse with human LRH-1 resolves liver injury and improves glucose homeostasis.
(A) Percentage of liver weight and hepatic/plasma TGs in mice fed high-fat diet (HFD) (n ≥ 8 for Lrh-1AAV8-GFP, n ≥ 6 for Lrh-1AAV8-Cre, n ≥ 13 for Lrh-1AAV8-Cre+hLrh1 and Lrh-1AAV8-Cre+hPM, n ≥ 7). (B) Representative images (original magnification, ×20) of H&E- and Sirius red–stained livers from mice fed HFD from different experimental groups. Arrows highlight periportal collagen staining. (C) Fasting plasma glucose in male mice fed HFD for 6 weeks (n = 11 for Lrh-1AAV8-GFP, n = 9 for Lrh-1AAV8-Cre, n = 13 for Lrh-1AAV8-Cre+hLrh1, n = 11 for Lrh-1AAV8-Cre+hPM). Glucose tolerance test (GTT) results (n = 12 for Lrh-1AAV8-Cre, n = 6 for Lrh-1AAV8-Cre+hLrh1, and n = 7 for Lrh-1AAV8-Cre+hPM). (D) Schematic showing that loss of LRH-1 decreases arachidonic acid phospholipids (AA PLs) and leads to hepatic lipid accumulation via regulation of arachidonic acid biosynthesis. Error bars represent ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, unpaired Student’s t test (A and C) or 2-way ANOVA with Sidak (C).
Discussion
Here, using a viral-mediated approach to acutely eliminate LRH-1 in the adult hepatocyte, our study has uncovered an unanticipated role for the phospholipid-binding nuclear receptor LRH-1 in maintaining normal phospholipid composition and lipid homeostasis in the liver, as shown schematically (Figure 7D). Loss of LRH-1 greatly expanded lipid droplet size and increased lipid accumulation in hepatocytes. Eliminating LRH-1 in the Lrh-1AAV8-Cre model also leads to stark periportal macrovesicular steatosis and poorer glucose tolerance in male mice following a HFD challenge. The liver phenotype observed in Lrh-1AAV8-Cre adult male mice fed a HFD displayed features commonly observed with human NAFLD, including macrovesicular steatosis and moderate liver injury, as evidenced by increases in αSma and Col1a1 and periportal Sirius red staining. The lipid imbalance in Lrh-1AAV8-Cre mice is not driven by changes in de novo lipogenesis or lipid secretion but rather by increases in circulating FFAs, a known contributor to hepatic TG accumulation (45). These phenotypic changes are associated with decreased hepatic AA phospholipids owing to repression of Fads2 and Elovl5. We also show for the first time to our knowledge that wild-type hLRH-1, but not a ligand-binding deficient mutant, fully complements mLRH-1 in restoring normal hepatic lipid metabolism, while having beneficial effects on glucose homeostasis in diet-induced obesity.
We posit that alterations in the AA phospholipid pools in LRH-1–deficient livers help drive the lipid storage defect and fatty liver phenotype. Reduction of AA phospholipids is associated with abnormal hepatic lipid accumulation and NASH (5). In this study, loss of LRH-1 reduced AA phospholipids by 25%, and in ELOVL5-null mice, AA phospholipids are further decreased by 37% (9). Targeting the primary hepatic acyl transferase, LPCAT3, that transfers arachidonic acid on to lysophospholipid acceptors reduces hepatic AA phospholipids even further by 70% (7, 8). All three of these mutant mouse models result in fatty liver but clearly impinge on different pathways in lipid synthesis or flux. Nonetheless, these studies in aggregate establish that tampering with phospholipid remodeling and lowering cellular AA phospholipids below a critical threshold disrupts hepatic lipid homeostasis.
Although AA phospholipids decrease after deleting LRH-1 in adult livers, the total PC pool increases in Lrh-1AAV8-Cre mice, including LPC. Whether any of these many PC or LPC lipid species serve as endogenous ligands that can regulate LRH-1 remains an open question. Despite an increase in PC pools, levels of the rate-limiting enzyme in PC biosynthesis, Pcyt1a, as well as Pemt and Gnmt, two methyltransferases that convert PE to PC, were all suppressed in Lrh-1AAV8-Cre livers. The global KO of GNMT, a direct gene target of LRH-1 (46), also results in spontaneous fatty liver, with a paradoxical increase in hepatic PC levels (47). Given that both Lrh-1AAV8-Cre and GNMT-null mice exhibit a similar increase in PC (~15%), we conclude that, while LRH-1 regulates methyltransferases in the one carbon pool, LRH-1 is not essential for maintaining cellular PC levels.
Differences documented here between the developmental Lrh-1Alb-Cre and acute Lrh-1AAV8-Cre models are quite pronounced. Directly comparing these two KO models, which are equally effective in ablating mLrh-1, implies that early compensatory mechanisms in lipid flux successfully mask the true effect of LRH-1 on liver function and phospholipid metabolism when LRH-1 is deleted prior to birth. Lowered plasma cholesterol is the single significant phenotype shared by both the acute Lrh-1AAV8-Cre and developmental Lrh-1Alb-Cre models, as reported here and in ref. 20. Indeed, lowered plasma LDL-C in Lrh-1AAV8-Cre mice is consistent with decreased hepatic expression of Pcsk9, whose gene product regulates clearance of the LDL receptor at the plasma membrane (30, 32). While the drop in Pcsk9 expression and plasma cholesterol occurs early (2 weeks after infection, Supplemental Figure 8), the precise link to LRH-1 remains to be explored.
That hLRH-1 mitigates liver steatosis in a ligand-dependent manner and improves liver function in the face of a dietary challenge underscore the need to identify new effective chemical probes for this enigmatic subclass of nuclear receptor. Despite the fact that both mouse and hLRH-1 possess a large hydrophobic ligand-binding pocket that easily accommodates C16:0,C18:0 acyl chains (23, 24), a high-affinity efficacious lipid mimetic that occupies this cavity has remained elusive. Indeed, while RJW100 (48, 49) and DLPC (19) will bind hLRH-1, neither ligand fully occupies the pocket nor mimics the exposed head group of signaling phosphatidylinositols that appear to help stabilize solvent exposed residues at the pocket entrance (22, 25). In this regard, we found that RJW100 is relatively inert at altering expression of the top hepatic DEGs identified in this study, as well as known LRH-1 targets in humanized hepatocytes (data not shown). Our results also suggest some species-specific benefits of hLRH-1 with respect to glucose homeostasis. Replacing hLRH-1 with mLRH-1 improved fasting plasma glucose in these “humanized mice” over that of control mice. Work by Oosterveer and colleagues using the developmental Lrh-1Alb-Cre model suggests that mLRH-1 might regulate hepatic glucose flux through Gck (50). Although Gck expression was unchanged in glucose-intolerant Lrh-1AAV8-Cre mice fed HFD, Gckr, a key regulator of GCK activity (51), is significantly suppressed in Lrh-1AAV8-Cre mice (Supplemental Data Set 1). Interestingly, expressing hLRH-1 does not restore Gckr expression (data not shown), implying that hLRH-1 must modulate other critical pathways in hepatic glucose homeostasis. Regardless of the precise mechanisms by which LRH-1 mediates these antidiabetic effects (19), the improved plasma lipid profile and resolution of macrovesicular steatosis reported in our study rationalize renewed efforts to “drug” hLRH-1.
Methods
Generation of hepatic LRH-1 KO mouse models.
Mice harboring the Lrh-1 floxed allele (Lrh-1fl/fl) were generated as previously described (20) and acquired from O. Kline (UCSF) with permission from S. Kliewer. Liver-specific LRH-1–KO mice were generated by crossing Lrh-1fl/fl mice with albumin-Cre (Alb-Cre) mice (The Jackson Laboratory) to generate Lrh-1fl/+; Alb-Cre mice. Lrh-1fl/+; Alb-Cre mice were then crossed with Lrh-1fl/fl to generate Lrh-1fl/fl; Alb-Cre mice (Lrh-1Alb-Cre mice). Lrh-1fl/fl were genotyped as previously described (20) using the following primer pairs: 1F-CGATGTCCCTACTGTCGA and 1R-CGCAGCATTCTTCGGCAG; 2F-CATAAGGGCTCAGTGGCAC and 2R-CTTCACTGGCTGCCAAGCTG. The presence of Alb-Cre transgene was accessed using the following primers: 1F-ACCTGAAGATGTTCGCGATTA and 1R-ACCGTCAGTACGTGAGATATC.
Adult KO of mLRH-1 and expression of hLRH-1 in liver was accomplished using viral vectors acquired or generated at the University of Pennsylvania Gene Therapy Vector Core. Briefly, adenoassociated virus 8–TBG vectors (AAV8-TBG) expressing wild-type hLRH-1 (AAV8-hLRH-1) or a ligand-binding deficient mutant (AAV8-hPM) were generated as previously described (44). Six-week-old Lrh-1fl/fl male mice were infected with either AAV8-GFP (Lrh-1AAV8-GFP) or AAV8-Cre (Lrh-1AAV8-Cre) at a concentration of 1 × 1011 genome copies/ml via retro-orbital injection. Replacing hepatic mLRH-1 with hLRH-1 or hPM was achieved by simultaneously infecting 6-week-old Lrh-1fl/fl male mice with AAV8-Cre and AAV8-hLRH-1 (Lrh-1AAV8-Cre+hLRH-1) at concentrations of 1 × 1011 and 5 × 1010 genome copies/ml, respectively. All subsequent studies were initiated 2 weeks after infection.
Animals studies.
Mice were housed in the UCSF Barrier Facility and euthanized in accordance with the UCSF Institutional Animal Care and Use Committee under the Ingraham lab protocol (AN109147-03E). Adult male mice were used for all studies. Mice were weaned onto SD (LabDiet, 5058) or placed on HFD (Research Diets Inc., D12492) composed of 60 kcal% fat for 6 weeks. Body weights were monitored weekly during the study period. Prior to GTT or ITT, mice were fasted for 6 hours with ad libitum access to water and then administered glucose (1 g/kg) or insulin (1 U/kg) i.p. Blood glucose was measured from blood samples collected from tail vein using a glucometer (Accu-Check Compact, Roche). Mice were fasted for 6 hours prior to euthanasia and perfused with PBS prior to collection of liver tissue used for all subsequent histological, biochemical, and gene expression analyses. Harvested livers were immediately frozen in liquid nitrogen and stored at –80°C.
Blood parameters and hepatic VLDL secretion.
Mice were fasted for 6 hours prior to blood collection via submandibular bleed. All plasma TG and cholesterol were measured using colorimetric assay (Infinity Triglyceride or Cholesterol Assay, Thermo Scientific). Plasma free FAs and alanine transaminase were measured by the Mouse Metabolic Phenotyping Core at University of California, Davis. Plasma lipoprotein profiles by fast protein liquid chromatography were performed at the Lipoprotein Analysis Laboratory at Wake Forest University School of Medicine from pooled plasma samples from 5 Lrh-1AAV8-GFP or Lrh-1AAV8-Cre mice. For VLDL secretion, mice were fasted for 4 hours prior to injection with Pluronic F-127 (1 mg/g BW i.p.) in PBS. Blood was collected 1, 2, and 3 hours after injection.
Histology.
Mouse livers were perfused with PBS and fixed in 4% PFA overnight, equilibrated in 10% and 20% sucrose, and maintained in 70% ethanol. Livers were then embedded in paraffin, sectioned, and stained with Sirius red or H&E at the Gladstone Histology Core. The percentage Sirius red–stained area was calculated from 3 independent fields using ImageJ (NIH; https://imagej.nih.gov/ij/docs/examples/stained-sections/index.html).
Primary hepatocyte analyses.
Primary hepatocytes were isolated from nonfasted mice as previously described (44). Hepatocytes were seeded at 4.5 × 105 cells per well on type 1 collagen–coated 6-well plates. For lipid staining, primary hepatocytes were seeded at 2 × 104 cells per well on a type 1 collagen–coated 8-well Lab-Tek chamber slide (Nunc). After 4 hours, hepatocytes were washed with PBS and fixed with 4% PFA for 15 minutes at room temperature. Lipids were stained by incubating hepatocytes with BODIPY 494 (1 μg/ml) for 10 minutes at room temperature and analyzed by fluorescent microscopy. Lipid droplet volume and number were quantified from confocal images using ImageJ. For measurement of TGs, hepatocytes were allowed to adhere for 4 hours prior to treating with or without 500 μM BSA-oleate for 16 hours. Lipid extraction was performed by incubating with 500 μl 3:2 hexane/isopropanol (repeated 2 times) for 30 minutes at room temperature. The solvent was then transferred to a 1.5-ml Eppendorf tube and dried down under a nitrogen gas stream. The remaining lipid film was solubilized in 100 μl of 1% Triton X-100 in absolute ethanol for 4 hours at room temperature. Total protein was extracted with 300 μl of 0.1% SDS/0.1 M NaOH solution and measured using Protein Assay Dye reagent concentrate (Bio-Rad). TGs were measured by colorimetric assay (Infinity Triglyceride Assay, Thermo Fisher) and normalized to total protein.
In vitro FA transport was determined in primary hepatocytes isolated from nonfasted mice and seeded at 3 × 104 cells per well on a type 1 collagen–coated 96-well black clear-bottom plates (Corning) containing DMEM 10% FBS. Hepatocytes were serum starved in DMEM for 2 hours prior to fluorescent C1-BODIPY-FA uptake assay. DMEM was replaced with prewarmed (37°C) QBT uptake solution (HBSS, 0.1% BSA, 20 mM glucose, 2 mM Trypan Blue, and 2 μM BODIPY 3823). Fluorescence was measured at excitation 488/emission 515 for 2 hours with measurements taken every minute using a SpectraMax i3 plate reader. Oxygen consumption rate was measured as previously described (52) on primary hepatocytes isolated from nonfasting mice and seeded at 2 × 104 cells per well on a type 1 collagen–coated 24-well Seahorse XF24 Microplates containing DMEM with 5% FCS. After 4 hours, cells were washed twice with PBS and allowed to equilibrate in assay buffer (111 mM NaCl, 4.7 mM KCl, 2 mM MgSO4, 1.2 mM Na2HPO4, 2.5 mM glucose, 0.5 mM carnitine) for 60 minutes at 37°C and placed in Seahorse XF24 microplate reader. Etomoxir (40 μM) was added 15 minutes prior to Seahorse analysis. For measuring exogenous FA oxidation, palmitic acid (1 mM) was added to hepatocytes immediately prior to Seahorse analysis.
Lipid analyses.
Lipid extraction from frozen livers was carried out as previously described (19). Briefly, frozen liver pieces were homogenized in 9 volumes of PBS. 200 μl lysate was then transferred to a 2-ml Eppendorf tube containing 1.2 ml of 2:1 chloroform/methanol and mixed vigorously for 30 seconds. An additional 100 μl PBS was then added and mixed vigorously on a table top Vortex Mixer (VWR) for 15 seconds. Samples were then centrifuged at 4,200 g for 10 minutes at 4°C. The organic phase (200 μl) was transferred to a 1.5-ml Eppendorf tube and dried down under a nitrogen gas stream. Dried lipids were solubilized in 100 μl of 1% Triton X-100 in absolute methanol and rotated at room temperature for 4 hours. TG and total cholesterol were quantified by colorimetric assay (Infinity, Thermo Scientific) and normalized to grams of tissue.
For lipidomics analysis, snap frozen liver pieces were homogenized in 10 volumes of 2:1 chloroform/methanol using an Ika Ultra homogenizer. 500 μl homogenate was then transferred to a 2-ml Eppendorf tube and mixed with an equal volume of double deionized water followed by the addition of 500 μl chloroform. The resulting mixture was vortexed vigorously for 1 hours at room temperature using a Vortex Mixer. Samples were then centrifuged at 18,000 g for 5 minutes at room temperature. 500 μl of the organic phase was then transferred to a 1.5-ml Eppendorf tube and dried down under a nitrogen gas stream. Dried samples were overlaid with argon gas and sealed with parafilm. Phospholipid species quantification and analysis were carried out as previously described (11) at the Singapore Lipidomics Incubator at National University of Singapore. The percentage of individual phospholipid species was calculated by dividing by the sum of the most abundant phospholipids from our analysis.
In vivo hepatic de novo lipogenesis was carried out using a modified protocol (53). Briefly, 2 weeks following AAV8 infection, 14-week-old male mice were injected i.p. with 99.8% 2H2O (heavy water) in 0.9% NaCl to achieve 5% enrichment of their body water. This was followed by ad libitum administration of 8.0% 2H2O into drinking water for 4 days. After lipid extraction, newly synthesized palmitate was measured from TG pools as follows. Total lipids were extracted from approximately 100 mg frozen liver tissue by overnight incubation in chloroform/methanol (v/v, 2:1). Aqueous and organic phases were separated by the addition of 1 M NaCl and TGs were isolated from total lipid fraction by thin-layer chromatography using silica gel G plates (20 × 20 cm, 250 μm, Analtech Inc.) in hexane/ethylether/acetic acid (80:20:1). Scraped TG bands were hydrolyzed and FA methyl esters were formed by incubating in 3 N HCl in methanol (MilliporeSigma) at 65°C for 1 hours. FA methyl esters were extracted and derivatives were analyzed for deuterium enrichment by gas chromatography/mass spectrometry as described previously (53, 54).
qPCR analyses and Western blotting.
Total RNA was extracted from cells and tissues using TRIzol Reagent (MilliporeSigma). DNAse-treated (1 Ug) total RNA was used to generate cDNA using the High-Capacity cDNA Reverse Transcription kit (Life Technologies). RT-qPCR was performed using SYBR green, High ROX (Quanta), and data were analyzed as described previously (44). For Western blotting, livers were lysed in Tris-SDS buffer (2% SDS, 0.6 M Tris-Cl, pH 8.0, and 0.1 M DTT) supplemented with protease inhibitors (Calbiochem) and 20 mM N-ethylmaleimide (MilliporeSigma) and then sonicated using a Diagenode Bioruptor. Lysates were clarified by centrifugation and protein concentration was measured using Protein Assay Dye Reagent (Bio-Rad). Antibodies were used at the following concentrations: anti-Flag M2 (1:2,000, MilliporeSigma, F1804), anti-LRH-1 (1:2,000, R&D Systems, PPH2325-00), and anti–β-actin (1:5,000, Cell Signaling Technology, 5125).
RNA-seq and analysis.
Total RNA was extracted from livers of Lhr-1AAV8-GFP and Lhr-1AAV8-Cre mice fed SD or HFD 8 weeks after infection using the PureLink RNA mini kit (Life Technologies). Prior to generating cDNA, the quality of DNAse-treated total RNA was assessed by a bioanalyzer using an Agilent RNA 6000 Pico chip (Agilent). DNAse treatment was performed on total RNA using a DNA-free DNA removal kit (ThermoFisher Scientific). cDNA libraries were generated from 100 ng DNAse-treated total RNA using the Ovation RNA-Seq System V2 (Nugen). cDNA libraries were sonicated to generate 150-bp fragments using a Covaris M220 sonicator. cDNA fragment size and quality was assessed by a bioanalyzer using a High-Sensitivity DNA Analysis kit (Agilent). Barcoded sequencing libraries were generated using the Ovation Ultralow Library System V2 (Nugen) from 100 ng fragmented cDNA. Single-end 50-bp reads were sequenced on the HiSeq4000 (Illumina) at the Center for Advanced Technology at UCSF. Sequences were mapped to the mouse genome (mm10) using TopHat2. Reads that mapped to exons in annotated genes were counted using HTSeq. Final quantification and statistical testing of differentially expressed (adjusted P < 0.01 or < 0.05) genes were performed using DESeq2.
Statistics.
All statistical analyses are indicated in figure legends. For unpaired Student’s t test, 2-tailed analysis was used. For 2-way ANOVA, Sidak correction was done for multiple comparisons. For all dot plots, error bars for data represent mean ± SEM. For all box-and-whisker plots, maximum and minimum values are shown with median.
Study approval.
All procedures making use of animal subjects were reviewed and approved by the UCSF Institutional Animal Care and Use Committee under Ingraham lab protocol (AN109147-03E).
Author contributions
DAM and HAI designed experiments, analyzed data, and wrote the paper. WCK performed RNA-Seq data analysis and edited paper. DSS performed the FA transport assay with supervision from AS. MS and HE performed hepatocyte isolation and qPCR analyses of liver tissue. ACG and JCF performed lipidomic analyses with oversight from MRW and DLS. MH, MF, and EN performed de novo lipogenesis analyses.
Supplementary Material
Acknowledgments
We thank K. Ashrafi, S. Koliwad, A. Pierce, and J. Maher and members of the Ingraham lab for helpful discussions and review of the paper. We also thank the Gladstone Institute Histology Core for technical assistance and Christoph Paillart in the UCSF Mouse Metabolic Core (Nutrition Obesity Research Centers National Institute of Diabetes and Digestive and Kidney Diseases [NIDDK] P30 DK098722). This research was supported by grants to HAI (NIH NIDDK R01 DK099722, American Diabetes Association 1-15-MI-08, NIDDK Nuclear Receptor Signaling Atlas–NURSA Data Source Project, and UCSF Liver Pilot and Feasibility Program Award from NIDDK P30 026743), DAM (American Heart Association [AHA] Postdoctoral Fellowship 14POST20130048), WCK (AHA Postdoctoral Fellowship 16POST27260361), DSS (National Research Foundation Singapore NRF2016-NRFI001-15), and MRW and ACG (National Research Foundation Singapore NRFI2015-05). We acknowledge the UCSF Diabetes and Endocrinology Research Center (NIDDK P30 DK063720) for SeaHorse Analyses, UCSF Liver Center (NIDDK P30 026743), and the University of California, Davis, Mouse Metabolic Phenotyping Center (NIDDK U24 092993).
Version 1. 03/08/2018
Electronic publication
Funding Statement
All other funding sources for cores are listed in acknowledgements
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(4):e96151. https://doi.org/10.1172/jci.insight.96151.
Contributor Information
Amaury Cazenave-Gassiot, Email: bchacgt@nus.edu.sg.
Miyuki Suzawa, Email: miyuki.suzawa@gmail.com.
Hazel Escusa, Email: hazelanne.escusa@ucsf.edu.
Juat Chin Foo, Email: lsifjc@nus.edu.sg.
Andreas Stahl, Email: AStahl@berkeley.edu.
Mark Fitch, Email: mark.fitch@berkeley.edu.
Edna Nyangau, Email: edna.n@berkeley.edu.
David L. Silver, Email: david.silver@duke-nus.edu.sg.
References
- 1.Haas JT, Francque S, Staels B. Pathophysiology and mechanisms of nonalcoholic fatty liver disease. Annu Rev Physiol. 2016;78:181–205. doi: 10.1146/annurev-physiol-021115-105331. [DOI] [PubMed] [Google Scholar]
- 2.Vassilatou E. Nonalcoholic fatty liver disease and polycystic ovary syndrome. World J Gastroenterol. 2014;20(26):8351–8363. doi: 10.3748/wjg.v20.i26.8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McPherson S, Hardy T, Henderson E, Burt AD, Day CP, Anstee QM. Evidence of NAFLD progression from steatosis to fibrosing-steatohepatitis using paired biopsies: implications for prognosis and clinical management. J Hepatol. 2015;62(5):1148–1155. doi: 10.1016/j.jhep.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 4.Pais R, et al. A systematic review of follow-up biopsies reveals disease progression in patients with non-alcoholic fatty liver. J Hepatol. 2013;59(3):550–556. doi: 10.1016/j.jhep.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Puri P, et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46(4):1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 6.Krahmer N, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14(4):504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rong X, et al. Lpcat3-dependent production of arachidonoyl phospholipids is a key determinant of triglyceride secretion. Elife. 2015;4:e06557. doi: 10.7554/eLife.06557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashidate-Yoshida T, et al. Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. Elife. 2015;4:e06328. doi: 10.7554/eLife.06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon YA, Hammer RE, Horton JD. Deletion of ELOVL5 leads to fatty liver through activation of SREBP-1c in mice. J Lipid Res. 2009;50(3):412–423. doi: 10.1194/jlr.M800383-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang B, et al. Intestinal phospholipid remodeling is required for dietary-lipid uptake and survival on a high-fat diet. Cell Metab. 2016;23(3):492–504. doi: 10.1016/j.cmet.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hishikawa D, Shindou H, Kobayashi S, Nakanishi H, Taguchi R, Shimizu T. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc Natl Acad Sci USA. 2008;105(8):2830–2835. doi: 10.1073/pnas.0712245105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rong X, et al. LXRs regulate ER stress and inflammation through dynamic modulation of membrane phospholipid composition. Cell Metab. 2013;18(5):685–697. doi: 10.1016/j.cmet.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shindou H, Shimizu T. Acyl-CoA:lysophospholipid acyltransferases. J Biol Chem. 2009;284(1):1–5. doi: 10.1074/jbc.R800046200. [DOI] [PubMed] [Google Scholar]
- 14.Cinti DL, Cook L, Nagi MN, Suneja SK. The fatty acid chain elongation system of mammalian endoplasmic reticulum. Prog Lipid Res. 1992;31(1):1–51. doi: 10.1016/0163-7827(92)90014-A. [DOI] [PubMed] [Google Scholar]
- 15.Devlin AM, Singh R, Wade RE, Innis SM, Bottiglieri T, Lentz SR. Hypermethylation of Fads2 and altered hepatic fatty acid and phospholipid metabolism in mice with hyperhomocysteinemia. J Biol Chem. 2007;282(51):37082–37090. doi: 10.1074/jbc.M704256200. [DOI] [PubMed] [Google Scholar]
- 16.Sahini N, Borlak J. Genomics of human fatty liver disease reveal mechanistically linked lipid droplet-associated gene regulations in bland steatosis and nonalcoholic steatohepatitis. Transl Res. 2016;177:41–69. doi: 10.1016/j.trsl.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin B, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6(3):517–526. doi: 10.1016/S1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 18.del Castillo-Olivares A, Gil G. Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha -hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J Biol Chem. 2000;275(23):17793–17799. doi: 10.1074/jbc.M000996200. [DOI] [PubMed] [Google Scholar]
- 19.Lee JM, et al. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474(7352):506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee YK, et al. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol. 2008;22(6):1345–1356. doi: 10.1210/me.2007-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mataki C, et al. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol Cell Biol. 2007;27(23):8330–8339. doi: 10.1128/MCB.00852-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blind RD, et al. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proc Natl Acad Sci USA. 2014;111(42):15054–15059. doi: 10.1073/pnas.1416740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120(3):343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 24.Ortlund EA, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12(4):357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 25.Sablin EP, et al. Structure of liver receptor homolog-1 (NR5A2) with PIP3 hormone bound in the ligand binding pocket. J Struct Biol. 2015;192(3):342–348. doi: 10.1016/j.jsb.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sablin EP, et al. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Mol Endocrinol. 2009;23(1):25–34. doi: 10.1210/me.2007-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisend CM, Kundert JA, Suvorova ES, Prigge JR, Schmidt EE. Cre activity in fetal albCre mouse hepatocytes: Utility for developmental studies. Genesis. 2009;47(12):789–792. doi: 10.1002/dvg.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cole LK, Jacobs RL, Vance DE. Tamoxifen induces triacylglycerol accumulation in the mouse liver by activation of fatty acid synthesis. Hepatology. 2010;52(4):1258–1265. doi: 10.1002/hep.23813. [DOI] [PubMed] [Google Scholar]
- 29.Yan Z, Yan H, Ou H. Human thyroxine binding globulin (TBG) promoter directs efficient and sustaining transgene expression in liver-specific pattern. Gene. 2012;506(2):289–294. doi: 10.1016/j.gene.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 30.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci USA. 2004;101(18):7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SW, Moon YA, Horton JD. Post-transcriptional regulation of low density lipoprotein receptor protein by proprotein convertase subtilisin/kexin type 9a in mouse liver. J Biol Chem. 2004;279(48):50630–50638. doi: 10.1074/jbc.M410077200. [DOI] [PubMed] [Google Scholar]
- 32.Rashid S, et al. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9. Proc Natl Acad Sci USA. 2005;102(15):5374–5379. doi: 10.1073/pnas.0501652102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao J, Sportsman R, Harris J, Stahl A. Real-time quantification of fatty acid uptake using a novel fluorescence assay. J Lipid Res. 2005;46(3):597–602. doi: 10.1194/jlr.D400023-JLR200. [DOI] [PubMed] [Google Scholar]
- 34.Stein S, et al. Impaired SUMOylation of nuclear receptor LRH-1 promotes nonalcoholic fatty liver disease. J Clin Invest. 2017;127(2):583–592. doi: 10.1172/JCI85499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bazinet RP, Layé S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat Rev Neurosci. 2014;15(12):771–785. doi: 10.1038/nrn3820. [DOI] [PubMed] [Google Scholar]
- 36.Gong J, et al. Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J Cell Biol. 2011;195(6):953–963. doi: 10.1083/jcb.201104142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolins NE, Skinner JR, Schoenfish MJ, Tzekov A, Bensch KG, Bickel PE. Adipocyte protein S3-12 coats nascent lipid droplets. J Biol Chem. 2003;278(39):37713–37721. doi: 10.1074/jbc.M304025200. [DOI] [PubMed] [Google Scholar]
- 38.Goh VJ, Silver DL. The lipid droplet as a potential therapeutic target in NAFLD. Semin Liver Dis. 2013;33(4):312–320. doi: 10.1055/s-0033-1358521. [DOI] [PubMed] [Google Scholar]
- 39.Tan JS, Seow CJ, Goh VJ, Silver DL. Recent advances in understanding proteins involved in lipid droplet formation, growth and fusion. J Genet Genomics. 2014;41(5):251–259. doi: 10.1016/j.jgg.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 40.Pagac M, et al. SEIPIN regulates lipid droplet expansion and adipocyte development by modulating the activity of glycerol-3-phosphate acyltransferase. Cell Rep. 2016;17(6):1546–1559. doi: 10.1016/j.celrep.2016.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilfling F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24(4):384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwanten WJ, Martinet W, Michielsen PP, Francque SM. Role of autophagy in the pathophysiology of nonalcoholic fatty liver disease: a controversial issue. World J Gastroenterol. 2014;20(23):7325–7338. doi: 10.3748/wjg.v20.i23.7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Musille PM, Pathak M, Lauer JL, Griffin PR, Ortlund EA. Divergent sequence tunes ligand sensitivity in phospholipid-regulated hormone receptors. J Biol Chem. 2013;288(28):20702–20712. doi: 10.1074/jbc.M113.472837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzawa M, et al. A gene-expression screen identifies a non-toxic sumoylation inhibitor that mimics SUMO-less human LRH-1 in liver. Elife. 2015;4:e09003. doi: 10.7554/eLife.09003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnelly KL, Smith CI, Schwarzenberg SJ, Jessurun J, Boldt MD, Parks EJ. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J Clin Invest. 2005;115(5):1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner M, et al. Liver receptor homolog-1 is a critical determinant of methyl-pool metabolism. Hepatology. 2016;63(1):95–106. doi: 10.1002/hep.28124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martínez-Chantar ML, et al. Loss of the glycine N-methyltransferase gene leads to steatosis and hepatocellular carcinoma in mice. Hepatology. 2008;47(4):1191–1199. doi: 10.1002/hep.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mays SG, et al. Crystal structures of the nuclear receptor, liver receptor homolog 1, bound to synthetic agonists. J Biol Chem. 2016;291(49):25281–25291. doi: 10.1074/jbc.M116.753541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whitby RJ, et al. Small molecule agonists of the orphan nuclear receptors steroidogenic factor-1 (SF-1, NR5A1) and liver receptor homologue-1 (LRH-1, NR5A2) J Med Chem. 2011;54(7):2266–2281. doi: 10.1021/jm1014296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oosterveer MH, et al. LRH-1-dependent glucose sensing determines intermediary metabolism in liver. J Clin Invest. 2012;122(8):2817–2826. doi: 10.1172/JCI62368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Farrelly D, et al. Mice mutant for glucokinase regulatory protein exhibit decreased liver glucokinase: a sequestration mechanism in metabolic regulation. Proc Natl Acad Sci USA. 1999;96(25):14511–14516. doi: 10.1073/pnas.96.25.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li JZ, et al. Chronic overexpression of PNPLA3I148M in mouse liver causes hepatic steatosis. J Clin Invest. 2012;122(11):4130–4144. doi: 10.1172/JCI65179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hellerstein MK, et al. Measurement of de novo hepatic lipogenesis in humans using stable isotopes. J Clin Invest. 1991;87(5):1841–1852. doi: 10.1172/JCI115206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pierce AA, et al. Isocaloric manipulation of macronutrients within a high-carbohydrate/moderate-fat diet induces unique effects on hepatic lipogenesis, steatosis and liver injury. J Nutr Biochem. 2016;29:12–20. doi: 10.1016/j.jnutbio.2015.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.