Summary
Root knot nematodes (RKNs) penetrate into the root vascular cylinder triggering morphogenetic changes to induce galls, de novo formed ‘pseudo-organs’ containing several giant cells (GCs). Distinctive gene repression events observed in early gall/GCs development is thought to be mediated by post-transcriptional silencing via-miRNAs, a process far from being fully characterized.
Arabidopsis backgrounds with altered activities based on target MIMICRY (35S::MIM172), 35S::TOE1-miR172-resistant (35S::TOE1R) and mutant (ft-10) lines, were used for functional analysis of nematode infective and reproductive parameters. GUS-reporter lines, MIR172A-E::GUS, auxin (IAA) and auxin-inhibitor (PEO-IAA) treatments, as well as a MIR172CAuxRE−::GUS line with two mutated auxin responsive elements (AuxREs) were assayed for nematode-dependent gene expression.
Arabidopsis backgrounds with altered expression of either MIRNA172, TARGET OF EAT1 (TOE1), and FLOWERING LOCUS T (FT), showed lower susceptibility to the RKNs and smaller feeding sites, galls/GCs. MIR172C-D::GUS showed restricted activity in galls/GCs that is regulated by auxins through auxin responsive factors. IAA induced their activity in galls while PEO-IAA treatment and mutations in AuxRe motifs abolished it.
The results show that the activity of the regulatory module miRNA172/TOE1/FT plays an important role in correct GCs and gall development where miRNA172 is modulated by auxins.
Keywords: AP2, Arabidopsis, flowering, giant cells, Meloidogyne spp., miRNA172, repression
Introduction
Plant parasitic nematodes that include two major groups, the root-knot nematodes (RKNs; Meloidogyne spp.) and the cyst nematodes (CNs; Heterodera spp. and Globodera spp.), constitute one of the major threats for plant productivity given the progressive ban of the most effective chemical nematicides (reviewed in Escobar et al., 2015; European Parliament Directive 2009/128/EC). Juvenile RKNs invade the root through the elongation zone and then migrate intercellularly through the cortex to the root apex where they penetrate into the vascular cylinder and get established (reviewed in Escobar et al., 2015). Once in the root vascular cylinder they select five to eight vascular cells to differentiate into their feeding cells, giant cells (GCs hereafter), as they present a much greater volume than adjacent cells in the vascular cylinder. The cells around the GCs proliferate and eventually a knot in the root, called gall is formed. This de novo structure is considered as a newly formed pseudo-organ in the roots (reviewed in Escobar et al., 2015).
In the last decade, major advances have been made to understand the molecular mechanisms underlying the formation of the galls/GCs. Interestingly, genes previously defined to have key functions in plant developmental pathways also showed a role during galls/GCs development in Arabidopsis (reviewed in Cabrera et al., 2015b). Some of them are transcription factors participating in root development, for example LBD16 or WRKY23 that are also crucial during the CNs and RKNs interaction in syncytia and gall and/or GCs development (Grunewald et al., 2008; Cabrera et al., 2014, respectively). Moreover, small signaling peptides such as CLAVATA3/ESR (CLE)-like or C-TERMINALLY ENCODED PEPTIDEs (CEP) peptides that participate in root and shoot apical meristem maintenance, vascular development and lateral root formation (Mohd-Radzman et al., 2016; Yamaguchi et al., 2016), were identified in RKNs secretions (Betsuyaku et al., 2011; Mitchum et al., 2012; Bobay et al., 2013). The role of CLE-like peptides have been studied more deeply in CNs than in RKNs. One of the most recent studies indicates a clear link between nematode B-type CLE signaling and the WOX4-mediated cell proliferation pathway for feeding cell formation (Guo et al., 2017). In this respect, 16D10 is a secretory peptide with a CLE-Like sequence identified in Meloidogyne spp., with a role during the RKN interaction (Huang et al., 2006). Hence, one of the putative strategies used by RKNs to induce the formation of their specialized feeding sites is to hijack or interfere with established plant developmental pathways to induce the formation of their feeding sites (Cabrera et al., 2015b).
Several transcriptomic analyses of galls and/or GCs induced by Meloidogyne spp. in a number of plant species have been performed, delivering extensive lists of misregulated genes (upregulated or downregulated) in the nematode galls and GCs (reviewed in Cabrera et al., 2016a). Interestingly, a conspicuous trait that prevails in all these studies is the large number of significantly downregulated genes, particularly at early infection stages (Barcala et al., 2010; Damiani et al., 2012; Portillo et al., 2013). Therefore, gene repression can be considered as a signature for proper nematode establishment and feeding site formation. In line with this, the overexpression of the repressed gene TPX1 in tomato caused a reduction in the infectivity and in the size of the GCs (Portillo et al., 2013). Similarly, in CNs, the overexpression of the repressed gene RAP2.6 led to an enhanced resistance against Heterodera schachtii and to smaller feeding cells in Arabidopsis (Ali et al., 2013). Bearing in mind the putative role of gene repression, changes in the sRNAs population in the feeding sites induced by RKNs and CNs have been studied by high-throughput sequencing (Hewezi et al., 2008; Li et al., 2012; Xu et al., 2014; Zhao et al., 2015; Cabrera et al., 2016b). However, we have just started to delineate the functional role of sRNAs and their targets during the nematode infection. Evidence showed that miR396 and its GRFs target genes participate in the regulation of a large number of genes in the CNs nematode feeding cells (Hewezi et al., 2012; Hewezi & Baum, 2015). Similarly, Hewezi et al. (2016) demonstrated that the miR827 and its target gene NLA (NITROGEN LIMITATION ADAPTATION) had a role in mediating the infectivity of H. schachtii. Furthermore, Arabidopsis loss of function lines for the module miR390/TAS3/ARFs displayed a decrease in the infectivity and smaller galls during RKN infection (Cabrera et al., 2016b) and in situ localization of miR390 in tomato indicated accumulation of miR390 in galls/GCs (Díaz-Manzano et al., 2016a) what suggests conserved roles in tomato. Additionally, miR319 and its target gene TCP4 regulate the systemic defense response during RKN infection in tomato (Zhao et al., 2015).
The riboregulator miRNA172 post-transcriptionally targets a small group of regulatory repressor genes (APETALA2 (AP2) and AP2-like transcription factors, such as TARGET OF EARLY ACTIVATION TAGGED 1 (TOE1), TOE2, and TOE3, SCHLAFMÜTZE (SMZ), and SCHNARCHZAPFEN (SNZ)) that encode transcription factors belonging to the evolutionarily conserved AP2/Ethylene Responsive Factor (ERF) plant family. This group of proteins comprises members of the AP2 class, ERF type (subgroup B1-B6), related to ABI3/VP1 (RAV), the DREB class (subgroup A1–A6), and others (Sakuma et al., 2002). These proteins regulate various developmental and stress responsive pathways (Licausi et al., 2013; Müller & Munné-Bosch, 2015) and bear at least one conserved AP2 domain of c. 60–70 amino acid residues, which recognizes particular DNA cis-elements (Nakano et al., 2006). When overexpressed some AP2/ERF type transcription factors enhance biotic and abiotic stress tolerance (Jisha et al., 2015; Mishra et al., 2015).
Regarding the members of miRNA172-targeted AP2-like transcription factors, they have been involved in controlling plant aging, flowering time, tuber formation, fruit growth, and nodulation (Martin et al., 2009; Zhu & Helliwell, 2010; Yan et al., 2013; Wang et al., 2014; Ripoll et al., 2015). Interestingly, miRNA172 is regulated by different auxin response factors (ARFs) during fruit development in Arabidopsis (Ripoll et al., 2015). In Arabidopsis, AP2-like transcription factors act as flowering repressors and at least TOE1 (Zhang et al., 2015) and SMZ (Mathieu et al., 2009), negatively regulate FT expression as for example TOE1 binds to its promoter region and represses its expression in the vascular tissue of the leaves during the floral transition (Zhang et al., 2015).
Hence, TOE1 functions in different developmental processes, but has also a role described only during a biotic interaction, that is, nodulation (Yanz et al., 2013). In line with this, previous studies showed that the repression of a subclade within the AP2-like transcription factor family in Arabidopsis has a crucial functional role during the establishment and development of the feeding cells induced by the plant parasitic cyst nematode H. schachtii (Ali et al., 2013). Moreover, TOE1 was downregulated in the transcriptomes of the RKNs GCs induced by M. javanica in Arabidopsis (Barcala et al., 2010).
Our investigations show that the miRNA172-dependent regulatory module miRNA172/TOE1/FT is active and de-regulated in the feeding sites induced by M. javanica in Arabidopsis roots, and also participates in gall and GCs development. This study adds knowledge on how RKNs interfere with endogenous developmental pathways and contributes to a better understanding of the molecular signatures associated with feeding cells development.
Materials and Methods
Nematode population
Meloidogyne javanica Treub, (1885) was maintained in vitro on cucumber etiolated seedlings (Cucumis sativus cv Hoffmanns Giganta). Egg hatching was stimulated in sterile tap water 4 d before inoculation as described in Díaz-Manzano et al. (2016b).
Plant material, growth conditions and nematode inoculation
Arabidopsis thaliana (L.) Heynh Columbia-0 (Col-0) was the background of all the transgenic lines used. Reporter GUS promoter lines MIR172A-E::GUS and MIR172CAuxRE−::GUS (Ripoll et al., 2015) were used for the expression analyses. Four independent transgenic lines, homozygous and harbouring one T-DNA insertion for 35S:TOE1R and two independent lines for MIM172 (Supporting Information Fig. S1a,b; Ripoll et al., 2015) were selected for the infection tests with M. javanica.
For histochemical analysis, three independent experiments were performed each with 30 individual plants per line. Galls were hand-dissected at the selected infection stages and the histochemical analysis and GUS staining was carried out according to Cabrera et al. (2014). Roots from MIR172C::GUS and MIR172D::GUS plants were incubated for 2 d on medium containing 300 μM α-(phenyl ethyl-2-one)-indole-3-acetic acid (PEO-IAA) as described in Cabrera et al. (2014).
For the infection tests in vitro, three to six independent infection tests, with at least 30 plants per experiment and line were performed using Col-0 (as control), 35S:TOE1R, MIM172 and ft-10 lines. Seeds were sterilized, grown vertically and homogeneously inoculated following the protocol set up in Olmo et al. (2017) at long day (LDs;16 h : 8 h, day : night) or short days (SDs;8 h : 16 h, day : night) regime.
For infection tests in soil, seedlings were planted singly into 15 ml clay pots containing steam-sterilized river sand. Arabidopsis plants were grown at 25 ± 2°C, 60% RH, at SDs for 2 months and inoculated with 200 J2 per pot. Nematode inoculum's was obtained as described in Andrés et al. (2012). Plants were watered as needed and fertilized with 1 ml of a 0.5% solution of 20-20-20 (N-P-K) every 15 d per pot. Each infection test was replicated at least three times with 10 plants per experiment. Reproductive parameters were evaluated 2 months after inoculation (i.e. 4 months after planting). The number of galls/egg masses per fresh root gram was counted from each root system. Egg production was obtained by extracting the eggs from the entire root system (Hussey & Barker, 1973).
Galls and GCs phenotyping
The volume or the estimation of the size of the GCs (n≥ 10 per line tested) induced by M. javanica in Col-0, 35S:TOE1R, MIM172 and ft-10 were obtained after 3D reconstruction following the method described in Cabrera et al. (2015a) by using the plugin TrakEM2 (Cardona et al., 2012) for FIJI (Schindelin et al., 2012). Gall diameters were measured from micrographs from each gall (n ≥ 20 per line tested) by using the straight line and measurement tools from FIJI (Schindelin et al., 2012).
Plant transgenic construction and selection
The 35S:TOE1R constructs were generated by introducing three mutations in the miRNA172 pairing sequence of the TOE1 transcription factor, avoiding alterations of the amino acid sequence of the protein. Specific oligos TOE1miR172 (Table S1; Fig. 1a) and the Quikchange lightning site-directed mutagenesis protocol (Agilent technologies) were used to generate the TOE1 miRNA172172 resistant version, which was fully sequenced and mobilized into the pGW2 vector under the control of a CaMV 35S promoter. This construct was transformed into Arabidopsis Col-0 plants, and several independent transgenic lines expressing the miRNA172-resistant version of TOE1 (35S:TOE1R) were selected on hygromycin plates. These miRNA172-resistant lines were developed under the framework of the TRANSPLANTA project (Coego et al., 2014).
Fig. 1.
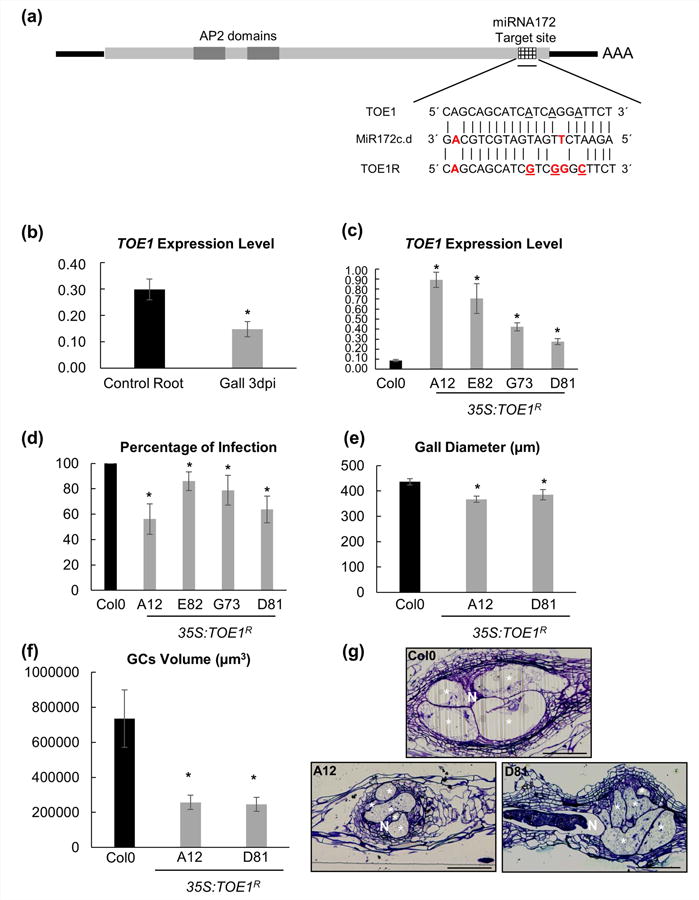
TARGET OF EAT1 (TOE1) is involved in the root-knot nematode infection. (a) Schematic representation of miRNA712c-d (GACGTCGTAGTAGTTCTAAGA) target sequence for TOE1 (CAGCAGCATCATCAGGATTCT) and TOE1 resistant (CAGCAGCATCGTCGGGCTTCT). Red bold text, nonmatching base pairs; underlined text, mutated TOE1 resistant base pairs respect to TOE1. (b) Q-PCR analysis of TOE1 abundance in galls as compared to control uninfected root segments. TOE1 decreased two-fold in galls induced by Meloidogyne javanica. (c) Q-PCRs of 35S:TOE1R lines of uninfected roots. The four lines showed higherTOE1 transcript abundance compared to their endogenous control Columbia 0 (Col-0). (d) In vitro infection tests of four independent 35S:TOE1R lines challenged with M. javanica. The percentage of galls per main root was lower in all TOE1 lines than in Col-0. (e) Galls diameter of 35S:TOE1R-A12 and 35S:TOE1R-D81. Both lines showed a smaller gall diameter than their control Col-0 at 14 d post infection (dpi) (P < 0.05). (f) Giant cells (GCs) volume in 35S:TOE1R-A12 and 35S:TOE1R-D81 lines was smaller than that of Col-0 at 14 dpi (P < 0.05). (g) Representative pictures of 35S:TOE1R lines and Col-0 at 7 dpi Araldite® gall sections (2 μm). Statistical analysis was performed with three independent experiments per line using T-test, significant differences with Col-0 or corresponding controls are indicated by asterisks, P < 0.05; values are means ± SE. N, nematodes; bars, 100 μm. GCs are labelled with a white asterisk.
Arabidopsis thaliana 35S:MIMICRY172-7 and 35S:MIMICRY172-23 plants (MIM172) are resistant to BASTA (Todesco et al., 2010). For the homozygous lines selection, we performed a double check. A selection according to the expected late flowering phenotype (Fig. S1d,f) as compared to Col-0 and a selection based on BASTA resistance, with 120 mg l-1 (DL-PHOSPHINOTHRICIN, Duchefa® Biochemie, Ref.: P0159 0250 and Silwet-77 surfactant 500 μl l-1 (Lehle Seeds) as described by Bouchez et al. (1993). Plants were grown in soil (a mixture of peat substrate, Kekkilä PROJAR 70L 50/50, and vermiculite, 3 : 1) at 21°C, 60% humidity and 16 h : 8 h, light : darkness for 6 wk.
Lines 35S:TOE1R were grown on Murashige and Skoog (MS) agar medium supplemented with 1% sucrose plus hygromycin 20 μl l-1 (Sigma®, Ref.: H3274) and compared with Col-0 for selection.
Flowering time assays
Flowering time was scored as the total leaf number, including rosette and cauline leaves, just before the first flower opened (Martin-Trillo et al., 2006).
RNA Isolation and quantitative PCR analysis
Total RNA was extracted from 25 hand-dissected root plant tissues (control root segments and/or galls from infected plants) using the miRNeasy Micro Kit (Qiagen, Hilden, Germany) following the manufacturer's instructions. An amount (0.75 mg) RNA from each sample (control root segments and/or galls) was used for cDNA synthesis with the High Capacity cDNA Reverse Transcription Kit with random primers (Applied Biosystems, Foster City, California, USA) according to the company's guidelines (35 cycles). The cDNA was diluted up to 1/10 and 1 ng was used from each sample as a template for the subsequent qPCR transcript analysis for TOE1 (TARGET OF EAT 1, AT2G28550); and 45 ng for FT (FLOWERING LOCUS T, AT1G65480) as its expression level is very low in most tissues.
Quantitative PCR analysis was done with SYBR-Green technology (SYBR Green Master Mix 2X, no ROX, Thermo Scientific, Waltham, MA, USA) in a LightCycler® 480 II machine (Roche, Indianapolis, USA). Quantification of the relative changes in gene expression levels was determined using the E-Method (Tellmann, 2006). In all cases, at least three independent experiments each with three technical replicates of each reaction were performed. Arabidopsis thaliana GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE C2 (GAPC2, AT1G13440) was used as internal control to normalize gene expression levels. Primers used for PCRs and qPCRs are described in Table S1.
Pharmacological treatments
For auxin treatments and MIR172C-D::GUS expression, plantlets were germinated and grown on 0.5 MS medium, as described in Chapman et al. (2012), transferred to media containing the corresponding auxin (IAA) concentration or DMSO (for the mock controls). The tissue was incubated for 30 min and total RNA was extracted using Trizol (Life Technologies) and treated for cDNA synthesis as previously described (Ripoll et al., 2015). Five micrograms of total RNA was used for cDNA synthesis with an oligo (dT) primer and Superscript III reverse transcriptase (Life Technologies, Carlsbad, CA, USA). After 1/10 dilution of the cDNA, 1 μl was used as a template for the subsequent qPCR reactions. Relative changes in gene expression levels were determined using the 2ΔΔ-CT method (Livak & Schmittgen, 2001) RNA levels were normalized to the constitutively expressed gene ACTIN2 as previously reported (Ripoll et al., 2011). Each experiment was executed using three biological replicates. Primers used for this set of experiments can be found in Ripoll et al. (2015).
Data analysis
Data obtained were represented with histograms with mean values and/or percentages per line/treatment and ± SE. Statistical analysis of the infection and reproduction parameters; and GCs volumes and areas were performed using the T-test in the SPSS package (IBM, Armonk, NY, USA). The corresponding confidence intervals (CI) were calculated with a significance level of 5% (P <0.05) which was indicated with an asterisk.
Moreover, data from q-PCRs were represented with histograms with pairing-fold change values (line/treatment vs control) and ± SE. Statistical analysis were performed using the T-test in the SPSS package (IBM, Armonk, NY, USA). The corresponding confidence intervals (CI) were calculated with a significance level of 5% (P <0.05) which was indicated with an asterisk.
Blast analysis
We searched for miRNA172 sequences described in tomato and pea on miRbase (http://www.mirbase.org), finding two sequences described in tomato (sly-miRNA172a and sly-miRNA172b). To find homologue sequences to TOE1 from Arabidopsis in other crop species, we selected the miRNA172c Arabidopsis sequence (http://www.mirbase.org) and performed a blastn (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE_TYPE=BlastSearch) against tomato (Solanum lycopersicum; taxid: 4081) and pea (Pisum sativum; taxid: 3888). We obtained SlAP2d (NM_001247718.2) and AP2-like (AF325506.1) genes with a 100% cover query for tomato and pea, respectively.
Results
Repression of TOE1 in galls and GCs induced by M. javanica in Arabidopsis is necessary for the correct progression of the infection and the development of the feeding sites
In the present study, we further investigated the role of TOE1 (RAP2.7; At2g28550), during the formation of the GCs as it was downregulated in the transcriptomes of the GCs induced by M. javanica in Arabidopsis (Barcala et al., 2010).
First, we corroborated by qPCR that TOE1 expression was two-fold down-regulated in galls induced by M. javanica in Arabidopsis at 3 d post infection (dpi; Fig. 1b; P < 0.05), which is in line with the results obtained in previous microarray experiments (Barcala et al., 2010). TOE1 expression is regulated by miRNA172 during the floral transition (Zhu & Helliwell, 2010). Consistent with this observation and the downregulation of TOE1, pri-miRNA172d expression was clearly induced in the same microarrays of micro-dissected Arabidopsis GCs as compared to vascular control cells (Table S2; Barcala et al., 2010). Hence, we generated four independent transgenic lines overexpressing a modified version of TOE1 being resistant to the degradation by miRNA172 (35S:TOE1R; Fig. 1a). All these lines (A12, E82, G73, D81) showed a noticeable increase in TOE1 transcript abundance as compared to the Col-0 wild type (WT) line, although there were differences in the expression levels among lines (Fig. 1c; P < 0.05). All four 35S:TOE1R lines showed a conspicuous delay in flowering time (P < 0.05) under SD conditions, while two of them (D81 and A12) flowered significantly late under LDs (Fig. S1). When we challenged these 35S:TOE1R plants with juveniles of M. javanica, all independent lines showed a significant reduction in the percentage of galls per plant in the range of 15–44% (Fig. 1d). Remarkably, the flowering phenotype was correlated with the nematode resistant traits, as those lines with the strongest flowering phenotypes also showed the highest reduction in nematode infection rates (see lines A12 and D81 in Figs 1d, S1a,c,e). Our next step was to characterize in more detail the gall and GCs phenotypes in the 35S:TOE1R lines with the most extreme phenotypic alterations (Fig. 1e–g). Galls of 35S:TOE1R lines were c. 15% smaller than those formed in Col-0 at 14 dpi (Fig. 1e; P < 0.05). In addition, the GCs formed in these galls at 14 dpi were reduced in size to two-thirds compared to those formed in Col-0 galls at the same infection stage (Fig. 1f,g; P < 0.05), measured after 3D reconstruction following Cabrera et al. (2015a). Thus, we corroborated that downregulation of TOE1 by miRNA172 in the GCs and galls induced by M. javanica in Arabidopsis is important for proper nematode establishment, and gall and GCs development.
FT, a defined target of TOE1, has a crucial role during gall development
It has been recently identified that the TOE1 regulatory protein directly binds to the promoter region of the gene FT and represses its expression in the vascular tissue of the leaves during the floral transition (Zhang et al., 2015).
Because TOE1 expression was repressed in 3 dpi galls induced by M. javanica in Arabidopsis (Fig. 1b) we checked by qPCR whether the expression levels of FT were altered after nematode infection in Col-0 roots (Fig. 2a). Interestingly, FT transcript levels were noticeable increased in the galls while they were almost undetectable in the uninfected Col-0 control roots (Fig. 2a; P < 0.05). By contrast, FT expression in 35S:TOE1R lines were barely detectable either in control roots or in galls, similarly to the uninfected control Col-0 roots (Fig. 2a). The levels of FT in control root tissue are extremely low, this might explain the large error bars in the 35S:TOE1R lines. This is consistent with TOE1R repressing the expression of FT in galls, what suggests that the downregulation of TOE1 in galls probably mediates the accumulation of FT transcripts in those cells.
Fig. 2.
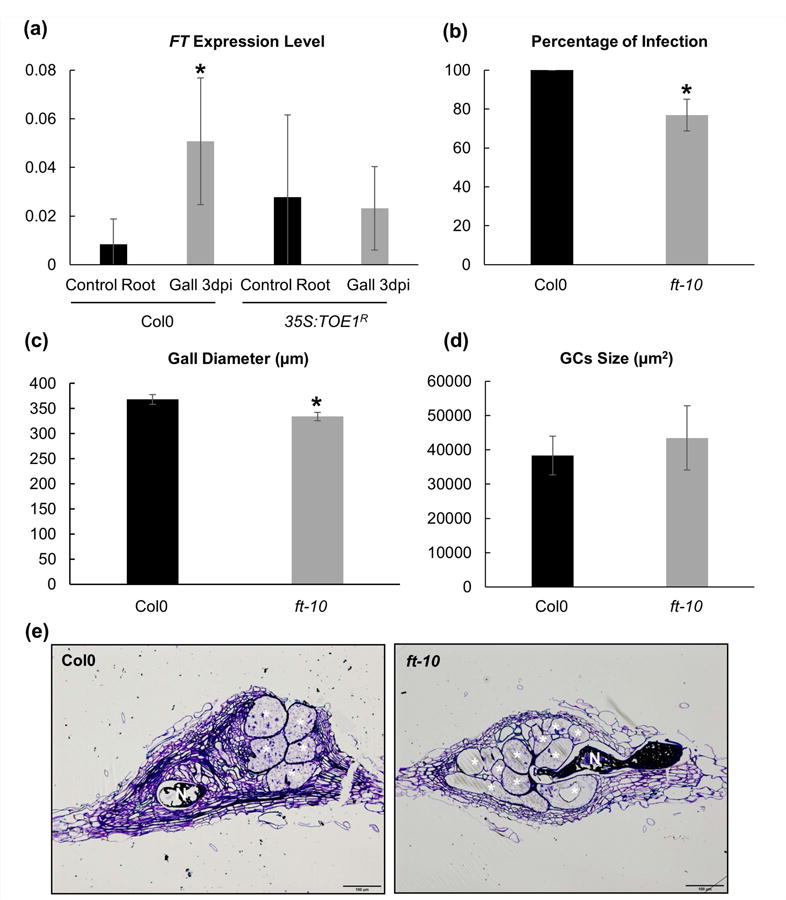
FLOWERING LOCUS T (FT) is functional during root-knot nematode interaction. (a) q-PCR analysis of the FT transcript in galls at 3 d post infection (dpi) compared to uninfected roots in background Col-0 and 35S:TOE1R-A12. FT was induced five-fold in galls formed by Meloidogyne javanica in the ecotype Col-0 as compared to uninfected control root samples, but not in 35S:TOE1R lines. (b) In vitro infection tests of ft-10 mutant line. The percentage of galls per main root was significantly lower in the ft-10 line than in the ecotype Col-0. (c) Galls diameter of ft-10 mutant line showing smaller values than their control Col-0 at 14 dpi (P < 0.05). (d) Giant cells (GCs) volume measurement compared to Col-0 at 14 dpi. The volume occupied by the pool of GCs within the gall showed no significant differences between ft-10 and Col-O. (e) Representative images of Araldite® gall sections (2 μm) of ft-10 line and Col-0 at 14 dpi. Asterisks, significant differences (T-student; P < 0.05) from three independent experiments. Values are means ±S E. N, nematodes; bars, 100 μm. GCs are labelled with a white asterisk.
To decipher the role of FT during the gall and GC formation we performed six independent infection tests using a previously characterized complete loss-of-function FT mutant allele (ft-10) of Arabidopsis that displayed a late flowering time phenotype (Yoo et al., 2005). Mutant plants for FT showed a significant reduction in the number of galls, 24% less when compared to the WT plants (Fig. 2b), suggesting that FT function is required for the proper establishment of M. javanica in Arabidopsis. This idea is reinforced by the fact that the galls formed in the ft-10 mutant plants were smaller than those formed in Col-0 at 14 dpi (Fig. 2c; P < 0.05).
MiRNA172 is upregulated in the galls induced by M. javanica in Arabidopsis and other crop species
While TOE1 downstream regulation is at least partially mediating FT transcript accumulation, TOE1 is regulated upstream by the action of the miRNA172 during Arabidopsis flowering (Aukerman & Sakai, 2003). As mentioned earlier, from the 83 pre-miRNAs that were detected in the microarray of developing GCs (Barcala et al., 2010), the pre-miRNA172d was induced, being the only one differentially expressed in GCs as compared to non-infected vascular cells (P < 0.05; Table S2). To investigate the expression pattern of the gene family that generate the mature miRNA172 in the feeding sites induced by M. javanica in Arabidopsis, we assayed the GUS-reporter lines for the promoters for all five miRNA172-encoding loci (MIR172a, MIR172b, MIR172c, MIR172d and MIR172e), fused to the coding sequence of the GUS marker gene (Ripoll et al., 2015). In roots of uninfected plants, the promoters of the five precursors for the miRNA172 showed differential activity patterns. MIR172A::GUS, MIR172B::GUS and MIR172E::GUS were activated along the vascular cylinder of the root (Fig. 3a,b,e), except in the elongation zone and root apex in the case of MIR172A::GUS and MIR172E::GUS (Fig. 3a1,e1). By contrast, the activity of the promoters for the precursors of miRNA172c and miRNA172d was not detectable along the vascular cylinder in the elongation or differentiation zones, being a specific signal only noticeable in the root tip (Fig. 3c,d,c1,d1). Hence, MIR172C::GUS and MIR172D::GUS showed a restricted staining pattern in root tips and both originate identical mature miRNA172 (Table S3a).
Fig. 3.
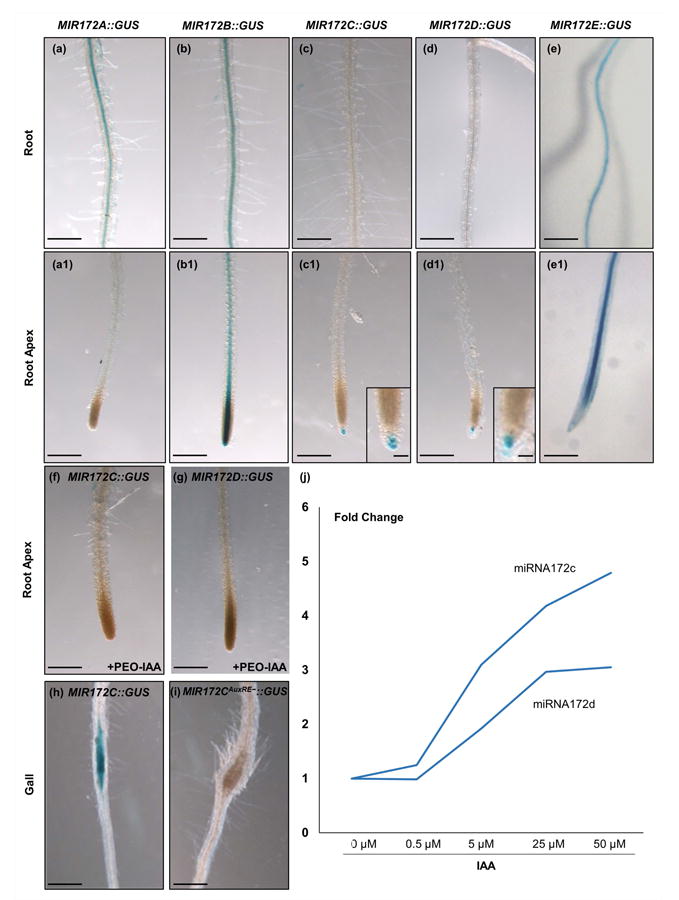
MIR172::GUS constructs are active in feeding sites and activity of miRNA172 promoter is modulated by auxin. (a–e) Basal GUS activity in roots and (a1–e1) zoom in detailed image of the elongation zone and root apex. Only C and D lines showed specific staining in the root apex (c1 and d1, respectively). The lines A (a, a1), B (b, b1) and E (e, e1) presented either no signal or it was centred in the vascular cylinder of the root. (f, g) GUS expression of MIR172C::GUS and D lines under auxin-inhibitor (PEO-IAA) treatment showing no GUS signal. (h, i) MIMR172C::GUS galls induced by Meloidogyne javanica at 5 d post infection showing restricted GUS signal in the center of the gall (h); by contrast, the MIR172CAuxRE−::GUS line with two mutated AuxRe elements displayed no GUS signal (i). (j) Expression changes of MIR172 C and D after treatments with exogenous auxin (up to 50 μM). Values are means ± SE. Bars, 200 μm (except in magnifications, 50 μm).
Because pri-miRNA172d was induced in the transcriptome of 3 dpi GCs (Table S2), we infected these two lines MIR172C::GUS and MIR172D::GUS with similar expression patterns in roots, with RKNs to check their activation patterns during gall development (Fig. 4). At 4 dpi the GUS staining for both reporters was localized specifically in the vascular cylinder inside the galls induced by M. javanica in Arabidopsis (Fig. 4a,d). The signal remained visible and specific in the centre of the galls at early-medium stages of development (7 dpi; Fig. 4b,e). The number of galls with positive GUS signal decreased at 11dpi (Fig. 4h,j) and eventually, at 14 dpi, >90% of the galls in the lines MIR172C::GUS and MIR172D::GUS did not express GUS (Fig. 4c,f,h,j). Moreover, MIR172C::GUS and MIR172D::GUS were clearly expressed in the GCs induced by the nematode and in adjacent vascular cells inside the galls at 4 dpi as shown in semi-thin sections (Fig. 4g,i). These results indicate a specific induction of MIR172C::GUS and MIR172D::GUS within the galls and GCs in roots upon nematode infection. However, no specific GUS pattern in galls of MIR172A::GUS, MIR172B::GUS and MIR172E::GUS lines was observed as they are highly activated along the vascular cylinder of the non-infected roots and their expression pattern was similar within the galls (Fig. 3a,b,a1,b1; data not shown). Although, it cannot be ruled out that these three miRNAs also might contribute, to some extent, to the total amount of miRNA172 molecules in the gall and GCs, no differences in the pattern of expression between uninfected roots and galls, or during the infection were observed (data not shown).
Fig. 4.
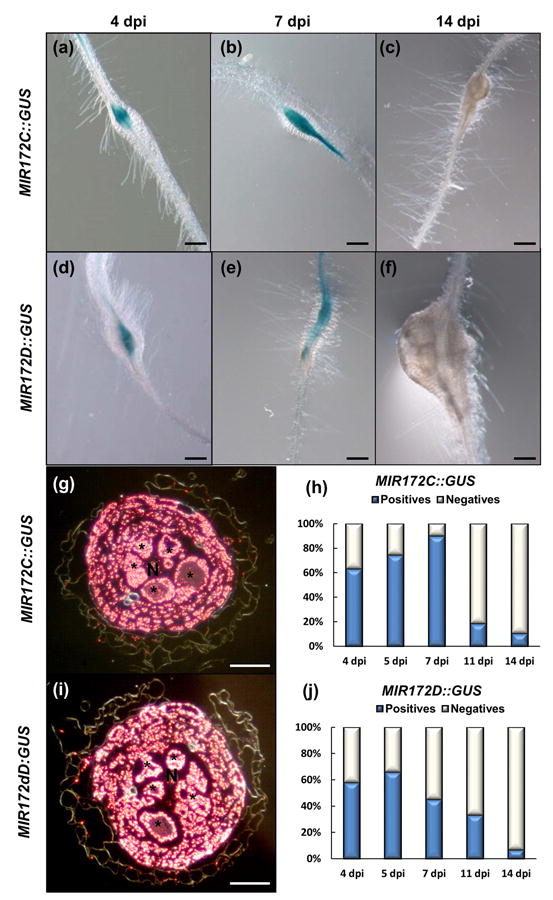
A timeline of miRNA172c and miRNA172d promoters indicated early activation patterns after root-knot nematode infection. (a–f) Representative pictures of GUS assays at 4, 7 and 14 d post infection (dpi), respectively of (a–c) MIR172C ::GUS and (d–f) MIR172D -::GUS lines in Arabidopsis galls induced by Meloidogyne javanica. (g, i) Dark field images of Araldite® cross sections of GUS stained galls at 7 dpi showing signal in the giant cells and adjacent cell layers within the vascular cylinder. Asterisks, giant cells; N, nematode. (h, j) Percentage of blue galls for (h) MIR172C ::GUS and (j) MIR172D ::GUS. Bars: (a–f) 200 μm; (g–i) 50 μm.
miRNA172 is highly conserved in the plant kingdom in angiosperms, gymnosperms, ferns, and across all tracheophytes (Luo et al., 2013), that is, the sly-miRNA172a and sly-miRNA172b from tomato showed a 100% homology across 20 out of 21 nucleotides to miRNA172c from Arabidopsis (Table S3b). Although the sequence for miRNA172 from pea has not been yet identified, miRNA172 is described as an active molecular partner during nodulation in legumes, a process with some molecular similarities to gall formation (see discussion). Hence, we performed in situ hybridisation in galls of tomato and pea with the Arabidopsis miRNA172c probe. Our results confirm accumulation of miRNA172 in galls and particularly in GCs of tomato and pea at 7 dpi (Fig. S2c,f, respectively). No signal was observed in either non-infected roots or galls of tomato or pea with a negative control probe (Scramble; Fig. S2a,d,b,e, respectively). Interestingly, BLAST analysis of the miRNA172c from Arabidopsis against the tomato and pea available genomes identified putative AP2-like gene targets with a high complementarity to miRNA172, similar to TOE1 in Arabidopsis (Fig. S2g). Those results suggest that induction of miRNA172 in galls is probably a mechanism conserved in these crop species.
MiRNA172 interferes with gall and GC development after nematode infection
To further investigate the putative role of miRNA172 in galls and GCs development, we tested two independent target mimicry lines for miRNA172 (MIM172; Franco-Zorrilla et al., 2007; Fig. 5e) in which the function of this miRNA was impaired. The two mimicry lines largely reproduced the late flowering phenotype showed by the 35S:TOE1R plants (Fig. S1b), either at SD or LD (Fig. S1d,f), as expected when TOE1 is not repressed by miRNA172. When the mimicry lines MIM172 were inoculated with M. javanica, both presented a conspicuous reduction in the infection level (at least 45%; Fig. 5a; P < 0.05), resembling again the phenotype displayed by the 35S:TOE1R lines after nematode infection (Fig. 1d). Moreover, the galls and GCs formed in the MIM172 lines were smaller than those formed in Col-0 (Fig. 5b–d; P < 0.05). These results further support a role for miRNA172 during gall and GC development.
Fig. 5.
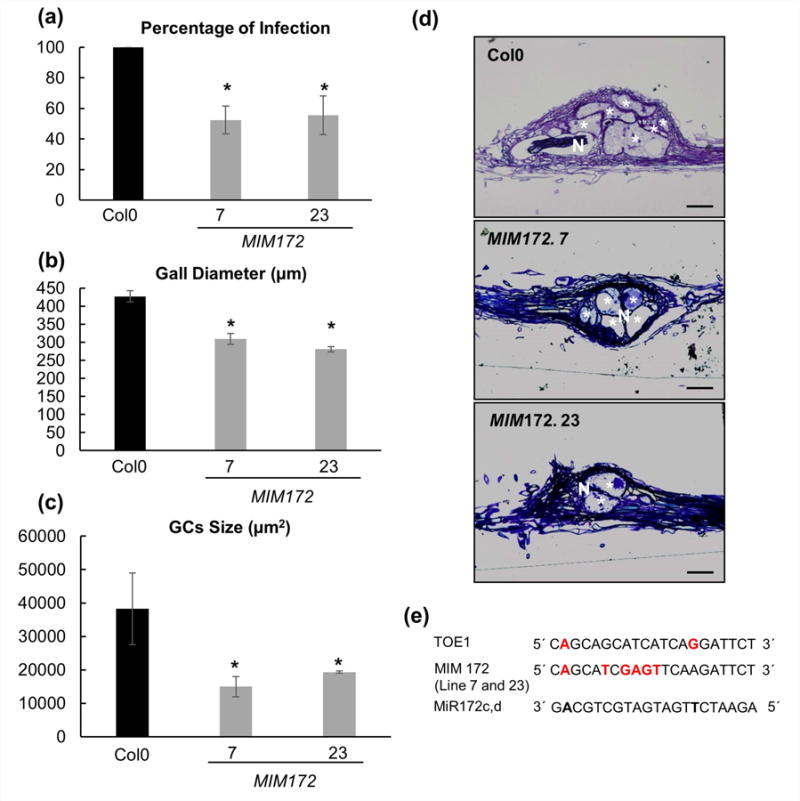
Target mimicry lines for miR172 show increased resistance to root-nematode infection. (a) In vitro infection tests of two independent mimicry (MIM) lines 7 and 23 showing significant differences (P < 0.05) between MIM lines and Col-0, as the percentage of galls is 40–50% lower in MIM lines as compared to controls. (b) Galls diameter of MIM lines and (c) volume of giant cells (GCs), were significantly lower than in Col-0 at 21 d post infection (dpi). (d) Representative pictures of Araldite® gall sections (2 μm) of MIM lines and Col-0 at 21 dpi. (e) Schematic comparison of miRNA172c-d, MIMICRY (MIM172) and miR172-TOE1 target sequences. In red bold, nonmatching base pairs with respect to miR172c-d. Asterisks, significant differences (T-Student; P < 0.05). Values are means ± SE. N, nematode; bars, 100 μm. GCs are labelled with a white asterisk.
Auxin regulation of miRNA172 function
The importance of an auxin response maxima during the formation of the nematode feeding cells has been demonstrated (reviewed in Cabrera et al., 2015b; Kyndt et al., 2016). On the other hand, the ARFs orchestrate auxin responses, by targeting the cis-motifs called auxin response elements (AuxREs) to regulate gene expression (Chapman & Estelle, 2009). Interestingly, miRNA172 function is directly modulated by the auxin signaling pathway via the regulation of miRNA172c. The promoter of miRNA172c contains two canonical AuxREs and it is already known that the miRNA172 is regulated by different ARFs during fruit development in Arabidopsis (Ripoll et al., 2015).
We analyzed the first 1000 bps of the promoter region upstream of miRNA172b, miRNA172c and miRNA172d loci, revealing the presence of canonical AuxREs (Table S3). To test whether auxin was influencing miRNA172c and miRNA172d expression in root tissues, we first treated seedlings with different auxin concentrations and measured transcript abundance for both loci. In both cases, auxin treatments increased transcript abundance when compared to mock treated samples (Fig. 3j). We next tested the activity of the GUS reporters for miRNA172c and miRNA172d after treatment with the auxin signaling inhibitor PEO-IAA. The signal for the reporters was largely abolished in the root zones where GUS activity was present in the mock treated roots (Fig. 3, compare c1, d1 to f, g, respectively). Moreover, when we challenged MIR172CAuxRE−::GUS plants bearing two mutated AuxREs motifs (Ripoll et al., 2015) with M. javanica juveniles, no induction was observed in the galls at 5 dpi as compared with the control reporter line MIR172C::GUS where a strong signal was detected in the center of the gall (Fig. 3h,i). Altogether, these results indicate that the miRNA172 is regulated by auxins both in uninfected roots and in the galls induced by M. javanica in Arabidopsis.
A role for miRNA172 and TOE1 in the RKN–plant interaction at different day length regimes and plant developmental stages
The previously shown infection tests for the miRNA172 and the 35S:TOE1R lines were carried out in vitro under LD photoperiods (16 h : 8 h, light : dark; Figs 1, 5). However, it has been reported that miRNA172 expression changes with the length of the light period, being higher under longer photoperiods (Jung et al., 2007). Besides, the expression of miRNA172 is regulated temporally, with no miRNA172 transcripts detected 2 d after germination, and progressively more steady state transcript accumulation was seen with age (Aukerman & Sakai, 2003; Chuck et al., 2007). With this in mind, we performed similar infection assays to those described above but under SD (8 h : 16 h, light : dark) and we also analyzed reproductive parameters in soil-grown plants under these conditions (Fig. 6). The reduction in the infection parameters for the MIM172 and 35S:TOE1R plants was also observed and even enhanced in some cases under SD as compared to LD (Fig. 6a; P < 0.05). Interestingly, a reproductive parameter as the number of eggs per root weight was severely impaired in most of the lines grown in soil and infected c. 2 months after germination (Fig. 6b; P < 0.05) which indicate that the module miRNA172-TOE1 is crucial for nematode infection and also for reproduction. This is in agreement with the reduction of galls and GCs size observed in the MIM172 and 35S:TOE1R lines in LDs (Figs 1, 5). These results confirmed that the miRNA172 performs an important function during the gall and GC development affecting also the reproductive cycle of the nematode. Therefore, the miRNA172 /TOE1 module play a role in galls and GCs development under different photoperiods and plant developmental stages, yet it seems to be independent of the day light regime.
Fig. 6.
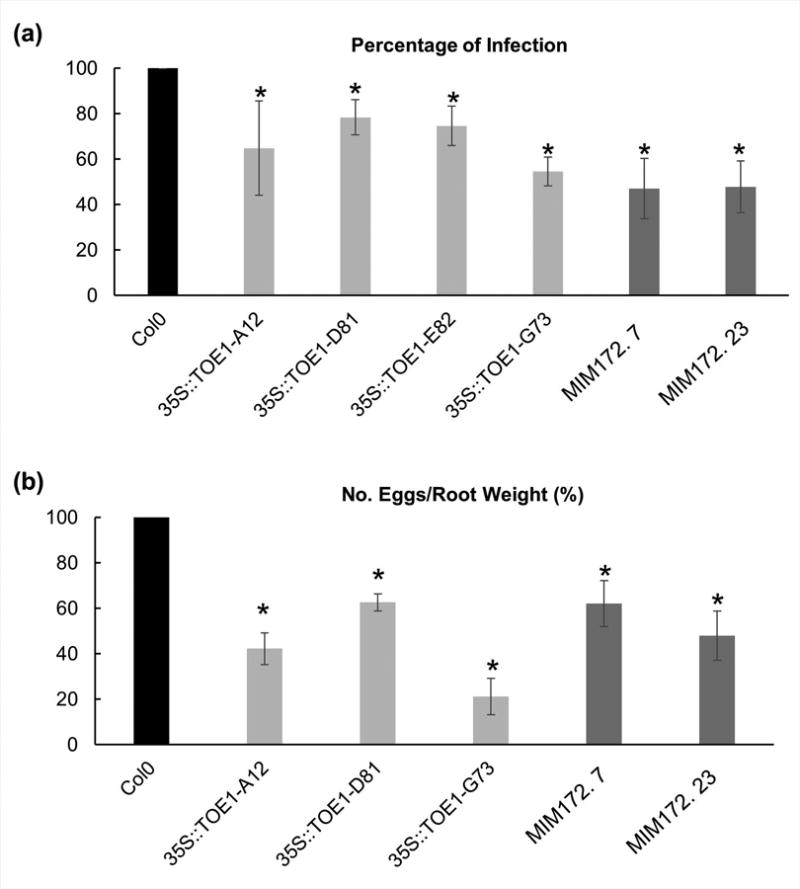
TOE1-miRNA172 resistant lines, 35S:TOE1R and target mimicry lines for miRNA172 show resistance to root-nematode infection under short days. (a) In vitro grown analysis of 35S:TOE1R lines A12, D81, E82 and G73 (in light grey); and mimicry (MIM) lines 7 and 23 (dark grey). The percentage of galls per main root was significantly lower in all lines studied than ecotype Col-0 (black). At least 80 plants per line were analyzed. (b) Soil reproduction tests for 35S:TOE1R lines A12, D81 and G73; and MIM lines 7 and 23. The percentage of number of eggs per root weight was lower in all lines than in Col-0 (P < 0.05). At least 30 plants per line were analyzed. Asterisks, significant differences (T-Student; P < 0.05). Three independent experiments were performed for each treatment. Values are means ± SE.
Discussion
RKNs constitute a major problem for the agriculture, exacerbated in recent years due to the ban of effective but contaminant chemical nematicides (Directives 91/414/EEC or 2009/128/EU). Meloidogyne spp. nematodes establish an obligate and endoparasitic interaction with a broad spectrum of valuable agronomic crops by inducing their feeding cells, GCs, inside the gall, a de novo developed structure (pseudo-organ) within the host roots (Escobar et al., 2015). Increasing knowledge of the molecular mechanisms orchestrating GCs and galls formation could assist in the development of new biotechnological tools against the RKNs (Fosu-Nyarko & Jones, 2015). Our results reveal the activation in the nematode feeding sites (GCs and galls) of a well-documented gene regulatory module (miRNA172/AP2-like), implicated in other developmental processes directing de novo emerging organs in plants like flowers (Aukerman & Sakai, 2003), tubers (Martin et al., 2009), fruit (Ripoll et al., 2015) or nodules (Yan et al., 2013; Wang et al., 2014).
The riboregulator miRNA172 regulates the abundance of a number of AP2-like transcription factors, like TOE1, at both transcriptional and translational levels (Chen, 2004). Yet, in our transcriptomic study of microdissected GCs compared to non-infected cells, the only flowering related AP2-like gene repressed was TOE1 (Barcala et al., 2010). Therefore, an increase in the level of expression of miRNA172 in the cell would result in a reduction of TOE1 transcript abundance and/or protein accumulation. Our expression analyses by q-PCR confirmed TOE1 down-regulation in galls (Fig. 1) while the promoters of the precursor genes for the miRNA172c and miRNA172d (Fig. 4) were strongly activated in galls and GCs induced by M. javanica in Arabidopsis. As mentioned, this is in agreement with previous results of microarrays experiments of laser-capture microdissected GCs at 3 dpi in Arabidopsis (Barcala et al., 2010; Table S2). Our previous work showed a massive and conserved down-regulation of genes in the transcriptome of early developing GCs in Arabidopsis and tomato (Barcala et al., 2010; Portillo et al., 2013) also evident in galls (Jammes et al., 2005; Barcala et al., 2010). The repression of TOE1 expression seems to have a role for proper nematode establishment as infection was reduced in overexpressing lines resistant to the miRNA172 mediated silencing (Fig. 1). Similarly, TPX1, a peroxidase coding gene was repressed in tomato GCs and in Arabidopsis GCs (Portillo et al., 2013). Overexpression of TPX1 in the plant conferred not only a higher resistance to the infection but impaired nematode feeding site development, showing smaller GCs. Likewise, RAP2.6, a transcription factor containing an AP2 domain, was down-regulated in syncytia induced by H. schachtii in Arabidopsis (Szakasits et al., 2009) and its overexpression enhanced the resistance against the nematode, resulting in smaller syncytia (Ali et al., 2013).
In our study we have observed lower infection and reproductive levels in genetic backgrounds with either impaired miRNA172 function or misexpressed TOE1-miRNA172-resistant (MIM172 and 35S:TOE1R, respectively). Moreover, in those lines the growth of galls and GCs was dramatically impaired when compared to the corresponding controls (Figs 1, 5, 6). These results suggest a role for the repression of TOE1 in the morphogenetic processes leading to gall/GCs development. Our results are in line with previous publications showing that AP2-like target repression by miR172 is essential for correct tuberization and fruit morphogenesis (Licausi et al., 2013; Ripoll et al., 2015). This is in accordance with previous studies showing the interference caused by the nematodes in different plant developmental pathways, hijacking regulatory circuits to use them for their own benefit (reviewed in Cabrera et al., 2015b).
MiRNA172 regulates the expression of a small group of AP2-like transcription factors during flowering, including TOE1 (Zhu & Helliwell, 2010). In this respect, we demonstrated the activation of the promoters of the precursor genes for miRNA172c and miRNA172d at early infection stages within the galls, with a high specific pattern in GCs and the adjacent vascular cell layers (Fig. 4) as well as the accumulation of miRNA172 homologues to Arabidopsis miRNA172c in galls from tomato and pea (Fig. S2). These observations are in line with a role for miRNA172 in the nematode feeding site. The regulation of TOE1 by miRNA172 is reinforced by the fact that mimicry lines for miRNA172 showed late flowering phenotype either at SDs or LDs similar to 35S:TOE1R lines (Fig. S1) and a significant reduction in the infection parameters and size of the galls and GCs (Figs 1, 5), somehow mimicking the phenotype observed for 35S:TOE1R also during nematode infection.
The relevance of the down-regulation of a gene for the proper nematode feeding site development has been recently demonstrated, that is, miRNA827 was induced and its target gene NLA, repressed, being necessary for the correct establishment of CNs feeding sites (Hewezi et al., 2016). Moreover, ARF3 is downregulated by the induction of the auxin responsive miRNA390 in Arabidopsis (Cabrera et al., 2016b) and mutants for the miRNA390 activity showed a decrease in the infection parameters and a reduction in the galls size (Cabrera et al., 2016b). This latter example suggests a connection between the hormone auxin and the regulation of genes by miRNAs in galls. Here we demonstrated the regulation of miRNA172 by auxins in RKN-induced galls by two independent assays, one after exposure to a TIR inhibitor, PEO-IAA, and the second by mutating two AuxREs in the miRNA172 promoter (MIR172CAuxRE− ::GUS). In both cases the specific activation of MIR172C::GUS in galls disappeared (Fig. 3). The regulation of miRNA172 expression by auxins has also been demonstrated during the Arabidopsis fruit development, where miRNA172 is induced by different auxin response factors (ARF6 and ARF8) and targets the expression of AP2 (TOE3), mediating in this way the proper fruit growth (Ripoll et al., 2015). The presence of RKNs in the roots alters the auxin levels, showing a high accumulation during early stages of feeding site development and it is a key signal for the formation of GCs and galls (Karczmarek et al., 2004; Kyndt et al., 2013; Cabrera et al., 2014). Auxins also drive fruit development and flowering (Ripoll et al., 2011; 2015). Moreover, nodulation also shows some parallels with gall formation (Mathesius, 2003), such as the accumulation of auxins at early stages (Grunewald et al., 2009). In addition, the regulatory module miRNA172-AP2 also plays a role during the formation of nodules in legumes (Yan et al., 2013; Wang et al., 2014). In parallel, miRNA172c expression induced in the nodules negatively regulates an AP2-like transcription factor, NNC1, allowing in this way the proper formation of the new organ (Wang et al., 2014). Interestingly, MIM172 mimicry lines showed abnormal phenotype of the GCs, as they were smaller than in wild type plants (Fig. 5). Similarly, during fruit development in MIM172 lines smaller valve cells are formed (Ripoll et al., 2015). All these suggest common regulatory networks mediated by miRNA172 in apparently distant processes of plant development and during biotic interactions.
Zhang et al. (2015) showed that TOE1 binds the FT promoter in leaves and that the expression levels of FT increase in an overexpressing line for the miRNA172 and in the double mutant line toe1/toe2, suggesting that TOE1 acts as a negative regulator of FT expression. We showed that FT transcripts accumulate in galls formed by M. javanica in Arabidopsis roots (Fig. 2). These data together with the fact that the phenotype of MIMR172 lines as well as of 35S:TOE1R lines was maintained at different day length regimes, suggest, that RKNs might induce local changes in FT abundance. Accordingly, mutant ft-10 lines showed a decrease in the infection by RKNs and in the gall size (Fig. 2). FT is a mobile molecule broadly characterized as a positive regulator of flowering (Turck et al., 2008). This data in addition to the down-regulation of TOE1 (Fig. 1) and the up-regulation of miRNA172 (Fig. 4) supports a scenario in which the expression of these three genes could be coordinated in nematode feeding sites (Fig. 7b). Belowground, FT orthologous genes have been identified to positively regulate tuber formation in potato (Navarro et al., 2011) or bulb formation in onion (Lee et al., 2013). These results correlate well with the up-regulation and positive role demonstrated for miRNA172 repressing the expression of a TOE1 relative, the AP2-like transcription factor RAP1, during the tuber formation in potato (Martin et al., 2009). Thus, taking into consideration all of the above, it is reasonable to postulate that all those developmental programs (tuber formation, nodule formation, gall development and fruit growth) share a common gene regulatory architecture mediated by miRNA172.
Fig. 7.
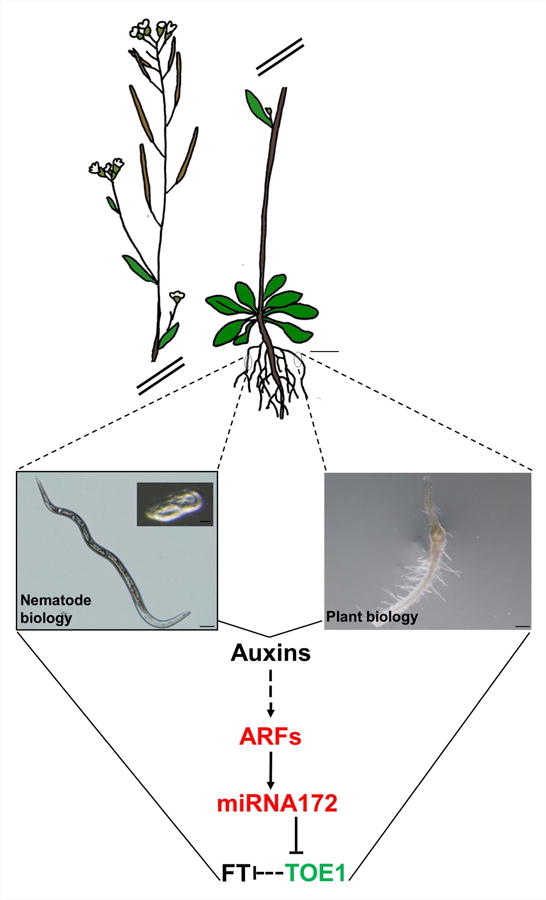
Schematic model of the miRNA172/TOE1/FT function and regulation during the plant–nematode interaction. As a result of the cross talk between Arabidopsis thaliana and Meloidogyne spp. hormonal signaling pathways such as auxins should be altered. Auxins are a positive signal for miRNA172d and c accumulation through auxin response factors (ARFs) activation that induce the miRNA172c promoter, and probably through intermediate partners still not characterized. miRNA172 mediates gene silencing of TOE1 that strongly correlates with the accumulation of FT transcripts, thus TOE1 should regulates FT either directly or indirectly through other partners, also still not known. Bars: egg picture, 20 μm; nematode, 50 μm; gall, 200 μm; Arabidopsis, 1 cm. Red text, induced genes in the microarray of microdissected giant cells (GCs) according to Barcala et al. (2010); green text, repressed; black text, nondifferentially expressed.
Our data support a model where the miRNA172/TOE/FT regulatory module is shared by seemingly unrelated and distant processes such as flowering in the aerial parts and responses to pathogens in roots (Fig. 7). Hence, the initial cells leading to de novo formation of GCs might share some characteristics. For instance, the establishment of floral organ founder cells precedes an auxin response maxima, providing local competence for G1-S cell cycle progression (Chandler, 2011; Seeliger et al., 2016). In this respect, although the founder cells of RKN feeding cells, GCs, are still undetermined, an auxin maxima occurs early during nematode establishment (Karczmarek et al., 2004; Cabrera et al., 2014; Kyndt et al., 2016) preceding successive mitosis and endoreduplication events during their differentiation (De Almeida-Engler et al., 2015; Coelho et al., 2017).
In conclusion, here we show the participation of the regulatory module miRNA172/TOE1/FT in GC and gall development induced by M. javanica in Arabidopsis (Fig. 7). Our data show that the activity of this module plays an important role during RKN parasitism, as several genotypes affected in the activities of this module show lower susceptibility to nematode infection and smaller galls/GCs. The regulation of miRNA172 by auxin response factors together with previous results highlights the key role of auxin signaling during early GC and gall development, strongly suggesting that this regulatory module is in turn regulated by auxins during feeding cells development. Moreover, FT, a gene encoding a mobile molecule that positively regulates flowering, is induced in galls and its loss of function compromised nematode infection levels. Further studies should be performed to understand these common regulatory networks. Yet, common regulatory molecular partners at the cellular level should be controlling different organogenetic processes triggered either by developmental cues, or by biotic interactions, as plant/nematodes, probably coordinated by internal hormonal signals.
Supplementary Material
Fig. S1 Delayed flowering time phenotype of 35S:TOE1R and mimicry lines as compared to Col-0 control.
Fig. S2 In situ detection of mature miRNA172c expression in crops.
Table S1 List of primers used for qPCR analysis of GAPC2, TOE1, TOE1R and FT
Table S3 Auxin responsive elements (AuxRE) present in the promoter regions of five genes that transcribe to miRNA172 and their sequence
Table S2 List of the pre-miRNAs present in the microarray of micro-dissected Arabidopsis GCs Barcala et al. (2010) in Arabidopsis thaliana
Acknowledgments
We thank to Dr G. Engler for their technical advices during in situ procedure. We thank Mr Iñaki Velasco, Mr Jose Perez and Ms Ana Bombín for his technical help. Work supported by the Spanish Government (grants AGL2013-48787; AGL2016-75287-R to C.E., CSD2007-057 and PCIN-2013-053 to C.F. and BIO2013-43098R and BIO2016-77559R to M.P. and J.A.J.; FPU-AP2009-1577 fellowship to F.E.D-M.) and by the Castilla-La Mancha government (PEII-2014-020-P to C.F. and Cátedra Fundación Enresa UCLM grant to F.E.D-M.). J.C. is supported by a Cytema-Santander contract from UCLM. NIH (grant 1R01GM112976-01A1) and the Paul D. Saltman Endowed Chair in Science Education (J-J.R. and M.F.Y.).
Footnotes
Author contributions: Conceptualization: F.E.D-M., J.C., C.E.; methodology: F.E.D-M., J.C., J-J.R., I.d.O., M.F.A., A.C.S., M.B., M.S., V.R-F., J.d.A-E.; investigation: all authors; writing – original draft: F.E.D-M., J.C., C.E., J-J.R., M.P., J.A.J., C.F.; writing – review and editing: all authors; funding acquisition: C.E., C.F., J-J.R., M.F.Y., M.P., J.A.J.; resources: C.E., C.F., M.F.Y., M.P., J.A.J.; supervision: C.E.
Supporting Information: Additional Supporting Information may be found online in the Supporting Information tab for this article:
References
- Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y, Ichinoki H, Notaguchi M, Goto K, Araki T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science. 2005;309(5737):1052–1056. doi: 10.1126/science.1115983. [DOI] [PubMed] [Google Scholar]
- Ali MA, Abbas A, Kreil DP, Bohlmann H. Overexpression of the transcription factor RAP2.6 leads to enhanced callose deposition in syncytia and enhanced resistance against the beet cyst nematode Heterodera schachtii in Arabidopsis roots. BMC Plant Biol. 2013;13:47. doi: 10.1186/1471-2229-13-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés MF, González-Coloma A, Sanz J, Burillo J, Sainz P. Nematicidal activity of essential oils: a review. Phytochemistry Reviews. 2012;11(4):371–390. [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15(11):2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcala M, Garcia A, Cabrera J, Casson S, Lindsey K, Favery B, Garcia-Casado G, Solano R, Fenoll C, Escobar C. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010;61(4):698–712. doi: 10.1111/j.1365-313X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- Betsuyaku S, Takahashi F, Kinoshita A, Miwa H, Shinozaki K, Fukuda H, Sawa S. Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant and Cell Physiology. 2011;52(1):14–29. doi: 10.1093/pcp/pcq157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobay BG, DiGennaro P, Scholl E, Imin N, Djordjevic MA, Mck Bird D. Solution NMR studies of the plant peptide hormone CEP inform function. FEBS Letters. 2013;587(24):3979–3985. doi: 10.1016/j.febslet.2013.10.033. [DOI] [PubMed] [Google Scholar]
- Bouche F, D'Aloia M, Tocquin P, Lobet G, Detry N, Perilleux C. Integrating roots into a whole plant network of flowering time genes in Arabidopsis thaliana. Scientific Reports. 2016;6:29042. doi: 10.1038/srep29042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchez D, Camilleri C, Caboche M. A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. Comptes Rendus de l'Académie des Sciences. Sciences de la Vie. 1993;316(10):1188–1193. [Google Scholar]
- Cabrera J, Barcala M, Fenoll C, Escobar C. The power of omics to identify plant susceptibility factors and to study resistance to root-knot nematodes. Current Issues in Molecular Biology. 2016a;19:53. [PubMed] [Google Scholar]
- Cabrera J, Barcala M, Garcia A, Rio-Machin A, Medina C, Jaubert-Possamai S, Favery B, Maizel A, Ruiz-Ferrer V, Fenoll C, et al. Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: a functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol. 2016b;209(4):1625–1640. doi: 10.1111/nph.13735. [DOI] [PubMed] [Google Scholar]
- Cabrera J, Díaz-Manzano FE, Barcala M, Arganda-Carreras I, de Almeida-Engler J, Engler G, Fenoll C, Escobar C. Phenotyping nematode feeding sites: three-dimensional reconstruction and volumetric measurements of giant cells induced by root-knot nematodes in Arabidopsis. New Phytologist. 2015a;206(2):868–880. doi: 10.1111/nph.13249. [DOI] [PubMed] [Google Scholar]
- Cabrera J, Díaz-Manzano FE, Fenoll C, Escobar C. Chapter seven – Developmental pathways mediated by hormones in nematode feeding sites. In: Escobar C, Fenoll C, editors. Advances in botanical research. Elsevier Academic Press; 2015b. pp. 167–188. [Google Scholar]
- Cabrera J, Diaz-Manzano FE, Sanchez M, Rosso MN, Melillo T, Goh T, Fukaki H, Cabello S, Hofmann J, Fenoll C, et al. A role for LATERAL ORGAN BOUNDARIES-DOMAIN 16 during the interaction Arabidopsis–Meloidogyne spp. provides a molecular link between lateral root and root-knot nematode feeding site development. New Phytol. 2014;203(2):632–645. doi: 10.1111/nph.12826. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Schindelin J, Arganda-Carreras I, Preibisch S, Longair M, Tomancak P, Hartenstein V, Douglas RJ. TrakEM2 software for neural circuit reconstruction. PLoS ONE. 2012;7(6):e38011. doi: 10.1371/journal.pone.0038011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JW. Founder cell specification. Trends in Plant Science. 2011;16(11):607–613. doi: 10.1016/j.tplants.2011.08.005. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annual Review of Genetics. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M. Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and-independent pathways. PLoS ONE. 2012;7(5):e36210. doi: 10.1371/journal.pone.0036210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Irish E, Sakai H, Hake S. The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nature Genetics. 2007;39(12):1517–1521. doi: 10.1038/ng.2007.20. [DOI] [PubMed] [Google Scholar]
- Coego A, Brizuela E, Castillejo P, Ruíz S, Koncz C, Del Pozo JC, Piñeiro M, Jarillo JA, Paz-Ares J, León J. The TRANSPLANTA collection of Arabidopsis lines: a resource for functional analysis of transcription factors based on their conditional overexpression. Plant Journal. 2014;77(6):944–953. doi: 10.1111/tpj.12443. [DOI] [PubMed] [Google Scholar]
- Coelho RR, Vieira P, de Souza Júnior JDA, Martin-Jimenez C, De Veylder L, Cazareth J, Engler G, Grossi-de-Sa MF, de Almeida-Engler J. Exploiting cell cycle inhibitor genes of the KRP family to control root-knot nematode induced feeding sites in plants. Plant, Cell & Environment. 2017;40(7):1174–1188. doi: 10.1111/pce.12912. [DOI] [PubMed] [Google Scholar]
- Damiani I, Baldacci-Cresp F, Hopkins J, Andrio E, Balzergue S, Lecomte P, Puppo A, Abad P, Favery B, Hérouart D. Plant genes involved in harbouring symbiotic rhizobia or pathogenic nematodes. New Phytologist. 2012;194(2):511–522. doi: 10.1111/j.1469-8137.2011.04046.x. [DOI] [PubMed] [Google Scholar]
- De Almeida-Engler J, Vieira P, Rodiuc N, Grossi-de-Sa MF, Gilbert E. Chapter four – The plant cell cycle machinery: usurped and modulated by plant-parasitic nematodes. In: Escobar C, Fenoll C, editors. Advances in botanical research. Elsevier Academic Press; 2015. pp. 91–118. [Google Scholar]
- Díaz-Manzano FE, Barcala M, Engler G, Fenoll C, de Almeida-Engler J, Escobar C. A reliable protocol for in situ microRNAs detection in feeding sites induced by root-knot nematodes. Front Plant Sci. 2016a;7:966. doi: 10.3389/fpls.2016.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz-Manzano FE, Olmo R, Cabrera J, Barcala M, Escobar C, Fenoll C. Long-term in vitro system for maintenance and amplification of root-knot nematodes in Cucumis sativus roots. Front Plant Sci. 2016b;7:124. doi: 10.3389/fpls.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [accessed 5 April 2017];Directive 91/414/EEC of 15 July 1991 concerning the placing of plant protection products on the market. [WWW document] URL http://eur-lex.europa.eu/legal-content/EN/TXT/?uri=uriserv%3Al13002a.
- [accessed 5 April 2017];Directive 2009/128/EC of the European Parliament and of the Council of 21 October 2009 establishing a framework for Community action to achieve the sustainable use of pesticides. [WWW document] URL http://eur-lex.europa.eu/legal-ontent/EN/TXT/?uri=CELEX%3A32009L0128.
- Escobar C, Barcala M, Cabrera J, Fenoll C. Chapter one – Overview of root-knot nematodes and giant cells. In: Escobar C, Fenoll C, editors. Advances in botanical research. Elsevier Academic Press; 2015. pp. 1–32. [Google Scholar]
- Fosu-Nyarko J, Jones MGK. Chapter fourteen - Application of biotechnology for nematode control in crop plants. In: Escobar C, Fenoll C, editors. Advances in botanical research. Elsevier Academic Press; 2015. pp. 339–376. [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics. 2007;39(8):1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Grunewald W, van Noorden G, Van Isterdael G, Beeckman T, Gheysen G, Mathesius U. Manipulation of auxin transport in plant roots during Rhizobium symbiosis and nematode parasitism. Plant Cell. 2009;21(9):2553–2562. doi: 10.1105/tpc.109.069617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang J, Gardner M, Fukuda H, Kondo Y, Etchells JP, Wang X, Mitchum MG. Identification of cyst nematode B-type CLE peptides and modulation of the vascular stem cell pathway for feeding cell formation. PLoS Pathogens. 2017;13(2):e1006142. doi: 10.1371/journal.ppat.1006142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewezi T, Piya S, Qi M, Balasubramaniam M, Rice JH, Baum TJ. Arabidopsis miR827 mediates post-transcriptional gene silencing of its ubiquitin E3 ligase target gene in the syncytium of the cyst nematode Heterodera schachtii to enhance susceptibility. Plant Journal. 2016;88(2):179–192. doi: 10.1111/tpj.13238. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Baum TJ. Chapter nine – Gene silencing in nematode feeding sites. In: Escobar C, Fenoll C, editors. Advances in Botanical Research. Elsevier Academic Press; 2015. pp. 221–239. [Google Scholar]
- Hewezi T, Howe P, Maier TR, Baum TJ. Arabidopsis small RNAs and their targets during cyst nematode parasitism. Molecular Plant–Microbe Interactions. 2008;21(12):1622–1634. doi: 10.1094/MPMI-21-12-1622. [DOI] [PubMed] [Google Scholar]
- Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiology. 2012;159(1):321–335. doi: 10.1104/pp.112.193649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Allen R, Davis EL, Baum TJ, Hussey RS. Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proceedings of the National Academy of Sciences, USA. 2006;103(39):14302–14306. doi: 10.1073/pnas.0604698103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey RS, Barker KR. A comparison of methods of collecting inocula of Meloidogyne spp., including a new technique. Plant Disease Reporter. 1973;57:1025–1028. [Google Scholar]
- Jammes F, Lecomte P, de Almeida-Engler J, Bitton F, Martin-Magniette ML, Renou JP, Abad P, Favery B. Genome-wide expression profiling of the host response to root-knot nematode infection in Arabidopsis. Plant Journal. 2005;44(3):447–458. doi: 10.1111/j.1365-313X.2005.02532.x. [DOI] [PubMed] [Google Scholar]
- Jisha V, Dampanaboina L, Vadassery J, Mithofer A, Kappara S, Ramanan R. Overexpression of an AP2/ERF Type Transcription Factor OsEREBP1 Confers Biotic and Abiotic Stress Tolerance in Rice. PLoS ONE. 2015;10(6):e0127831. doi: 10.1371/journal.pone.0127831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Seo YH, Seo PJ, Reyes JL, Yun J, Chua NH, Park CM. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell. 2007;19(9):2736–2748. doi: 10.1105/tpc.107.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczmarek A, Overmars H, Helder J, Goverse A. Feeding cell development by cyst and root-knot nematodes involves a similar early, local and transient activation of a specific auxin-inducible promoter element. Mol Plant Pathol. 2004;5(4):343–346. doi: 10.1111/j.1364-3703.2004.00230.x. [DOI] [PubMed] [Google Scholar]
- Kyndt T, Goverse A, Haegeman A, Warmerdam S, Wanjau C, Jahani M, Engler G, de Almeida-Engler J, Gheysen G. Redirection of auxin flow in Arabidopsis thaliana roots after infection by root-knot nematodes. Journal of Experimental Botany. 2016;67(15):4559–4570. doi: 10.1093/jxb/erw230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndt T, Vieira P, Gheysen G, de Almeida-Engler J. Nematode feeding sites: unique organs in plant roots. Planta. 2013;238(5):807–818. doi: 10.1007/s00425-013-1923-z. [DOI] [PubMed] [Google Scholar]
- Lee R, Baldwin S, Kenel F, McCallum J, Macknight R. FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat Commun. 2013;4:2884. doi: 10.1038/ncomms3884. [DOI] [PubMed] [Google Scholar]
- Li X, Wang X, Zhang S, Liu D, Duan Y, Dong W. Identification of soybean microRNAs involved in soybean cyst nematode infection by deep sequencing. PLoS ONE. 2012;7(6):e39650. doi: 10.1371/journal.pone.0039650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F, Ohme-Takagi M, Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 2013;199(3):639–649. doi: 10.1111/nph.12291. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo Y, Guo Z, Li L. Evolutionary conservation of microRNA regulatory programs in plant flower development. Developmental Biology. 2013;380(2):133–144. doi: 10.1016/j.ydbio.2013.05.009. [DOI] [PubMed] [Google Scholar]
- Martin A, Adam H, Diaz-Mendoza M, Zurczak M, Gonzalez-Schain ND, Suarez-Lopez P. Graft-transmissible induction of potato tuberization by the microRNA miR172. Development. 2009;136(17):2873–2881. doi: 10.1242/dev.031658. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M, Lázaro A, Poethig RS, Gómez-Mena C, Piñeiro MA, Martinez-Zapater JM, Jarillo JA. EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;133(7):1241–1252. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- Mathesius U. Conservation and divergence of signalling pathways between roots and soil microbes – the Rhizobium–legume symbiosis compared to the development of lateral roots, mycorrhizal interactions and nematode-induced galls. Plant and Soil. 2003;255(1):105–119. [Google Scholar]
- Mishra S, Phukan UJ, Tripathi V, Singh DK, Luqman S, Shukla RK. PsAP2 an AP2/ERF family transcription factor from Papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol Biol. 2015;89(1-2):173–186. doi: 10.1007/s11103-015-0361-7. [DOI] [PubMed] [Google Scholar]
- Mitchum MG, Wang X, Wang J, Davis EL. Role of nematode peptides and other small molecules in plant parasitism. Annual Review of Phytopathology. 2012;50:175–195. doi: 10.1146/annurev-phyto-081211-173008. [DOI] [PubMed] [Google Scholar]
- Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid DE, Stougaard J, Frugier F, Imin N, Djordjevic MA. Different pathways act downstream of the peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiology. 2016;171(4):2536–2548. doi: 10.1104/pp.16.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Munne-Bosch S. Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 2015;169(1):32–41. doi: 10.1104/pp.15.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oro E, Cuellar CA, Tamaki S, Silva J, Shimamoto K, Prat S. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature. 2011;478(7367):119–122. doi: 10.1038/nature10431. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140(2):411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo R, Silva AC, Díaz-Manzano FE, Cabrera J, Fenoll C, Escobar C. A standardized method to assess infection rates of root-knot and cyst nematodes in Arabidopsis thaliana mutants with alterations in root development related to auxin and cytokinin signaling. Auxins and Cytokinins in Plant Biology: Methods and Protocols. 2017:73–81. doi: 10.1007/978-1-4939-6831-2_5. [DOI] [PubMed] [Google Scholar]
- Portillo M, Cabrera J, Lindsey K, Topping J, Andres MF, Emiliozzi M, Oliveros JC, Garcia-Casado G, Solano R, Koltai H, et al. Distinct and conserved transcriptomic changes during nematode-induced giant cell development in tomato compared with Arabidopsis: a functional role for gene repression. New Phytol. 2013;197(4):1276–1290. doi: 10.1111/nph.12121. [DOI] [PubMed] [Google Scholar]
- Ripoll JJ, Bailey LJ, Mai QA, Wu SL, Hon CT, Chapman EJ, Ditta GS, Estelle M, Yanofsky MF. MicroRNA regulation of fruit growth. Nature Plants. 2015;1:15036. doi: 10.1038/nplants.2015.36. [DOI] [PubMed] [Google Scholar]
- Ripoll JJ, Roeder AH, Ditta GS, Yanofsky MF. A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development. 2011;138(23):5167–5176. doi: 10.1242/dev.073031. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger I, Frerichs A, Glowa D, Velo L, Comelli P, Chandler JW, Werr W. The AP2-type transcription factors DORNRÖSCHEN and DORNRÖSCHEN-LIKE promote G1/S transition. Molecular Genetics and Genomics. 2016;291(5):1835–1849. doi: 10.1007/s00438-016-1224-x. [DOI] [PubMed] [Google Scholar]
- Szakasits D, Heinen P, Wieczorek K, Hofmann J, Wagner F, Kreil DP, Sykacek P, Grundler FM, Bohlmann H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 2009;57(5):771–784. doi: 10.1111/j.1365-313X.2008.03727.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellmann G. The E-Method: a highly accurate technique for gene-expression analysis. Nature Methods Application Notes. 2006:i–ii. [Google Scholar]
- Todesco M, Rubio-Somoza I, Paz-Ares J, Weigel D. A collection of target mimics for comprehensive analysis of microRNA function in Arabidopsis thaliana. PLoS Genet. 2010;6(7):e1001031. doi: 10.1371/journal.pgen.1001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Van Helden J. Regulatory sequence analysis tools. Nucleic Acids Research. 2003;31(13):3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang L, Zou Y, Chen L, Cai Z, Zhang S, Zhao F, Tian Y, Jiang Q, Ferguson BJ, et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell. 2014;26(12):4782–4801. doi: 10.1105/tpc.114.131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Li Y, Zhang Q, Xu T, Qiu L, Fan Y, Wang L. Novel miRNA and phasiRNA biogenesis networks in soybean roots from two sister lines that are resistant and susceptible to SCN race 4. PLoS ONE. 2014;9(10):e110051. doi: 10.1371/journal.pone.0110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi YL, Ishida T, Sawa S. CLE peptides and their signaling pathways in plant development. J Exp Bot. 2016;67(16):4813–4826. doi: 10.1093/jxb/erw208. [DOI] [PubMed] [Google Scholar]
- Yan Z, Hossain MS, Wang J, Valdes-Lopez O, Liang Y, Libault M, Qiu L, Stacey G. MiR172 regulates soybean nodulation. Mol Plant–Microbe Interact. 2013;26(12):1371–1377. doi: 10.1094/MPMI-04-13-0111-R. [DOI] [PubMed] [Google Scholar]
- Yoo SK, Chung KS, Kim J, Lee JH, Hong SM, Yoo SJ, Yoo SY, Lee JS, Ahn JH. CONSTANS activates SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 through FLOWERING LOCUS T to promote flowering in Arabidopsis. Plant Physiology. 2005;139(2):770–778. doi: 10.1104/pp.105.066928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang L, Zeng L, Zhang C, Ma H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015;29(9):975–987. doi: 10.1101/gad.251520.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Li Z, Fan J, Hu C, Yang R, Qi X, Chen H, Zhao F, Wang S. Identification of jasmonic acid-associated microRNAs and characterization of the regulatory roles of the miR319/TCP4 module under root-knot nematode stress in tomato. Journal of Experimental Botany. 2015;66(15):4653–4667. doi: 10.1093/jxb/erv238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Helliwell CA. Regulation of flowering time and floral patterning by miR172. J Exp Bot. 2010;62(2):487–495. doi: 10.1093/jxb/erq295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Delayed flowering time phenotype of 35S:TOE1R and mimicry lines as compared to Col-0 control.
Fig. S2 In situ detection of mature miRNA172c expression in crops.
Table S1 List of primers used for qPCR analysis of GAPC2, TOE1, TOE1R and FT
Table S3 Auxin responsive elements (AuxRE) present in the promoter regions of five genes that transcribe to miRNA172 and their sequence
Table S2 List of the pre-miRNAs present in the microarray of micro-dissected Arabidopsis GCs Barcala et al. (2010) in Arabidopsis thaliana


