Abstract
Specialized renal epithelial cells known as podocytes are essential components of the filtering structures within the kidney that coordinate the process of removing waste from the bloodstream. Podocyte loss initiates many human kidney diseases as it triggers subsequent damage to the kidney, leading to progressive loss of function that culminates with end stage renal failure. Podocyte morphology, function and gene expression profiles are well conserved between zebrafish and humans, making the former a relevant model to study podocyte development and model kidney diseases. Recently, we reported that whole genome sequencing of the zeppelin (zep) zebrafish mutant, which exhibits podocyte abrogation, revealed that the causative lesion for this defect was a splicing mutation in the breast cancer 2, early onset (brca2) gene. This was a surprising and novel discovery, as previous research on brca2/BRCA2 in a number of vertebrate animal models had not implicated an explicit role for this gene in kidney mesoderm development. Interestingly, the abrogation of the podocyte lineage in zep mutants was also accompanied by the formation of a larger interrenal (IR) gland, which is analogous to the adrenal gland in mammals, and suggested a fate switch between the renal and inter renal mesodermal derivatives. Mirroring these findings, knockdown of brca2 also recapitulated the loss of podocytes and increased IR population. In addition, brca2 overexpression was sufficient to partially rescue podocytes in zep mutants, and induced ectopic podocyte formation in wild-type embryos. Interestingly, immunofluorescence studies indicated that zep mutants had elevated P-h2A.X levels, suggesting that DNA repair is dysfunctional in these animals and contributes to the zep phenotype. Moving forward, this unique zebrafish mutant provides a new model to further explore how brca2 contributes to the development of tissues including the kidney mesoderm—roles which may have implications for renal diseases as well.
Keywords: brca2, kidney, development, podocyte, interrenal gland, zebrafish, fanconi anemia
Introduction
The classic tumor suppressor genes BRCA1, DNA repair associated and BRCA2, DNA repair associated (BRCA1 and BRCA2) have risen to notoriety as they are two of the most frequently mutated genes found in hereditary breast cancer tumors[1, 2]. The BRCA1/2 proteins play central roles in repairing double-stranded breaks in DNA through mediating homologous recombination in somatic cells and meiotic recombination in germ cells, where they prevent the accumulation of genetic errors[3, 4]. People with severe heterozygous genetic mutations in BRCA1 and BRCA2 have increased susceptibility to cancer because the dysfunction of these proteins has widespread and catastrophic results that promote genome instability[1–4]. While genetic mutations of BRCA1 and BRCA2 are widely recognized to increase the risk of breast and ovarian cancers, mutations in these critical genes have also been linked to tumor formation in dozens of other cancer types including prostate, melanoma, pancreatic, and kidney among others[5].
Additionally, the inheritance of homozygous copies of mild mutations in BRCA2, which is also known as FANCD1, results in Fanconi anemia (FA), a rare recessive disease that has been associated with 22 genes to date[6–8]. The products of the FA genes all participate in DNA damage repair[6–8]. FA is a phenotypically heterogenous condition, which is characterized by anemia and congenital malformations that range in severity and can involve any of the organ systems, though approximately one-third of patients do not exhibit major malformations[9–13]. FA patients also have increased risk for cancer due to their inherent genome instability: 20% or more develop malignancies, where acute leukemia is by far the most common pathology[9–13]. While the effects of BRCA2 mutations have been extensively studied in the cancer field, the impact of these mutations in the early developmental context of FA has been less scrutinized. Thus, there is an unmet need to understand why FA patients present with pleiotropic abnormalities of the skeletal, cardiac, nervous, gastrointestinal and renal systems, and how these phenotypes could be prevented, lessened, or cured[11]. In part, this has been hampered because of limitations in existing animal models, as homozygous null BRCA2 mutations in mice are embryonic lethal, highlighting the need for alternative models like tissue-specific conditional knockouts[14–16]. Further, as tumors are capable of activating long-dormant developmental pathways, exploring the roles of tumor suppressors in developmental models can provide unique insights that may highlight new therapy options.
One vertebrate animal model that has risen in popularity for developmental genetics and disease research, including cancer and the FA genes, is the zebrafish, Danio rerio[17–22]. This small freshwater fish has a number of attributes that have fostered its husbandry in a laboratory setting and use as an experimental system[17]. These traits include high fecundity, as large clutches (100-200 embryos or even several hundred more) can be obtained weekly from a single pair of healthy breeding adults. The embryos are transparent and develop externally from the parents, making them tractable for research at all stages of ontogeny, and ideal for high-throughput genetic and chemical screens due to their size and ability to obtain large sample sizes. Organogenesis occurs rapidly in zebrafish[23], and the ease of visualization of the developing tissues is a unique advantage that is unrivaled in comparison to other vertebrate models. As there is considerable genetic conservation between the zebrafish and humans[24], many biological aspects are highly similar or even have direct relevance, which has opened many possibilities for translating zebrafish research findings into new biomedical applications[25].
The embryonic kidney is just one essential organ that can be visualized in the zebrafish embryo at 24 hours post fertilization (hpf), known at this stage as the pronephros[26]. The pronephros has a simple anatomy, with two nephron functional units that perform excretory tasks beginning by the 48 hpf stage[27]. The rapid formation of the pronephros has facilitated a steadily expanding literature in which many fundamental aspects of renal ontogeny have been examined along with the establishment of powerful kidney disease models[28–31]. Recent work on zebrafish kidney development has revealed insights ranging from the control of cell fate decisions to the mechanics underlying tubulogenesis, and morphological processes involving collective cell migration[32–43]. These exciting advances have been fostered by the formulation of advanced cell, molecular, and imaging techniques along with methods of transgenesis and genome editing[44–48].
The adult human kidney is comparatively much more complex, being composed of several hundred thousand to upwards of over 1 million nephron functional units[49]. Fundamentally, however, human and zebrafish nephrons exhibit a shared organization wherein a variety of specialized epithelial cells form three distinct functional zones, known as the blood filter, tubule and duct[49, 50]. Each nephron part accomplishes explicit tasks that regulate excretion and osmoregulation, such as the various tubule segments wherein groups of different epithelial cells perform the absorption and secretion of metabolites[49, 50]. The blood filter, also termed the renal corpuscle, is comprised of a glomerulus housed within the Bowman’s capsule[51, 52]. The glomerulus forms an intricate sieve that sends filtrate into the capsule and then into the nephron tubule where it will be refined to recover essential nutrients before excretion of the solution. The glomerulus is a capillary tuft where fenestrated endothelial cells make up the capillary walls, and mesangial cells, a type of modified smooth muscle cell, lie between the capillaries. As the blood passes into the capillaries, a filtrate solution from the circulation traverses the capillaries and is percolated through a surrounding glomerular basement membrane (GBM) on which the specialized epithelial cells known as a podocytes are situated. Podocytes are characterized by an octopus-like morphology with a cell body that has long, intricate cell extensions known as foot processes, in which protein complexes form interlocking structures that connect with neighboring podocytes, termed the slit diaphragm. The integrity of the slit diaphragm and maintenance of podocyte attachment to the GBM are among the essential components of each nephron filtration apparatus.
As such, damage to podocytes results in a compromised filtration system and can initiate a cascade of phenotypes involving damage and/or death to nephron cells, leading to the destruction of entire nephrons and chronic kidney disease[53, 54]. In stark contrast to their high importance in the kidney, mechanisms to regenerate podocytes are currently thought to be quite limited. Indeed, there is strong evidence that adult podocytes are mitotically quiescent, where podocyte loss can be compensated for through hypertrophy of remaining cells. Medical options to reverse significant podocyte losses are unknown at present. Therefore, it is of critical importance to learn the genetic pathways that promote the formation of this cell type to gain a better understanding of kidney diseases and options to treat them. Because of the well-demonstrated conservation in genetic ontogeny and cellular morphology, our lab has chosen to use the zebrafish embryo to discover novel regulators of podocyte development.
The zebrafish pronephros has been a particularly useful model for the in vivo study of podocyte and renal corpuscle development, real-time physiology assessment of filtration, and how acute damage to the various components leads to regeneration or permanent defects[55]. With regard to development of the cellular components in the zebrafish pronephros, the podocyte field is derived from the anterior portion of intermediate mesoderm as marked by genes that include wilms tumor 1a and 1b (wt1a, wt1b) (Figure 1A)[27,56, 57]. By 24 hpf these podocyte progenitors appear as two bilateral cell clusters situated rostral to the tubules[27, 56, 57]. Over the next day of development, the podocytes undergo maturation wherein the clusters migrate toward the midline of the trunk and recruit vascular cells by secreting the growth factor encoded by vascular endothelial growth factor Aa (vegfaa) (Figure 1A)[58–62]. The podocytes and capillaries form a central renal corpuscle that connects laterally to the pair of nephrons, which provide renal function for the first several weeks of embryonic life. Subsequently, a kidney with several hundred nephrons (each with their own blood filter) forms, which is termed the mesonephros, and provides renal function for the remainder of juvenile and adult life[63]. Although a number of key milestones of renal corpuscle development are known, the genetic regulatory networks that control the patterning and differentiation of the podocyte lineage are not yet fully understood[64]. Thus, forward genetic approaches with zebrafish are uniquely situated to facilitate discovery of the conserved molecules responsible for podocyte emergence, and renal corpuscle development as well, because of their rapid genesis and the other features of the zebrafish embryo as outlined above.
Figure 1. Zebrafish zeppelin (zep) mutant embryos exhibit edema, a decrease in kidney podocyte (P) cells, and an increased interrenal gland (IR).
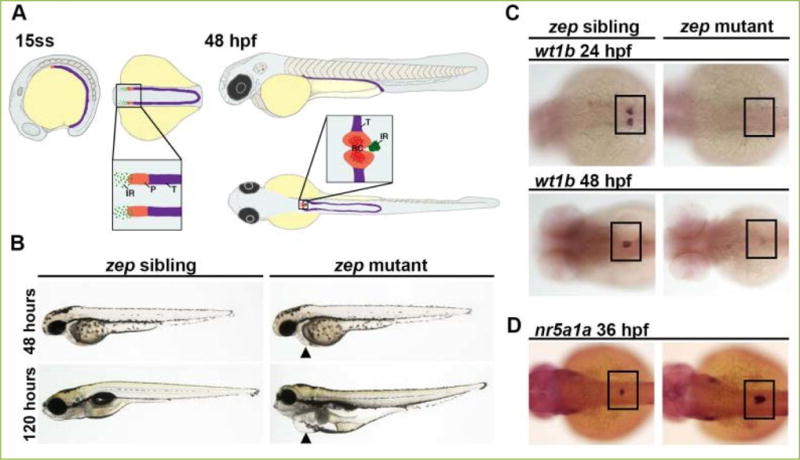
A) The zebrafish kidney arises from the intermediate mesoderm (IM, purple) and by the 15 somite stage (ss) the anteriormost region of the IM is patterned by two fields of P progenitors, as distinguished by the marker wt1a, whereas non-wt1a expressing cells are predestined as kidney tubule (T). It is hypothesized that IR precursors arise from the wt1a-expressing portion of IM. The P progenitor fields begin to migrate towards the midline after 24 hpf, and by 48 hpf this field is fused at the midline, has become innervated with capillaries, and begins to function as the renal corpuscle (RC). The IR cells also exhibit a migration toward the midline and fuse to become a single organ by the 36 hpf time point. B) Brightfield imaging showing live morphology: zep mutants appeared similar to siblings at the 48 hpf time point, but by 120 hpf they exhibited severe edema including around the pericardial region (black arrowheads). C) zep mutants exhibited a severe decrease in podocytes at 24 hpf and 48 hpf compared to siblings, demonstrated by whole mount in situ hybridization (WISH) using the podocyte marker wt1b. D) WISH on zep mutants at 36 hpf with the IR marker nr5a1a showed an increased IR in mutants compared to siblings. C–D) Black boxes demarcate the embryonic region where P or IR areas are located. Figure adapted from[66], and reprinted as allowed by the terms of the CC BY 4.0 Creative Commons License of the Authors.
Results
Utilizing the advantages of the zebrafish as a model, a forward genetic screen to identify novel genetic regulators of kidney development was performed using the mutagen ethylnitrosourea (ENU)[65]. Hits were determined by the presence of pericardial edema, which is a hallmark of fluid imbalance and can be an indicator of kidney dysfunction. Hits were also assessed by using whole mount in situ hybridization (WISH) at 24 hpf to examine changes in genetic territories of developing nephron segment cells. One such mutant line exhibited edema that became so dramatic by 120 hpf that it was dubbed zeppelin (zep) (Figure 1B)[66]. Tissue sections of these embryos revealed dilated pronephric tubules but an absent glomerulus. Interestingly, WISH revealed that despite having an intact nephron tubule at 24 hpf, zep homozygous (−/−) mutant embryos, hereafter referred to simply as zep mutants, exhibited a complete or nearly complete loss of podocytes at 24 hpf onwards as evidenced by several podocyte-specific markers including wt1b (Figure 1C). The filtration ability of these embryos also appeared to be debilitated as the mutants were unable to absorb and clear injected fluorescent dextran compared to siblings.
While the podocyte cell type was missing in zep mutants, there were no significant differences in cell proliferation or death events at several developmental time points, as seen using immunofluorescence assays to evaluate phosphorylated histone H3, which marks cells in G2/M phase, or Caspase-3 which marks cells undergoing apoptosis, as well as acridine orange which also labels apoptotic cells[66]. In exploring related tissue types, we noticed a significant increase in area and volume of the interrenal gland (IR) in zep mutants, the teleost equivalent of the adrenal cortex, which is marked by the transcripts of nuclear receptor subfamily 5, group A, member 1a (nr5a1a) at 36 hpf (Figure 1D). This expansion was verified by 3β-hydroxysteroid dehydrogenase (3β-HSD) chromogenic staining, which indicates an enzymatic property of differentiated IR cells[67]. In previous studies, it has been shown that podocytes and IR cells arise from the wt1a progenitor field, and that knockdown of wt1a leads to a decrease in IR size[67, 68]. Because there was no distinguishable difference in proliferation and cell death, we hypothesized that the cells that would normally be predestined for the podocyte lineage in the wt1a-expressing renal progenitor field undergo a fate change to the IR lineage in the zep mutants.
To directly identify the causal lesion underlying the zep phenotype, we undertook a whole-genome sequencing (WGS) strategy[66]. In recent years, advances in using next-generation sequencing (NGS) have made it possible to perform genetic mapping of spontaneous and ENU-induced mutations by WGS in both zebrafish and mouse[69]. This approach takes advantage of the fact that examination of mutant DNA pools (i.e. bulk segregant analysis) is typified by a lack of genetic diversity of single nucleotide polymorphisms (SNPs) in the region harboring the mutation compared to wild-type pools, while diversity increases as the distance from the mutation position increases. We performed WGS as previously described, invoking a hidden Markov model to define the likely mutation area, along with the web based software SNPtrack to assess SNPs in the critical region[69, 70].
WGS localized the zep lesion to zebrafish chromosome 15[66]. Next, our SNP analysis revealed a T to C mutation at the splice acceptor site between exons 20 and 21 in the brca2 gene within the zep mutant pools, where alleles were also detected within wild-type genomic DNA pools, consistent with heterozygous animals in that group (Figure 2A). The brca2 gene is ubiquitously expressed in zebrafish from early stages through the 24 hpf time point, but expression had not been closely scrutinized in kidney development (Figure 2B)[71–73]. Our WISH analyses to assess brca2 expression in zep mutants and wild-type embryos recapitulated these results and tantalizingly suggested that the mesodermal region from which podocytes arise possibly exhibited an elevated number of brca2 transcripts (Figure 2B). Tissue-specific quantification of this observation is technically challenging, but warrants further study and would likely require the development of transgenic lines to enable isolation of kidney mesodermal precursors at very early stages. Interestingly, the level and spatial distribution of brca2 transcripts were unchanged based on our WISH studies of zep mutants and wild-type embryos.
Figure 2. brca2 is necessary and sufficient to promote podocyte development in the embryonic zebrafish kidney.
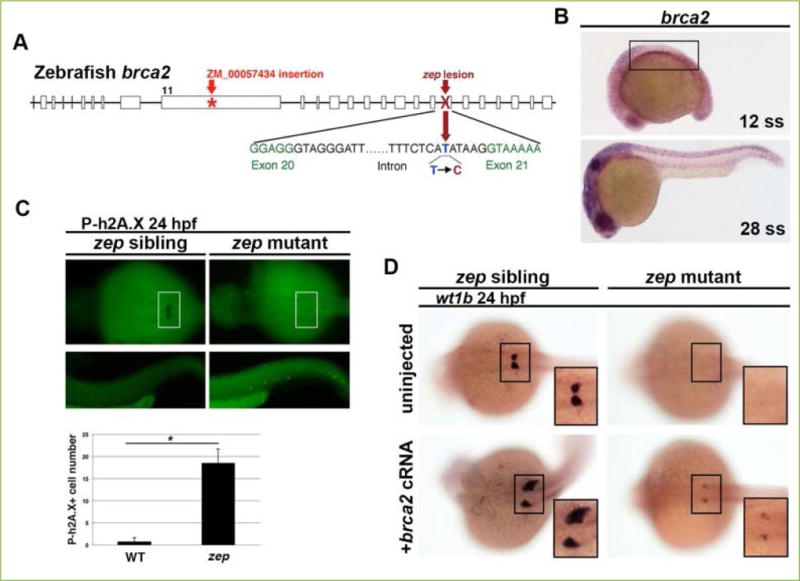
A) Whole genome sequencing (WGS) revealed that zep mutants contain a T to C transversion in the brca2 gene that we found resulted in aberrant splicing. This schematic map also indicates the location of the lesion in brca2ZM_00057434 insertional mutants used in our study. B) WISH on wild-type fish at a variety of stages showed that brca2 transcripts (purple stain) were expressed ubiquitously throughout the organism, including the intermediate mesoderm (IM) which gives rise to kidney (boxed area). C) zep siblings and mutants at the 24 hpf time point were subjected to WISH with wt1b to demarcate podocytes (boxed area), then immunofluorescence to detect phosphorylated h2A.X (P-h2A.X). There was a significant increase in cells positive for P-h2A.X in zep mutants, which indicates an accumulation of double stranded DNA breaks, and suggested that zep mutants have dysfunctional brca2 protein (* = p<0.05). D) Both zep siblings and zep mutants were injected with brca2 capped RNA (cRNA) and then subjected to WISH with wt1b. Interestingly, some injected wild-type and heterozygous sibling embryos had drastically increased areas of podocytes (frequency of 13.5%) compared to uninjected controls. Podocyte development was partially rescued in zep mutants (frequency of 55%) following brca2 overexpression. Figure adapted from[66], and reprinted as allowed by the terms of the CC BY 4.0 Creative Commons License of the Authors.
Next, we found that knockdown of brca2 using two independently verified morpholinos led to a loss of podocytes and an expanded IR that resembles the phenotypes seen in zep mutants[66]. Interestingly, WISH with the podocyte marker wt1b conducted concomitantly with immunohistochemistry with anti-phospho-H2AX antibody (P-h2A.X) to detect double-stranded DNA breaks revealed a marked increase in damaged DNA in podocyte-absent zep mutants compared to siblings (Figure 2C). This suggested that the Brca2 protein was not functioning in DNA repair in mutant animals.
Previous reports have isolated two different brca2 mutant alleles in zebrafish, one being a C to T point mutation that produces a glutamine (Q) to stop (X) change, termed the brca2Q658X allele[71], the other being an insertional mutation in exon 11, termed the brca2ZM_00057434 allele[72]. Neither mutant allele has been associated with renal defects, but instead with a constellation of phenotypes including defects in ovary development, gametogenesis (both oogenesis and spermatogenesis), sex determination, and genome instability leading to gonadal tumors[71–74]. With our collaborators, we explored kidney and IR development in brca2ZM_00057434 −/− mutants, and interestingly did not detect alterations in podocyte precursors during early embryogenesis[66]. However, we observed a modest but significant increase in IR area at the 36 hpf stage in brca2ZM_00057434 −/− mutant embryos. Similarly, when performing complementation crosses between zep+/− adults and brca2ZM_00057434 +/− adults, we observed a modest, but significant, increase in the IR area in compound heterozygous zep+/−; brca2ZM_00057434 +/− embryos, which resembled the IR area that developed in the brca2ZM_00057434 −/− mutant embryos. We concluded that these two lines failed to complement each other, and this provides further genetic evidence that Brca2 is essential for mesoderm development.
We next examined the gain of function brca2 phenotype, to test whether Brca2 was sufficient to ameliorate the podocyte deficiency in zep mutants[66]. zep mutants injected with brca2 capped mRNA (cRNA) exhibited a partial podocyte rescue, where 55% of mutant embryos developed wt1b+ podocytes (Figure 2D). Furthermore, some wild-type siblings injected with a similar dose of brca2 cRNA had an expanded podocyte field and/or ectopic podocyte formation, at a frequency of 13.5%, indicating that brca2 is sufficient for podocyte formation. This overexpression result was particularly astonishing to us, as is the first time to our knowledge that Brca2 has been shown to be sufficient to promote formation of a particular embryonic lineage. This surprising result warrants further study so we can understand the mechanisms by which Brca2 function influences mesoderm ontogeny. Additional studies to examine the simultaneous outcomes of brca2 overexpression for podocyte and IR would be very interesting, and could be accomplished with more sophisticated gene expression analyses such as the implementation of fluorescent WISH.
While these studies had validated that brca2 acts as an intriguing regulator of podocyte and IR formation in the embryonic zebrafish, how it coordinates this task with the greater podocyte genetic network was unclear[66]. Retinoic acid (RA) has been shown to act to promote the formation of podocytes, in part through direct transcriptional regulation of wt1a[75, 76]. We performed exogenous RA treatments at a range of concentrations on zep mutants and control wild-type siblings, which revealed that RA was not sufficient to rescue podocyte formation in zep mutants[66]. This suggests that brca2 acts downstream of RA signaling in podocyte development, or in an alternative essential pathway.
We also investigated the relationship between Notch signaling and brca2[66], as Notch is a major pathway that promotes podocyte formation in zebrafish[68] and also amphibians[77]. We utilized the transgenic line Tg (hsp70::GAL4, UAS::NICD) that upon heat-shock induces the ubiquitous ectopic activation of Notch via expression of the transcriptional regulator, known as the Notch intracellular signaling domain (NICD). NICD overexpression led to a slight, though not significant, increase in podocytes in wild-type control embryos. In contrast, heat-shocked brca2 morphants continued to show an absence of podocytes, indicating a failure of NICD to rescue podocyte genesis. This suggests that Notch signaling acts upstream of brca2 in podocyte development, or in an alternative pathway. Inhibition of Notch signaling via chemical treatments, using the gamma-secretase inhibitor known as N-[N-(3,5-difluoro-phenactyl)-Lalanyl]-S-phenylglycine t-butyl ester (DAPT), led to a reduction of the podocyte field in wild-type control embryos and absent podocytes in brca2 MO injected animals. With regard to IR development, inhibition of Notch led to an expanded IR field in wild-type embryos, analogous to the brca2-deficient embryos. Overexpression of Notch was sufficient to decrease the enlarged IR typically seen in brca2 morphants, while DAPT had no additional effects in brca2 deficient embryos. While further studies are needed to discern these tissue difference, our data suggest that Notch may act downstream of brca2 to inhibit IR formation, but upstream of brca2, to promote podocyte formation.
Conclusions and perspectives
We have shown in this study that zebrafish can be used to identify novel genetic regulators of podocyte formation[66]. While we collected compelling evidence that brca2 is both necessary and sufficient to promote podocyte development at the expense of the IR, the molecular mechanisms by which Brca2 influences kidney mesoderm and related intermediate mesoderm lineages like the IR have yet to be elucidated. For example, further work is needed to assess whether Brca2 expression is autonomously required in mesodermal precursors, along with the renal and IR precursors, or works cell-autonomously in neighboring tissues. Our expression data in the mid-somitogenesis stages suggest that Brca2 could have roles in the mesoderm directly, but our analysis may be misleading for any number of reasons. For example, the time window when Brca2 exerts its effects is unknown at present and therefore the expression we noted may not be linked to kidney mesoderm development. Manipulations of both the spatial and temporal activity of Brca2 would be necessary to tease apart these nuances for its courses of action in development.
Like its mammalian counterpart, zebrafish Brca2 has previously been implicated to act as a tumor suppressor through a complex it forms to repair double-stranded DNA breaks. Deficiency of another critical member of this complex, rad51, has recently been studied in zebrafish and shown to display symptoms of FA and smaller kidneys[78]. In light of this, other gene members of the FA pathway should be more closely scrutinized in the context of podocyte and kidney development to assess whether diminished DNA repair by other FA genes is causative of the fate switch seen between podocytes and IR in zep mutants. Continued study of FA has accumulated a list of 22 genes, to date, whose mutations result in the disease, and the current list of these published genes is curated on the Fanconi Anemia Mutation Database, (http://www.rockefeller.edu/fanconi/), a publicly available resource. Something else to consider is the variation of IR and podocyte phenotypes seen in zep and brca2ZM_00057434 homozygous mutants, as this possibly suggests that only a portion of the Brca2 protein may be critical in regulating mesodermal development.
Interestingly, there is clinical link between Fanconi anemia patients that have bi-allelic BRCA2 mutations and kidney diseases such as Wilms’ tumor and clear cell carcinoma that have been appreciated but never fully understood[11, 79–86]. There have also been clinical links between BRCA2 mutations and adrenal cortical carcinoma[87]. Our observations that brca2 mutations cause expanded interrenal glands in a developmental context suggest that adrenal gland neoplasms and the prevalence of kidney disorders in Fanconi anemia patients could be caused by mutations in BRCA2.
Future studies are needed to determine if BRCA2 is capable of influencing renal progenitors during mammalian kidney development. In mammalian development, three successive kidney forms develop and degrade; the pronephros, the mesonephros and the terminal kidney, the metanephros[88, 89]. While the major cell components of the renal corpuscle and nephron are conserved between the mammalian and teleost system, zebrafish only form a pronephros followed by a mesonephros. Related to this difference, it is possible that BRCA2 only plays a role in the earliest kidney forms in mammalian development and may be less effective in controlling podocyte fate than in zebrafish. In opposition to this notion, it is also possible that brca2 is a more potent factor in regulating kidney development than we have reported, as we have not exhaustively characterized all the nephron cell types or each time point critical to kidney development in zep mutants.
Looking forward, given the strengths of the zebrafish system for studying glomerular development and amenability to chemical genetics[90, 91], our brca2 model affords new opportunities to identify modulators of Brca2 that influence mesoderm formation and other tissues during ontogeny. Our findings indicate that brca2 can be considered in a new light, as a participant in aspects of both development and cancer that may have important ramifications and biomedical significance to humans as well.
Acknowledgments
This work was supported in part by National Institutes of Health Grant R01DK100237 to R.A.W., and a National Science Foundation Graduate Research Fellowship DGE-131583 to B.E.D. We thank the Gallagher and Hiller Families for their generous support of stem cell and developmental biology research at the University of Notre Dame, which provided funding to the brca2 research team that included a summer undergraduate research fellowship to our co-author R. Miceli[66]. We are indebted to our lab for useful discussions, the Center for Zebrafish Research at the University of Notre Dame for their care of our vivarium, and the Department of Biological Sciences office staff for their ongoing administrative support. R.A.W. dedicates this work to her mentor, B.H.P., who was a beloved friend and gifted scientist who used zebrafish genetics to make long-lasting contributions to the world.
Abbreviations
- 3β-Hsd
3β-hydroxysteroid dehydrogenase
- P-h2A.X
anti-phospho-H2AX antibody
- brca2
breast cancer 2, early onset
- BRCA1
BRCA DNA repair associated
- BRCA2
BRCA2 DNA repair associated
- cRNA
capped mRNA
- ENU
ethylnitrosourea
- FA
Fanconi anemia
- GBM
glomerular basement membrane
- hpf
hours post fertilization
- nr5a1a
nuclear receptor subfamily 5, group A, member 1a
- IR
interrenal gland
- SNPs
single nucleotide polymorphisms
- vegfaa
vascular endothelial growth factor Aa
- WGS
whole genome sequencing
- WISH
whole mount in situ hybridization
- wt1a
wilms tumor 1a
- wt1b
wilms tumor 1b
- zep
zeppelin.
Footnotes
Conflicting interests
The authors have declared that no conflicts of interest exist.
Author contributions
B.E.D. and R.A.W. wrote and revised the manuscript.
References
- 1.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16:110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 2.Godet I, Gilkes DM. BRCA1 and BRCA2 mutations and treatment strategies for breast cancer. Integr Cancer Sci Ther. 2017;4 doi: 10.15761/ICST.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen EM, Pishvaian MJ. Targeting the BRCA1/2 tumor suppressors. Curr Drug Targets. 2014;15:17–31. doi: 10.2174/1389450114666140106095432. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes WD, Shuen AY. In brief: BRCA1 and BRCA2. J Pathol. 2013;230:347–349. doi: 10.1002/path.4205. [DOI] [PubMed] [Google Scholar]
- 5.Mersch J, Jackson MA, Park M, Nebgen D, Peterson SK, Singletary C, Arun BK, Litton JK. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer. 2015;2:269–275. doi: 10.1002/cncr.29041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2008;668:11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Stecklein SR, Jensen RA. Identifying and exploiting defects in the Fanconi anemia/BRCA pathway in oncology. Trans Res. 2012;160:178–197. doi: 10.1016/j.trsl.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 8.Dong H, Nebert DW, Bruford EA, Thompson DC, Joenje H, Vasiliou V. Update of the human and mouse Fanconi anemia genes. Human Genomics. 2015;9:32. doi: 10.1186/s40246-015-0054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D’Andrea AD, Grompe M. The Fanconi anemia/BRCA pathway. Nat Rev Cancer. 2002;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy RD, D’Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;24:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 11.Auerbach AD. Fanconi anemia and its diagnosis. Mutat Res. 2009;668:4–10. doi: 10.1016/j.mrfmmm.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung RS, Taniguchi T. Recent insights into the molecular basis of Fanconi anemia: genes, modifiers, and drivers. Int J Hematol. 2017;106:335–344. doi: 10.1007/s12185-017-2283-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malric A, Defachelles AS, Leblanc T, Lescoeur B, Lacour B, Peuchmaur M, et al. Fanconi anemia and solid malignancies in childhood: a national retrospective study. Pediatr Blood Cancer. 2014;9999:1–8. doi: 10.1002/pbc.25303. [DOI] [PubMed] [Google Scholar]
- 14.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki A, de la Pompa JL, Hakem R, Elia A, Yoshida R, Mo R, et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 16.Tischkowitz M, Winqvist R. Using mouse models to investigate the biological and physiological consequences of defects in the Fanconi anaemia/breast cancer DNA repair signaling pathway. J Pathol. 2011;224:301–305. doi: 10.1002/path.2903. [DOI] [PubMed] [Google Scholar]
- 17.Lieschke GJ, Currie PD. Animal models of human disease: zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 18.Etchin J, Kanki JP, Look AT. Zebrafish as a model for the study of human cancer. Methods Cell Biol. 2011;105:309–337. doi: 10.1016/B978-0-12-381320-6.00013-8. [DOI] [PubMed] [Google Scholar]
- 19.van Rooijen E, Fazio M, Zon LI. From fish bowl to bedside: The power of zebrafish to unravel melanoma pathogenesis and discover new therapeutics. Pigment Cell Melanoma Res. 2017;30:402–412. doi: 10.1111/pcmr.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idilli AI, Precazzini F, Mione MC, Anelli V. Zebrafish in translational cancer research: insight into leukemia, melanoma, glioma and endocrine tumor biology. Genes (Basel) 2017;8 doi: 10.3390/genes8090236. pii:E236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titus TA, Selvig DR, Qin B, Wilson C, Starks AM, Roe BA, et al. The Fanconi anemia gene network is conserved from zebrafish to human. Gene. 2006;371:211–223. doi: 10.1016/j.gene.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 22.Titus TA, Yan YL, Wilson C, Starks AM, Frohnmayer JD, Bremiller RA, et al. The Fanconi anemia/BRCA gene network in zebrafish: embryonic expression and comparative genomics. Mutat Res. 2009;668:117–132. doi: 10.1016/j.mrfmmm.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 24.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chao HT, Liu L, Bellen HJ. Building dialogues between clinical and biomedical research through cross-species collaborations. Semin Cell Dev Biol. 2017;70:49–57. doi: 10.1016/j.semcdb.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond BE, Wingert RA. Insights into kidney stem cell development using zebrafish. World J Stem Cells. 2016;8:22–31. doi: 10.4252/wjsc.v8.i2.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drummond IA, Majumdar A, Hentschel H, Elger M, Solnica-Krezel L, Schier AF, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 28.Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:559–585. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poureetezadi SJ, Wingert RA. Little fish, big catch: zebrafish as a model of kidney disease. Kidney Int. 2016;89:1204–1210. doi: 10.1016/j.kint.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jerman S, Sun Z. Using zebrafish to study kidney development and disease. Curr Top Dev Biol. 2017;124:43–79. doi: 10.1016/bs.ctdb.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Schenk H, Müller-Deile J, Kinast M, Schiffer M. Disease modeling in genetic kidney diseases: zebrafish. Cell Tissue Res. 2017;369:127–141. doi: 10.1007/s00441-017-2593-0. [DOI] [PubMed] [Google Scholar]
- 32.Naylor RW, Przepiorski A, Ren Q, Yu J, Davidson AJ. HNF1B is essential for nephron segmentation during nephrogenesis. J Am Soc Nephrol. 2013;24:77–87. doi: 10.1681/ASN.2012070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerlach GF, Wingert RA. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota and zeta. Dev Biol. 2014;396:183–200. doi: 10.1016/j.ydbio.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKee R, Gerlach GF, Jou J, Cheng CN, Wingert RA. Temporal and spatial expression of tight junction genes during zebrafish pronephros development. Gene Expr Patterns. 2014;16:104–113. doi: 10.1016/j.gep.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marra AN, Wingert RA. Roles of Iroquois transcription factors in kidney development. Cell Dev Biol. 2014;3:131. doi: 10.4172/2168-9296.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng CN, Wingert RA. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish. Dev Biol. 2015;399:100–116. doi: 10.1016/j.ydbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ott E, Wendik B, Srivastava M, Pacho F, Töchterle S, Salvenmoser W, Meyer D. Pronephric tubule morphogenesis in zebrafish depends on Mnx mediated repression of irx1b within the intermediate mesoderm. Dev Biol. 2016;411:101–14. doi: 10.1016/j.ydbio.2015.10.014. [DOI] [PubMed] [Google Scholar]
- 38.Marra AN, Wingert RA. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev Biol. 2016;411:231–245. doi: 10.1016/j.ydbio.2016.01.035. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naylor RW, Dodd RC, Davidson AJ. Caudal migration and proliferation of renal progenitors regulates early nephron segment size in zebrafish. Sci Rep. 2016;19:35647. doi: 10.1038/srep35647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poureetezadi SJ, Cheng CN, Chambers JM, Drummond BE, Wingert RA. Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. Elife. 2016;5 doi: 10.7554/eLife.17551. pii:e17551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond BE, Li Y, Marra AN, Cheng CN, Wingert RA. The tbx2a/b transcription factors direct pronephros segmentation and corpuscle of Stannius formation in zebrafish. Dev Biol. 2017;421:52–66. doi: 10.1016/j.ydbio.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naylor RW, Han HI, Hukriede NA, Davidson AJ. Dev Biol. Wnt8a expands the pool of embryonic kidney progenitors in zebrafish. 2017;425:130–141. doi: 10.1016/j.ydbio.2017.03.027. [DOI] [PubMed] [Google Scholar]
- 43.Kuechlin S, Schoels M, Slanchev K, Lassmann S, Walz G, Yakulov TA. EpCAM controls morphogenetic programs during zebrafish pronephros development. Biochem Biophys Res Commun. 2017;487:209–215. doi: 10.1016/j.bbrc.2017.04.035. [DOI] [PubMed] [Google Scholar]
- 44.Vasilyev A, Drummond IA. Live imaging kidney development in zebrafish. Methods Mol Biol. 2012;886:55–70. doi: 10.1007/978-1-61779-851-1_6. [DOI] [PubMed] [Google Scholar]
- 45.Drummond IA, Davidson AJ. Zebrafish kidney development. Methods Cell Biol. 2016;134:391–429. doi: 10.1016/bs.mcb.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 46.Perner B, Englert C. Immunofluorescence staining of Wt1 on sections of zebrafish embryos and larvae. Methods Mol Biol. 2016;1467:129–132. doi: 10.1007/978-1-4939-4023-3_11. [DOI] [PubMed] [Google Scholar]
- 47.Perner B, Schnerwitzki D, Graf M, Englert C. Analysis of zebrafish Kidney Development with time-lapse imaging using a dissecting microscope equipped for optical sectioning. J Vis Exp. 2016;110:e53921. doi: 10.3791/53921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marra AN, Ulrich M, White A, Springer M, Wingert RA. Visualizing multiciliated cells in the zebrafish through a combined protocol of whole mount fluorescent in situ hybridization and immunofluorescence. J Vis Exp. 2017;129:e56261. doi: 10.3791/56261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McMahon AP. Development of the mammalian kidney. Curr Top Dev Biol. 2016;117:31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wingert RA, Davidson AJ. The zebrafish pronephros: a model to study nephron segmentation. Kid Int. 2008;73:1120–1127. doi: 10.1038/ki.2008.37. [DOI] [PubMed] [Google Scholar]
- 51.Quaggin SE, Kreidberg JA. Development of the renal glomerulus: good neighbors and good fences. Development. 2008;135:609–620. doi: 10.1242/dev.001081. [DOI] [PubMed] [Google Scholar]
- 52.Scott RP, Quaggin SE. Review series: The cell biology of renal filtration. J Cell Biol. 2015;209:199–210. doi: 10.1083/jcb.201410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reiser J, Sever S. Podocyte biology and pathogenesis of kidney disease. Annu Rev Med. 2013;1:357–366. doi: 10.1146/annurev-med-050311-163340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagata M. Podocyte injury and its consequences. Kidney Int. 2016;89:1221–1230. doi: 10.1016/j.kint.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 55.Kroeger PT, Wingert RA. Using zebrafish to study podocyte genesis during kidney development and regeneration. Genesis. 2014;9:771–792. doi: 10.1002/dvg.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bollig F, Mehringer R, Perner B, Hartung C, Schafer M, Schartl M, Volff JN, Winkler C, Englert C. Identification and comparative expression analysis of a second wt1 gene in zebrafish. Dev Dyn. 2006;235:554–561. doi: 10.1002/dvdy.20645. [DOI] [PubMed] [Google Scholar]
- 57.Hsu HJ, Lin G, Chung BC. Parallel early development of zebrafish interrenal glands and pronephros: Differential control by wt1 and ff1b. Development. 2003;130:2107–2116. doi: 10.1242/dev.00427. [DOI] [PubMed] [Google Scholar]
- 58.Majumdar A, Drummond IA. Podocyte differentiation in the absence of endothelial cells as revealed in the zebrafish avascular mutant, cloche. Dev Genet. 1999;24:220–229. doi: 10.1002/(SICI)1520-6408(1999)24:3/4<220::AID-DVG5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 59.Majumdar A, Drummond IA. The zebrafish floating head mutant demonstrates podocytes play an important role in directing glomerular differentiation. Dev Biol. 2000;222:147–157. doi: 10.1006/dbio.2000.9642. [DOI] [PubMed] [Google Scholar]
- 60.Majumdar A, Lun K, Brand M, Drummond IA. Zebrafish no isthmus reveals a role for pax2.1 in tubule differentiation and patterning events in the pronephric primordia. Development. 2000;127:2089–2098. doi: 10.1242/dev.127.10.2089. [DOI] [PubMed] [Google Scholar]
- 61.Serluca FC, Drummond IA, Fishman MC. Endothelial signaling in kidney morphogenesis: A role for hemodynamic forces. Curr Biol. 2002;12:492–497. doi: 10.1016/s0960-9822(02)00694-2. [DOI] [PubMed] [Google Scholar]
- 62.Zhu X, Chen Z, Zeng C, Wang L, Xu F, Hou Q, et al. Ultrastructural characterization of the pronephric glomerulus development in zebrafish. J Morphol. 2016;277:1104–1112. doi: 10.1002/jmor.20560. [DOI] [PubMed] [Google Scholar]
- 63.Diep CQ, Peng Z, Ukah TK, Kelly PM, Daigle RV, Davidson AJ. Development of the zebrafish mesonephros. Genesis. 2015;53:257–269. doi: 10.1002/dvg.22846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Miceli R, Kroeger PT, Jr, Wingert RA. Molecular mechanisms of podocyte development revealed by zebrafish kidney research. Cell Dev Biol. 2014;3:138. doi: 10.4172/2168-9296.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wingert RA, Davidson AJ. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev Dyn. 2011;240:2011–2027. doi: 10.1002/dvdy.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kroeger PT, Drummond BE, Miceli R, McKernan M, Gerlach GF, Marra AN, et al. The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev Biol. 2017;428:148–163. doi: 10.1016/j.ydbio.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu YW. Interrenal organogenesis in the zebrafish model. Organogenesis. 2007;3:44–48. doi: 10.4161/org.3.1.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Brien LL, Grimaldi M, Kostun Z, Wingert RA, Selleck R, Davidson AJ. Wt1a, Foxc1a, and the Notch mediator Rbpj physically interact and regulate the formation of podocytes in zebrafish. Dev Biol. 2011;2:318–330. doi: 10.1016/j.ydbio.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leshchiner I, Alexa K, Kelsey P, Adzhubei I, Austin-Tse CA, Cooney JD, et al. Mutation mapping and identification by whole-genome sequencing. Genome Res. 2002;22:1541–1548. doi: 10.1101/gr.135541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ryan S, Willer J, Marjoram L, Bagwell J, Mankiewicz J, Leshchiner I, et al. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development. 2013;140:4445–4451. doi: 10.1242/dev.101170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shive HR, West RR, Embree LJ, Azuma M, Sood R, Liu P, et al. brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:19350–19355. doi: 10.1073/pnas.1011630107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodríguez-Marí A, Wilson C, Titus TA, Cañestro C, BreMiller RA, Yan YL, et al. Roles of brca2 (fancd1) in oocyte nuclear architecture, gametogenesis, gonad tumors, and genome stability in zebrafish. PLoS Genet. 2011;7:e1001357. doi: 10.1371/journal.pgen.1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodríguez-Marí A, Postlethwait JH. The role of Fanconi anemia/BRCA genes in zebrafish sex determination. Methods Cell Biol. 2011;105:461–490. doi: 10.1016/B978-0-12-381320-6.00020-5. [DOI] [PubMed] [Google Scholar]
- 74.Shive HR, West RR, Embree LJ, Golden CD, Hickstein DD. BRCA2 and TP53 collaborate in tumorigenesis in zebrafish. PLoS One. 2014;9:e87177. doi: 10.1371/journal.pone.0087177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wingert RA, Selleck R, Yu J, Song HD, Chen Z, Song A, et al. cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;10:1922–1938. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bollig F, Perner B, Besenbeck B, Köthe S, Ebert C, Taudien S, et al. A highly conserved retinoic acid responsive element controls wt1a expression in the zebrafish pronephros. Development. 2009;136:2883–2892. doi: 10.1242/dev.031773. [DOI] [PubMed] [Google Scholar]
- 77.White JT, Zhang B, Cerqueira DM, Tran U, Wessely O. Notch signaling, wt1 and foxc2 are key regulators of the podocyte gene regulatory network in Xenopus. Development. 2010;137:1863–1873. doi: 10.1242/dev.042887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Botthof JG, Bielczyk-Maczyńska E, Ferreira L, Cvejic A. Loss of the homologous recombination gene rad51 leads to Fanconi anemia-like symptoms in zebrafish. Proc Natl Acad Sci U S A. 2017;22:E4461. doi: 10.1073/pnas.1620631114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Chadarévian JP, Vekemans M, Bernstein M. Fanconi’s anemia, medulloblastoma, Wilms’ tumor, horseshoe kidney, and gonadal dysgenesis. Arch Pathol Lab Med. 1985;109:367–369. [PubMed] [Google Scholar]
- 80.Bissig H, Staehelin F, Tolnay M, Avoledo P, Richter J, Betts D, Bruder E, Kühne T. Co-occurrence of neuroblastoma and nephroblastoma in an infant with Fanconi’s anemia. Hum Pathol. 2002;33:1047–1051. doi: 10.1053/hupa.2002.128062. [DOI] [PubMed] [Google Scholar]
- 81.Reid S, Renwick A, Seal S, Baskcomb L, Barfoot R, Jayatilake H, et al. Biallelic BRCA2 mutations are associated with multiple malignancies in childhood including familial Wilms tumour. J Med Genet. 2005;42:147–151. doi: 10.1136/jmg.2004.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Berrebi D, Lebras MN, Belarbi N, Couturier J, Fattet S, Faye A, Peuchmaur M, de Lagausie P. Bilateral adrenal neuroblastoma and nephroblastoma occurring synchronously in a child with Fanconi’s anemia and VACTERL syndrome. J Pediatr Surg. 2006;41:e11–14. doi: 10.1016/j.jpedsurg.2005.10.087. [DOI] [PubMed] [Google Scholar]
- 83.Sari N, Akyuz C, Aktas D, Gumruk F, Orhan D, Alikasifoglu M, Aydin B, Alanay Y, Buyukpamukcu M. Wilms tumor, AML and medulloblastoma in a child with cancer prone syndrome of total premature chromatid separation and Fanconi anemia. Pediatr Blood Cancer. 2009;53:208–210. doi: 10.1002/pbc.21966. [DOI] [PubMed] [Google Scholar]
- 84.Compostella A, Toffolutti T, Soloni P, Dall’Igna P, Carli M, Bisogno G. Multiple synchronous tumors in a child with Fanconi anemia. J Pediatr Surg. 2010;45:e5–8. doi: 10.1016/j.jpedsurg.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 85.Rizk T, Taslakian B, Torbey PH, Issa G, Hourani R. Sequential development of Wilms tumor and medulloblastoma in a child: an unusual presentation of fanconi anemia. Pediatr Hematol Oncol. 2013;30:400–402. doi: 10.3109/08880018.2013.788593. [DOI] [PubMed] [Google Scholar]
- 86.Trejo Bittar HE, Radder JE, Ranganathan S, Srinivasan A, Madan-Khetarpal S, Reyes-Múgica M. Clear cell sarcoma of the kidney in a child with Fanconi anemia. Pediatr Dev Pathol. 2014;17:297–301. doi: 10.2350/14-03-1450-CR.1. [DOI] [PubMed] [Google Scholar]
- 87.El Ghorayeb N, Grunenwald S, Nolet S, Primeau V, Côté S, Maugard CM, Lacroix A, Gaboury L, Bourdeau I. First case report of an adrenocortical carcinoma caused by a BRCA2 mutation. Medicine (Baltimore) 2016;95:e4756. doi: 10.1097/MD.0000000000004756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saxen L. Organogenesis of the kidney. Cambridge: Cambridge University Press; 1987. [Google Scholar]
- 89.Romagnani P, Lasagni L, Remuzzi G. Renal progenitors: an evolutionary conserved strategy for kidney regeneration. Nat Rev Nephrol. 2013;9:137–146. doi: 10.1038/nrneph.2012.290. [DOI] [PubMed] [Google Scholar]
- 90.Hanke N, Staggs L, Schroder P, Litteral J, Fleig S, Kaufeld J, Pauli C, Haller H, Schiffer M. “Zebrafishing” for novel genes relevant to the glomerular filtration barrier. Biomed Res Int. 2013;2013:658270. doi: 10.1155/2013/658270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.MacRae CA, Peterson RT. Zebrafish as tools for drug discovery. Nat Rev Drug Discov. 2015;10:721–731. doi: 10.1038/nrd4627. [DOI] [PubMed] [Google Scholar]


