Genetically identical bacteria possess varying numbers of surface-adhering patches.
Abstract
Understanding and controlling the surface adhesion of pathogenic bacteria is of urgent biomedical importance. However, many aspects of this process remain unclear (for example, microscopic details of the initial adhesion and possible variations between individual cells). Using a new high-throughput method, we identify and follow many single cells within a clonal population of Escherichia coli near a glass surface. We find strong phenotypic heterogeneities: A fraction of the cells remain in the free (planktonic) state, whereas others adhere with an adhesion strength that itself exhibits phenotypic heterogeneity. We explain our observations using a patchy colloid model; cells bind with localized, adhesive patches, and the strength of adhesion is determined by the number of patches: Nonadherers have no patches, weak adherers bind with a single patch only, and strong adherers bind via a single or multiple patches. We discuss possible implications of our results for controlling bacterial adhesion in biomedical and other applications.
INTRODUCTION
Bacterial colonization of myriad niches, both natural and man-made, begins with adhesion to surfaces (1, 2). The colonization of man-made surfaces (catheters, surgical implants, etc.) causes infection (3) because adhering biofilms resist physical and chemical assaults (4), and it contributes to the emergence of antimicrobial resistance (5). Understanding and minimizing bacterial adhesion is a cross-disciplinary “grand challenge” (6), in which it is key to understand the forces involved. Besides generic electrostatics and dispersion forces (7), there are also forces of biological origin associated with various adhesins and sticky organelles (for example, fimbriae) (6, 8–10).
Biophysical studies of bacterial adhesion typically fall into two categories (11): high-throughput, population-level work giving little single-cell information or low-throughput single-cell studies. We report a robust, generic, and high-throughput tracking and big-data analysis technique that reveals detailed single-cell information in a large population. Applying this technique to Escherichia coli on glass, we find substantial variability in the propensity for adhesion and the postadhesion dynamics. Some cells do not adhere despite repeated encounters, whereas others adhere rapidly upon contact. Among the adherers, a fraction pivot freely around their own attachment points and can be removed from the surface by gravity, whereas the remainder spend part of their time in a more strongly bound, rotationally constrained state. This variability remains in flagella- and fimbriae-deletion mutants.
From the biological perspective, such variability exemplifies phenotypic heterogeneity in a clonal population (12) and offers a new model for its study (13, 14). Given the variety of surfaces that bacteria may encounter, such adhesive phenotypic heterogeneity may be an instance of “bet hedging” (15), allowing survival when the environment changes unpredictably (for example, following fecal excretion for Enterobacteriaceae).
In soft matter terms, genetically monodisperse E. coli cells are adhesively polydisperse and behave as “living patchy colloids” bearing a variable number of sticky patches on their cell bodies. Thus, the considerable knowledge about patchy colloids accumulated over the past decade (16–18) can be deployed in the design of abiotic surfaces to minimize bacterial adhesion. Bacterial adhesion to engineered patchy soft surfaces has been studied before (19), but the possibility of patchiness on the bacteria themselves has not been considered in much detail.
We studied E. coli strain AB1157. Like all K-12 derivatives, it has mutations in the rfb gene cluster [here, in rfbD (20)] preventing O-antigen production (21). Thus, one major cause of variability and molecular roughness on the cell surface is absent. Our cells display a highly conserved layer of core oligosaccharides anchored to the outer membrane by lipid A (22), although the precise terminal sugars in AB1157 are unknown. Besides the wild-type (WT), to further simplify the surface, we also used a mutant (AD19 = AB1157 fimA−, fliF−; hereafter ΔFF) defective in producing type 1 fimbriae and flagella. In transmission electron micrographs (fig. S1), these cells appear smooth down to ≲ 10 nm. Any heterogeneities, or “patchiness,” on the surface of this mutant are likely due to membrane proteins.
RESULTS
A high-throughput method reveals complex adhesion behavior
To monitor surface adhesion of E. coli, we loaded cell suspensions into 400-μm-high borosilicate glass capillaries at ~ 4.5 × 107 cells/ml (WT) or ~ 1.5 × 107 cells/ml (ΔFF) in phosphate motility buffer (MB) and observed them in an inverted microscope at ca. 22°C using a 60× phase-contrast objective (focal depth ~ 3.5 μm) (Fig. 1A). From measurements at the lower surface at the start of the experiment, we estimate an initial motile fraction between 25 and 35% [nonadhering motile cells swim at an average speed of ≲ 20 μm s−1 (23)]; the remaining cells are nonmotile and diffuse as passive colloids. With 0.72 μM glucose included in the MB, the swimmers maintain their speed for at least 20 hours (23). Atomic force microscopy (AFM) measurements reveal that the glass surface is smooth, with a root-mean-squared roughness of 0.25 ± 0.003 nm (fig. S2).
Fig. 1. Tracking and classification of bacteria.
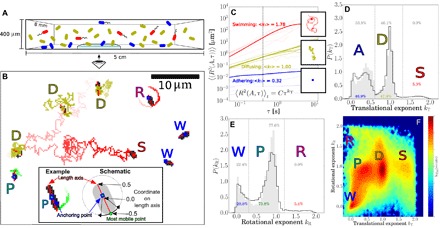
(A) Schematic of adhering (blue), diffusing (olive), and swimming (red) bacteria in a capillary. (B) Trajectories tracing the most mobile point on a cell. Straight red lines are fits of the length axis, squares mark anchoring points, and letters mark the type: diffuser (D), swimmer (S), wobbler (W), pivoter (P), and active rotator (R). The image is black/white-inverted and thresholded to be clear in print. Inset: Adhering cell and schematic depicting the anchoring coordinate and most mobile coordinate ∈ [−0.5, 0.5] on its length axis. (C) MSDs as a function of time interval τ from trajectories of swimming (red), diffusing (olive), and adhering (blue) bacteria; thin lines show randomly selected individual trajectories and thick lines represent the average 〈R2(A, τ)〉i,t of each category. The insets show an example trajectory for each category; squares denote the position in the last frame. (D) Normalized distribution of the translational exponent kT for WT cells on glass during the first 2 hours of the experiment, showing distinct peaks for adhering (A), diffusing (D), and swimming (S) subpopulations. (E) Normalized distribution of the rotational exponent kR for adhering cells, showing three peaks for wobblers (W), pivoters (P), and active rotators (R). Data in in panels D and E for the nonflagellated mutant ΔFF are shown in gray. (F) Two-dimensional histogram (logarithmic scaling) of the translational (kT) and rotational (kR) exponents for WT cells, showing three peaks for adherers (W, P, and R), one for diffusers (D), and one for swimmers (S). The distributions in panels D to F are based on 139,335 cells for WT and 10,582 cells for ΔFF, and data are weighted for trajectory durations.
Time-lapse images of the bottom and top glass surfaces were taken automatically at multiple locations to track arriving cells and their subsequent fate. We adapted algorithms used for colloidal rods (24) to determine the position r, orientation θ, and projected length lP of each cell (movie S1) and track these in time. Typical analyzed trajectories are shown in Fig. 1B and movie S2. Quantifying these trajectories allows us to classify cells’ adhesive phenotypes.
Focusing first on translation, we identified for each cell the point of least motion along its length axis (fig. S3), corresponding roughly to the center of attachment for adhered cells and to the hydrodynamic center for free cells, and determined its mean-squared displacement (MSD) = 〈(r(t + τ) − r(t))2〉. At short delay time (τ ≤ 0.4 s), all cells show , with a range of {CT, kT} (Fig. 1C), exhibiting constrained, diffusive, or ballistic (directed) motion with values peaked around kT = 0, 1, or 2, respectively (25, 26).
The probability distribution of kT, P(kT), shows three well-separated peaks (Fig. 1D), classifying cells into adherers (A), diffusers (D), or swimmers (S). Cells with kT ≲ 0.6 and kT ≳ 1.3 correspond to adherers and swimmers, respectively. Diffusers show 0.6 ≲ kT ≲ 1.3; the spread and skewness of the distribution in this range come from the finite, stochastic trajectories used in tracking.
The initial analysis misidentified some diffusers as adherers and vice versa, for example, because of the width of the distributions and a few adherers that pivot around their poles and swing out of plane, showing up as rapid short-time motion. To resolve this issue, we further required that adherers be visible for at least 6 s. Trajectories with kT ≲ 0.6 and shorter than 6 s were labeled as ambiguous. For trajectories greater than 6 s, we also calculated the translational exponents for the longer delay time of τ = 4 s; we then used this exponent, , as a cell’s translational exponent if both and and identified the trajectory on this basis.
Visual inspection of trajectories (movies S3 to S7) revealed correct automated classification of ≳ 95 % of all cells using this algorithm. The shallow minimum in P(kT) demarcating the D and S subpopulations coincides with the cutoff in P(kT) measured for the ΔFF mutant (Fig. 1E, gray), which does not swim.
Similarly, we fitted the mean-squared orientational displacement (MSOD) for τ < 0.4 s. The probability distribution of kR, P(kR), for WT adherers shows two peaks falling off to a long “tail” at kR ~ 2 (Fig. 1E). We interpret kR ~ 0 as corresponding to adhering cells that “wobble” (W) around a fixed orientation; kR ~ 1 as corresponding to cells that “pivot” (P) around a fixed attachment point and undergo apparently free rotational diffusion; and kR ~ 2 as corresponding to cells that “rotate ballistically” (R).
Only adherers show three rotational modes. Diffusing and swimming cells both appear only as a single mode in kR. This pattern is clear in the two-dimensional heat map of {kT, kR} (Fig. 1F), which shows three features for adherers (W, P, and R), one for diffusers (D), and one for swimmers (S). Of the three different adhering modes, the active rotators observed for WT strains were not found in ΔFF cells, which is reflected by the missing tail at kR > 1.3 for ΔFF (gray area in Fig. 1E). We attribute ballistic rotation to the presence of active flagella (27), which we further verified using a strain modified to permit flagella staining (movie S8) (28). We observed that most rotating cells were found adhering with a point on or close to the body. From the movies, it appears that active rotation originated from direct adhesion by a rotating motor or short filament or, indirectly, by freely moving flagella rotating an otherwise adhering cell. We also observed cells “tethered” by a flagellum with the cell body not attached to the surface, but these occurrences were rare. Flagella-mediated adhesion is implicated in biofilm formation and pathogenicity (29, 30). It may be electrostatically mediated (31–33), so that its rarity (≤ 6 % of cells; cf. Fig. 1E) under our high-ionicity conditions (Debye screening length κD−1 ≈ 1 nm) is perhaps unsurprising. Hereafter, we mostly neglect active rotators and focus on wobblers and pivoters.
The pivoting state is particularly intriguing: The diffusive angular motion implies that the cell is attached at a single, freely rotating locus. We observed many cells pivoting more than 2π within a single movie. Virtually no pivoters were observed in the adhesion of sterically stabilized synthetic bacterium-shaped hollow silica particles (fig. S1) (34), so that such free rotation is not a generic feature of colloidal rods attached to surfaces. Neither is it due to surface organelles, because pivoters were well represented in WT and ΔFF strains and in further mutants ΔFH and ΔFFH, which additionally lacked the hook protein (fig. S4). We later examine this pivoting behavior in more detail.
In sum, tracking the translational and rotational motion of E. coli at a borosilicate glass surface has revealed a variety of adhesive behavior: Some cells adhere, others do not; of the adherers, a small number rotate ballistically, whereas most either wobble or undergo pivoted rotational diffusion. The question naturally arises: Is this variety due to cells switching between different modes of behavior within our observational time scale or due to the presence of different kinds of cells? In statistical physics terms, is the “disorder” (presence of multiple modes of behavior) annealed or quenched? To answer this question, we turn our attention to the adhesion dynamics.
Adhesive propensity is phenotypically heterogeneous
We followed the arrival of cells on the lower capillary surface from the bulk (shown schematically in Fig. 2A) and observed the buildup of the different subpopulations to a steady state (Fig. 2B). The time between sealing the capillary and beginning observations (≲ 10 min) was long enough for the near-surface swimmers to achieve dynamic equilibrium with the bulk (35), so that we observed a roughly constant number of near-surface swimmers. Meanwhile, the number of adherers and diffusers both increased steadily before saturating after ~ 2 hours, which is consistent with nonswimmers sedimenting through the h = 400 μm capillary at an independently measured speed of vs = 0.06 ± 0.01 μm s−1. There was no detectable delay between the arrival of the first diffusers and the first adherers (Fig. 2B), so that adhesion is rapid on the scale of 5 min. Repeating with ΔFF mutants gave qualitatively similar dynamics except for the absence of swimmers and rotators (Fig. 2C). From this, we infer that flagella and fimbriae are not essential to the observed WT phenomenology.
Fig. 2. Capillary inversion assay.
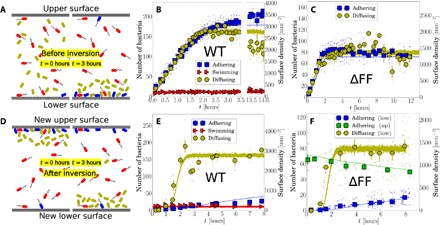
(A) Schematic of adhering (blue), diffusing (olive), and swimming (red) cells in the capillary at loading (0 hours) and 3 hours after, when nonmotile cells have sedimented, and motile cells have reached the top and bottom surfaces. (B) Number of adherers, diffusers, and swimmers on the lower surface as a function of time for WT and (C) for ΔFF. (D) Schematic immediately (0 hours) and 3 hours after capillary inversion. (E) Number of cells on the lower surface after inversion for WT and (F) for ΔFF, where the number of adhering cells on the upper surface is also given. Small points are average values for single movies, and large points are weighted averages over groups of multiple movies, with vertical bars as the SE and horizontal bars as the time window for each point. The lines in panels B, C, E, and F are results from simulations with a minimal kinetic model.
Figure 2B shows that more than half of the population does not adhere to the surface and that this fraction stays approximately constant over 14 hours. We will argue that this is primarily due to inherent, phenotypic heterogeneity between the cells. However, we must first exclude the alternative possibility that the incomplete adhesion is maintained by a dynamic equilibrium between surface binding and unbinding of otherwise identical cells. For this, we use an inversion assay (36), in which we inverted the capillary and observed that nonadhering diffusing cells sediment toward the new lower surface, which, at this point, is virtually empty (Fig. 2D, left panel). When the sedimenting cells reach the new lower surface around 1 to 2 hours after inversion (Fig. 2, E and F), almost none of them adhere. This is inconsistent with a dynamic equilibrium between otherwise identical cells, for which we would expect the same ratio of diffusers to adherers on the lower surface before and after inversion. Therefore, we reason that there are differences between the adhesive properties of individual cells. In statistical physics terms, the disorder in adhesive propensity is quenched. This inversion assay also rules out a third possibility, that the number of adherers is limited by the availability of binding sites on the glass surface. In this unobserved scenario, we would expect phenotypically sticky cells, which were unable to find a binding site before inversion, to adhere post-inversion to the new, empty lower surface.
Over longer time scales, there is a slight increase in the number of adhering cells on the new lower surface for both ΔFF and WT cells. This increase is due to previously adhering cells detaching slowly from the new upper surface and sedimenting downward and reattaching to the new lower surface. We can show this by monitoring the decay in adherer numbers remaining on the (new) upper surface (green squares in Fig. 2F). Fitting the adherer number N to N(t) = N(0)e− γt returns a detachment rate of γ ≈ 0.032 hour−1. After the time it takes for detached adherers to sediment through the capillary (less than 2 hours), all of these cells appear at and re-adhere to the (new) lower surface, with the rate of this attachment being approximately equal to the detachment rate, as one would expect from our picture. This shows that the adhesion properties of individual cells are preserved in time: Cells that were previously adhering on the (new) upper surface, detach, and sediment downward are capable of reattaching on the (new) lower surface.
Postadhesion dynamics are also phenotypically heterogeneous
We next focus on adhering cells and distinguish between wobblers and pivoters on the basis of their MSOD. We report data for ΔFF mutants, for which there are no active rotators. As cells arrived on the lower capillary surface, the number of wobblers and pivoters increased linearly before saturating, showing a constant P/W ratio of 3:1 throughout (Fig. 3A). After capillary inversion, the dynamics of these two subpopulations diverged: Pivoters left the (new) upper surface at least an order of magnitude faster than wobblers, whose number remained almost constant (Fig. 3B). Thus, wobblers appear more tightly bound.
Fig. 3. Polydispersity in adhering cells for ΔFF.
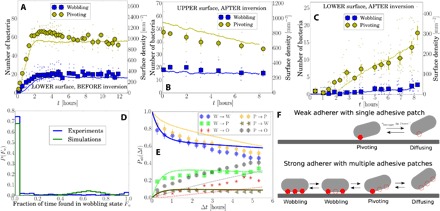
(A) Number of wobbling and pivoting cells on the lower surface before capillary inversion and (B and C) on the upper and lower surface after capillary inversion. Solid lines in panels A to C are from simulations with a minimal kinetic model. Data correspond to the inversion experiments in Fig. 2 (C and F). (D) Histogram showing the fraction of time cells are identified as wobbling on the lower surface before inversion, as in experiments (from a total of 117 movies over six different positions recorded between 4 and 12 hours before capillary inversion) and simulations. (E) Probabilities for a cell in a wobbling (W) or pivoting (P) state at time t to be found wobbling, pivoting, or off the surface (O) at a later time t + Δt (lines are for the kinetic model). (F) Schematic of weakly and strongly adhering cells: filled red circle, bound adhesive patch; empty circles, unbound adhesive patch.
The rate of arrival of pivoters on the (new) lower surface (Fig. 3C) equaled the rate of their disappearance from the (new) upper surface (Fig. 3B), with a delay in arrival consistent with sedimenting through 400 μm. The buildup of wobblers on the (new) lower surface commenced ≳ 1 hour after the arrival of the first pivoters, suggesting that pivoters may become wobblers. To confirm this, we monitored adherers on the lower surface before inversion over 8 hours in the steady state and measured the fraction of time Fw, each cell spent as a wobbler (Fig. 3D): ~ 70% of cells were always pivoting, whereas the remainder switched between wobbling and pivoting.
The data in Fig. 3D are not consistent with a single population of adherers stochastically switching between wobbling and pivoting states, for which we expect a broad distribution peaked around some intermediate Fw. The sharp peak at Fw = 0 (always pivoting) implies at least two populations: a stochastically switching fraction and a pure pivoting fraction.
The interstate switching dynamics can be quantified (Fig. 3E) by a time-dependent transition matrix, Pab(Δt), giving the probability of a cell in state “a” at time t being in state “b” at time t + Δt, where {a} = {P, W} and {b} = {P, W, O}, with P, W, and O standing for pivoting, wobbling, and off, respectively, with the latter denoting a cell detaching from the surface altogether. There is significant P ↔ W switching on a time scale of several hours. However, some cells are mostly in either the P state or the W state: Only ~ 10 % of originally P cells become W cells and ~ 30 % of originally W cells become P cells. The discernible rate of W → O transitions explains the slow buildup of wobblers on the (new) lower surface after inversion (Fig. 3C). Direct visual analysis of the trajectories confirms that W → O transitions occurred via the sequence W ↔ P → O. This implies that wobbling cells detaching from the upper surface first become pivoters before they detach and that they reattach as pivoters before becoming wobblers. On the lower surface before inversion, detachments and reattachments of cells occurred at a steady rate over time scales of hours. Detached cells were rarely observed to reattach at the same site but almost always at alternative locations on the surface (fig. S5 and movie S9), which again demonstrates that adhesion is not limited by a low number of binding sites on the glass substrate.
In sum, we conclude that adhering cells are also phenotypically heterogeneous and can either be purely pivoting or switch between pivoting and wobbling states. Moreover, a pivoting cell may relatively easily detach from the surface, whereas wobbling cells can only do so by first becoming pivoters.
A patchy-colloid model explains E. coli adhesion dynamics
In a simple model consistent with our observations (Fig. 3), cells may have 0, 1, or ≥ 2 adhesive patches available for simultaneous surface binding. Cells without patches are permanent nonadherers. Cells with 1 patch are “pure pivoters”—such a cell bound to a surface using its single adhesive patch may detach stochastically because of thermal motion. Cells with ≥ 2 patches may be actually bound to a surface at ≥ 2, 1, or 0 of these patches and appear as a wobbler, pivoter, or a free diffuser. Note that our ΔFF mutant data show that these patches are not necessarily (if at all) associated with flagella or fimbriae. Moreover, in our model, cells with at least one available adhesive patch will eventually encounter the surface with this patch because of rotational diffusion. This mimics reality, where the rotational self-diffusion time in bulk τself,rot ≈ (2π)2/4Drot ≈ 1 min for a free cell [using typical measured values for Drot for deflaggelated E. coli (37)], so that it can explore many different orientations during an experiment.
The observed rate at which pivoters leave the upper capillary surface after inversion (γ ≈ 0.032 hour−1; Fig. 3B) allows us to estimate the adhesive strength of a single patch. A particle confined to a potential of depth ΔU escapes at rate (38)
| (1) |
Here, kB is Boltzmann’s constant, T is the temperature, and D is the particle’s diffusivity inside a potential well of stiffness κS. Dimensional analysis suggests that κS ≈ ΔU/L2, where L is the interaction range. From AFM, we estimate 20 ≲ L ≲ 100 nm for bacteria adhering to a variety of surfaces (39, 40). Separately, from the typical maximum MSD of the anchoring coordinate of pivoting cells, we estimate L ≲ 200 nm (fig. S6). ΔU is rather insensitive to the exact value used. Taking a conservatively broad range of 5 nm < L < 150 nm and D = 0.4 μm2 s−1 (41), we obtain 15 kBT < ΔU < 23 kBT. This is well within the range of, for example, electrostatic and hydration effects (42) and hydrophobic interactions (43) between glass and proteins (44) or lipopolysaccharides (45). We note that if electrostatic attractions were to play a role in adhesion, these are likely very local because of the smallness of the Debye screening length (κD−1 ≈ 1 nm) and the fact that the bacterial surface bears a net negative charge (zeta potential = − 16 mV measured for our WT strain). The latter observation means that any electrostatic attraction with a negative glass surface (33) must be due to localized minority positive charges on the bacterium arising from, for example, amine and hydroxyl groups at around neutral pH (9).
In a previous work where an inversion assay was used (36), it was suggested that gravitational pull might contribute to the detachment of cells from the upper surface (after inversion). The potential energy difference due to gravity for displacing a cell over a distance comparable to the interaction range, Ug ≈ 0.05 kBT, is much smaller than our estimated binding energy of an adhesive patch and therefore has a negligible effect on the detachment rate. Nonetheless, gravity causes a more subtle difference between the adhesion dynamics on the lower and upper surfaces. Once a cell on the upper surface detaches, it immediately starts sedimenting downward (away from the surface), making it highly improbable that it will reattach. On the lower surface, detaching cells remain close to the same surface and reattach to it eventually.
To deduce the likely maximum number of adhesive patches on our cells available for simultaneous surface binding, we consider the statistics of the point of minimum translation (“anchoring point,” − 0.5 ≤ A ≤ 0.5; Fig. 1B, inset). The distribution of anchoring points for wobblers (≥ 2 patches) is given by some weighted average over the individual patch locations (see the Supplementary Materials) and so must be narrower than that for a pivoter with a single patch. Figure 4 plots the observed normalized distribution of the anchoring coordinate |A| for pivoters (Fig. 4A) and wobblers (Fig. 4B), being narrower for the latter (SDs 0.27 and 0.22, respectively). We modeled the anchor-point distribution for adherers with n patches (Fig. 4B), assuming that these patches are placed along the cell axis according to the P(|A|) for pivoters, and that the anchoring coordinate for a wobbler is the average of the coordinates for these patches (see the Supplementary Materials). Our data suggest n = 2 or 3. This is as expected: Higher n would mean that pivoters and nonadherers should be very rare and that wobblers would almost never detach, contrary to observations.
Fig. 4. Distributions of anchoring coordinates for ΔFF.
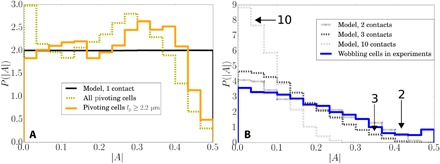
(A) The measured distribution of anchoring coordinates |A| for all pivoting cells (olive, 11,359 cells), only pivoting cells with lP > 2.2 μm (orange, 4338 cells) and for modeled cells with one contact point (black line). (B) The measured distribution of wobbling cells (blue, 4099 cells) fits best with models of cells with two or three patches. Modeled distribution of cells with 10 patches is also shown and peaks sharply close to zero. Modeled wobbler distributions were calculated using the P(|A|) for pivoters with artifacts removed.
Note that the experimental distribution of |A| for pivoters is slightly peaked near the cell center (|A| ≈ 0) and toward the cell pole (|A| ≈ 0.3) (Fig. 4A). The former is an artifact: A spherocylindrical cell adhering at one pole will appear as a small circle with an adhesion point identified in the cell center, artificially shifting the distribution toward |A| = 0. This can be removed by excluding cells with small projected lengths (orange line), although this does not significantly modify the predicted P(|A|) for wobblers. The other peak indicates that cells have a slight preference for polar adhesion, as previously found for other strains (36, 46). The observation that the peak is not at the pole itself is again due to the cell’s spherocylindrical shape.
We can simulate a minimal kinetic model based on the schematic in Fig. 3F. Bacteria are modeled as noninteracting point particles in a box with periodic boundary conditions in the horizontal (x, y) plane and impenetrable boundaries in the vertical (z) direction. Cells diffuse and sediment toward the lower surface. For ΔFF mutants, there are three independent subpopulations: nonadherers that do not bind to the surface; weak adherers that bind to the surface to become pivoters at rate kadh when within a distance Δzadh; and strong adherers that bind to the surface as pivoters, also at kadh, and can switch stochastically to a wobbling state or back, respectively, with rates kPW and kWP. Both weak and strong adherers can only detach from the surface, at rate kdet, when in the pivoting state.
The model uses values of the diffusivity and sedimentation speed based on independent measurements, and Δzadh = 100 nm, in line with the typical interaction range. Fitting the on-surface switching dynamics (Fig. 3E) gives estimates for kPW, kWP, and the ratio of weak to strong adherers. Varying the remaining parameters, kadh, kdet, and the total numbers of adherers and nonadherers credibly reproduces the main features of all our observations (curves in relevant parts of Figs. 2 and 3). We model the WT cells in the same way, except that a fraction of cells can swim and a subfraction of those can adhere as active rotators. The cell numbers are fitted, but the swimming speed is measured independently. Full model details are given in the Supplementary Materials, including used cell numbers (table S1) and movies depicting the simulations (movies S10 and S11).
DISCUSSION
We investigated the adhesion of E. coli AB1157 in buffer solution to glass. Using high-throughput and big-data analysis methods, we found strong heterogeneities between individual cells within a clonal population that are not related to type 1 fimbriae or flagella. These heterogeneities appear in the propensity for adhesion as well as the postadhesion dynamics. At least two types of adherers exist. Freely pivoted weak adherers are rotationally unconstrained and detach relatively easily. Strong adherers switch between this pivoting state and a rotationally constrained wobbling motion, in which they are more firmly bound to the surface.
A model of cells with adhesive patches that bind and unbind stochastically to the surfaces can explain our results. In this model, what differentiates subpopulations is the number of accessible patches per cell. Nonadhering cells have no patches and are always freely diffusing, weak adherers have one patch, and strong adherers have multiple adhesive patches that can bind simultaneously. These patches are estimated to have an interaction range of ≲ 100 nm and contact energy of 15 to 23 kBT. A very recent direct measurement of two Gram-positive species of Staphylococcus bacteria (47) shows that their adhesion to surfaces is mediated by sticky patches of radii ~ 100 to 300 nm, from which an interaction range of 20 ≲ L ≲ 120 nm is obtained. Thus, the physical mechanisms we have deduced from our experiments for E. coli may well be generic, even where specific details vary, for example, the interaction strengths measured in (47) are much larger, at >103 kBT.
However, the properties of the bacterial surface depend on many variables. Thus, for example, the acid-base properties of the surfaces of E. coli K-12 and Bacillus brevis cells are dependent on species, growth phase, as well as the composition of the growth media (48). These properties also change upon adhesion (9). When initial adhesion leads to the formation of a full-blown biofilm, different adhesive proteins are involved in binding cells to the substrate or to each other (10, 49). Both population variations in adhesion strength (50) and the location on the cell surface of adhesive interactions (51) are of importance. The study of a variety of organisms under diverse conditions will therefore be needed to establish a full picture.
Our work raises important questions about the biomolecular details of adhesion, most obviously the nature of the adhesive patches. Their rotational freedom implicates membrane proteins, which can rotate freely in lipid bilayers (52), whereas the interaction range of L ~ 100 nm suggests the involvement of cell surface macromolecules, consistent with a recent suggestion based on more direct, AFM measurements (47). Such rotational freedom raises the further question of the possible translational freedom of these patches, because individual proteins can also translate diffusively in the bacterial outer membrane (53). Both motions are strongly dependent on temperature because of lipidic phase transitions. If future work finds these adhesive patches to have temperature-dependent mobility, then bacteria will constitute a new class of patchy colloids. The interaction of mobile adhesive patches with rough (54) and patchy (19) substrates offers intriguing possibilities.
Another key question is the origin of the phenotypic heterogeneity we observed. This may be an instance of phase variation—the reversible switching on and off of gene expression (55), often of surface structures (56). On the other hand, adhesion may trigger a process of diversification in gene expression analogous to the process discovered recently in biofilm formation (57). Elucidating these issues using modern tools for time-dependent gene and protein expression assays is now an urgent task.
Finally, our work has implications for the design of antiadhesion surfaces, which is a key part of the ongoing fight against bacterial infections. First, it appears that we must now think in terms of designing antiadhesion surfaces that account for the range of adhesive phenotypes. Second, designing antiadhesion technologies will involve computer simulations, which, however, will never be fine-grained enough to include all details. Our finding that bacteria are patchy colloids vis-à-vis surface adhesion can help to design viable coarse-graining strategies and provide the necessary parameters for the computational study of this important phenomenon.
MATERIALS AND METHODS
Bacterial suspensions
All bacterial cells (E. coli AB1157 WT and various mutants; see the Supplementary Materials) were initially grown on LB agar plates, then inoculated and transferred to 10 ml of liquid LB, and incubated for 16 hours at 30°C. A fresh culture was inoculated as 1:100 dilution of overnight grown cells in 35 ml of tryptone broth and grown for 4 hours (to late exponential phase). Cells were washed three times with MB [6.2 mM K2HPO4 (Sigma-Aldrich), 3.8 mM KH2PO4 (Fisher Chemical), 67 mM NaCl (Fisher Chemical), and 0.1 mM EDTA (Sigma-Aldrich)] by careful filtration and were resuspended in MB at an optical density of ≈ 0.03 (WT) and ≈ 0.01 (ΔFF) at 600 nm, corresponding to ≈ 4.5 × 107 (WT) and ≈ 1.5 × 107 (ΔFF) cells/ml (23). Just before starting the adhesion assays, glucose solution in MB was added to the dispersion (dispersion contains 0.72 μM glucose after addition). We measured the electrophoretic mobility of the bacteria μE using a Malvern Zetasizer Nano Z.
Adhesion assays
Cell suspensions were loaded into borosilicate glass capillaries (Vitrocom, 0.4 × 8.0 × 50 mm) subsequently sealed with Vaseline. We recorded movies (1040 × 1024 pixels, 30 frames/s) focused on lower or upper capillary surfaces using a Mikrotron MC1362 camera on an inverted Nikon Ti-U microscope with a 60× phase-contrast objective (Nikon Plan Fluor, Ph2; numerical aperture, 0.7). Custom LabVIEW software controls the camera, a motorized stage (H117P21N4 with ProScan III, Prior Scientific), and a piezo objective actuator (P-725.4CD with E-753, Physik Instrumente) to automatically record movies at multiple locations on the inner capillary surface in time (23).
Analysis
For each movie, we filtered out background signals and noise, after which we determined the positions, orientations, and projected lengths of bacterial cells using an adapted algorithm specifically developed for rod-shaped colloids (24) and tracked bacteria in successive frames to obtain continuous trajectories. The suspension was so dilute that cells rarely collided, but those sections of trajectories crossing closer than 2.5 μm were omitted from the analysis. Other remaining misidentifications (for example, two neighboring cells recognized as one) were rare and did not affect the results significantly. Trajectories shorter than 0.8 s were not considered for classification. Trajectories displaying an adhering signature kT < 0.6 but shorter than 6 s were considered ambiguous.
Supplementary Material
Acknowledgments
We thank A. Jepson, A. Morozov, J. Thijssen, M. Cates, R. di Leonardo, F. Sciortino, A. Lips, T. Pilizota, A. Lepore, and H. Berg for useful discussions. Funding: T.V. was funded by FP7-PEOPLE-2013-IEF Marie Curie fellowships (LivPaC, 623364) and N.K. through H2020-MSCA-IF-2014 (ActiDoC, 654688). J.M.F. held an Engineering and Physical Sciences Research Council (EPSRC) studentship (EP/L015536/1 SOFI CDT), and A.T.B. received funding from a University of Edinburgh Chancellor’s Fellowship. Others were funded by EPSRC program grant EP/J007404/1 and European Research Council Advanced Grant GA 340877-PHYSAPS. We acknowledge the support of the Wellcome Trust Multi-user Equipment Grant (WT104915MA) for recording the transmission electron microscopy (TEM) images. Author contributions: T.V. and W.C.K.P. designed the research. A.D. constructed the mutants. A.D. and J.S.-L. prepared the bacteria. A.B.S. prepared the silica-coated bacteria and performed the scanning electron microscopy measurements. T.V. performed the adhesion experiments. V.A.M. assisted in the preliminary experiments, microscopy setup, and measurement protocols. J.M.F. and V.K. performed the AFM measurements. J.M.F. and T.V. performed TEM measurements. T.V., A.T.B., N.K., M.H., and W.C.K.P. analyzed the data. A.T.B. and T.V. performed the modeling. V.A.M., J.A., and N.K. assisted with the experiments. T.V., A.T.B., and W.C.K.P. wrote the paper. All authors discussed the results and commented on the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Data used in this publication are available on the Edinburgh DataShare repository (DOI: http://dx.doi.org/10.7488/ds/2315). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/4/eaao1170/DC1
section S1. Characterization
section S2. Synthesis and experimental details of bacterium-shaped hollow silica particles
section S3. Determining the anchoring point of a cell and classifying its dynamics
section S4. Adhesion for mutants without the flagellar hook
section S5. Pivoters, wobblers, and deserted adhesion sites
section S6. MSDs of anchoring point for wobblers and pivoters
section S7. Kinetic model details
fig. S1. Electron microscopy images of E. coli WT and ΔFF and bacterium-shaped hollow silica particles.
fig. S2. AFM images of the inner surface of the glass capillaries.
fig. S3. MSDs and anchoring points on the length axis of different types of bacteria.
fig. S4. Distributions of rotational exponents kR for mutants lacking essential hook proteins.
fig. S5. Pivoters, wobblers, and deserted binding sites for E. coli WT and ΔFF.
fig. S6. MSDs versus τ for pivoters and wobblers.
table S1. The cell numbers used for simulating the dynamics of bacterial adhesion.
movie S1. Determining positions and orientations of bacteria within the field of view.
movie S2. Lower surface before inversion for WT, categorized cells.
movie S3. Fifteen examples of trajectories for WT cells identified as diffusing.
movie S4. Fifteen examples of trajectories for WT cells identified as swimming.
movie S5. Fifteen examples of trajectories for WT cells identified as adhering and wobbling.
movie S6. Fifteen examples of trajectories for WT cells identified as adhering and pivoting.
movie S7. Fifteen examples of trajectories for WT cells identified as adhering and actively rotating.
movie S8. Examples of adhering cells with stained flagella (AD14) that are actively rotating.
movie S9. Illustration of deserted binding sites on the surface in time for ΔFF.
movie S10. Computer simulations of the inversion experiment for E. coli WT using a simple kinetic model.
movie S11. Computer simulations of the inversion experiment for ΔFF using a simple kinetic model.
REFERENCES AND NOTES
- 1.Petrova O. E., Sauer K., Sticky situations: Key components that control bacterial surface attachment. J. Bacteriol. 194, 2413–2425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunne W. M., Jr, Bacterial adhesion: Seen any good biofilms lately? Clin. Microbiol. Rev. 15, 155–166 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.J. Chan, S. Wong, Biofouling: Types, Impact, and Anti-Fouling (Nova Science Publishers, 2010). [Google Scholar]
- 4.Costerton J. W., Stewart P. S., Greenberg E. P., Bacterial biofilms: A common cause of persistent infections. Science 284, 1318–1322 (1999). [DOI] [PubMed] [Google Scholar]
- 5.Stewart P. S., Costerton J. W., Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138 (2001). [DOI] [PubMed] [Google Scholar]
- 6.D. Linke, A. Goldman, Bacterial Adhesion: Chemistry, Biology and Physics (Springer, 2011). [Google Scholar]
- 7.Poortinga A. T., Bos R., Norde W., Busscher H. J., Electric double layer interactions in bacterial adhesion to surfaces. Surf. Sci. Rep. 47, 1–32 (2002). [Google Scholar]
- 8.Tuson H. H., Weibel D. B., Bacteria–surface interactions. Soft Matter 9, 4368–4380 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong Y., Brown D. G., Alteration of bacterial surface electrostatic potential and pH upon adhesion to a solid surface and impacts to cellular bioenergetics. Biotechnol. Bioeng. 105, 965–972 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Beloin C., Houry A., Froment M., Ghigo J.-M., Henry N., A short–time scale colloidal system reveals early bacterial adhesion dynamics. PLOS Biol. 6, e167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katsikogianni M., Missirlis Y. F., Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur. Cell. Mater. 8, 37–57 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S., Stochastic gene expression in a single cell. Science 297, 1183–1186 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Ackermann M., A functional perspective on phenotypic heterogeneity in microorganisms. Nat. Rev. Microbiol. 13, 497–508 (2015). [DOI] [PubMed] [Google Scholar]
- 14.van Gestel J., Nowak M. A., Phenotypic heterogeneity and the evolution of bacterial life cycles. PLOS Comput. Biol. 12, e1004764 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philippi T., Seger J., Hedging one’s evolutionary bets, revisited. Trends Ecol. Evol. 4, 41–44 (1989). [DOI] [PubMed] [Google Scholar]
- 16.Bianchi E., Blaak R., Likos C. N., Patchy colloids: State of the art and perspectives. Phys. Chem. Chem. Phys. 13, 6397–6410 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Yi G. R., Pine D. J., Sacanna S., Recent progress on patchy colloids and their self-assembly. J. Phys. Condens. Matter 25, 193101 (2013). [DOI] [PubMed] [Google Scholar]
- 18.C. N. Likos, F. Sciortino, E. Zaccarelli, Soft Matter Self-Assembly (IOS Press, 2016), vol. 193. [Google Scholar]
- 19.Gon S., Kumar K.-N., Nüsslein K., Santore M. M., How bacteria adhere to brushy PEG surfaces: Clinging to flaws and compressing the brush. Macromolecules 45, 8373–8381 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.B. J. Bachmann, Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, in Escherichia coli and Salmonella, F. C. Neidhardt, Ed. (ASM Press, ed. 2, 1996), pp. 2460–2488. [Google Scholar]
- 21.Browning D. F., Wells T. J., França F. L. S., Morris F. C., Sevastsyanovich Y. R., Bryant J. A., Johnson M. D., Lund P. A., Cunningham A. F., Hobman J. L., May R. C., Webber M. A., Henderson I. R., Laboratory adapted Escherichia coli K-12 becomes a pathogen of Caenorhabditis elegans upon restoration of O antigen biosynthesis. Mol. Microbiol. 87, 939–950 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Prehm P., Schmidt G., Jann B., Jann K., The cell-wall lipopolysaccharide of Escherichia coli K-12. Structure and acceptor site for O-antigen and other substituents. Eur. J. Biochem. 70, 171–177 (1976). [DOI] [PubMed] [Google Scholar]
- 23.Schwarz-Linek J., Arlt J., Jepson A., Dawson A., Vissers T., Miroli D., Pilizota T., Martinez V. A., Poon W. C. K., Escherichia coli as a model active colloid: A practical introduction. Colloids Surf. B Biointerfaces 137, 2–16 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Besseling T. H., Hermes M., Kuijk A., de Nijs B., Deng T.-S., Dijkstra M., Imhof A., van Blaaderen A., Determination of the positions and orientations of concentrated rod-like colloids from 3D microscopy data. J. Phys. Condens. Matter 27, 194109 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Qian H., Sheetz M. P., Elson E. L., Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 60, 910–921 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saxton M. J., Jacobson K., Single-particle tracking: Applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26, 373–399 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Silverman M., Simon M., Flagellar rotation and the mechanism of bacterial motility. Nature 249, 73–74 (1974). [DOI] [PubMed] [Google Scholar]
- 28.Brown A. T., Vladescu I. D., Dawson A., Vissers T., Schwarz-Linek J., Lintuvuori J. S., Poon W. C. K., Swimming in a crystal. Soft Matter 12, 131–140 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Friedlander R. S., Vogel N., Aizenberg J., Role of flagella in adhesion of Escherichia coli to abiotic surfaces. Langmuir 31, 6137–6144 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Haiko J., Westerlund-Wikström B., The role of the bacterial flagellum in adhesion and virulence. Biology 2, 1242–1267 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vigeant M. A.-S., Ford R. M., Wagner M., Tamm L. K., Reversible and irreversible adhesion of motile Escherichia coli cells analyzed by total internal reflection aqueous fluorescence microscopy. Appl. Environ. Microbiol. 68, 2794–2801 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hermansson M., The DLVO theory in microbial adhesion. Colloids Surf. B Biointerfaces 14, 105–119 (1999). [Google Scholar]
- 33.Rijnaarts H. H. M., Norde W., Lyklema J., Zehnder A. J. B., DLVO and steric contributions to bacterial deposition in media of different ionic strengths. Colloids Surf. B Biointerfaces 14, 179–195 (1999). [Google Scholar]
- 34.Nomura T., Morimoto Y., Ishikawa M., Tokumoto H., Konishi Y., Synthesis of hollow silica microparticles from bacterial templates. Adv. Powder Technol. 21, 8–12 (2010). [Google Scholar]
- 35.Vladescu I. D., Marsden E. J., Schwarz-Linek J., Martinez V. A., Arlt J., Morozov A. N., Marenduzzo D., Cates M. E., Poon W. C. K., Filling an emulsion drop with motile bacteria. Phys. Rev. Lett. 113, 268101 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Agladze K., Wang X., Romeo T., Spatial periodicity of Escherichia coli K-12 biofilm microstructure initiates during a reversible, polar attachment phase of development and requires the polysaccharide adhesin PGA. J. Bacteriol. 187, 8237–8246 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tavaddod S., Charsooghi M. A., Abdi F., Khalesifard H. R., Golestanian R., Probing passive diffusion of flagellated and deflagellated Escherichia coli. Eur. Phys. J. E Soft. Matter 34, 16 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Hänggi P., Talkner P., Borkovec M., Reaction-rate theory: Fifty years after Kramers. Rev. Mod. Phys. 62, 251–341 (1990). [Google Scholar]
- 39.Razatos A., Ong Y.-L., Sharma M. M., Georgiou G., Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 95, 11059–11064 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dufrêne Y. F., Sticky microbes: Forces in microbial cell adhesion. Trends Microbiol. 23, 376–382 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Jepson A., Martinez V. A., Schwarz-Linek J., Morozov A., Poon W. C. K., Enhanced diffusion of nonswimmers in a three-dimensional bath of motile bacteria. Phys. Rev. E 88, 041002 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Trewby W., Livesey D., Voïtchovsky K., Buffering agents modify the hydration landscape at charged interfaces. Soft Matter 12, 2642–2651 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Chandler D., Interfaces and the driving force of hydrophobic assembly. Nature 437, 640–647 (2005). [DOI] [PubMed] [Google Scholar]
- 44.van Loosdrecht M. C. M., Norde W., Lyklema J., Zehnder A. J. B., Hydrophobic and electrostatic parameters in bacterial adhesion. Aquat. Sci. 52, 103–114 (1990). [Google Scholar]
- 45.Amro N. A., Kotra L. P., Wadu-Mesthrige K., Bulychev A., Mobashery S., Liu G.-y., High-resolution atomic force microscopy studies of the Escherichia coli outer membrane: Structural basis for permeability. Langmuir 16, 2789–2796 (2000). [Google Scholar]
- 46.Jones J. F., Velegol D., Laser trap studies of end-on E. coli adhesion to glass. Colloids Surf. B Biointerfaces 50, 66–71 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Spengler C., Thewes N., Jung P., Bischoff M., Jacobs K., Determination of the nano-scaled contact area of staphylococcal cells. Nanoscale 9, 10084–10093 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Hong Y., Brown D. G., Cell surface acid–base properties of Escherichia coli and Bacillus brevis and variation as a function of growth phase, nitrogen source and C:N ratio. Colloids Surf. B Biointerfaces 50, 112–119 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Berk V., Fong J. C. N., Dempsey G. T., Develioglu O. N., Zhuang X., Liphardt J., Yildiz F. H., Chu S., Molecular architecture and assembly principles of Vibrio cholerae biofilms. Science 337, 236–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schluter J., Nadell C. D., Bassler B. L., Foster K. R., Adhesion as a weapon in microbial competition. ISME J. 9, 139–149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duvernoy M.-C., Mora T., Ardre M., Croquette V., Bensimon D., Quilliet C., Ghigo J.-M., Balland M., Beloin C., Lecuyer S., Desprat N., Asymmetric adhesion of rod-shaped bacteria controls microcolony morphogenesis. bioRxiv 2017, 104679 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aisenbrey C., Bechinger B., Investigations of polypeptide rotational diffusion in aligned membranes by 2H and 15N solid-state NMR spectroscopy. J. Am. Chem. Soc. 126, 16676–16683 (2004). [DOI] [PubMed] [Google Scholar]
- 53.Nenninger A., Mastroianni G., Robson A., Lenn T., Xue Q., Leake M. C., Mullineaux C. W., Independent mobility of proteins and lipids in the plasma membrane of Escherichia coli. Mol. Microbiol. 92, 1142–1153 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharma S., Jaimes-Lizcano Y. A., McLay R. B., Cirino P. C., Conrad J. C., Subnanometric roughness affects the deposition and mobile adhesion of Escherichia coli on silanized glass surfaces. Langmuir 32, 5422–5433 (2016). [DOI] [PubMed] [Google Scholar]
- 55.van der Woude M. W., Phase variation: How to create and coordinate population diversity. Curr. Opin. Microbiol. 14, 205–211 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Jayaraman R., Phase variation and adaptation in bacteria: A ‘Red Queen’s Race’. Curr. Sci. 100, 1163–1171 (2011). [Google Scholar]
- 57.Besharova O., Suchanek V. M., Hartmann R., Drescher K., Sourjik V., Diversification of gene expression during formation of static submerged biofilms by Escherichia coli. Front. Microbiol. 7, 1568 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DeWitt S. K., Adelberg E. A., The occurrence of a genetic transposition in a strain of Escherichia coli. Genetics 47, 577–585 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.M. S. Guyer, R. R. Reed, J. A. Steitz, K. B. Low, Identification of a sex-factor-affinity site in E. coli as γδ, in Cold Spring Harbor Symposia on Quantitative Biology (Cold Spring Harbor Laboratory Press, 1981), vol. 45, pp. 135–140. [DOI] [PubMed] [Google Scholar]
- 60.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K. A., Tomita M., Wanner B. L., Mori H., Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/4/4/eaao1170/DC1
section S1. Characterization
section S2. Synthesis and experimental details of bacterium-shaped hollow silica particles
section S3. Determining the anchoring point of a cell and classifying its dynamics
section S4. Adhesion for mutants without the flagellar hook
section S5. Pivoters, wobblers, and deserted adhesion sites
section S6. MSDs of anchoring point for wobblers and pivoters
section S7. Kinetic model details
fig. S1. Electron microscopy images of E. coli WT and ΔFF and bacterium-shaped hollow silica particles.
fig. S2. AFM images of the inner surface of the glass capillaries.
fig. S3. MSDs and anchoring points on the length axis of different types of bacteria.
fig. S4. Distributions of rotational exponents kR for mutants lacking essential hook proteins.
fig. S5. Pivoters, wobblers, and deserted binding sites for E. coli WT and ΔFF.
fig. S6. MSDs versus τ for pivoters and wobblers.
table S1. The cell numbers used for simulating the dynamics of bacterial adhesion.
movie S1. Determining positions and orientations of bacteria within the field of view.
movie S2. Lower surface before inversion for WT, categorized cells.
movie S3. Fifteen examples of trajectories for WT cells identified as diffusing.
movie S4. Fifteen examples of trajectories for WT cells identified as swimming.
movie S5. Fifteen examples of trajectories for WT cells identified as adhering and wobbling.
movie S6. Fifteen examples of trajectories for WT cells identified as adhering and pivoting.
movie S7. Fifteen examples of trajectories for WT cells identified as adhering and actively rotating.
movie S8. Examples of adhering cells with stained flagella (AD14) that are actively rotating.
movie S9. Illustration of deserted binding sites on the surface in time for ΔFF.
movie S10. Computer simulations of the inversion experiment for E. coli WT using a simple kinetic model.
movie S11. Computer simulations of the inversion experiment for ΔFF using a simple kinetic model.


