Abstract
Recurrent chromosomal translocations leading to gene fusion formation have been described in uterine sarcomas, including low-grade endometrial stromal sarcoma (LG-ESS). Involvement of the PHF1 gene in chromosomal rearrangements targeting band 6p21 has been found in LG-ESS with different partners from JAZF1 mapping in 7p15, to EPC1 from 10p11, MEAF6 from 1p34, and BRD8 from 5q31.
In the present study, RNA sequencing of a LG-ESS showed a novel recombination of PHF1 with the Enhancer of Polycomb homolog 2 (EPC2). RT-PCR followed by Sanger sequencing and FISH analysis confirmed the EPC2-PHF1 fusion transcript.
Keywords: fusion gene, EPC2, PHF1, RNA sequencing, low-grade endometrial stromal sarcoma
INTRODUCTION
Recurrent chromosomal translocations leading to gene fusion formation have been described in uterine sarcomas [1, 2]. The discovery of such fusion transcripts provides not only fundamental knowledge about the pathogenetic mechanisms behind tumor formation and progression, but opens up for the fusions’ or fusion products’ use as potential molecular diagnostic markers and eventually as targets for smart drugs [3–5]. Many of the fusions identified are specific for distinct neoplastic entities.
The PHD finger protein-1 (PHF1) gene, mapping on chromosome band 6p21, was first identified as being rearranged in low-grade endometrial stromal sarcoma (LG-ESS) [6]. The gene is known to recombine with four partners through different chromosomal rearrangements within the same tumor type. It was found fused with JAZF1 (from 7p15; through an unbalanced 6;7-translocation) [6], with EPC1 (from 10p11; through a 6;10-translocation) [6], with MEAF6 (from 1p34; through a 1;6-translocation) [7], and, the so far latest addition to the list, with BRD8 (from 5q31, through a 5;6-rearrangement) [8]. The very same gene is also involved in the pathogenesis of non-ESS, non-endometrial stromal tumors (EST) such as cardiac ossifying sarcoma [9] as well as benign, atypical, and malignant ossifying fibromyxoid tumors (OFMT) [10–12]. In OFMT, PHF1 has been shown to generate a fusion transcript not only with EP400 (from 12q24) [13], but also with MEAF6 and EPC1 [10]. In cardiac ossifying sarcoma, a JAZF1-PHF1 fusion was demonstrated [9].
We report here a new fusion partner for PHF1 in an LG-ESS (Figure 1) detected by RNA sequencing and validated by RT-PCR followed by Sanger sequencing and Fluorescence In Situ Hybridization (FISH).
Figure 1. Histological examination of the LG-ESS.
(A) H&E – stained slide; tumor cells are spindle-shaped and monomorphic, with low-grade atypia. Multiple small thin-walled vessels are seen between tumor cells. The morphology is characteristic of LG-ESS; (B) Immunoexpression of caldesmon; (C) Immunoexpression of CD10; (D) Immunoexpression of desmin. The tumor has the characteristic IHC profile of LG-ESS, with expression of CD10 and absence of caldesmon and desmin.
RESULTS
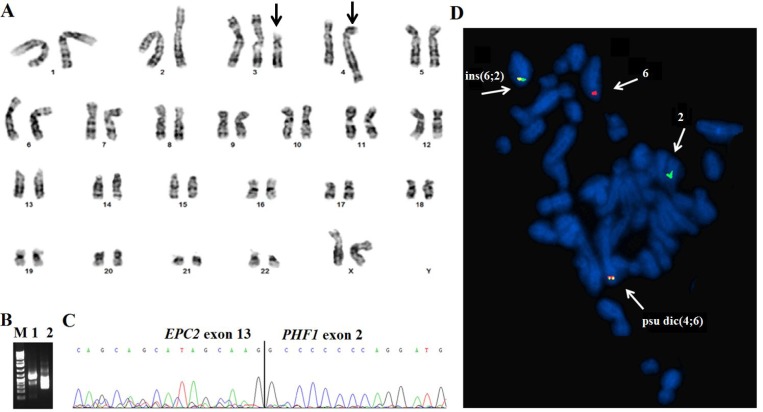
The cytogenetic investigation of the LG-ESS showed an abnormal karyotype described as 47,XX,+add(3)(p11),add(4)(q35) [15] (Figure 2A). PCR investigations for ESS-specific fusion transcripts did not show the presence of any known fusions. A total of five chimeric transcripts were obtained using the FusionCatcher algoritm searching for novel fusions (Table 1). These were tested using the BLAT command (https://genome-euro.ucsc.edu/cgi-bin/hgBlat?command=start the program) to identify those with 100% identity in the genome according to the UCSC Genome Browser (update Dec. 2013, GRCh38/hg38). Only one out of five detected transcripts showed such identity, involving the Enhancer of Polycomb homolog 2 (EPC2) gene with PHF1.
Figure 2. G-banding, RT-PCR, and FISH analysis of the low-grade endometrial stromal sarcoma.
(A) Karyogram of the LG-ESS. Derivative chromosomes, add(3p), and psu dic(4;6) are indicated by arrows. (B) Gel electrophoresis showing the amplified cDNA fragments. M, 1 Kb DNA ladder (GeneRuler, ThermoFisher); lane 1, amplification of cDNA fragment using the primers EPC2-2110F1 and PHF1-524R1; lane 2, Nested PCR using the primers EPC2-2266F2 and PHF1-376R1. (C) Partial sequence chromatogram of the amplified cDNA fragment showing the junction point of the EPC2-PHF1 fusion. (D) Metaphase FISH for the detection of the EPC2-PHF1 fusion gene. The green signal is the EPC2 probe from 2q23 whereas the red signal corresponds to the PHF1 gene from 6p21. The two fusion signals (yellow) were seen one on the psu dic(4;6) and ins(6;2).
Table 1. Fusion transcripts detected using FusionCatcher.
| 5′-Chr | 3′- Chr | 5′- Partner gene | 3′- Partner gene | Fusion sequence |
|---|---|---|---|---|
| 17 | 17 | C17orf107 | GP1BA | GAAGAGATGGTGATAAAGACGCAGTTCCTCGTTCTTCCCCACACCCCTGC*CTGGAGCTGCAG AGGGGACGGCAAGTGACAGTGCCCCGGGCCTGGCTGCT |
| 2 | 6 | EPC2 | PHF1 |
TGCCAAAGGTTACTCCCAGCAGTGCCATCAGCAGCATAGCAAG*GCCCCCCCAGGATGC AATGGCGCAGCCCCCCCGGCTGAGCCGC |
| 2 | 2 | MBD5 | EPC2 | TGAAGGCTTTATGGTTTACAGACAAGATCTTTAGAAGATAAGCACTAAAG*AGAGAACCACG AACCAGAAAGATTGGGCTTAAATGGAATAGCAGAGACAA |
| 19 | 19 | EIF3K | ACTN4 | AGAAGAGAGCATTAAACCCAAGAACATTGTGGAGAAGATTGACTTTGACA*ACCTTCACGGC ATGGTGCAACTCCCACCTGCGGAAGGCAGGCACACAGAT |
| X | 14 | MED12 | IRF2BPL | TCTTATAGCAGCAGCAGCAACAGCAACAGCAGCAGCAGCAGCAGCAGCAA*CAGCAGCAGC AGCAGCAGCAGCAGCAACAACAGCTCAACCACGTTGATGG |
RT-PCR with specific primer combinations confirmed an in-frame fusion between exon 13 of EPC2 (accession number NM_015630.3) and exon 2 of PHF1 (accession number NM_002636.4) (Figure 2B and 2C; Table 2).
Table 2. Primers used for PCR and sanger sequencing analyses.
| Name | Sequence | Position | Gene | Accession number |
|---|---|---|---|---|
|
EPC2-2110F1 PHF1-524R1 EPC2-2266F2 PHF1-376R1 |
5′ -cagcttgtaaggacagttggc - 3′ 5′-ctcagagcgacagacacaac- 3′ 5′ -aatacacggacttcagcacc- 3′ 5′ -tatagcagcccatcagtcca- 3′ |
2110-2139 505-524 2266-2285 357-376 |
EPC2 PHF1 EPC2 PHF1 |
NM_015630.3 NM_002636.4 NM_015630.3 NM_002636.4 |
Since the G-banding analysis did not show any rearrangements of 6p21, i.e., the band in which PHF1 is located, we performed metaphase FISH experiments to see the chromosomal location of the EPC2-PHF1 fusion. Two fusion signals (yellow color) were identified, one on the pseudo dicentric(4;6) and the other on the inserted(6;2) (Figure 2D). The revised karyotype incorporating the FISH and RNA-sequencing data thus became 47,XX, +add(3)(p11),psu dic(4;6)(q31;q15)ins(6;2)(p21;q23q23), +6, ins(6;2)(p21;q23q23).ish psu dic(4;6)(PHF1+, EPC2+), ins(6;2)(PHF1+, EPC2+;EPC2-)[4].
DISCUSSION
Rearrangements of PHF1 from chromosomal band 6p21 were first reported in ESS in 2006 when the gene was found recombined with two different partners, JAZF1 (from 7p15) and EPC1 (from 10p11), through unbalanced 6p;7p- and 6p;10p-rearrangements, respectively [6]. Subsequent cytogenetic and molecular studies detected additional partners for PHF1: MEAF6, recombined through a t(1;6)(p34;p21) [14] and, recently, BRD8 from 5q31 [8]. We now report the fifth recombination partner, namely EPC2 from 2q23.
The EPC2 gene is a paralog of Enhancer of Polycomb homolog 1 of Drosophila (EPC1), a member of the Polycomb Group of Genes (PcG). EPC2 is conserved from yeast to man. It is a component of an essential chromatin regulatory complex which has a potential oncogenic role as it contributes to cellular processes such as induction of apoptotic death [15]. Little is known about EPC2 though different genetic studies suggest that deletion of its yeast homolog Ep11 [16] causes accumulation of cells in G2M, increases sensitivity to DNA damaging agents, leads to defective telomeric silencing, and may result in global loss of histone H2A as well as H4 acetylation. The gene also has an important role in homeotic gene silencing in Drosophila. Interestingly, a target knockdown screen of EPC1 and EPC2 in AML cell lines caused apoptosis and loss of stem cell potential [17]. Mutation of EP400 complex components EPC1 and EPC2 has been identified as pathogenetic events in AML [18].
The fusion of EPC2 and PHF1 led to a chimeric transcript retaining the entire coding regions from both genes interlocked in an open reading frame. The resulting putative protein would consist of 855 amino acid residues from EPC2 (AAH93818) and 662 amino acids from PHF1 (AAC52062.1); thus, the predicted protein sequence would consist of 1,517 amino acids in total.
The conserved domains from EPC2 include EPL1 (Enhancer of polycomb-like1) and –E-Pc-C (Enhancer of Polycomb C-terminus), complexes involved in transcriptional activation and heterocromatin formation, respectively. From the PHF1 protein are included its Tudor and PHD zinc finger as well as MTF2 domains, as was also the case in other PHF1-fusions previously described [6, 8, 14].
It appears that genes fused with PHF1 through various translocations are involved in regulation of gene expression through formation of zing finger motifs or acetylation of histone proteins [14, 19]. They would therefore have the possibility of deregulating the transcription of a number of genes in LG-ESS as well as in seemingly unrelated tumors such as OFMT and cardiac ossifying sarcoma [9, 10]. This suggests a common neoplastic mechanism, namely rearrangement of the same gene, in these tumors that show little or no morphologic or immunophenotypic overlap. Indeed, the very same EPC1-PHF1 and MEAF6-PHF1 chimeric transcripts have been identified in both LG-ESS and OFMT [6, 12, 14], making these entities genocopies. In this regard, PHF1 and the tumors characterized by its rearrangement do not differ principally from Soft Tissue Tumors in general where similar situations are known to be common [5].
As seen in the LG-ESS, OFMT, and cardiac ossifying sarcomas analyzed so far, the rearrangements of PHF1 as the 3′- partner have breakpoints in exon 2 leading to retention of most of the gene sequence [6–10, 13, 14]. As a result of the rearrangements, the gene comes under the influence of a new promoter belonging to its 5′ partners. The fact that all detected alterations of PHF1 are similar makes it likely that the gene is a key player in the tumorigenesis of these sarcoma subgroups [6, 10, 11]. Furthermore, the chromosomal aberrations behind the gene recombinations were always unbalanced allowing PHF1 to retain its 5′-3′ orientation [6]. It is finally worthy of note that the rearrangement of 6p21, the chromosomal band where PHF1 is located, was cryptic in the LG-ESS we report, hinting that it may be cytogenetically invisible also in other tumors. It seems certain that the use of FISH or molecular methods is necessary to detect this and related gene-level rearrangements in tumors with seemingly normal or complex karyotypes, perhaps especially when the lesions are histologically unusual.
The finding underscores the complex pathogenetic interplay that exists among ESS. Since EPC2 is a paralogue of EPC1, one could speculate that the EPC2-PHF1 fusion, too, will soon to be found in the rare OFMT and other non-ESS tumors.
MATERIALS AND METHODS
Case history
A 49-year-old woman presented with a tumor of the uterus whereupon total hysterectomy with left-sided salpingo-oophorectomy and right-sided salpingectomy was performed. Gross evaluation showed a 10 cm tumor with heterogeneous cut section in the uterine corpus, as well as several smaller nodules which were presumed to be leiomyomas. Morphological assessment of the large lesion showed a spindle cell tumor with low-grade atypia, 3 mitoses/10 HPF, but no necrosis. The histological diagnosis was LG-ESS. This was confirmed by immunostaining showing positivity for CD10, ER, PR, and SMA but negativity for desmin and caldesmon (Figure 1). The left ovary and both fallopian tubes were without tumor involvement as was the right ovary, which was also subsequently removed.
G-banding and karyotyping
Fresh tissue from a representative area of the tumor was analyzed cytogenetically as part of our diagnostic routine [20]. The karyotype was written following the recommendations of the International System for Human Cytogenomic Nomenclature (ISCN) [21].
The study was approved by the Regional Committee for Medical and Health Research Ethics, South-East Norway (REK Sør-Øst; http://helseforskning.etikkom.no). Written informed consent was obtained from the patient. The consent included acceptance that the clinical details be published. The ethics committee’s approval included a review of the consent procedure. All patient information has been de-identified.
Molecular genetic analyses
Total RNA was extracted from fresh frozen tumor tissue using miRNeasy (Qiagen, Hilden, Germany) and QIAcube (Qiagen). The RNA quality was evaluated using 2100 Bioanalyzer (Agilent, Santa Clara, California, USA) according to the manufacturer’s instructions. One µg of total RNA was reverse-transcribed in a 20 µL reaction volume using iScript Advanced cDNA synthesis Kit for RT-PCR according to the manufacturer’s instructions (Bio-Rad Laboratories, Oslo, Norway).
RT-PCR was used to investigate whether known ESS-specific fusion transcripts were present. The primers and PCR cycles are listed in previous publications [8, 22].
One µg of total RNA was sent for RNA-sequencing at the Genomics Core Facility, Oslo University Hospital and University of Oslo (http://oslo.genomics.no/). The sequencing was performed using an Illumina HiSeq 2000 instrument and the Illumina software pipeline. FusionCatcher (version 0.99.3a beta-April 15, 2014) with the associated ENSEMBL, UCSC, and RefSeq databases automatically downloaded by FusionCatcher (https://code.google.com/p/fusioncatcher/) were used for the discovery of fusion transcripts.
The primers used for PCR reactions and Sanger sequencing are listed in Table 2. The 25 μl PCR volume contained 12.5 μl Premix Ex Taq DNA Polymerase Hot Start Version (Takara Bio Europe/SAS, Saint-Germain-en-Laye, France), 1 μl of cDNA, and 1 µL of each of the forward and reverse primers. The primer combinations were EPC2-2110F1 and PHF1-524R1 for the first PCR reaction and EPC2-2266F2 and PHF1-376R1 for Nested PCR. The PCR amplifications were run on a C-1000 Thermal cycler (Bio-Rad Laboratories) with an initial denaturation at 94° C for 30 sec, followed by 35 cycles at 98° C for 7 sec, 55° C for 30 sec, 1 min at 72° C, and a final extension at 72° C for 5 min. Three µL of the PCR product were stained with GelRed (Biotium, Hayward, CA, USA), analyzed by electrophoresis through 1.0% agarose gel, and photographed. The remaining 22 µl PCR product were purified using the QIAquick PCR Purification Kit (Qiagen) and sequenced using 3500 Genetic Analyzer (Applied Biosystems). The BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and BLAT (http://genome.ucsc.edu/cgi-bin/hgBlat) softwares were used for computer analysis of sequence data.
Fluorescence in situ hybridization (FISH)
BAC probes were retrieved from the Human ‘32K’ BAC Re-Array library (BACPAC Resources, https://bacpacresources.org/). They were selected according to physical and genetic mapping data on chromosomes 2 and 6 (see below) as reported on the Human Genome Browser at the University of California, Santa Cruz website (May 2004, http://genome.ucsc.edu/). The clones used were RP11-899B15, mapping to 2q23 and containing the EPC2 gene (labelled in green), and RP11-436J22 and RP11-600P03 mapping to 6p21 and overlapping with the PHF1 locus (labelled in red). FISH was performed as described elsewhere [23]. Fluorescent signals were captured and analyzed using the CytoVision system (Leica Biosystems, Newcastle, UK).
Acknowledgments
The authors thank Hege Kilen Andersen and Kristin Andersen for excellent technical assistance. This study was supported by grants from the Norwegian Radium Hospital Foundation.
Footnotes
CONFLICTS OF INTEREST
The author(s) declare that they have no competing interests.
REFERENCES
- 1.Mitelman F, Johansson B, Mertens F. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2017 [Google Scholar]
- 2.Mertens F, Johansson B, Fioretos T, Mitelman F. The emerging complexity of gene fusions in cancer. Nat Rev Cancer. 2015;15:371–81. doi: 10.1038/nrc3947. https://doi.org/10.1038/nrc3947. [DOI] [PubMed] [Google Scholar]
- 3.Conklin CM, Longacre TA. Endometrial stromal tumors: the new WHO classification. Adv Anat Pathol. 2014;21:383–93. doi: 10.1097/PAP.0000000000000046. https://doi.org/10.1097/PAP.0000000000000046. [DOI] [PubMed] [Google Scholar]
- 4.Micci F, Heim S. Tumors of the female genital organs. Cancer Cytogenetics. 2015:447–80. [Google Scholar]
- 5.Mertens F, Antonescu CR, Mitelman F. Gene fusions in soft tissue tumors: recurrent and overlapping pathogenetic themes. Genes Chromosomes Cancer. 2016;55:291–310. doi: 10.1002/gcc.22335. https://doi.org/10.1002/gcc.22335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Micci F, Panagopoulos I, Bjerkehagen B, Heim S. Consistent rearrangement of chromosomal band 6p21 with generation of fusion genes JAZF1/PHF1 and EPC1/PHF1 in endometrial stromal sarcoma. Cancer Res. 2006;66:107–12. doi: 10.1158/0008-5472.CAN-05-2485. https://doi.org/10.1158/0008-5472.CAN-05-2485. [DOI] [PubMed] [Google Scholar]
- 7.Micci F, Gorunova L, Gatius S, Matias-Guiu X, Davidson B, Heim S, Panagopoulos I. MEAF6/PHF1 is a recurrent gene fusion in endometrial stromal sarcoma. Cancer Lett. 2014;347:75–78. doi: 10.1016/j.canlet.2014.01.030. https://doi.org/10.1016/j.canlet.2014.01.030. [DOI] [PubMed] [Google Scholar]
- 8.Micci F, Brunetti M, Dal Cin P, Nucci MR, Gorunova L, Heim S, Panagopoulos I. Fusion of the genes BRD8 and PHF1 in endometrial stromal sarcoma. Genes Chromosomes Cancer. 2017;56:841–45. doi: 10.1002/gcc.22485. https://doi.org/10.1002/gcc.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoolmeester JK, Sukov WR, Maleszewski JJ, Bedroske PP, Folpe AL, Hodge JC. JAZF1 rearrangement in a mesenchymal tumor of nonendometrial stromal origin: report of an unusual ossifying sarcoma of the heart demonstrating JAZF1/PHF1 fusion. Am J Surg Pathol. 2013;37:938–42. doi: 10.1097/PAS.0b013e318282da9d. https://doi.org/10.1097/PAS.0b013e318282da9d. [DOI] [PubMed] [Google Scholar]
- 10.Antonescu CR, Sung YS, Chen CL, Zhang L, Chen HW, Singer S, Agaram NP, Sboner A, Fletcher CD. Novel ZC3H7B-BCOR, MEAF6-PHF1, and EPC1-PHF1 fusions in ossifying fibromyxoid tumors—molecular characterization shows genetic overlap with endometrial stromal sarcoma. Genes Chromosomes Cancer. 2014;53:183–93. doi: 10.1002/gcc.22132. https://doi.org/10.1002/gcc.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebre-Medhin S, Nord KH, Möller E, Mandahl N, Magnusson L, Nilsson J, Jo VY, Vult von Steyern F, Brosjö O, Larsson O, Domanski HA, Sciot R, Debiec-Rychter M, et al. Recurrent rearrangement of the PHF1 gene in ossifying fibromyxoid tumors. Am J Pathol. 2012;181:1069–77. doi: 10.1016/j.ajpath.2012.05.030. https://doi.org/10.1016/j.ajpath.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 12.Graham RP, Weiss SW, Sukov WR, Goldblum JR, Billings SD, Dotlic S, Folpe AL. PHF1 rearrangements in ossifying fibromyxoid tumors of soft parts: A fluorescence in situ hybridization study of 41 cases with emphasis on the malignant variant. Am J Surg Pathol. 2013;37:1751–55. doi: 10.1097/PAS.0b013e31829644b4. https://doi.org/10.1097/PAS.0b013e31829644b4. [DOI] [PubMed] [Google Scholar]
- 13.Endo M, Kohashi K, Yamamoto H, Ishii T, Yoshida T, Matsunobu T, Iwamoto Y, Oda Y. Ossifying fibromyxoid tumor presenting EP400-PHF1 fusion gene. Hum Pathol. 2013;44:2603–08. doi: 10.1016/j.humpath.2013.04.003. https://doi.org/10.1016/j.humpath.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Panagopoulos I, Micci F, Thorsen J, Gorunova L, Eibak AM, Bjerkehagen B, Davidson B, Heim S. Novel fusion of MYST/Esa1-associated factor 6 and PHF1 in endometrial stromal sarcoma. PLoS One. 2012;7:e39354. doi: 10.1371/journal.pone.0039354. https://doi.org/10.1371/journal.pone.0039354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. https://doi.org/10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–28. doi: 10.1101/gad.1056603. https://doi.org/10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinclair DA, Clegg NJ, Antonchuk J, Milne TA, Stankunas K, Ruse C, Grigliatti TA, Kassis JA, Brock HW. Enhancer of Polycomb is a suppressor of position-effect variegation in Drosophila melanogaster. Genetics. 1998;148:211–20. doi: 10.1093/genetics/148.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang X, Spencer GJ, Lynch JT, Ciceri F, Somerville TD, Somervaille TC. Enhancers of Polycomb EPC1 and EPC2 sustain the oncogenic potential of MLL leukemia stem cells. Leukemia. 2014;28:1081–91. doi: 10.1038/leu.2013.316. https://doi.org/10.1038/leu.2013.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avvakumov N, Côté J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–407. doi: 10.1038/sj.onc.1210608. https://doi.org/10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 20.Mandahl N. Methods in solid tumor cytogenetics. 2001 [Google Scholar]
- 21.McGowan-Jordan J, Simons A, Schmid M. An International System for Human Cytogenomic Nomenclature (ISCN 2016) 2016 [Google Scholar]
- 22.Micci F, Gorunova L, Agostini A, Johannessen LE, Brunetti M, Davidson B, Heim S, Panagopoulos I. Cytogenetic and molecular profile of endometrial stromal sarcoma. Genes Chromosomes Cancer. 2016;55:834–46. doi: 10.1002/gcc.22380. https://doi.org/10.1002/gcc.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panagopoulos I, Thorsen J, Gorunova L, Haugom L, Bjerkehagen B, Davidson B, Heim S, Micci F. Fusion of the ZC3H7B and BCOR genes in endometrial stromal sarcomas carrying an X;22-translocation. Genes Chromosomes Cancer. 2013;52:610–18. doi: 10.1002/gcc.22057. [DOI] [PubMed] [Google Scholar]