Abstract
Traditionally, reflux esophagitis was assumed to develop as a caustic, chemical injury inflicted by refluxed acid. Recently, however, studies in rats and humans suggest that reflux esophagitis develops as a cytokine-mediated inflammatory injury, with hypoxia inducible factor (HIF)-2α playing a major role. In response to the reflux of acid and bile, HIF-2α in esophageal epithelial cells becomes stabilized, thereby increasing production of pro-inflammatory cytokines that attract T lymphocytes and other inflammatory cells to damage the esophagus. Recent studies have identified small molecule inhibitors of HIF-2α that demonstrate exquisite isoform selectivity, and clinical trials for treatment of HIF-2α-driven kidney cancers are ongoing. It is conceivable that a HIF-2α-directed therapy might be a novel approach to prevention and treatment of reflux esophagitis.
Reflux esophagitis: acid burn or cytokine sizzle?
Acid Burn
Traditionally, reflux esophagitis was assumed to develop as an acid burn in which esophageal squamous epithelial cells succumbed to the caustic chemical effects of refluxed gastric acid [1]. The acid-induced death of esophageal surface cells was thought to incite an acute, granulocytic inflammatory response that started in the epithelium and later progressed into the lamina propria and, with ulceration, into the submucosa. The loss of esophageal surface cells also was assumed to stimulate hyperplasia of progenitor cells in the basal layer of the squamous epithelium, which is a characteristic histologic feature of gastroesophageal reflux disease (GERD) [2,3].
Cytokine sizzle
In 2009, the acid burn concept of reflux esophagitis was challenged in a report describing the histologic progression of GERD in rats that had reflux induced by creating an esophago-duodenostomy [4]. Reflux esophagitis in these animals began, not with the death of surface cells and epithelial infiltration by granulocytes, but rather with T lymphocytes infiltrating the esophageal submucosa first, and later progressing into the lamina propria and epithelium. Surface cell erosions did not appear until weeks after esophago-duodenostomy, and basal cell hyperplasia developed well before the loss of surface cells. The report also described in vitro studies showing that acid and bile salts caused human esophageal epithelial cells in culture to secrete pro-inflammatory and pro-proliferative cytokines such as interleukin (IL)-8. These findings suggested an alternative hypothesis for reflux esophagitis pathogenesis in which refluxed gastric juice did not kill esophageal epithelial cells directly, but rather stimulated them to secrete cytokines that induce epithelial proliferative changes and attract the T lymphocytes and other inflammatory cells that ultimately damage the mucosa. Thus, reflux esophagitis appears to develop as a cytokine sizzle.
A recent clinical study explored this hypothesis that reflux esophagitis develops as a cytokine-mediated injury rather than as an acid burn [5]. In 12 patients with reflux esophagitis healed by proton pump inhibitors (PPIs), the investigators induced acute esophagitis by interrupting PPI therapy. Endoscopic examinations performed at 1 and 2 weeks after stopping PPIs showed that all patients had redeveloped reflux esophagitis within two weeks, and esophageal biopsies confirmed that, as in the rat model, acute reflux esophagitis in humans begins with T lymphocyte-predominant inflammation and with basal cell hyperplasia developing before the loss of surface cells. These findings support a cytokine-mediated pathogenesis for reflux esophagitis.
Hypoxia-inducible factor-2α: a key mediator of the cytokine sizzle
HIF-2α stabilization and signaling
The same group subsequently explored a role for hypoxia inducible factors (HIFs) in mediating the GERD-induced release of pro-inflammatory cytokines by the esophagus. HIFs are transcription factors that enable cells to respond to hypoxic stress, and HIFs are known to mediate some inflammatory processes [6–9]. HIF proteins are heterodimers comprising a HIF-1β subunit that is expressed constitutively and a HIF-α subunit that can be regulated by oxygen [9]. Under normal oxygen conditions, prolyl hydroxylases (PHDs) in the cytoplasm catalyze the hydroxylation of proline residues in the oxygen-dependent degradation domain of HIF-α, and this hydroxylation initiates the rapid degradation of HIF-α by proteasomes. Hypoxia decreases PHD activity, thereby preventing proteasomal degradation of the HIF-α subunit. Thus, hypoxia stabilizes HIFs and enables them to accumulate in the cytoplasm and translocate to the nucleus to induce transcription of their target genes, which can include pro-inflammatory cytokines [10]. Like hypoxia, reactive oxygen species (ROS) also can decrease PHD activity and stabilize HIF proteins, and esophageal squamous cells exposed to acid and bile salts in vitro have been shown to produce ROS [11]. Based on these observations, the investigators hypothesized that refluxed acid and bile salts cause esophageal squamous epithelial cells to produce ROS that inactivate PHDs, enabling HIFs to accumulate and to stimulate the production of pro-inflammatory molecules [12].
HIF-2α in human reflux esophagitis
Using esophageal biopsy specimens taken from GERD patients in the aforementioned clinical study at 1 and 2 weeks after stopping PPIs, the investigators noted that the development of acute reflux esophagitis was associated with a significant increase in epithelial immunostaining for HIF-2α and phosphorylated NF-kB subunit p65 (phospho-p65), and with increased mRNA expression of a number of pro-inflammatory mediators including IL-8, IL-1β, tumor necrosis factor (TNF)-α, cyclooxygenase (COX)-2, and intercellular adhesion molecule (ICAM)-1 [12]. Using the statistical technique of computing eta2 values for non-linear correlations, large associations were found among levels of HIF-2α and phospho-p65 and mRNA expression of the pro-inflammatory mediators. These findings suggest that the development of reflux esophagitis is associated with increases in HIF-2α that appear to contribute to increased NF-kB/p65 activity, which in turn appears to contribute to increased expression of pro-inflammatory mediators in esophageal squamous epithelium.
Reflux-induced HIF-2α stabilization and signaling in esophageal squamous cells in vitro
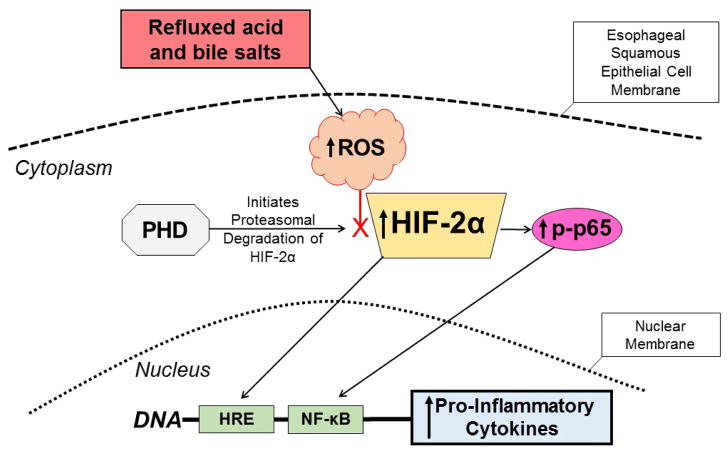
The investigators also performed in vitro studies in human esophageal squamous cell lines showing that acidic bile salts decrease PHD function by generating intracellular ROS through the squamous cell’s NADPH oxidase system (Figure 1) [12]. This ROS-induced decrease in PHD function caused a sustained increase in squamous cell levels of HIF-2α, which mediated an NF-κB/p65-dependent inflammatory response with an increase in expression of pro-inflammatory molecule mRNAs. In addition, conditioned media from esophageal cells that were treated with acidic bile salts were found to increase T cell migration rates in a transwell system. Finally, HIF-2α knockdown by shRNA and HIF-2α inhibition using a selective small molecule antagonist (S,R)-4 (Figure 2) [13] blocked the increases in pro-inflammatory molecule expression and T cell migration induced by acidic bile salts [12]. [Note that (S,R)-4 was named (S,R)-37 Peak 2 in Reference 12] These observations support the hypothesis that reflux esophagitis develops through a cytokine-mediated process in which HIF-2α plays a central role (Figure 1).
Figure 1.
Proposed mechanism for the pathogenesis of reflux esophagitis. Esophageal squamous epithelial cells exposed to refluxed acid and bile salts generate intracellular ROS that decrease the activity of PHD, the enzyme that initiates proteasomal degradation of HIF-2α. The decreased PHD activity stabilizes HIF-2α and enables it to accumulate in the cytoplasm. The stabilized HIF-2α then can translocate to the nucleus to induce transcription of pro-inflammatory cytokine genes that have hypoxia responsive elements (HREs) in their promoter regions. The stabilized HIF-2α in the cytoplasm also can increase levels of phospho-p65, enabling it to translocate to the nucleus and bind the NF-κB gene promoter, which further stimulates production of the pro-inflammatory cytokines that mediate the development of reflux esophagitis. ROS=reactive oxygen species, PHD=prolyl hydroxylase, HIF=hypoxia inducible factor, HRE= hypoxia responsive element, NF-κB=nuclear factor κB.
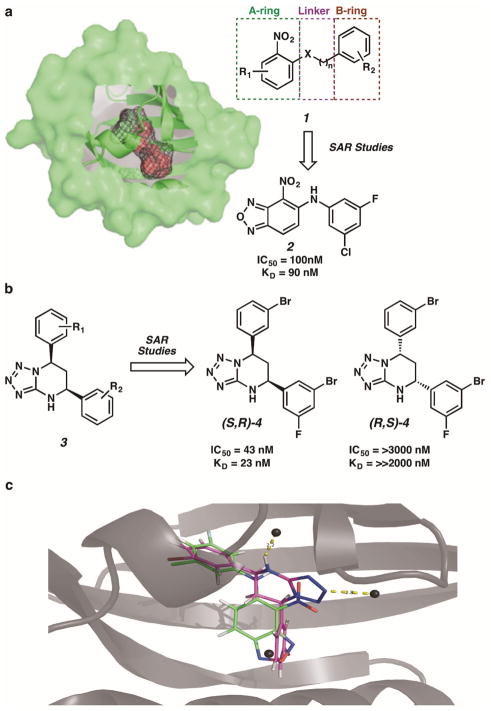
Figure 2.
Direct inhibition of HIF-2α with small molecule antagonists. (a) A large 290 Å3 cavity buried within the HIF-2α PAS-B domain was identified as a target for small molecule binding. The general class of inhibitor 1 was optimized via structure-activity-relationship studies to compound 2. (b) The general class of inhibitor 3 was optimized to compound (S,R)-4. The enantiomer (R,S)-4 was not an effective inhibitor of HIF-2α. (c) Both inhibitors 2 and (S,R)-4 bind within the same buried cavity of the HIF-2α PAS-B domain.
Isoform-selective HIF-2α inhibitors
Small molecule inhibition of the HIF family of transcription factors is recognized as a potentially valuable therapeutic approach to treat HIF-dependent diseases. Although several high throughput screen-derived synthetic small molecules and natural products have been reported to inhibit the HIF pathway [14–17], most of these directly interact with targets other than the HIF proteins themselves, limiting their utility in testing HIF-centric therapeutic hypotheses.
In one study, Bruick, Gardner, MacMillan, and Tambar exploited an unusually large 290 Å3 cavity buried within the Per-ARNT-Sim (PAS)-B domain of the HIF-2α isoform to demonstrate allosteric inhibition via direct small molecule binding (Figure 2a) [18,19]. An extensive high throughput screen of more than 200,000 small molecules was performed to detect direct small molecule interaction with HIF-2α PAS-B [20,21]. “Hits” within the screening process were determined based on a compound’s ability to inhibit HIF-2α–HIF-β heterodimerization in a homogenous, bead-based luminescence proximity assay (AlphaScreen). The most effective small molecule disrupters of HIF-2α–HIF-β heterodimerization possessed the general structure of compound 1, which served as a starting point for structure-activity-relationship (SAR) studies. Diversification of 1 to a variety of HIF-2α antagonists focused on three major regions: (1) modification of the left-hand A-ring portion characterized by a nitro-bearing oxadiazole, (2) variation of the central heteroatom-containing linker, and (3) diversification of the right hand aromatic B-ring region. Ultimately, analogue 2 exhibited the strongest inhibitory activity in this structural class.
To better understand the nature of the interaction of HIF-2α and inhibitor 2, NMR-based structural analysis was paired with X-ray crystallographic studies [20]. A co-crystal structure of inhibitor 2 shows the compound bound within the HIF-2α PAS-B internal cavity, favoring a weak electrostatic interaction between the nitro functionality of the A-ring and the H248 imidazole side chain. The specificity in positioning required to accommodate this interaction situates the B-ring of the molecule adjacent to the β-sheet, inducing a series of notable conformational changes throughout the domain as observed by 15N/1H HSQC NMR studies of a 15N-enriched HIF-2α PAS-B-2 complex. While these conformational changes perturb interactions mediated through the β-sheet observed between isolated PAS domains [20], additional effects on residues residing on the opposing face of the domain may interfere with key dimer contacts observed in structures of the complete HIF heterdimerization units [22,23].
Isoform selective HIF-2α inhibitors-the next generation
Subsequent to this original study, Bruick, Gardner, MacMillan, and Tambar identified a more potent class of inhibitors of HIF-2α–HIF-β heterodimerization, which are based on a tetrazolo-tetrahydropyrimidine ring system (3, Figure 2b) [13]. This general pharmacophore is preferred over compound 2 for several reasons. While the nitro functionality in compound 2 proved to be necessary for activity, it also posed pharmacokinetic disadvantages that are averted in this second scaffold. Additionally, tetrazole moieties increase compound solubility and have been widely studied as pharmacophores for carboxylic acids [24,25]. Finally, these compounds are unique in that they are chiral molecules and thus may prove to exhibit interesting selectivity.
After extensive structure-activity-relationship studies, tetrazolo-tetrahydropyrimidine (S,R)-4 was identified as high affinity binding, effective, isoform-selective inhibitor of HIF-2α in cells. Further examination revealed stereo-preferential binding of this compound, where (S,R)-4 alone presented inhibitory activity (IC50 = 43 nM, KD = 23 nM) and its enantiomer (R,S)-4 was entirely inactive (IC50 = >30,000 nM, KD = ≫2,000 nM). Co-crystal structures of (S,R)-4 bound within the HIF-2α PAS-B cavity were instrumental in defining the absolute stereochemistry of the active enantiomeric series and providing insight into the structural features that govern binding affinity.
Comparison of ternary complex (S,R)-4-HIF-2α PAS-B to 2-HIF-2α PAS-B revealed that both small molecules bind within the same buried cavity of the PAS-B domain and share many structural similarities as well as some dissimilarities that may account for the enhanced binding witnessed with inhibitor (S,R)-4 (Figure 2c). The more pronounced binding interactions observed with (S,R)-4-HIF-α PAS-B lead to a series of conformational changes within the binding domain, allowing for an expanded internal cavity 40–65% larger in volume than that seen with inhibitor 2 and increasing the disruption of important intermolecular contacts with the HIF-β PAS-B domain.
These improved analogs feature sufficient potency and pharmacologic properties to demonstrate efficacy in cultured cells [13,20]. Both compounds 2 and (S,R)-4 antagonize HIF-2α–HIF-β heterodimer formation. The diminished HIF-2 DNA-binding activity results in decreased expression of HIF-2α target genes. These effects are not observed with the inactive (R,S)-4 enantiomer. Moreover, these compounds are selective for HIF-2α; no inhibition of HIF-1α target gene expression was observed. Despite the >70% identity between the HIF-1α and -2α PAS-B domains, several bulkier residues face the internal HIF-1α cavity to preclude inhibitor binding. Effectively, nature has provided an endogenous internal control to assess the on-target effects of these compounds. These “tool” inhibitors are suitable for proof-of-concept studies in cultured cells.
Clinical application of HIF-2α small molecule inhibitors
Excitingly, insights gleaned from these lead compounds served as the launching point for a comprehensive medicinal chemistry effort by Peloton Therapeutics, Inc. to improve inhibitor potency, selectivity, pharmacokinetic, and toxicity profiles. These efforts have led to the successful development of well-tolerated HIF-2α inhibitors capable of selectively antagonizing HIF-2α in vivo in preclinical animal models [23,26,27]. Clinical trials of these inhibitors for the treatment of HIF-2α driven kidney cancers are ongoing (ClinicalTrials.gov Identifiers: NCT02553356, NCT02293980, NCT03108066, and NCT02974738). These results support the feasibility of targeting the HIF-2α transcription factor in therapeutic settings.
Conclusion: implications for HIF-2α inhibitors in reflux esophagitis
Instead of the traditional notion that refluxed gastric acid causes a chemical burn in the esophagus, the studies discussed above suggest that refluxed acid and bile salts cause esophageal epithelial cells to produce ROS that decrease PHD activity, thereby stabilizing HIF-2α and enabling it to accumulate in the cytoplasm and translocate to the nucleus. In the nucleus, HIF-2α stimulates the production of pro-inflammatory cytokines. HIF-2α in the cytoplasm also mediates the activity of phospho-p65, enabling it to translocate to the nucleus and bind the NF-κB gene promoter, which further stimulates production of the pro-inflammatory cytokines that mediate the development of reflux esophagitis. Medicinal chemistry efforts have led to the development of well-tolerated, exquisitely selective HIF-2α inhibitors that are in ongoing clinical trials. Since HIF-2α appears to play such a central role in the esophageal epithelium’s inflammatory response to gastroesophageal reflux, a HIF-2α-directed therapy might be on the horizon as a novel approach to the prevention and treatment of reflux esophagitis.
Highlights.
Reflux esophagitis (RE) traditionally was thought to develop as a caustic chemical injury.
Recent studies show that RE develops as a cytokine-mediated injury initiated by HIF-2α.
Refluxed material stabilizes esophageal HIF-2α, which increases inflammatory cytokine levels.
Highly selective HIF-2α small molecule inhibitors have been developed and are in clinical trials.
Investigational opportunities are open for HIF-2α-directed therapies in RE.
Acknowledgments
Funding: This work was supported by the National Institutes of Health (R01-DK63621and R01-DK103598 to R.F.S. and S.J.S.; R01-GM102604 to U.K.T.); W.W. Caruth, Jr. Endowed Scholarship and the Welch Foundation (I-1748 to U.K.T.); Sloan Research Fellowship (U.K.T.); R.K.B. is the Michael L. Rosenberg Scholar in Medical Research and was supported by the Cancer Prevention and Research Institute of Texas (RP130513) and the Welch Foundation (I-1568 to R.K.B.).
Footnotes
COI: U.K.T. and R.K.B. own equity in Peloton Therapeutics, Inc. as licensors (U.K.T. and R.K.B.) or advisors (R.K.B.).
References
- 1.Winkelstein A. Peptic esophagitis: a new clinical entity. Jama. 1935;104:906–909. [Google Scholar]
- 2.Ismail-Beigi F, Horton PF, Pope CE., 2nd Histological consequences of gastroesophageal reflux in man. Gastroenterology. 1970;58:163–174. [PubMed] [Google Scholar]
- 3.Frierson HF., Jr Histology in the diagnosis of reflux esophagitis. Gastroenterol Clin North Am. 1990;19:631–644. [PubMed] [Google Scholar]
- 4.Souza RF, Huo X, Mittal V, Schuler CM, Carmack SW, Zhang HY, Zhang X, Yu C, Hormi-Carver K, Genta RM, et al. Gastroesophageal reflux might cause esophagitis through a cytokine-mediated mechanism rather than caustic acid injury. Gastroenterology. 2009;137:1776–1784. doi: 10.1053/j.gastro.2009.07.055. [DOI] [PubMed] [Google Scholar]
- 5**.Dunbar KB, Agoston AT, Odze RD, Huo X, Pham TH, Cipher DJ, Castell DO, Genta RM, Souza RF, Spechler SJ. Association of Acute Gastroesophageal Reflux Disease With Esophageal Histologic Changes. Jama. 2016;315:2104–2112. doi: 10.1001/jama.2016.5657. This study reports the clinical, endoscopic, and histologic findings of acute reflux esophagitis induced by interrupting PPI therapy in patients with severe reflux esophagitis that had been healed by PPIs. The investigators found that acute reflux esophagitis is a T-lymphocyte predominant form of esophageal inflammation, and that esopahgeal basal cell hyperplasia precedes the development of surface cell erosions. These findings suggest that reflux esophagitis develops as a cytokine-mediated injury rather than as an acid burn (a caustic, chemical injury) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor CT. Interdependent roles for hypoxia inducible factor and nuclear factor-kappaB in hypoxic inflammation. J Physiol. 2008;586:4055–4059. doi: 10.1113/jphysiol.2008.157669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eltzschig HK, Carmeliet P. Hypoxia and inflammation. N Engl J Med. 2011;364:656–665. doi: 10.1056/NEJMra0910283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zinkernagel AS, Johnson RS, Nizet V. Hypoxia inducible factor (HIF) function in innate immunity and infection. J Mol Med (Berl) 2007;85:1339–1346. doi: 10.1007/s00109-007-0282-2. [DOI] [PubMed] [Google Scholar]
- 9.Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors--similar but not identical. Mol Cells. 2010;29:435–442. doi: 10.1007/s10059-010-0067-2. [DOI] [PubMed] [Google Scholar]
- 10.Shah YM, Ito S, Morimura K, Chen C, Yim SH, Haase VH, Gonzalez FJ. Hypoxia-inducible factor augments experimental colitis through an MIF-dependent inflammatory signaling cascade. Gastroenterology. 2008;134:2036–2048. 2048.e2031–2033. doi: 10.1053/j.gastro.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feagins LA, Zhang HY, Zhang X, Hormi-Carver K, Thomas T, Terada LS, Spechler SJ, Souza RF. Mechanisms of oxidant production in esophageal squamous cell and Barrett’s cell lines. Am J Physiol Gastrointest Liver Physiol. 2008;294:G411–417. doi: 10.1152/ajpgi.00373.2007. [DOI] [PubMed] [Google Scholar]
- 12*.Huo X, Agoston A, Dunbar KB, Cipher DJ, Zhang X, Yu C, Cheng E, Zhang Q, Pham TH, Tambar UK, et al. Hypoxia-Inducible Factor 2a Plays a Role in Mediating Oesophagitis in Gastro-Oesophageal Reflux Disease. Gut. 2016 doi: 10.1136/gutjnl-2016-312595. Epub Using non-neoplastic, human eosphageal squamous cell lines and esophageal biopsies taken from patients with acute GERD, this study demonstrates how the gastroesophageal reflux of acid and bile stabilizes HIF-2α, which plays a major role in triggering the release of the pro-inflammatory cytokines that mediate the development of reflux esophagitis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheuermann TH, Stroud D, Sleet CE, Bayeh L, Shokri C, Wang H, Caldwell CG, Longgood J, MacMillan JB, Bruick RK, et al. Isoform-Selective and Stereoselective Inhibition of Hypoxia Inducible Factor-2. J Med Chem. 2015;58:5930–5941. doi: 10.1021/acs.jmedchem.5b00529. [DOI] [PubMed] [Google Scholar]
- 14.Nagle DG, Zhou YD. Natural product-derived small molecule activators of hypoxia-inducible factor-1 (HIF-1) Curr Pharm Des. 2006;12:2673–2688. doi: 10.2174/138161206777698783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenza GL. Evaluation of HIF-1 inhibitors as anticancer agents. Drug Discov Today. 2007;12:853–859. doi: 10.1016/j.drudis.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Onnis B, Rapisarda A, Melillo G. Development of HIF-1 inhibitors for cancer therapy. J Cell Mol Med. 2009;13:2780–2786. doi: 10.1111/j.1582-4934.2009.00876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K, Zhang H, Qian DZ, Rey S, Liu JO, Semenza GL. Acriflavine inhibits HIF-1 dimerization, tumor growth, and vascularization. Proc Natl Acad Sci U S A. 2009;106:17910–17915. doi: 10.1073/pnas.0909353106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Key J, Scheuermann TH, Anderson PC, Daggett V, Gardner KH. Principles of ligand binding within a completely buried cavity in HIF2alpha PAS-B. J Am Chem Soc. 2009;131:17647–17654. doi: 10.1021/ja9073062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheuermann TH, Li Q, Ma HW, Key J, Zhang L, Chen R, Garcia JA, Naidoo J, Longgood J, Frantz DE, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nat Chem Biol. 2013;9:271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers JL, Bayeh L, Scheuermann TH, Longgood J, Key J, Naidoo J, Melito L, Shokri C, Frantz DE, Bruick RK, et al. Development of inhibitors of the PAS-B domain of the HIF-2alpha transcription factor. J Med Chem. 2013;56:1739–1747. doi: 10.1021/jm301847z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 23**.Wallace EM, Rizzi JP, Han G, Wehn PM, Cao Z, Du X, Cheng T, Czerwinski RM, Dixon DD, Goggin BS, et al. A Small-Molecule Antagonist of HIF2alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma. Cancer Res. 2016;76:5491–5500. doi: 10.1158/0008-5472.CAN-16-0473. This study describes the characterization of PT2385, a potent and selective HIF-2α antagonist that binds within the internal cavity of the PAS-B domain. PT2385 disrupts HIF-2α heterodimerization in vivo to inhibit the expression of HIF-2α target genes. PT2385 treatment resulted in tumor regression in preclinical mouse xenograft models initiated from VHL-null cells expressing HIF-2α. PT2385 was well-tolerated with no adverse effects on cardiovascular performance as is often observed with other standard of care agents for the treatment of clear cell Renal Cell Carcinoma. Though transcription factors such as HIF-2α are often considered “undruggable”, these results provide the first demonstration of in vivo efficacy for this class of HIF-2α selective inhibitors now in clinical trials. [DOI] [PubMed] [Google Scholar]
- 24.Butler RN. Comprehensive Herterocyclic Chemistry. Vol. 4. Oxford, U.K: Pergamon; 1996. [Google Scholar]
- 25.Herr RJ. 5-Substituted-1H-tetrazoles as carboxylic acid isosteres: medicinal chemistry and synthetic methods. Bioorg Med Chem. 2002;10:3379–3393. doi: 10.1016/s0968-0896(02)00239-0. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, Homayoun F, Ma Y, Patel N, Yell P, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539:112–117. doi: 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho H, Du X, Rizzi JP, Liberzon E, Chakraborty AA, Gao W, Carvo I, Signoretti S, Bruick RK, Josey JA, et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature. 2016;539:107–111. doi: 10.1038/nature19795. [DOI] [PMC free article] [PubMed] [Google Scholar]