Abstract
BACKGROUND
Vascular anomalies currently are classified according to their clinical and histological characteristics. Recent advances in molecular genetics have enabled the identification of somatic mutations in most types of vascular anomalies. The purpose of this study was to collate information regarding the genetic basis of vascular anomalies.
METHODS
The PubMed literature was reviewed for all citations that identified a mutation in a vascular anomaly between 1994–2017. Search words included “vascular anomaly”, “mutation”, “gene”, “hemangioma”, “pyogenic granuloma”, “kaposiform hemangioendothelioma”, “capillary malformation”, “venous malformation”, lymphatic malformation”, “arteriovenous malformation”, and “syndrome”. Articles that identified both germline as well as somatic mutations in vascular anomalies were analyzed. Mutations were categorized by type (germline or somatic), gene, signaling pathway, and cell(s) enriched for the mutation.
RESULTS
The majority of vascular anomalies had associated mutations that commonly affected tyrosine kinase receptor signaling through the RAS or PIK3CA pathways. Mutations in PIK3CA and G-protein coupled receptors were most frequently identified. Specific types of vascular anomalies usually were associated with a single gene. However, mutations in the same gene occasionally were found in different vascular lesions, and some anomalies had a mutation in >1 gene. Mutations were most commonly enriched in endothelial cells.
CONCLUSIONS
Identification of somatic mutations in vascular anomalies is changing the paradigm by which lesions are diagnosed and understood. Mutations and their pathways are providing potential targets for the development of novel pharmacotherapy. In the future, vascular anomalies will be managed based on clinical characteristics as well as molecular pathophysiology.
Keywords: vascular, anomaly, malformation, hemangioma, mutation, gene, classification, molecular, tumor
INTRODUCTION
Vascular anomalies currently are classified by their clinical and histologic characteristics into 2 broad categories: tumors or malformations (Table 1).1 Vascular tumors have proliferating endothelium while vascular malformations are structural anomalies. Since the proposal of the clinicohistological classification in 1982 by Mulliken and Glowacki (Level of Evidence III),2 minimal insight into the pathophysiology of vascular anomalies has been achieved and pharmacotherapy for many lesions does not exist. Beginning in 1994, the genetic basis of familial forms of vascular anomalies began to be identified. Because almost all lesions are sporadic, the cause of most vascular anomalies has remained unknown. The development of next generation sequencing has enabled the identification of low-level somatic mosaic mutations in sporadic lesions; in 2011 this technology was applied to vascular anomalies.3 As a result, over the past few years mutations associated with many lesions in the field has expanded significantly. The purpose of this manuscript was to evaluate the field of vascular anomalies based on new information regarding the molecular basis of these lesions.
Table 1.
Current Classification of Vascular Anomalies*
| TUMORS | MALFORMATIONS | |||
|---|---|---|---|---|
|
| ||||
| Simple | Combined | Major Named Vessels | Associated With Other Anomalies | |
|
| ||||
| Benign | Capillary | Capillary-venous | Aneurysm | CLOVES |
| Infantile hemangioma | Cutis marmorata telangiectatica congenita | Capillary-lymphatic | Atresia | Klippel-Trenaunay |
| Congenital hemangioma | Capillary-arteriovenous | Ectasia | Megalencephaly-capillary malformation | |
| Pyogenic granuloma | Fading stain | Lymphatic-venous | Stenosis | Mafucci |
| Infantile myofibroma | Capillary-lymphatic-venous | Parkes Weber | ||
| Enzinger hemangioma | Lymphatic | Capillary-lymphatic-arteriovenous | Proteus | |
| Macrocystic | Capillary-venous-arteriovenous | PTEN hamartoma | ||
| Intermediate | Microcystic | Capillary-lymphatic-venous-arteriovenous | Sturge-Weber | |
| Kaposiform hemangioendothelioma | Generalized | |||
| Tufted angioma | Gorham-Stout | |||
| Primary lymphedema | ||||
| Malignant | ||||
| Angiosarcoma | Venous | |||
| Epitheliod hemangioendothelioma | Cutaneoumucosal | |||
| Glomuvenous | ||||
| Cerebral cavernous | ||||
| Blue rubber bleb nevus | ||||
| Fibro adipose | ||||
| Verrucous | ||||
| Arteriovenous | ||||
| Capillary malformation-arteriovenous malformation | ||||
| Hereditary hemorrhagic telangiectasia | ||||
Adapted from “Vascular Anomalies Classification: Recommendations From the International Society for the Study of Vascular Anomalies,” Wassef et al., 2015, Pediatrics, 136, p. 203–214.
METHODS
The PubMed literature was reviewed for all citations that identified a mutation in a vascular anomaly between 1994–2017. Search words included “vascular anomaly”, “mutation”, “gene”, “hemangioma”, “pyogenic granuloma”, “kaposiform hemangioendothelioma”, “capillary malformation”, “venous malformation”, lymphatic malformation”, “arteriovenous malformation”, and “syndrome”. Articles were analyzed that identified either somatic or germline mutations. A somatic mutation is a randomly acquired alteration to the genetic sequence of a cell any time after fertilization that can be passed to the affected cell’s progeny but not to the organism’s offspring. A germline mutation is an alteration to the genetic sequence that occurs in gametes and can be inherited by the patient’s children.
Mutations were categorized by type (germline or somatic), gene, signaling pathway, and cell(s) enriched for the mutation. Mutant allele frequency (MAF) associated with somatic mutations were recorded when possible. An allele is one of two alternatives of a gene found at the same location along a chromosome that codes for a specific protein. Because each cell has two alleles (one inherited from each parent) it is generally assumed that a random somatic mutation would only affect one of the alleles. The MAF is calculated by measuring the number of mutant alleles and dividing them by the total number of alleles present (mutant alleles / mutant alleles + wild-type alleles). The higher the MAF, the greater number of affected cells in the lesion.
RESULTS
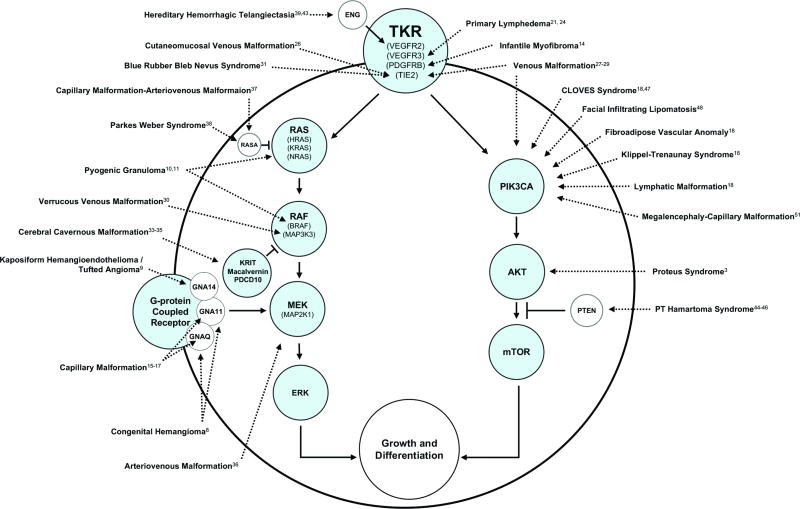
Forty-three publications identifying mutations in vascular anomalies were identified. Mutations most commonly involved tyrosine kinase receptor signaling through the RAS or PIK3CA pathways (Figure 1). Mutations in the tyrosine kinase receptors TIE2, PDGF, and ENG were associated with venous malformation, infantile myofibroma, and hereditary hemorrhagic telangiectasia, respectively. Mutations affecting RAS were found in pyogenic granuloma and capillary malformation-arteriovenous malformation. Pyogenic granuloma and venous malformation were associated with mutations affecting RAF. Alterations affecting MEK, including G-protein coupled receptors, were noted in arteriovenous malformation, capillary malformation, congenital hemangioma, and kaposiform hemangioendothelioma. PIK3CA mutations were identified in: lymphatic malformation, venous malformation, fibroadipose vascular anomaly, and overgrowth syndromes (CLOVES, Klippel-Trenaunay, megalencephaly-capillary malformation). Mutations affecting AKT were found in proteus syndrome and PTEN hamartoma-tumor syndrome. Most vascular anomalies were associated with a single gene. However, mutations in the same gene occasionally were noted in different types of vascular lesions, and some anomalies had a mutation in >1 gene. Mutations were most commonly enriched in endothelial cells.
Figure 1.
Mutations in vascular anomalies most commonly affect genes associated with tyrosine kinase signaling through the RAS or PIK3CA pathways.
DISCUSSION
Current Clinicohistologic Framework
Most vascular anomalies are identified by history and physical examination. Imaging can aid the diagnosis and histopathology rarely is necessary. Although lesions usually can be identified clinically, significant heterogeneity and response to treatment exists. Successful management of a vascular anomaly is based on an accurate diagnosis. Tumors have dividing endothelium and thus may be treated with pharmacotherapy. In contrast, vascular malformations have minimal cellular turnover and have not shown efficacy to drugs (an exception is lymphatic malformation which can respond to oral sirolimus).4 Management of vascular malformations includes resection, laser, sclerotherapy, and/or embolization. Many vascular malformations are unable to be cured and the goal of treatment is to control the lesion. Progression and recurrence after intervention is common. Consequently, drug development is important to potentially regress the anomaly and/or prevent its growth and recurrence after treatment. A limitation of the clinicohistologic classification is that it does not incorporate the molecular basis of vascular anomalies. For example, venous malformations can be caused by mutations in eight different genes.
Genetic Basis of Vascular Anomalies
Infantile Hemangioma (IH)
IH is the most common tumor of infancy and has a unique growth cycle: it enlarges rapidly during the first few months of life and then regresses in early childhood. A mutation has yet to be identified. Explanations that have been proposed for its etiopathology include placental, stem cell, and follicle stimulating hormone hypotheses.5–7
Congenital Hemangioma (CH)
Two types of CHs exist: rapidly involuting congenital hemangioma (RICH) and non-involuting congenital hemangioma (NICH). Unlike infantile hemangioma, these lesions are fully-formed at birth. Mutations in GNAQ or GNA11 have been found in 12 of 16 specimens; the mutant allele frequency (MAF) is 1%–11%.8 GNAQ and GNA11 mutations are present in both RICH and NICH specimens. Partially involuting congenital hemangioma (PICH) is a rare subtype of RICH that does not involute fully and persists as a NICH-type lesion; its somatic mutation has not been identified (Nasseri 2014).9
Kaposiform Hemangioendothelioma (KHE)
KHE is a vascular neoplasm that usually is present at birth, enlarges during infancy, and then partially involutes. Most patients are treated with systemic pharmacotherapy for associated Kasabach-Merritt phenomenon (thrombocytopenia, petechiae, bleeding) or to minimize fibrosis. A mutation in GNA14 was found in 1/3 KHE specimens (MAF 10%) and in 1/4 closely related tufted angiomas.10
Pyogenic Granuloma (PG)
Pyogenic granuloma is a postnatal lesion with a mean age of onset of 6 years. It averages 6mm in diameter and often bleeds. Mutations in KRAS, NRAS, or HRAS have been identified in 6/42 specimens in one study (MAF 16%–40%).11 Another investigation of 25 PGs found mutations in BRAF in 4 specimens and KRAS in 1 specimen.12 PGs that arise from capillary malformations show mutations in both GNAQ and BRAF in 7/10 specimens.12
Rare Vascular Tumors
Angiosarcoma is a malignant lesion that rarely affects children; mutations in PTPRB and PLCG have been found in 10/39 and 3/34 specimens, respectively.13 Epithelioid hemangioendothelioma is a malignant endothelial tumor with variable clinical behavior. Less than 10% involve the pediatric population. Lesions usually are multifocal and may be stable, slowly enlarge, or rapidly progress and metastasize. A WWTR1-CAMTA1 gene fusion has been identified in 17/17 samples.14 Infantile myofibroma is a fibrous tumor of early childhood that may be solitary, multifocal, or generalized. Solitary lesions can regress, while multifocal or generalized disease affecting the viscera can be life-threatening. The lesion is associated with mutations in PDGFRB in 7/16 specimens.15
Capillary Malformation (CM)
CM is the most common type of vascular malformation. Lesions are present at birth and affect the skin. The pink stain darkens over time and patients can develop soft-tissue and bony hypertrophy underneath the integument. Mutations in GNAQ have been found in 45 of 52 patients with sporadic or syndromic (Sturge-Weber syndrome) CM (MAF 1%–22%).16, 17 A mutation in GNA11 has been identified in the extremities of 3/8 specimens with a diffuse CM including overgrowth (MAF 0.3%–5.0%).18 GNAQ mutations in CM are enriched in endothelial cells (3%–43%), compared to pericytes (0%), stromal cells (0.4%–11%), or hematopoietic cells (0.3%).17
Lymphatic Malformation (LM)
LMs are cystic structures that can be macrocystic, microcystic, or combined. Mutations in PIK3CA have been found in 16/17 specimens (MAF 0.8%–10%).19 Primary lymphedema also is a type of vascular malformation that can be hereditary. Only 8% of sporadic and 36% of familial cases have an identifiable mutation (e.g.,VEGFR3/FLT-4, FOXC2, SOX18, CCBE1).20–26
Venous Malformation (VM)
VMs usually are sporadic, but can be familial. Cutaneomucosal autosomal dominant VMs were found to be caused by TIE2 mutations (61/61 samples).27 Mutations in TIE2 then were identified in sporadic VMs as well (80/130 specimens; MAF 4%–48%)28, 29 PIK3CA mutations also are found in sporadic VMs in 27/130 specimens (MAF 1%–18%); lesions usually involve the subcutis.30 Verrucous venous malformations are hyperkeratotic anomalies typically affecting the skin of an extremity; mutations in MAP3K3 have been identified in 6/10 specimens (MAF 6%–19%).31 Blue rubber bleb nevus syndrome is a non-hereditary condition with multifocal VMs in the skin, soft tissue, and gastrointestinal tract; a TIE2 mutation has been found in 32/35 samples.32 Fibroadipose vascular anomaly is a lesion that previously was thought to be an intramuscular VM but exhibits more fibroadipose tissue and smaller, nonspongiform vessels. Mutations in PIK3CA were identified in 4/8 samples (MAF 5%–20%).19 Glomuvenous malformation is an autosomal dominant condition that usually has multiple small lesions caused by mutations in Glomulin in 6/7 families.33 Cerebral cavernous malformation can be sporadic or autosomal dominant with anomalies usually in the brain; approximately 10% of patients have hyperkeratotic skin lesions. Cerebral cavernous malformation results from mutations in CCM1/KRIT1 (12/20 samples),34 CCM2/malcavernin (29/35 samples),35 or CCM3/PDCD10 (8/20 samples).36
Arteriovenous Malformation (AVM)
AVMs are abnormal connections between arteries and veins without a normal capillary bed through either a nidus or fistula. AVMs are associated with mutations in the MAP2K1 gene in 16/25 specimens (MAF 1%–13%).37 MAP2K1 mutations in AVM are exclusive to endothelial cells (MAF 31%–53%).37 Capillary malformation-arteriovenous malformation is an autosomal dominant disease characterized by multifocal cutaneous lesions and a 1/3 risk of having an AVM, including Parkes Weber syndrome (an overgrown extremity with a diffuse AVM). Capillary malformation-arteriovenous malformation is caused by a mutation in RASA1 (6/17 families);38 13/16 patients with Parkes Weber syndrome exhibited a RASA1 mutation.39 Hereditary hemorrhagic telangiectasia is an autosomal dominant disease that exhibits epistaxis, mucocutaneous telangiectasias, and/or visceral AVMs (pulmonary, cerebral, hepatic, gastrointestinal). Mutations in endoglin (ENG) or activating A receptor type 2-like 1 (ACVRL1/ALK1) are responsible for almost all cases.40, 41 SMAD4 (associated with juvenile polyposis) or GDF2 mutations affect 2% of patients.42, 43 Recently, endoglin has been shown to affect VEGFR2 signaling.44 PTEN-associated vascular anomaly is present in approximately 50% of patients with the autosomal dominant PTEN hamartoma-tumor syndrome.45–47 Lesions can be multiple, are often intramuscular, and have excess adipose tissue and disproportionate dilatation of draining veins.
Overgrowth Syndromes Associated With Vascular Anomalies
Vascular anomalies are major components of several types of syndromes that cause enlargement of soft-tissue and/or bone. Proteus syndrome (progressive, asymmetrical overgrowth of the skeleton, cerebriform nevi of the hands or feet, epidermal nevi, vascular malformations, cerebral anomalies, skull hyperostosis, and megaspondylodysplasia) is caused by a mutation in AKT1 (26/29 patients, MAF 2%–39%).3 CLOVES syndrome (congenital lipomatosis, overgrowth, vascular malformations, epidermal nevi, and skeletal anomalies) was found to have a PIK3CA mutation in 36/38 patients (MAF 1%–32%).19, 48 Individuals with isolated facial infiltrating lipomatosis also exhibit PIK3CA mutations in the subcutaneous adipose (6/6 samples, MAF 9%–31%).49 The mutation is present throughout all structures of the face and is enriched in non-endothelial cells (MAF 28%–49%) compared to endothelial cells (1%–5%)50 Klippel-Trenaunay syndrome is a capillary-lymphatic-venous malformation of an extremity causing overgrowth; PIK3CA mutations have been found in 19/21 patients (MAF 3%–12%).19 Maffuci Syndrome is characterized by multiple enchondromas and soft-tissue VMs. The condition is caused by a somatic mutation in isocitrate dehydrogenase (IDH) in 37/40 individuals; 98% have an IDH1 mutation and 2% exhibit an IDH2 error.51 Megalencephaly-capillary malformation commonly causes neurologic abnormalities and patients typically have a CM involving the upper lip, trunk, and/or extremities; patients have mutations in PIK3CA.52
Evolving Genetic Framework
Most mutations found in vascular anomalies involve tyrosine kinase receptor signaling through the RAS or PIK3CA pathways; the function of some mutations remains unknown. Mutations in the same gene can be associated with different lesions. While PIK3CA mutations result in relatively similar types of vascular malformations, GNAQ/GNA11/GNA14 mutations are found in both tumors and malformations. It is unclear how mutations in the same gene can lead to different phenotypes, but it may be due to the location of the mutation in the gene, cell type(s) affected, and/or stage of development when the error occurred. RAS and PIK3CA signaling have broad effects on cell proliferation, differentiation, and survival.53, 54 Consequently, mutations can result in both tumors and malformations. For example, germline mutations in MAP2K1 cause cardio-facio-cutaneous syndrome which includes malformations,55 while different somatic MAP2K1 mutations result in arteriovenous malformation37 or cancers (melanoma, lung, hematopoietic).56–58
Occasionally lesions reviewed in our vascular anomalies center have an unclear diagnosis. We now have the capability of testing specimens with droplet digital PCR (ddPCR) against known vascular anomaly mutations. Mutation discovery in vascular anomalies also has allowed the formulation of genotype-phenotype relationships. For example, VMs have been classified as one entity, but it is now recognized that different mutations are associated with unique phenotypes: cutaneous VM (TIE2), subcutaneous VM (PIK3CA), hyperkeratotic VM (MAP3K3), painful VM (glomulin). Similarly, AVMs can be caused by mutations in MAP2K1 (most common), PTEN (more adipose), or RASA1 (Parkes Weber syndrome). Molecular findings also have confirmed clinical observations that certain lesions were closely related (e.g., verrucous “hemangioma” and VM).30
Targeted treatments based on the specific pathway that is affected for the anomaly can be developed, similar to focused therapy of malignant neoplasms based on their mutation.59 For example, vascular anomalies caused by PIK3CA mutations have been grouped under the umbrella term PROS (PIK3CA related overgrowth spectrum) and are no longer viewed as independent diseases.60 Instead, they are considered a spectrum of conditions caused by the same mutation. Similarly, capillary malformation, congenital hemangioma, and kaposiform hemangioendothelioma may be affiliated under the category of G-protein receptor mutations affecting the closely related GNAQ/GNA11/GNA14 genes [(e.g., GNA-vascular anomaly (GNAVA)].
The genetic framework of vascular anomalies is still rapidly evolving (Table 2). Somatic mutations are not found in all patient specimens subjected to whole exome sequencing (WES) or ddPCR. Possible explanations include: (1) the mutant allele frequency is below the detection limit of the assay; (2) a mutation is present in another part of the gene; (3) the mutation is in a different gene. Other mutations causing vascular anomalies will continue to be discovered, likely affecting components of the same signaling pathway. The cost of mutation identification is inexpensive if the mutation is already known using ddPCR (approximately $4 per sample); the expense of mutation discovery using WES continues to decrease (approximately $2700 per sample). Further investigation will show the specific cell type(s) harboring the mutation, as well as how the mutation affects cell biology. Understanding the mutations associated with vascular anomalies is the first step to developing targeted pharmacotherapy for these lesions. In the future, the framework by which vascular anomalies are diagnosed and managed will become more comprehensive by incorporating genetic findings with clinical information.
Table 2.
Molecular Framework of Vascular Anomalies
| TKR | RAS | RAF | MEK | PIK3CA | AKT | Other | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Anomaly | Gene | Anomaly | Gene | Anomaly | Gene | Anomaly | Gene | Anomaly | Gene | Anomaly | Gene | Anomaly |
| TIE2 | VM CMVM BRBNS | HRAS | PG | BRAF | PG | MAP2K1 | AVM | PIK3CA | LM CLOVES KTS FIL FAVA MCAP VM | AKT1 | PS | ACVRL SMAD4 GDF2 SMAD4 | HHT |
|
| |||||||||||||
| Glomulin | GVM | ||||||||||||
|
|
|
||||||||||||
| PDGF | IM | KRAS | PG | MAP3K3 | VVM | GNAQ | CM CH | PTEN | PTENAVA | PTPRB PLCG | AS | ||
|
|
|
||||||||||||
| ENG | HHT | NRAS | PG | KRIT Macalvernin PDCD10 | CCM | GNA11 | CM CH | WWTR1-CAMTA1 | EHE | ||||
|
|
|
|
|||||||||||
| RASA | CM-AVM PWS | GNA14 | KHE TA | IDH1 IDH2 | MS | ||||||||
The major headings (bold) represent principal signaling pathway components. Mutations listed below are either subtypes of the heading or affect its signaling pathway (see Figure 1).
CONCLUSIONS
Next generation sequencing has allowed the recent discovery of somatic mosaic mutations in vascular anomalies. Most mutations affect the RAS or PIK3CA tyrosine kinase signaling pathways. Identification of the molecular basis of vascular anomalies has facilitated the identification of lesions with an equivocal clinicohistologic diagnosis, as well as enabled genotype-phenotype correlations. Further investigation will determine how specific mutations affect the etiopathogenesis of the vascular anomaly. Pharmacotherapy targeting altered signaling pathways may prove efficacious in the future.
Acknowledgments
This manuscript was supported by the National Institutes of Health Awards NICHD-081004 (AKG), NICHD-082606 (AKG), NHLBI-127030 (AKG), and the Translational Research Program Boston Children’s Hospital (AKG). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
INDEX OF TERMS
Vascular Anomalies
- AS
Angiosarcoma
- AVM
Arteriovenous malformation
- BRBNS
Blue rubber bleb nevus syndrome
- CM
Capillary malformation
- CM-AVM
Capillary malformation-arteriovenous malformation
- CCM
Cerebral cavernous malformation
- CLOVES
Congenital lipomatosis overgrowth, vascular malformations, epidermal nevus, spinal/skeletal anomalies/scoliosis
- CH
Congenital hemangioma
- CMVM
Cutaneomucosal venous malformation
- EHE
Epithelioid hemangioendothelioma
- FAVA
Fibroadipose vascular anomaly
- FIL
Facial infiltrating lipomatosis
- GVM
Glomovenous malformation
- HHT
Hereditary hemorrhagic telangiectasia
- IM
Infantile myofibroma
- KHE
Kaposiform hemangioendothelioma
- KTS
Klippel-Trenaunay syndrome
- LM
Lymphatic malformation
- MCAP
Megalencephaly-capillary malformation
- MS
Maffucci syndrome
- PTENAVA
PTEN associated vascular anomaly
- PWS
Parkes Weber syndrome
- PS
Proteus syndrome
- PG
Pyogenic granuloma
- TA
Tufted angioma
- VM
Venous malformation
- VVM
Verrucous venous malformation
GENES
- ACVRL
Activin receptor-like kinase
- AKT
Ak-thymoma
- BRAF
B-rapidly accelerated fibrosarcoma
- CAMTA1
Calmodulin binding transcription activator 1
- ENG
Endoglin
- GDF2
Growth differentiation factor 2
- GNA 11
Guanine nucleotide-binding protein subunit alpha 11
- GNA 14
Guanine nucleotide-binding protein subunit alpha 14
- GNAQ
Guanine nucleotide-binding protein subunit alpha Q
- HRAS
Harvey rat sarcoma proto-oncogene
- IDH1
Isocitrate dehydrogenase 1
- IDH2
Isocitrate dehydrogenase 2
- KRAS
Kirsten rat sarcoma proto-oncogene
- KRIT
Krev interaction trapped protein 1
- MAP2K1
Mitogen-activated protein kinase kinase 1
- MAP3K3
Mitogen-activated protein kinase kinase kinase 3
- MEK
Mitogen activating pathway kinase-extracellular signal-regulated kinase
- NRAS
Neuroblastoma rat sarcoma proto-oncogene
- PDGF
Platelet-derived growth factor
- PDCD10
Programmed cell death protein 10
- PIK3CA
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- PLCG
Phospholipase C, gamma 1
- PTEN
Phosphatase and tensin homolog
- PTPRB
Protein tyrosine phosphatase, receptor type B
- RAF
rapidly accelerated fibrosarcoma
- RAS
Rat sarcaoma
- RASA
Rat sarcoma p21 protein activator 1
- SMAD
Small body size - mothers against decapentaplegic, Drosophila) homolog 4
- TIE2
Tyrosine Kinase With Ig And EGF Homology Domains-2
- TKR
Tyrosine kinase receptor
- WWTR1
WW domain-containing transcription regulator protein 1
Footnotes
Presented at: 2017 New England Society for Plastic and Reconstructive Surgery (NESPRS) meeting in Falmouth, MA.
Disclosures: None of the authors has a financial interest in any of the products, devices, or drugs mentioned in this manuscript.
AUTHOR ROLE AND PARTICIPATION
AKG and J.A.G. contributed to overall design, data analysis, writing manuscript, and revising manuscript. Both authors gave final approval.
References
- 1.Wassef M, Blei F, Adams D, et al. Vascular Anomalies Classification: Recommendations From the International Society for the Study of Vascular Anomalies. Pediatrics. 2015;136:e203–214. doi: 10.1542/peds.2014-3673. [DOI] [PubMed] [Google Scholar]
- 2.Mulliken JB, Glowacki J. Hemangiomas and vascular malformations in infants and children: a classification based on endothelial characteristics. Plast Reconstr Surg. 1982;69:412–422. doi: 10.1097/00006534-198203000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Lindhurst MJ, Sapp JC, Teer JK, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hammill AM, Wentzel M, Gupta A, et al. Sirolimus for the treatment of complicated vascular anomalies in children. Pediatr Blood Cancer. 2011;57:1018–1024. doi: 10.1002/pbc.23124. [DOI] [PubMed] [Google Scholar]
- 5.Barnes CM, Christison-Lagay EA, Folkman J. The placenta theory and the origin of infantile hemangioma. Lymphat Res Biol. 2007;5:245–255. doi: 10.1089/lrb.2007.1018. [DOI] [PubMed] [Google Scholar]
- 6.Khan ZA, Boscolo E, Picard A, et al. Multipotential stem cells recapitulate human infantile hemangioma in immunodeficient mice. J Clin Invest. 2008;118:2592–2599. doi: 10.1172/JCI33493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maclellan RA, Vivero MP, Purcell P, et al. Expression of follicle-stimulating hormone receptor in vascular anomalies. Plast Reconstr Surg. 2014;133:344e–351e. doi: 10.1097/01.prs.0000438458.60474.fc. [DOI] [PubMed] [Google Scholar]
- 8.Ayturk UM, Couto JA, Hann S, et al. Somatic Activating Mutations in GNAQ and GNA11 Are Associated with Congenital Hemangioma. Am J Hum Genet. 2016;98:789–795. doi: 10.1016/j.ajhg.2016.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nasseri E, Piram M, McCuaig CC, Kokta V, Dubois J, Powell J. Partially involuting congenital hemangiomas: a report of 8 cases and review of the literature. Journal of the American Academy of Dermatology. 2014;70:75–79. doi: 10.1016/j.jaad.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Lim YH, Bacchiocchi A, Qiu J, et al. GNA14 Somatic Mutation Causes Congenital and Sporadic Vascular Tumors by MAPK Activation. Am J Hum Genet. 2016;99:443–450. doi: 10.1016/j.ajhg.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim YH, Douglas SR, Ko CJ, et al. Somatic Activating RAS Mutations Cause Vascular Tumors Including Pyogenic Granuloma. J Invest Dermatol. 2015;135:1698–1700. doi: 10.1038/jid.2015.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groesser L, Peterhof E, Evert M, Landthaler M, Berneburg M, Hafner C. BRAF and RAS Mutations in Sporadic and Secondary Pyogenic Granuloma. J Invest Dermatol. 2016;136:481–486. doi: 10.1038/JID.2015.376. [DOI] [PubMed] [Google Scholar]
- 13.Behjati S, Tarpey PS, Sheldon H, et al. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat Genet. 2014;46:376–379. doi: 10.1038/ng.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Errani C, Zhang L, Sung YS, et al. A novel WWTR1-CAMTA1 gene fusion is a consistent abnormality in epithelioid hemangioendothelioma of different anatomic sites. Genes Chromosomes Cancer. 2011;50:644–653. doi: 10.1002/gcc.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arts FA, Sciot R, Brichard B, et al. PDGFRB gain-of-function mutations in sporadic infantile myofibromatosis. Hum Mol Genet. 2017;26:1801–1810. doi: 10.1093/hmg/ddx081. [DOI] [PubMed] [Google Scholar]
- 16.Shirley MD, Tang H, Gallione CJ, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couto JA, Huang L, Vivero MP, et al. Endothelial Cells from Capillary Malformations Are Enriched for Somatic GNAQ Mutations. Plast Reconstr Surg. 2016;137:77e–82e. doi: 10.1097/PRS.0000000000001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couto JA, Ayturk UM, Konczyk DJ, et al. A somatic GNA11 mutation is associated with extremity capillary malformation and overgrowth. Angiogenesis. 2017 doi: 10.1007/s10456-016-9538-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luks VL, Kamitaki N, Vivero MP, et al. Lymphatic and other vascular malformative/overgrowth disorders are caused by somatic mutations in PIK3CA. J Pediatr. 2015;166:1048–1054. e1041–1045. doi: 10.1016/j.jpeds.2014.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferrell RE, Levinson KL, Esman JH, et al. Hereditary lymphedema: evidence for linkage and genetic heterogeneity. Hum Mol Genet. 1998;7:2073–2078. doi: 10.1093/hmg/7.13.2073. [DOI] [PubMed] [Google Scholar]
- 21.Fang J, Dagenais SL, Erickson RP, et al. Mutations in FOXC2 (MFH-1), a forkhead family transcription factor, are responsible for the hereditary lymphedema-distichiasis syndrome. Am J Hum Genet. 2000;67:1382–1388. doi: 10.1086/316915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Irrthum A, Karkkainen MJ, Devriendt K, Alitalo K, Vikkula M. Congenital hereditary lymphedema caused by a mutation that inactivates VEGFR3 tyrosine kinase. Am J Hum Genet. 2000;67:295–301. doi: 10.1086/303019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irrthum A, Devriendt K, Chitayat D, et al. Mutations in the transcription factor gene SOX18 underlie recessive and dominant forms of hypotrichosis-lymphedema-telangiectasia. Am J Hum Genet. 2003;72:1470–1478. doi: 10.1086/375614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alders M, Hogan BM, Gjini E, et al. Mutations in CCBE1 cause generalized lymph vessel dysplasia in humans. Nat Genet. 2009;41:1272–1274. doi: 10.1038/ng.484. [DOI] [PubMed] [Google Scholar]
- 25.Mendola A, Schlogel MJ, Ghalamkarpour A, et al. Mutations in the VEGFR3 signaling pathway explain 36% of familial lymphedema. Mol Syndromol. 2013;4:257–266. doi: 10.1159/000354097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brouillard P, Boon L, Vikkula M. Genetics of lymphatic anomalies. J Clin Invest. 2014;124:898–904. doi: 10.1172/JCI71614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vikkula M, Boon LM, Carraway KL, 3rd, et al. Vascular dysmorphogenesis caused by an activating mutation in the receptor tyrosine kinase TIE2. Cell. 1996;87:1181–1190. doi: 10.1016/s0092-8674(00)81814-0. [DOI] [PubMed] [Google Scholar]
- 28.Limaye N, Wouters V, Uebelhoer M, et al. Somatic mutations in angiopoietin receptor gene TEK cause solitary and multiple sporadic venous malformations. Nat Genet. 2009;41:118–124. doi: 10.1038/ng.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soblet J, Limaye N, Uebelhoer M, Boon LM, Vikkula M. Variable Somatic TIE2 Mutations in Half of Sporadic Venous Malformations. Mol Syndromol. 2013;4:179–183. doi: 10.1159/000348327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limaye N, Kangas J, Mendola A, et al. Somatic Activating PIK3CA Mutations Cause Venous Malformation. Am J Hum Genet. 2015;97:914–921. doi: 10.1016/j.ajhg.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couto JA, Vivero MP, Kozakewich HP, et al. A somatic MAP3K3 mutation is associated with verrucous venous malformation. Am J Hum Genet. 2015;96:480–486. doi: 10.1016/j.ajhg.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Soblet J, Kangas J, Natynki M, et al. Blue Rubber Bleb Nevus (BRBN) Syndrome Is Caused by Somatic TEK (TIE2) Mutations. J Invest Dermatol. 2017;137:207–216. doi: 10.1016/j.jid.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 33.Brouillard P, Boon LM, Mulliken JB, et al. Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations ("glomangiomas") Am J Hum Genet. 2002;70:866–874. doi: 10.1086/339492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laberge-le Couteulx S, Jung HH, Labauge P, et al. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- 35.Liquori CL, Berg MJ, Siegel AM, et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergametti F, Denier C, Labauge P, et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Couto JA, Huang AY, Konczyk DJ, et al. Somatic MAP2K1 Mutations Are Associated with Extracranial Arteriovenous Malformation. Am J Hum Genet. 2017;100:546–554. doi: 10.1016/j.ajhg.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eerola I, Boon LM, Mulliken JB, et al. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet. 2003;73:1240–1249. doi: 10.1086/379793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Revencu N, Boon LM, Mulliken JB, et al. Parkes Weber syndrome, vein of Galen aneurysmal malformation, and other fast-flow vascular anomalies are caused by RASA1 mutations. Hum Mutat. 2008;29:959–965. doi: 10.1002/humu.20746. [DOI] [PubMed] [Google Scholar]
- 40.McAllister KA, Grogg KM, Johnson DW, et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat Genet. 1994;8:345–351. doi: 10.1038/ng1294-345. [DOI] [PubMed] [Google Scholar]
- 41.Johnson DW, Berg JN, Baldwin MA, et al. Mutations in the activin receptor-like kinase 1 gene in hereditary haemorrhagic telangiectasia type 2. Nat Genet. 1996;13:189–195. doi: 10.1038/ng0696-189. [DOI] [PubMed] [Google Scholar]
- 42.Gallione CJ, Repetto GM, Legius E, et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- 43.Wooderchak-Donahue WL, McDonald J, O'Fallon B, et al. BMP9 mutations cause a vascular-anomaly syndrome with phenotypic overlap with hereditary hemorrhagic telangiectasia. Am J Hum Genet. 2013;93:530–537. doi: 10.1016/j.ajhg.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jin Y, Muhl L, Burmakin M, et al. Endoglin prevents vascular malformation by regulating flow-induced cell migration and specification through VEGFR2 signalling. Nat Cell Biol. 2017;19:639–652. doi: 10.1038/ncb3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelen MR, Padberg GW, Peeters EA, et al. Localization of the gene for Cowden disease to chromosome 10q22–23. Nat Genet. 1996;13:114–116. doi: 10.1038/ng0596-114. [DOI] [PubMed] [Google Scholar]
- 46.Marsh DJ, Kum JB, Lunetta KL, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 47.Tan WH, Baris HN, Burrows PE, et al. The spectrum of vascular anomalies in patients with PTEN mutations: implications for diagnosis and management. J Med Genet. 2007;44:594–602. doi: 10.1136/jmg.2007.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurek KC, Luks VL, Ayturk UM, et al. Somatic mosaic activating mutations in PIK3CA cause CLOVES syndrome. Am J Hum Genet. 2012;90:1108–1115. doi: 10.1016/j.ajhg.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maclellan RA, Luks VL, Vivero MP, et al. PIK3CA activating mutations in facial infiltrating lipomatosis. Plast Reconstr Surg. 2014;133:12e–19e. doi: 10.1097/01.prs.0000436822.26709.7c. [DOI] [PubMed] [Google Scholar]
- 50.Couto JA, Konczyk DJ, Vivero MP, Kozakewich HPW, Upton J, Fu Xi, Padwa BL, Mulliken JB, Warman ML, Greene AK. Somatic PIK3CA Mutations are Present in Multiple Tissues of Facial Infiltrating Lipomatosis. Pediatric Research. 2017 doi: 10.1038/pr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amary MF, Damato S, Halai D, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 52.Mirzaa GM, Riviere JB, Dobyns WB. Megalencephaly syndromes and activating mutations in the PI3K-AKT pathway: MPPH and MCAP. Am J Med Genet C Semin Med Genet. 2013;163C:122–130. doi: 10.1002/ajmg.c.31361. [DOI] [PubMed] [Google Scholar]
- 53.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harbor perspectives in biology. 2012;4:a011189. doi: 10.1101/cshperspect.a011189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15:577–592. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez-Viciana P, Tetsu O, Tidyman WE, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. [DOI] [PubMed] [Google Scholar]
- 56.Nikolaev SI, Rimoldi D, Iseli C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 57.Chakraborty R, Hampton OA, Shen X, et al. Mutually exclusive recurrent somatic mutations in MAP2K1 and BRAF support a central role for ERK activation in LCH pathogenesis. Blood. 2014;124:3007–3015. doi: 10.1182/blood-2014-05-577825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arcila ME, Drilon A, Sylvester BE, et al. MAP2K1 (MEK1) Mutations Define a Distinct Subset of Lung Adenocarcinoma Associated with Smoking. Clin Cancer Res. 2015;21:1935–1943. doi: 10.1158/1078-0432.CCR-14-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Druker BJ, Tamura S, Buchdunger E, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 60.Keppler-Noreuil KM, Rios JJ, Parker VE, et al. PIK3CA-related overgrowth spectrum (PROS): diagnostic and testing eligibility criteria, differential diagnosis, and evaluation. Am J Med Genet A. 2015;167A:287–295. doi: 10.1002/ajmg.a.36836. [DOI] [PMC free article] [PubMed] [Google Scholar]