Abstract
BACKGROUND. Cirrhosis is associated with gut microbial changes, but current 16S rDNA techniques sequence both dead and live bacteria. We aimed to determine the rRNA content compared with DNA from the same stool sample to evaluate cirrhosis progression and predict hospitalizations.
METHODS. Cirrhotics and controls provided stool for RNA and DNA analysis. Comparisons were made between cirrhotics/controls and within cirrhosis (compensated/decompensated, infected/uninfected, renal dysfunction/not, rifaximin use/not) with respect to DNA and RNA bacterial content using linear discriminant analysis. A separate group was treated with omeprazole for 14 days with longitudinal microbiota evaluation. Patients were followed for 90 days for hospitalizations. Multivariable models for hospitalizations with clinical data with and without DNA and RNA microbial data were created.
RESULTS. Twenty-six controls and 154 cirrhotics (54 infected, 62 decompensated, 20 renal dysfunction, 18 rifaximin) were included. RNA and DNA analysis showed differing potentially pathogenic taxa but similar autochthonous taxa composition. Thirty subjects underwent the omeprazole study, which demonstrated differences between RNA and DNA changes. Thirty-six patients were hospitalized within 90 days. In the RNA model, MELD score and Enterococcus were independently predictive of hospitalizations, while in the DNA model MELD was predictive and Roseburia protective. In both models, adding microbiota significantly added to the MELD score in predicting hospitalizations.
CONCLUSION. DNA and RNA analysis of the same stool sample demonstrated differing microbiota composition, which independently predicts the hospitalization risk in cirrhosis. RNA and DNA content of gut microbiota in cirrhosis are modulated differentially with disease severity, infections, and omeprazole use.
TRIAL REGISTRATION. NCT01458990.
FUNDING. VA Merit I0CX001076.
Keywords: Hepatology
Keywords: Bioinformatics, Clinical practice
RNA and DNA analysis of microbiota from the same stool sample in cirrhosis are complementary in predicting hosptialization risk.
Introduction
Cirrhosis is a leading cause of mortality and healthcare expenditure due to hospitalizations worldwide (1, 2). Bacterial products such as endotoxin play a key role in the development of a proinflammatory milieu and disease progression in cirrhosis (3, 4). Specifically, the development of hepatic encephalopathy (HE) and spontaneous bacterial peritonitis (SBP) have a strong gut-based origin (3). A growing body of literature has linked gut microbial DNA results with negative outcomes in cirrhosis (5–7). However, altered bacterial functionality characterized by endotoxemia and variability in secondary bile acids may influence outcomes rather than their composition (6, 8). These processes require live or highly active bacteria that are capable of interacting with the human host, mucosal immune system, and with other microbiota (9). This may not be discernible using conventional 16S rDNA sequencing since this technique assesses the presence of microbial DNA regardless of bacterial activity and their dead/alive status (9, 10). It has been reported that the ribosome content of bacteria reflects the microbial metabolic activity and assaying the RNA content of a community would reflect the metabolic activity of the species in the community (11–16). Therefore, our hypothesis is that sequencing of 16S rRNA using the RNA of the same samples that were performed for the DNA analysis would provide a different and better understanding of the nature of microbiota functional changes in cirrhosis and help predict 90-day hospitalizations.
Results
We included 180 subjects for the cross-sectional part of the study, of whom 26 were healthy controls with 154 cirrhotic patients. Within the cirrhosis group there were 62 patients with decompensation (controlled HE) (Table 1). There were also 54 patients who were infected and currently on antibiotics (Table 2). The leading infections were SBP (n = 15) followed by urinary tract infections (n = 13), bacteremia (n = 10), cellulitis (n = 5), Clostridium difficile (n = 6), and respiratory infections (n = 5).
Table 1. Comparison between cirrhotic patients with and without decompensation.
Table 2. Comparison between uninfected and infected cirrhotic patients.
Thirty additional subjects (15 controls and 15 compensated cirrhotic patients) were included for the before/after proton pump inhibitor (PPI) part of the study (17).
RNA alone, DNA alone, and RNA ratio microbiota comparison between groups
Controls versus cirrhosis comparison.
The genera that were different between controls and cirrhotics were different in RNA versus DNA. Using DNA, Veillonella was higher in cirrhosis, while for RNA it was higher in Lactobacillus and Enterococcus. While the taxa found in greater relative abundance in controls belonged largely to the same families (Lachnospiraceae and Ruminococcacaeae), the genera themselves were different when the RNA was compared with the DNA between the groups (Figure 1, A and B). RNA ratios (RNA/[RNA + DNA]) in controls versus cirrhosis comparison: A higher representation of RNA ratios of Enterobacteriaceae, Enterococcacae, and Veillonellaceae was seen in cirrhotic patients (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.98019DS1). In controls, on the other hand, there was a higher representation of Clostridiales members, families belonging to Bacteroidetes (Rikenelleaceae and Prevotellaceae) and Verrucomicrobiaceae. There was a significantly lower median cirrhosis dysbiosis ratio (CDR: [Lachnospiraceae + Ruminococaceae + ClostridialesXIV + Veillonellaceae]/[Enterobacteriaceae + Bacteroidaceae]) in cirrhotic patients on DNA (cirrhosis 0.24 vs. 1.1 controls, P < 0.0001) and on RNA (cirrhosis 0.32 vs. 0.98 controls, P < 0.001) compared with controls (6).
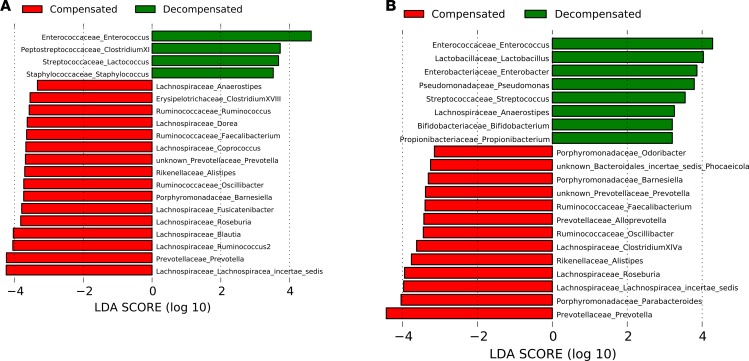
Figure 1. DNA comparison of controls versus cirrhosis.
Histograms of linear discriminant analysis (LDA) effect size (LEfSe) comparison between stool microbiota at the genus level between healthy controls (n = 26) and patients with cirrhosis (n = 154). Log-level changes in LDA score are displayed on the x axis. (A) DNA analysis comparison. (B) RNA analysis comparison. Green bars: taxa found in greater relative abundance in healthy controls. Red bars: taxa found in greater relative abundance in patients with cirrhosis.
Patients with and without decompensation.
In RNA analysis, there were higher potentially pathogenic genera (Enterococcus, Pseudomonas, and Enterobacter) in HE patients, while those belonging to autochthonous families were higher in the no-HE patients. On the other hand, DNA still showed Enterococcus at a higher level, while genera belonging to Gram-positive cocci (Peptostreptococcaceae, Streptococcaceae, and Staphylococcaceae) were higher in HE patients. The autochthonous family genera, however, were largely similar to the ones increased in the RNA results (Figure 2, A and B). RNA ratio of patients with and without decompensation: A higher Bacteroidetes taxa (Rikenelleaceae and Prevotellaceae) and lower Enterococceae and Peptostreptococcaceae were seen in patients without decompensation (Supplemental Figure 2). There was a significantly lower CDR by DNA analysis (median 0.32 decompensated vs. 0.54 compensated, P = 0.04) and RNA analysis (median 0.24 decompensated vs. 0.37 compensated, P = 0.02).
Figure 2. DNA comparison of compensated versus decompensated cirrhosis.
Histograms of linear discriminant analysis (LDA) effect size (LEfSe) comparison between stool microbiota at the genus level between compensated-cirrhosis patients (n = 92) and patients with decompensated cirrhosis (n = 2). Decompensation in cirrhosis is defined as hepatic encephalopathy, ascites, jaundice, and variceal bleeding. Log-level changes in LDA score are displayed on the x axis. (A) DNA analysis comparison. (B) RNA analysis comparison. Green bars: taxa found in greater relative abundance in decompensated cirrhosis. Red bars: taxa found in greater relative abundance in patients with compensated cirrhosis.
Infected with or without uninfected cirrhotic patients.
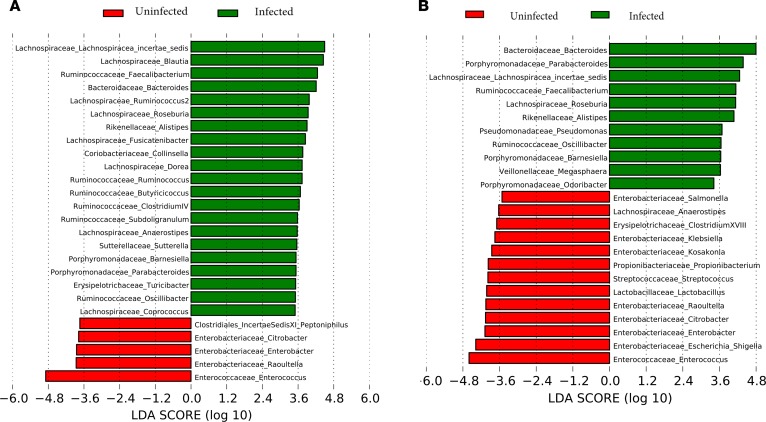
There were significant differences between these groups in both DNA and RNA analyses by linear discriminant analysis (LDA) effect size (LEfSe). By RNA analysis the infected groups had a relatively higher abundance of taxa belonging to Enterobacteriaceae, Lactobacillaceae, Erysipelothricaceae, and Propionibacteriaceae. Interestingly, very few of these were actually found in the DNA analysis differentiating infected from uninfected cirrhotic patients. On the other hand, regardless of RNA and DNA, similar autochthonous taxa were seen in uninfected patients (Figure 3, A and B). RNA ratio of patients with and without infection: Infected patients had a higher RNA ratio of Enterococcaceae and Actinomycetales, and a lower ratio of Bacteroidetes (Rikenellaceae and Porphyromonadaceae) and Acidaminococcaceae compared with uninfected cirrhotic patients (Supplemental Figure 3). There was a significantly lower CDR in infected patients in the DNA (median 0.31 infected vs. 0.55 uninfected, P < 0.0001) and in RNA (median 0.22 infected vs. 0.35 uninfected, P = 0.01) analyses compared with uninfected patients.
Figure 3. DNA comparison of infected versus uninfected cirrhotic patients.
Histograms of linear discriminant analysis (LDA) effect size (LEfSe) comparison between stool microbiota at the genus level between infected cirrhotic patients (n = 54) and uninfected patients with cirrhosis (n = 100). Infected patients were those with clinically determined bacterial infections. Log-level changes in LDA score are displayed on the x axis. (A) DNA analysis comparison. (B) RNA analysis comparison. Red bars: taxa found in greater relative abundance in infected cirrhosis. Green bars: taxa found in greater relative abundance in uninfected patients.
Renal dysfunction and its impact on microbiota.
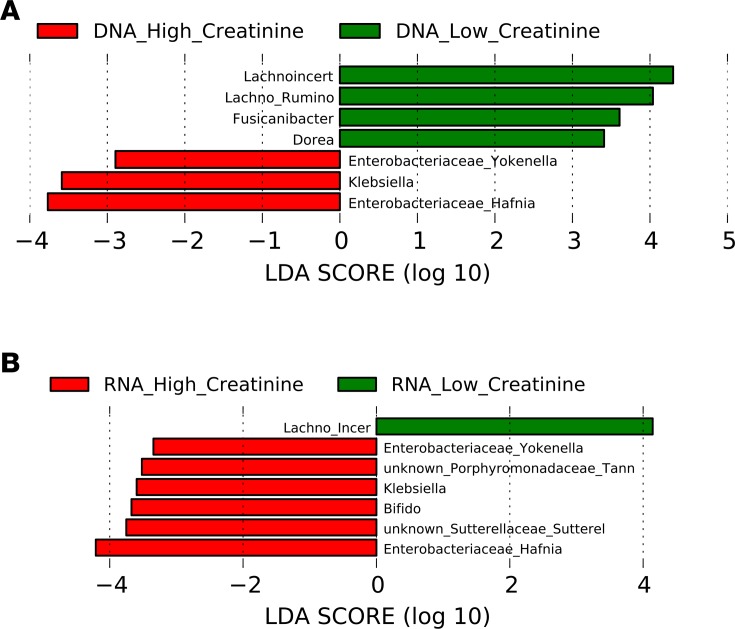
Since the model for end-stage liver disease (MELD) score consists of 2 primary liver-related laboratory variables (international normalized ratio [INR] and total serum bilirubin) and 1 renal variable (serum creatinine), we also defined the relative contribution of renal dysfunction (serum creatinine ≥ 1.5 mg/dl) to microbial composition. Twenty cirrhotic subjects had renal dysfunction and these patients had equivalent INR (1.7 ± 1.01 vs. 1.5 ± 0.5, P = 0.1) and bilirubin (2.6 ± 2.7 vs. 2.7 ± 4.2, P = 0.9) compared with cirrhotic patients without renal dysfunction. On LEfSe, renal dysfunction was associated with significantly higher relative abundances of Enterobacteriaceae taxa and lower Lachnospiraceae taxa (Figure 4, A and B).
Figure 4. DNA comparison of cirrhotic patients with and without renal dysfunction.
Histograms of linear discriminant analysis (LDA) effect size (LEfSe) comparison between stool microbiota at the genus level between cirrhotic patients with renal dysfunction (n = 20) and cirrhotic patients without renal dysfunction (n = 134). Renal dysfunction is defined as serum creatinine > 1.5 mg/dl on the day of sample collection. Log-level changes in LDA score are displayed on the x axis. (A) DNA analysis comparison. (B) RNA analysis comparison. Green bars: taxa found in greater relative abundance in those without renal dysfunction (low creatinine). Red bars: taxa found in greater relative abundance in patients with renal dysfunction (high creatinine) with compensated cirrhosis.
Comparison of patients with and without rifaximin.
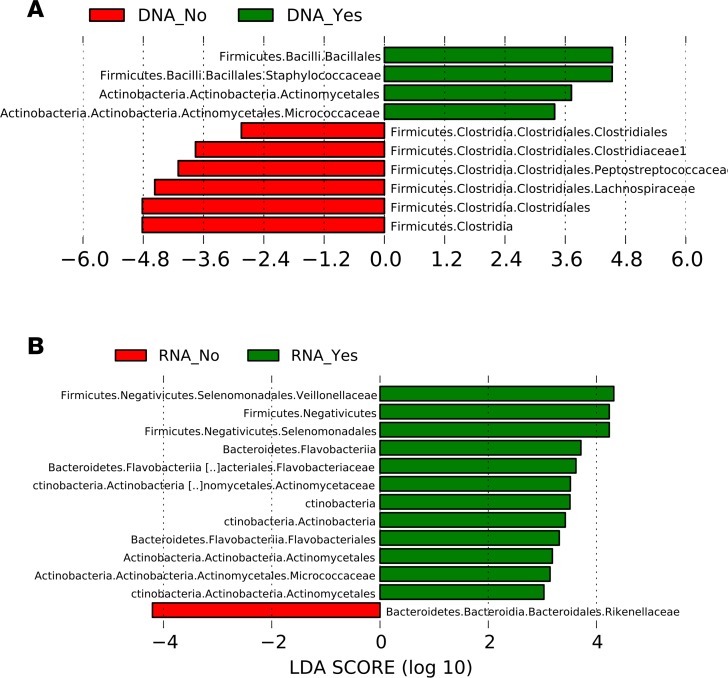
In the 85 cirrhotic patients who were not on absorbable antibiotics and were free of infection, 19 were on rifaximin. Using the DNA analysis, there was a higher relative abundance of Staphylococcaceae and members of Actinobacteria such as Micrococcaceae and lower relative abundance of Lachnospiraceae and other Clostridial taxa in patients on rifaximin. In the RNA analysis, there was again higher relative abundance of taxa belonging to Actinobacteria with also higher Veillonellaceae, while only Rikenelleaceae was lower in rifaximin users (Figure 5, A and B).
Figure 5. Comparison of cirrhotic patients on and not on rifaximin.
In the cirrhotic subgroup without infections or on other antibiotics (n = 85), histograms of linear discriminant analysis (LDA) effect size (LEfSe) comparison of stool microbiota at the genus level between patients on rifaximin (n = 19) and not on rifaximin (n = 66) are shown. Rifaximin is a nonabsorbable antibiotic used for hepatic encephalopathy. Log-level changes in LDA score are displayed on the x axis. (A) DNA comparison. (B) RNA comparison. Green bars: taxa found in greater relative abundance in patients on rifaximin. Red bars: taxa found in greater relative abundance in patients not on rifaximin.
Longitudinal study with omeprazole
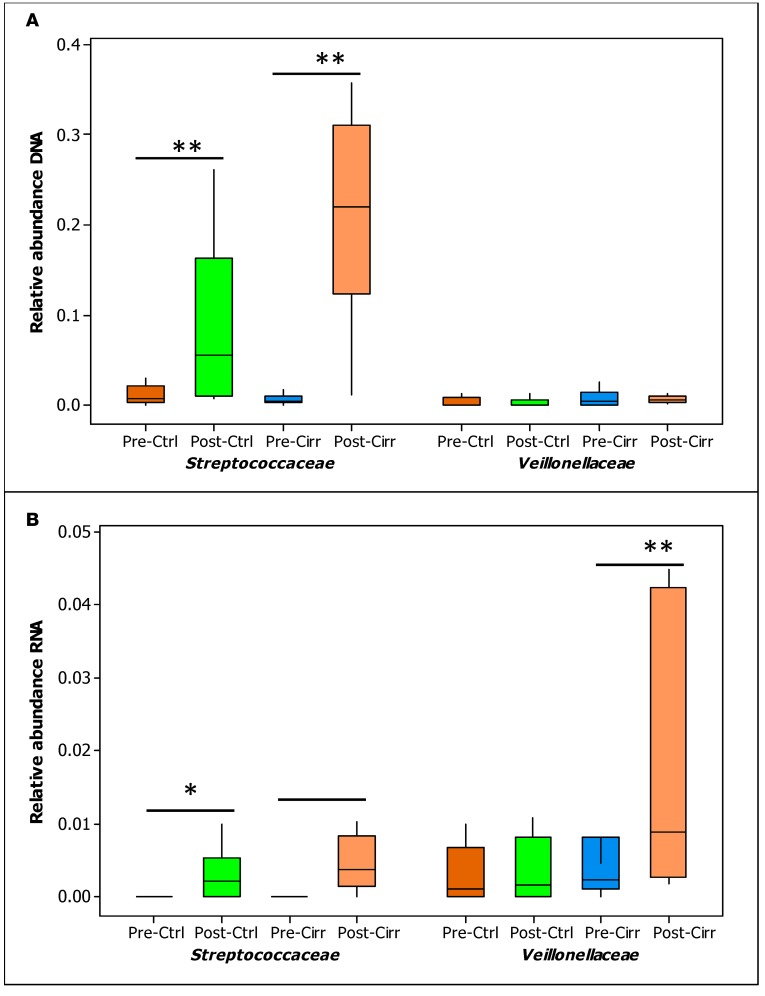
As part of a previous study, we analyzed samples of compensated cirrhotic patients and age-matched healthy controls before/after 40 mg omeprazole daily for 14 days (17). There was a significant change in 2 families related to oral microbiota, Streptococcoceae and Veillonellaceae, in varying degrees. While the relative abundance of Streptococcoceae increased in controls and cirrhotics after PPI, the magnitude of increase was higher in the DNA isolates, likely indicating a less metabolically active increase. On the other hand, Veillonellaceae were higher in cirrhotics by RNA but not DNA analysis, indicating a more metabolically active change after PPI (Figure 6, A and B).
Figure 6. Comparison of controls and cirrhotic patients before/after 14 days of omeprazole.
Healthy controls and compensated cirrhotic patients were administered 40 mg/day of omeprazole for 14 days. Stool microbiota were analyzed at baseline and after 14 days using Wilcoxon’s signed rank-sum test, focusing on relative abundance of oral-origin families Streptococcaceae and Veillonellaceae. (A) DNA comparison. (B) RNA comparison. Data shown as median and 95% CI. *P < 0.05, **P < 0.01. Ctrl, control; Cirr, compensated cirrhosis; Pre, before omeprazole; Post, after 14 days of 40 mg omeprazole daily.
Hospitalization prediction
Of the 154 patients included, 9 died within 90 days (3 during the index hospitalization and 6 were sent to hospice). Of the remaining 145, 36 (25%) were hospitalized within 90 days for nonelective reasons. The major causes were HE (n = 11), followed by ascites/hepatic hydrothorax management (n = 9), acute kidney injury (n = 8), infections (n = 4), and liver-unrelated reasons (n = 4). On univariate analyses, several clinical and bacterial variables were significantly associated with these hospitalizations (Supplemental Table 1). Because of the large number of variables under consideration, a screening process was conducted to determine variables to consider for a multivariate model. This screening was conducted by performing a series of univariate binary logistic regression models. Any variables that had a P < 0.20 level of significance in the univariate binary logistic regression models were used in a multivariable binary logistic regression model. The clinical variables that met this criterion for both RNA and DNA models (P < 0.20) are as follows: age, male gender, HE, infection, PPI, and MELD score. Additionally, the DNA microbiota that were significant were Bacteroides, Citrobacter, Enterococcus, Blautia, Lachnospiraceae incertae Sedis, Roseburia, Ruminococcus, Parabacteroides, Fecalibacterium, Subdoligranulum, and Veillonella. The RNA microbiota that were significant on univariate analysis were Bacteroides, Klebsiella, Raoultella, Enterococcus, Lachnospiraceae incertae Sedis, Roseburia, Parabacteroides, Prevotella, Alistipes, Fecalibacterium, Oscillispira, and Veillonella.
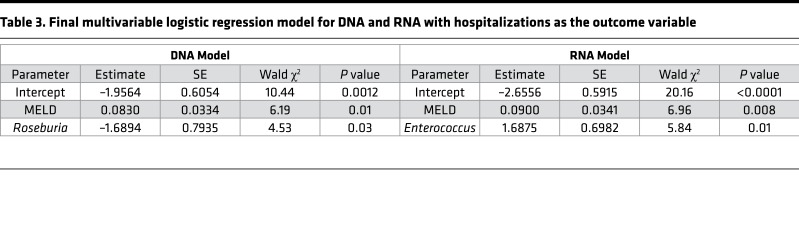
Once the variables to be considered in the multivariable binary logistic regression were determined, 2 models for hospitalizations were considered: 1 for DNA-linked microbiota and clinical variables and 1 for RNA-linked microbiota and clinical variables. The multivariable binary logistic regression models were fit using a backwards elimination procedure where all variables that remained in the model had to be significant at the P < 0.05 significance level, the results of which are shown in Table 3.
Table 3. Final multivariable logistic regression model for DNA and RNA with hospitalizations as the outcome variable.
On multivariable analysis in the RNA model, MELD score (odds ratio [OR] 1.1, 95%CI 1.02–1.17, P = 0.008) and Enterococcus (OR 5.41, 95% CI 1.38–19.31, P = 0.01) were associated with hospitalizations.
In the DNA model, MELD was again associated with hospitalizations (OR 1.1, 95% CI 1.02–1.16, P = 0.01), while Roseburia was protective against hospitalizations (OR 0.19, 95% CI 0.04–0.87, P = 0.03).
Clinical utility of hospitalization prediction beyond the MELD score
The final models for both DNA and RNA were compared to the model with MELD-only using the method proposed by DeLong et al. (18) for comparing the AUC under correlated receiver operating characteristic (ROC) curves. For the DNA model, the model with MELD-only had an AUC of 0.72 (95% CI [0.60, 0.82]), while the model with MELD and Roseburia had an AUC of 0.79 (95% CI [0.69, 0.89]). The increase in AUC with the addition of Roseburia was 0.07 (95% CI [0.0102, 0.1096]), which was statistically significant (P = 0.045) compared with the MELD model alone.
For the RNA model, the model with MELD-only had an AUC of 0.71 (95% CI [0.60, 0.83]), while the model with MELD and Enterococcus had an AUC of 0.77 (95% CI [0.66, 0.88]). The increase in AUC with the addition of Enterococcus was 0.06 (95% CI [0.018, 0.11]), which was statistically significant (P = 0.042) compared with MELD alone.
Discussion
Patients with cirrhosis have a poor prognosis that is often related to disease processes that are initiated and propagated by a proinflammatory gut and systemic milieu (19). Since bacterial functionality, including endotoxin production and bile acid modification, may play an important role in the disease progression, the delineation of living or metabolically active bacteria is relevant (3). In addition, current prognostic systems in cirrhosis are often inadequate in predicting relevant outcomes such as hospitalizations (2).
There have been several studies evaluating the role of alive versus dead bacteria, and qPCR of the DNA has been shown to detect bacterial cells several days to weeks after a loss of viability with antibiotics (20). This has also been corroborated with other techniques that focus on individual strains and bacteria (21–23). Alternatives such as viability PCR and molecular viability testing have been studied to definitively assess live versus dead bacteria but these are difficult to do in a microbiome-wide assay. Gut microbiota based on DNA have been used in cirrhotic patients to define disease severity, link with systemic inflammation, and also define outcomes such as death and hospitalizations (5–7). Hence, the linkages between dead and live bacteria in these samples are important since cadaver cells after antibiotic and other therapies may persist and confound interpretation of the 16S rRNA sequencing (20). Therefore, the current study employed bacterial RNA extracted from the same sample and the ultimate DNA was extracted with the hypothesis that active RNA synthesis implies active bacteria, while DNA can be a mixture of both live and dead bacteria or at the very least bacteria with a relatively low activity (9).
In cross-sectional analyses that differentiated between healthy controls and cirrhotic patients and between cirrhotic patients with and without decompensation or infection, we found significant changes in the DNA, as previously published (5, 6, 24). Most of these changes pointed towards a higher relative abundance of autochthonous taxa and lower potentially pathogenic taxa such as Enterobacteriaceae and Enterococcaceae. The interesting finding was that while the RNA changes were similar with respect to the autochthonous taxa, the taxa that were higher in the more diseased groups (cirrhosis, decompensated, and infected) were different in the RNA-only analysis compared with the DNA-only analysis. Therefore, there was a lack of concordance between taxa present in the DNA- versus RNA-only analyses. The taxa present in only DNA but not RNA could be potentially dead bacteria. It is also likely that the taxa present in RNA but not in DNA are metabolically active but did not reach the relative abundance cutoff (1%) that was used in the DNA analysis.
It is interesting that autochthonous taxa were overrepresented in both RNA-only and DNA-only analyses in the less severe groups. This likely demonstrates the ability of these taxa to continue producing their beneficial end-products such as short-chain fatty acids and their relative stability after analysis (25). This is encouraging in that these bacteria could continue to beneficially impact host-microbiota interactions. On the other hand, while potentially pathogenic families were higher in the groups with advanced disease or infections, specific taxa differed when compared between RNA and DNA analyses. This may be relevant because these taxa are widely used as markers of advanced disease, can predict hospitalizations and negative outcomes, and can also help determine who develops organ failure based on the DNA analysis. Many of these taxa are directly linked to infections and organ failure in cirrhosis, especially Klebsiella, Enterococcus, and Streptococcus (26, 27). These bacterial taxa in the past have only been characterized in cirrhosis using direct culture methods or DNA. However, our analysis using the RNA methodology examines a potentially novel aspect of bacterial taxa description in cirrhosis. In addition, we found that taxa normally found in the saliva, such as Streptococcaceae, were higher after PPI use in both controls and cirrhotics by DNA analysis, as published previously (17), indicating that PPI use allows these microbiota to be found in the stool. However the increase in Veilloneallaceae, another upper-GI microbial family, was nuanced, since it was higher in RNA, indicating potentially greater metabolic activity, but not in the DNA after PPI. This difference reflects another aspect of the discordance between the DNA and RNA assessment.
The additional contribution of renal dysfunction in cirrhosis progression has been published extensively (28). There are also studies demonstrating that renal dysfunction without cirrhosis is associated with dysbiosis (29). We found that additional renal dysfunction, regardless of cirrhosis severity on INR and bilirubin levels, was associated with further depletion of autochthonous taxa and increased relative abundance of Enterobacteriaceae taxa compared with those without renal dysfunction. This was similar in both RNA and DNA analysis, demonstrating the robust negative contribution of renal dysfunction towards this change.
A clinically relevant reason for microbiota analysis is their association with outcomes in patients, which has the potential to increase its use in clinical practice. In cirrhosis, most negative outcomes stem from complications such as infections that result in hospitalizations. The hospitalization risks in cirrhosis are incompletely predicted by purely clinical parameters, which is why the addition of microbiota variables could add a novel aspect (2). DNA analysis showed that Roseburia, a short-chain fatty acid–producing genus of Lachnospiraceae, was associated with protection from hospitalizations independent of the expected association with the MELD score. On the other hand, the RNA of Enterococcus was additive to MELD in the prediction of hospitalizations. Members of the genus Enterococcus have been associated with infections and also increase after PPI use in patients with cirrhosis and alcohol use (30, 31). Therefore, this pattern of differential modulation with Roseburia being protective and Enterococcus being predictive of hospitalizations lends biological plausibility to these results. Moreover, we found a significant increase in the predictive capability of the regression formula for both RNA- and DNA-related microbiota in addition to the current clinical variables. For an outcome that is as expensive and clinically significant as hospitalizations, we believe this significant increase in both these models is an important finding that substantiates our conclusions. The potential mechanisms regarding the occurrence of dysbiosis is related to overgrowth of potentially pathogenic taxa that would normally be suppressed by the existing barriers against this (gastric acid, bile secretion, and autochthonous taxa), all of which are impaired in cirrhosis and further worsened by superadded PPI use (6, 8, 17, 24).
Recent studies have suggested that the ribosome content of cells is not necessarily directly related to metabolic activity (32, 33) and it has been suggested that quorum sensing in complex communities also affects microbial community activity (9). Thus, more direct measures of metabolic activity need to be utilized, such as transcriptomics, proteomics, or metabolomics, to fully understand the microbial dynamics in the gut ecosystem. Further analyses before and after antibiotics are also needed to fully evaluate their role in suppressing the activity of bacteria.
We conclude that DNA and RNA analysis of the same stool sample demonstrated differing microbiota composition regardless of whether it was tested from healthy subjects or cirrhotic patients. Autochthonous taxa are relatively similar between DNA and RNA analyses and are modulated differentially with renal dysfunction and rifaximin and PPI use. These changes in the microbiota composition with RNA and DNA profiles predict the hospitalization risk in cirrhotic patients independently of clinical variables.
Methods
This study was carried out prospectively in the Virginia Commonwealth University and Richmond VA Medical Centers. Patients with cirrhosis (defined through biopsy, features of decompensation, endoscopic or radiological evidence of varices or cirrhosis in the setting of chronic liver disease) were recruited from inpatient and outpatient settings after informed consent. Stool collection for DNA and RNA was performed using Parapak and stool was stored in RNAlater until it was ultimately extracted. Concomitantly, blood was drawn for MELD score (validated logarithmic score for cirrhosis severity) (34). In the cross-sectional study, we enrolled 4 groups of subjects: (a) healthy controls, (b) compensated outpatient cirrhotics, (c) decompensated outpatients due to HE, and (d) inpatients with infections.
Ultimately, patients were followed for 90 days for nonelective hospitalizations and multivariable analysis predicting these outcomes was evaluated.
An additional 30 subjects (15 healthy controls and 15 compensated cirrhotic outpatients) were included for the before/after omeprazole study (17). All subjects underwent stool collection at baseline and were given 40 mg omeprazole once per day for 14 days, at which point stool was collected again.
Statistics.
RNA relative abundances, DNA relative abundances, and a separate variable, RNA/(RNA + DNA) (referred to here as the RNA ratio), were evaluated for each taxon present at the genus level. The RNA ratio could only be calculated if both RNA and DNA of a particular taxon were present. Therefore, this was calculated in the selected population with both RNA and DNA of a particular taxon. Analysis of DNA alone, RNA alone, and RNA ratios between groups was performed using LEfSe techniques that compared (a) all cirrhotic patients to healthy controls, (b) compensated patients to decompensated patients, and (c) infected patients to uninfected cirrhotic patients. We also studied the CDR ([Lachnospiraceae + Ruminococaceae + ClostridialesXIV + Veillonellaceae]/[Enterobacteriaceae + Bacteroidaceae]) between groups.
In addition, we used LEfSe to study changes in microbial composition on RNA and DNA analysis in cirrhotic patients who were on and not on rifaximin after excluding those with infections and those who were not on SBP prophylaxis. We also split the group of cirrhotic patients according to renal dysfunction (serum creatinine ≥ 1.5 mg/dl) and used LEfSe to compare microbial differences in RNA and DNA analyses depending on predominant renal or hepatic failure. In addition, we used Wilcoxon’s signed rank-sum test to evaluate before/after–PPI changes in taxa at the family level by RNA and DNA analyses for the trial analysis.
Hospitalizations: Due to the large number of variables under consideration, a screening process was conducted to determine variables to consider for a multivariate model. This screening was conducted by performing a series of univariate binary logistic regression models. Any variables that had a P < 0.20 level of significance in the univariate binary logistic regression models were used in a multivariable binary logistic regression model. Once the variables to be considered in the multivariable binary logistic regression were determined, 2 models for hospitalizations were created: 1 for DNA-linked microbiota and clinical variables and 1 for RNA-linked microbiota and clinical variables. The multivariable binary logistic regression models were fit using a backwards elimination procedure where all variables that remained in the model had to be significant at the P < 0.05 significance level.
We created ROC curves to define the relative AUC for RNA-containing versus DNA-containing models. AUCs were determined by analyzing the Youden index of the ROC curves. In order to define the utility of the ultimate clinical+microbiota model compared to clinical variables alone, we used the method proposed by DeLong et al. (18) for comparing the AUCs of correlated ROC curves.
Study approval.
The protocol was approved by the Institutional Review Boards of both hospitals and all participants provided written informed consent.
Detailed methods.
Stool samples, which were kept at –80°C in RNAlater, were thawed at 4°C. Two aliquots (approximately 100 μl each) were separated after mixing the sample for DNA and RNA extractions. The aliquoted samples were centrifuged for 10 minutes at 13,000 g to pellet the stool and separate RNAlater. Supernatant RNAlater was discarded and the stool pellet was used in further extractions according to the respective protocols. The RNA was extracted with an RNeasy Mini Kit from Qiagen and the cDNA was made with GoScript kit from Promega. The reverse bacterial 16S primer (1492R) was used to make the cDNA. A control PCR for RNA samples was run to detect any DNA contamination and RNA samples were treated with extra DNase treatment if any DNA contamination was detected. DNA was extracted with FastDNA Spin kit for Soil (MP Biomedicals) according to the manufacturer’s protocol. Both DNA and cDNA were used in PCRs with universal bacterial primers (please see detail below) for the first 2 variable regions (L27F and 355R) (35). DNA or cDNA was amplified by PCR for sequencing using Ion Torrent technology (36). The same products were also fingerprinted as a quality control (37). For PCR, a fusion forward primer 27F (5′-AGAGTTºTGATCCTGGCTCAG-3′), which contained different 8-bp tags and an adapter for sequencing with PGM (Ion Torrent technology), and a FAM-labeled reverse primer 355R′ (5′-GCTGCCTCCCGTAGGAGT-3′), which included the reverse adapter, were used in duplicate PCRs. Both primers are universal for bacteria and amplify the first 2 variable regions of the 16S rDNA. For fingerprinting, the PCR products were diluted according to their intensity based on agarose gel electrophoresis images and mixed with ILS-600 size standards (Promega) and HiDi Formamide (Applied Biosystems). The diluted samples were separated on an ABI 3130xl fluorescent capillary sequencer (Applied Biosystems) and analyzed using the Genemapper software package (Applied Biosystems). The quality of products was determined and duplicates were checked for confirmation and the best dilution was selected for pooling into one sample for sequencing. The sample was purified using Ampure magnetic beads (Beckman Coulter) and quantified using a DTX880 Multimode Fluorescent detector (Beckman Coulter) before sequencing. The sequencing was done on a Personal Genome Machine using Ion Torrent technology (Applied Biosystems) following the manufacturer’s protocols. The sequences were demultiplexed and uploaded into Galaxy portal for further analysis.
Author contributions
JSB and PMG conceptualized the study. JSB, AF, EAG, CA, MBW, DMH, and MF were involved in patient recruitment and clinical aspects of the study. PMG, RB, SD, and MS were involved in sample and data analysis. LRT performed the biostatistical analysis. PBH helped with critical revisions and drafting of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by VA Merit Review grant I0CX01076 to J.S. Bajaj.
Version 1. 03/08/2018
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(5):e98019. https://doi.org/10.1172/jci.insight.98019.
Contributor Information
Andrew Fagan, Email: andrew.fagan@va.gov.
Robert Brown, Email: rbrownf@gmu.edu.
Chathur Acharya, Email: chathur.acharya@vcuhealth.org.
Michael Fuchs, Email: mfuchs@vcu.edu.
Swati Dalmet, Email: sdalmet@masonlive.gmu.edu.
Masoumeh Sikaroodi, Email: msikaroo@gmu.edu.
References
- 1.Rehm J, Samokhvalov AV, Shield KD. Global burden of alcoholic liver diseases. J Hepatol. 2013;59(1):160–168. doi: 10.1016/j.jhep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Bajaj JS, et al. The 3-month readmission rate remains unacceptably high in a large North American cohort of patients with cirrhosis. Hepatology. 2016;64(1):200–208. doi: 10.1002/hep.28414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tandon P, Garcia-Tsao G. Bacterial infections, sepsis, and multiorgan failure in cirrhosis. Semin Liver Dis. 2008;28(1):26–42. doi: 10.1055/s-2008-1040319. [DOI] [PubMed] [Google Scholar]
- 4.Lin CY, et al. Endotoxemia contributes to the immune paralysis in patients with cirrhosis. J Hepatol. 2007;46(5):816–826. doi: 10.1016/j.jhep.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 5.Ahluwalia V, et al. The etiology of cirrhosis is a strong determinant of brain reserve: A multimodal magnetic resonance imaging study. Liver Transpl. 2015;21(9):1123–1132. doi: 10.1002/lt.24163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bajaj JS, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2014;60(5):940–947. doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, et al. Gut dysbiosis in acute-on-chronic liver failure and its predictive value for mortality. J Gastroenterol Hepatol. 2015;30(9):1429–1437. doi: 10.1111/jgh.12932. [DOI] [PubMed] [Google Scholar]
- 8.Kakiyama G, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58(5):949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J. 2013;7(11):2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gralla JD. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol. 2005;55(4):973–977. doi: 10.1111/j.1365-2958.2004.04455.x. [DOI] [PubMed] [Google Scholar]
- 11.Brettar I, Christen R, Höfle MG. Analysis of bacterial core communities in the central Baltic by comparative RNA-DNA-based fingerprinting provides links to structure-function relationships. ISME J. 2012;6(1):195–212. doi: 10.1038/ismej.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egert M, Schmidt I, Höhne HM, Lachnit T, Schmitz RA, Breves R. rRNA-based profiling of bacteria in the axilla of healthy males suggests right-left asymmetry in bacterial activity. FEMS Microbiol Ecol. 2011;77(1):146–153. doi: 10.1111/j.1574-6941.2011.01097.x. [DOI] [PubMed] [Google Scholar]
- 13.Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13(5):1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- 14.Hunt DE, et al. Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol. 2013;79(1):177–184. doi: 10.1128/AEM.02155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Männistö MK, Kurhela E, Tiirola M, Häggblom MM. Acidobacteria dominate the active bacterial communities of Arctic tundra with widely divergent winter-time snow accumulation and soil temperatures. FEMS Microbiol Ecol. 2013;84(1):47–59. doi: 10.1111/1574-6941.12035. [DOI] [PubMed] [Google Scholar]
- 16.Wüst PK, Horn MA, Drake HL. Clostridiaceae and Enterobacteriaceae as active fermenters in earthworm gut content. ISME J. 2011;5(1):92–106. doi: 10.1038/ismej.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajaj JS, et al. Systems biology analysis of omeprazole therapy in cirrhosis demonstrates significant shifts in gut microbiota composition and function. Am J Physiol Gastrointest Liver Physiol. 2014;307(10):G951–G957. doi: 10.1152/ajpgi.00268.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 19.Wiest R, Albillos A, Trauner M, Bajaj JS, Jalan R. Targeting the gut-liver axis in liver disease. J Hepatol. 2017;67(5):1084–1103. doi: 10.1016/j.jhep.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 20.Cangelosi GA, Meschke JS. Dead or alive: molecular assessment of microbial viability. Appl Environ Microbiol. 2014;80(19):5884–5891. doi: 10.1128/AEM.01763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aellen S, Que YA, Guignard B, Haenni M, Moreillon P. Detection of live and antibiotic-killed bacteria by quantitative real-time PCR of specific fragments of rRNA. Antimicrob Agents Chemother. 2006;50(6):1913–1920. doi: 10.1128/AAC.00869-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ben-Amor K, Heilig H, Smidt H, Vaughan EE, Abee T, de Vos WM. Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl Environ Microbiol. 2005;71(8):4679–4689. doi: 10.1128/AEM.71.8.4679-4689.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez G, González M, Isabal S, Blanc V, León R. Method to quantify live and dead cells in multi-species oral biofilm by real-time PCR with propidium monoazide. AMB Express. 2013;3(1):1. doi: 10.1186/2191-0855-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Y, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54(2):562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 25.Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2(2):99–104. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández J, Tandon P, Mensa J, Garcia-Tsao G. Antibiotic prophylaxis in cirrhosis: Good and bad. Hepatology. 2016;63(6):2019–2031. doi: 10.1002/hep.28330. [DOI] [PubMed] [Google Scholar]
- 27.Bajaj JS, et al. Second infections independently increase mortality in hospitalized patients with cirrhosis: the North American consortium for the study of end-stage liver disease (NACSELD) experience. Hepatology. 2012;56(6):2328–2335. doi: 10.1002/hep.25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nadim MK, et al. Management of the critically ill patient with cirrhosis: A multidisciplinary perspective. J Hepatol. 2016;64(3):717–735. doi: 10.1016/j.jhep.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Ramezani A, Raj DS. The gut microbiome, kidney disease, and targeted interventions. J Am Soc Nephrol. 2014;25(4):657–670. doi: 10.1681/ASN.2013080905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Llorente C, et al. Gastric acid suppression promotes alcoholic liver disease by inducing overgrowth of intestinal Enterococcus. Nat Commun. 2017;8(1):837. doi: 10.1038/s41467-017-00796-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajaj JS, et al. Second infections independently increase mortality in hospitalized patients with cirrhosis: the North American consortium for the study of end-stage liver disease (NACSELD) experience. Hepatology. 2012;56(6):2328–2335. doi: 10.1002/hep.25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell A, et al. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460(7252):220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- 33.Sukenik A, Kaplan-Levy RN, Welch JM, Post AF. Massive multiplication of genome and ribosomes in dormant cells (akinetes) of Aphanizomenon ovalisporum (Cyanobacteria) ISME J. 2012;6(3):670–679. doi: 10.1038/ismej.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamath PS, Kim WR, Advanced Liver Disease Study Group The model for end-stage liver disease (MELD) Hepatology. 2007;45(3):797–805. doi: 10.1002/hep.21563. [DOI] [PubMed] [Google Scholar]
- 35. Lane DJ. 16S/23S rRNA Sequencing. In: Stackebrandt E, Goodfellow M, eds, Nucleic Acid Techniques in Bacterial Systematic. New York, New York: John Wiley and Sons; 1991:115–175. [Google Scholar]
- 36.Kang DJ, et al. Rifaximin exerts beneficial effects independent of its ability to alter microbiota composition. Clin Transl Gastroenterol. 2016;7(8):e187. doi: 10.1038/ctg.2016.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sikaroodi M, Gillevet PM. Quality control in multi-tag pyrosequencing of microbial communities. BioTechniques. 2012;53(6):381–383. doi: 10.2144/000113967. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.