ABSTRACT
The mouse model of West Nile virus (WNV), which is a leading cause of mosquito-borne encephalitis worldwide, has provided fundamental insights into the host and viral factors that regulate viral pathogenesis and infection outcome. In particular, CD8+ T cells are critical for controlling WNV replication and promoting protection against infection. Here, we present the characterization of a T cell receptor (TCR)-transgenic mouse with specificity for the immunodominant epitope in the WNV NS4B protein (here referred to as transgenic WNV-I mice). Using an adoptive-transfer model, we found that WNV-I CD8+ T cells behave similarly to endogenous CD8+ T cell responses, with an expansion phase in the periphery beginning around day 7 postinfection (p.i.) followed by a contraction phase through day 15 p.i. Through the use of in vivo intravascular immune cell staining, we determined the kinetics, expansion, and differentiation into effector and memory subsets of WNV-I CD8+ T cells within the spleen and brain. We found that red-pulp WNV-I CD8+ T cells were more effector-like than white-pulp WNV-I CD8+ T cells, which displayed increased differentiation into memory precursor cells. Within the central nervous system (CNS), we found that WNV-I CD8+ T cells were polyfunctional (gamma interferon [IFN-γ] and tumor necrosis factor alpha [TNF-α]), displayed tissue-resident characteristics (CD69+ and CD103+), persisted in the brain through day 15 p.i., and reduced the viral burden within the brain. The use of these TCR-transgenic WNV-I mice provides a new resource to dissect the immunological mechanisms of CD8+ T cell-mediated protection during WNV infection.
IMPORTANCE West Nile Virus (WNV) is the leading cause of mosquito-borne encephalitis worldwide. There are currently no approved therapeutics or vaccines for use in humans to treat or prevent WNV infection. CD8+ T cells are critical for controlling WNV replication and protecting against infection. Here, we present a comprehensive characterization of a novel TCR-transgenic mouse with specificity for the immunodominant epitope in the WNV NS4B protein. In this study, we determine the kinetics, proliferation, differentiation into effector and memory subsets, homing, and clearance of WNV in the CNS. Our findings provide a new resource to dissect the immunological mechanisms of CD8+ T cell-mediated protection during WNV infection.
KEYWORDS: brain, CD8+ T cells, spleen, West Nile virus, neuroimmunology, viral pathogenesis
INTRODUCTION
West Nile virus (WNV) is a positive-sense single-stranded RNA virus that belongs to the family Flaviviridae and is transmitted by mosquito vectors (1). Since its introduction into the United States in 1999, WNV has remained the leading cause of mosquito-borne encephalitis (1, 2). WNV infection is generally asymptomatic in the vast majority of individuals; however, symptomatic individuals can present with arthralgia, myalgia, and cephalea. A small percentage of WNV-infected individuals may also progress to encephalitis that can be fatal or result in permanent neurologic deficits (3, 4). Neuroinvasive WNV infection is more prevalent among elderly and immunodeficient individuals (5). Currently, there are no antivirals or vaccines approved for use in humans to treat or prevent WNV infection.
Studies in humans infected with WNV have provided valuable insights into the correlates of protective immune responses. Postmortem central nervous system (CNS) tissues from individuals who have succumbed to WNV infection show generalized parenchymal infiltration of CD3+ T cells, which colocalize to areas of viral antigen (6, 7). Observations in the peripheral blood of symptomatic WNV-infected patients found that neuroinvasive disease was correlated with atypical CD4+ T cells that expressed Th1 and Th2 cytokines simultaneously (8). Additional studies have found a positive correlation between symptomatic WNV disease and increased T cell immunoglobulin domain-containing molecule 3 (Tim-3) expression on CD8+ T cells, strongly suggesting that WNV may induce T cell-inhibitory molecules as a mechanism to dampen T cell immune responses (9). Combined, the data show that T cells play an integral role in mediating clinical disease progression and infection outcome in humans.
WNV infection of mice recapitulates many aspects of viral pathogenesis observed in WNV-infected humans (10). Through the use of the murine model, several components of the innate and adaptive immune response have been found to control WNV replication, tissue tropism, and infection outcome. Following WNV infection, CD8+ T cells are activated and reach peak expansion in the periphery by day 7 postinfection (p.i.), followed by CXCR3-dependent trafficking to the CNS (11). There, CD8+ T cells control virus dissemination, limit neuronal injury, and mediate viral clearance through cytolytic (Fas, TRAIL, and perforin) and, potentially, noncytolytic mechanisms (12–15). Recently, the CD8+ T cell immunodominant epitope within WNV was identified, which has provided insight into the dynamics of virus-specific CD8+ T cell responses during WNV infection (16, 17). However, we still have a rudimentary understanding of the kinetics, differentiation, expansion, and trafficking of WNV-specific CD8+ T cells to the CNS during infection.
In this study, we present the generation and characterization of a novel T cell receptor (TCR)-transgenic mouse with specificity for the immunodominant epitope in the WNV NS4B protein (here referred to as transgenic WNV-I mice). Using an adoptive-transfer model, we found that WNV-I CD8+ T cells behave similarly to endogenous CD8+ T cell responses, with an expansion phase in the periphery beginning around day 7 p.i., followed by a contraction phase through day 15 p.i. Through the use of in vivo intravascular (i.v.) antibody immune cell staining, we determine that the kinetics, expansion, and differentiation into effector and memory subsets of WNV-I CD8+ T cells are influenced by anatomic location within the spleen. Within the CNS, we found that WNV-I CD8+ T cells are polyfunctional (gamma interferon [IFN-γ] and tumor necrosis factor alpha [TNF-α]); display a tissue-resident phenotype, characterized by increased CD69 and CD103 expression; and persist through day 15 p.i. Most importantly, WNV-I cells were able to reduce the viral burden within the brains of WNV-infected mice.
RESULTS
Titration of WNV-I T cells during WNV infection.
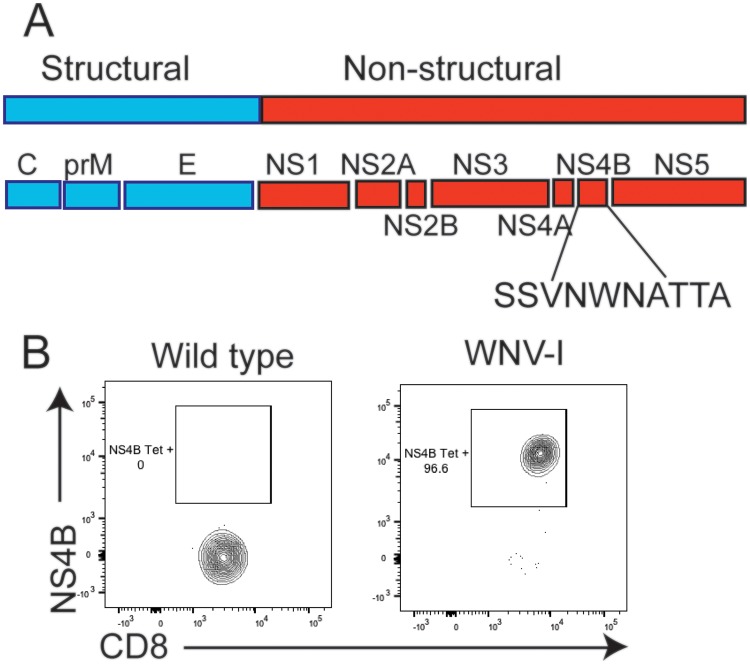
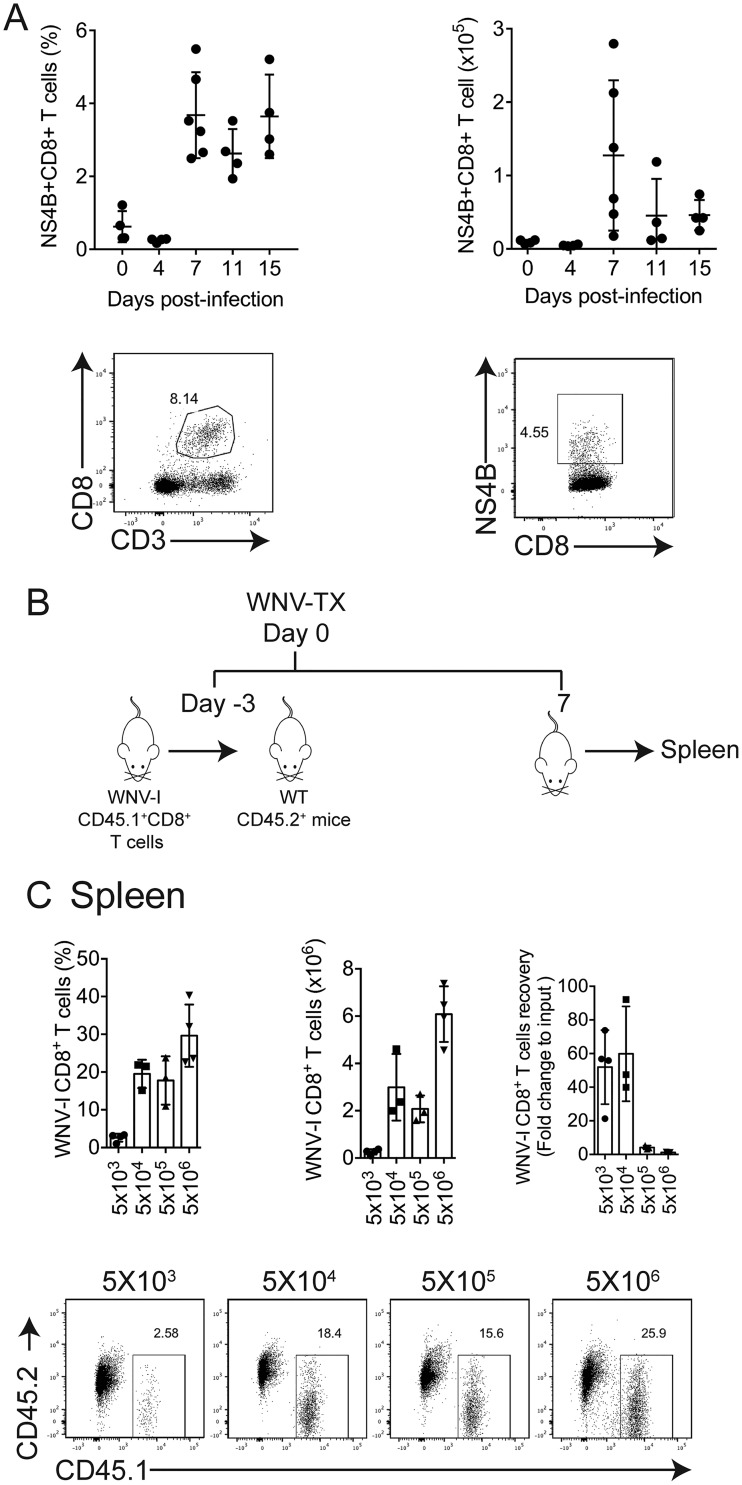
CD8+ T cells are essential for regulating protection against WNV infection (17). However, studying CD8+ T cells in the context of WNV infection has been challenging due to the lack of a T cell-transgenic system. Previous studies identified the Db binding immunodominant peptide epitope within WNV NS4b (SSVWNATTAI) following infection of C57BL/6 mice (16, 17). In this study, we report the generation of a TCR-transgenic mouse with specificity for this immunodominant epitope found within the WNV NS4B protein (here referred to as WNV-I mice) (Fig. 1A). We isolated and stained peripheral blood mononuclear cells (PBMCs) from naive WNV-I TCR-transgenic mice with a Db-NS4b tetramer (18) and found that more than 95% of the CD8+ T cells were WNV specific, which is consistent with other T cell-transgenic mouse models (Fig. 1B) (19, 20). Following WNV infection of C57BL/6 mice, WNV-specific CD8+ T cells peak on day 7 p.i. within the spleen (Fig. 2A). To determine an optimal number of WNV cells that could be tracked during infection, we adoptively transferred between 5 × 103 and 5 × 106 WNV-I CD8+ T cells into congenically marked mice and evaluated the expansion of WNV-I CD8+ T cells on day 7 p.i. (Fig. 2B). Three days following adoptive transfer, recipient mice were infected with 100 PFU of WNV isolate TX 2002-HC (WNV-TX) through the subcutaneous (s.c.) route. In the spleen, we observed a substantial increase in recovery/expansion of WNV-I CD8+ T cells following transfer of either 5 × 103 (52-fold) or 5 × 104 (59.8-fold) cells (Fig. 2C). Interestingly, we observed a dramatic reduction in this ratio in WNV-infected recipient mice that received either 5 × 105 (3.1-fold) or 5 × 106 (1.1-fold) WNV-I CD8+ T cells. Based on these findings, we proceeded to use 5 × 104 WNV-I CD8+ T cells as an optimal number of transgenic cells to transfer into recipient mice for analyzing CD8+ T cell responses during WNV infection.
FIG 1.
Generation of WNV-I CD8+ T cells. (A) WNV genome showing structural proteins (blue) and nonstructural proteins (red). The peptide sequence for the immunodominant epitope in the NS4B protein is shown. (B) Flow cytometry staining of peripheral blood obtained from naive C57BL/6J (left) or WNV-I (right) mice using a WNV-specific tetramer (NS4B) and anti-CD8 antibody.
FIG 2.
Titration of WNV-I-specific CD8+ T cells. (A) Expansion of endogenous WNV-specific CD8+ T cells during WNV-I infection. (B) Experimental design. Increasing amounts of WNV-I CD8+ T cells were transferred to congenically marked mice 3 days prior to infection. The mice were infected with WNV-TX (100 PFU) via the s.c. route. WNV-I CD8+ T cells in the spleen were evaluated on day 7 p.i. WT, wild type,. (C) (Top) (Left and middle) Frequency (left) and counts (middle) of WNV-I CD8+ T cells recovered from adoptive transfers on day 7 p.i. (Right) Fold change recovery calculated relative to the initial input of WNV-I CD8+ T cells. (Bottom) Representative flow cytometry plots for individual adoptive-transfer conditions. The data are representative of the results of two independent experiments (n = 3 or 4 mice per group). The error bars indicate standard deviations.
WNV-specific CD8+ T cells accumulate within the RP of the spleen.
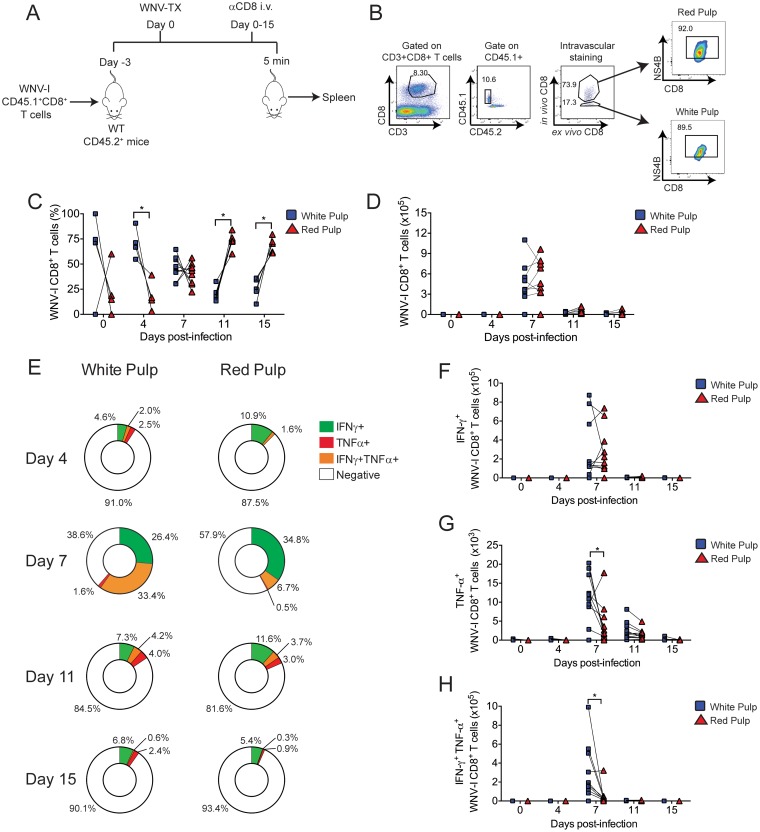
In vivo intravascular immune cell staining has been demonstrated to be a valuable technique for identifying tissue-resident immune cells (21, 22). Next, we combined adoptive transfer of WNV-I CD8+ T cells with in vivo intravascular immune cell staining to better understand the dynamics and composition of WNV-specific CD8+ T cell responses in the spleen during WNV infection (Fig. 3A and B). As a baseline, uninfected mice showed an accumulation of WNV-I-specific CD8+ T cells within the white pulp (WP) versus the red pulp (RP) of the spleen on day 3 posttransfer (Fig. 3C). This finding is consistent with previous studies using P14 CD8+ T cells (23). Following WNV infection, we observed strong skewing in the frequency of WNV-I CD8+ T cells within the RP compared to the WP of the spleen (Fig. 3C). On day 7 p.i., we observed a reduction in the frequency of WNV-I CD8+ T cells within the WP with a concomitant increase in the frequency of WNV-I CD8+ T cells within the RP. The frequency of WNV-I CD8+ T cells within the RP was higher than in the WP on day 15 p.i. (RP, 67.9% ± 3% versus WP, 27.1% ± 4%; P < 0.05). Interestingly, the total number of WNV-I CD8+ T cells expanded through day 7 p.i. and contracted through day 15 p.i. (Fig. 3D).
FIG 3.
Distribution of WNV-I CD8+ T cells in the spleen during WNV infection. (A) Experimental design. WNV-I CD8+ T cells (5 × 104) were transferred into congenically marked mice. Three days later, the recipients were infected with 100 PFU of WNV-TX s.c., and on the day of sacrifice, the mice received 10 μg of allophycocyanin-labeled anti-CD8 antibody for 5 min through the intravenous route. (B) Gating strategy to determine the anatomic location of WNV-I CD8+ T cells in the spleen. (C and D) Frequency (C) and cell counts (D) of WNV-I CD8+ T cells present in RP or WP compartments during WNV infection. (E) Frequencies of IFN-γ-single-positive, TNF-α-single-positive, and IFN-γ- TNF-α-double-positive cells in the spleen. (F to H) Cell counts of IFN-γ- and TNF-α-single- and double-positive WNV-I CD8+ T cells in the spleen. The data are representative of the results of two independent experiments (n = 4 to 9 mice per group). *, P < 0.05; paired t test.
We next evaluated the functional properties of WNV-specific CD8+ T cell responses within the spleen. Following ex vivo restimulation with a peptide carrying the immunodominant WNV CD8+ T cell epitope, we observed both single- and double-positive IFN-γ- and TNF-α-secreting cells through day 15 p.i. Interestingly, cytokine responses between RP and WP WNV CD8+ T cells were heterogeneous in nature, but peak responses occurred on day 7 p.i. (Fig. 3E to H). The frequency of IFN-γ single-positive (RP, 34% ± 5.2% versus WP, 26.4% ± 3.7%; P < 0.05) WNV-I CD8+ T cells were slightly, yet significantly, higher in the RP than the WP on day 7 p.i. In contrast, cell counts and frequencies of TNF-α single-positive (WP, 1.6% ± 0.7% versus RP, 0.5% ± 0.6%; P < 0.05) and IFN-γ and TNF-α double-positive (WP, 33.4% ± 6.7% versus RP, 6.7% ± 2.8%; P < 0.05) WNV-I CD8+ T cells were slightly, yet significantly, higher in the WP than in the RP at 7 and 11 days p.i., respectively. Combined, our findings demonstrate that the WNV-I CD8+ T cells are functional and respond during WNV infection. Furthermore, the anatomic location within the spleen has a strong influence on the distribution and cytokine potential of CD8+ T cell responses during WNV infection.
Effector and memory WNV-specific CD8+ T cell differentiation within the spleen.
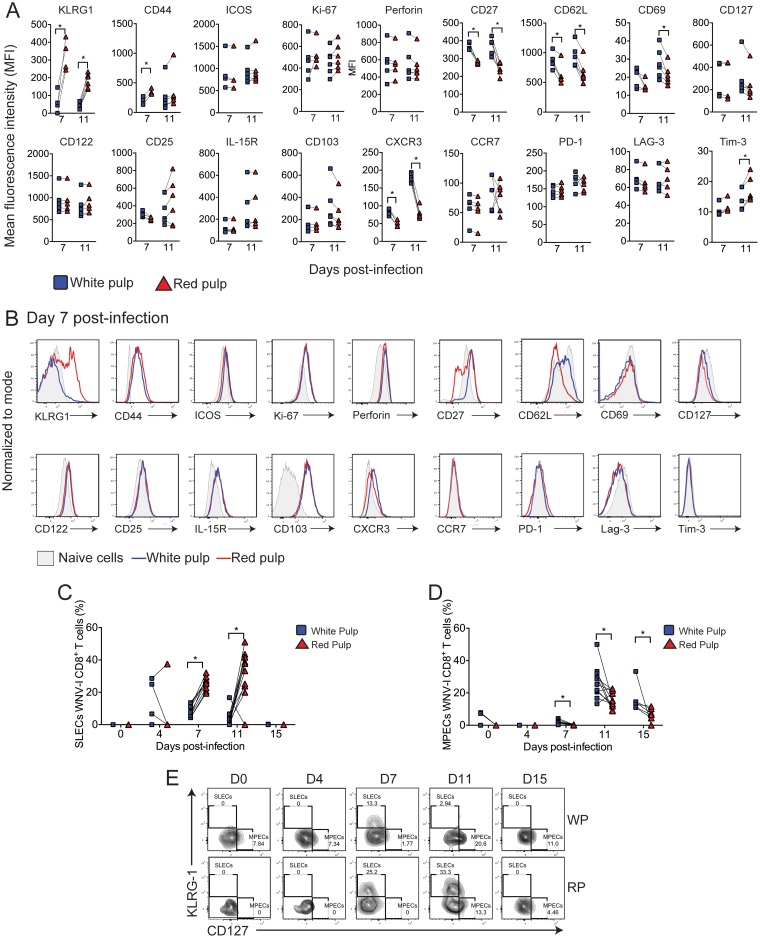
We next characterized differentiation into effector and memory WNV-specific CD8+ T cell subsets during infection (Fig. 4A and B). We observed that KLRG1 (RP, mean fluorescence intensity [MFI] of 325.8 ± 44.2, versus WP, 62.5 ± 30.3; P < 0.05) and CD44 (RP, MFI of 354 ± 30.9, versus WP, 205 ± 40; P < 0.05), two cell surface markers of effector T cell differentiation, showed significantly increased expression on WNV-I CD8+ T cells within the RP compared to the WP of the spleen on day 7 p.i. Interestingly, WP WNV-I CD8+ T cells on days 7 and 11 p.i. displayed KLRG1 expression similar to that of naive WNV-I CD8+ T cells. Compared to naive CD8+ T cells, ICOS, which is a marker typically found on activated CD8+ T cells (24); Ki-67, which is a molecule expressed on cells that have undergone recent proliferation; and perforin, which is a T cell effector molecule, were similarly upregulated on WNV-I CD8+ T cells within the RP or WP on day 7 or 11 p.i.
FIG 4.
Effector and memory CD8+ T cell differentiation during WNV infection. WNV-I CD8+ T cells (5 × 104) were transferred into congenically marked mice. Three days later, the recipients were infected with 100 PFU of WNV-TX s.c., and on the day of sacrifice, the mice received 10 μg of allophycocyanin-labeled anti-CD8 antibody for 5 min through the intravenous route. (A and B) Mean fluorescence intensity (MFI) (A) and representative histograms (B) on day 7 p.i. (C) Frequencies of WNV-I CD8+ T SLECs in the WP and RP. (D) Frequencies of WNV-I CD8+ T MPEC phenotypes in the WP and RP. (E) Representative flow cytometry plots of WNV-I CD8+ T SLECs and MPECs during WNV infection. The data are representative of the results of two independent experiments (n = 4 or 5 mice per group). *, P < 0.05; paired t test.
We next evaluated memory markers on WNV-I CD8+ T cells within the RP or WP during WNV infection. CD27, a marker typically found on central and effector memory T cells (25–27), was expressed at significantly higher levels on WP than RP WNV-I CD8+ T cells on days 7 (WP, MFI of 368.6 ± 21.2, versus RP, 280 ± 9.0; P < 0.05) and 11 (WP, MFI of 379 ± 49.4, versus RP, 239.3 ± 3; P < 0.05) p.i. In contrast, CD62L, a marker of memory CD8+ T cell differentiation, was downregulated to a greater extent on RP than WP WNV-I CD8+ T cells or naive T cells on days 7 (RP, MFI of 631 ± 186, versus WP, 869 ± 134.4; P < 0.05) and 11 (RP, MFI of 667.5 ± 189.6, versus WP, 932 ± 189.2; P < 0.05) p.i. Expression of CD69, a marker of tissue-resident memory CD8+ T cells, on WNV-I CD8+ T cells did not change relative to naive CD8+ T cells on day 7 p.i., but WP WNV-I CD8+ T cells did show increased expression on day 11 p.i. (WP, MFI of 27.6 ± 8.1, versus RP, 21.5 ± 5.5; P < 0.05). We next evaluated memory precursor effector cell (MPEC) (KLRG-1-CD127+) and short-lived effector cell (SLEC) (KLRG-1+CD127−) subsets on WNV-I CD8+ T cells found within the spleen (Fig. 4C to E). We observed a greater frequency of SLECs in the RP than in the WP of the spleen on days 7 (RP, 25.5% ± 1.3% versus WP, 8.4% ± 1.24%; P < 0.05) and 11 (RP, 32.5% ± 4.3% versus WP, 4% ± 1.5%; P < 0.05) p.i. (Fig. 4C). Conversely, we observed a significant enrichment of MPECs within the WP compared to the RP of the spleen on days 7 (2.1% ± 0.4% versus 0.17% ± 0.1%; P < 0.05), 11 (26.5% ± 3.0% versus 15.7% ± 1.5%; P < 0.05), and 15 (16.16% ± 3.4% versus 6.7% ± 1.7%; P < 0.05) p.i. (Fig. 4D). While CD122, CD25, interleukin 15 receptor (IL-15R), and CD103, which are additional markers of T cell memory, were upregulated on WNV-I CD8+ T cells compared to naive CD8+ T cells, no differences in expression were observed between WNV-I CD8+ T cells located within the RP and those in the WP.
We next evaluated homing molecules typically found on effector CD8+ T cells during virus infection. CXCR3, a chemokine receptor that has previously been implicated in promoting CD8+ T cell trafficking to the CNS during WNV infection (11), was expressed at significantly higher levels on WNV-I CD8+ T cells in the WP than on those in the RP on days 7 (WP, MFI of 83.5 ± 8.8, versus RP, 50.4 ± 7.7; P < 0.05) and 11 (WP, MFI of 178.1 ± 12.2, versus RP, 79.6 ± 15.4; P < 0.05) p.i. CCR7 was not expressed on WNV-I CD8+ T cells more than in naive controls at either time point.
Lastly, we measured the expression of the T cell inhibitor markers PD-1, Lag-3, and Tim-3 on WNV-I CD8+ T cells. We found that PD-1 and Lag-3 were not differentially expressed on RP or WP WNV-I CD8+ T cells compared to naive CD8+ T cells. However, we did observe a modest, yet significant, increase in Tim-3 expression on RP compared to WP WNV-I CD8+ T cells on day 11 p.i. (RP, MFI of 17.7 ± 1.9, versus WP, 14.3 ± 1.6; P < 0.05). Altogether, these data indicate that WNV-I CD8+ T cells display asymmetric phenotypic characteristics depending on their anatomic location within the spleen. Our data indicate that effector WNV-I CD8+ T cells tend to accumulate in the RP and memory precursor cells accumulate in the WP of the spleen.
Polyfunctional WNV-I CD8+ T cells infiltrate the brain on day 7 postinfection.
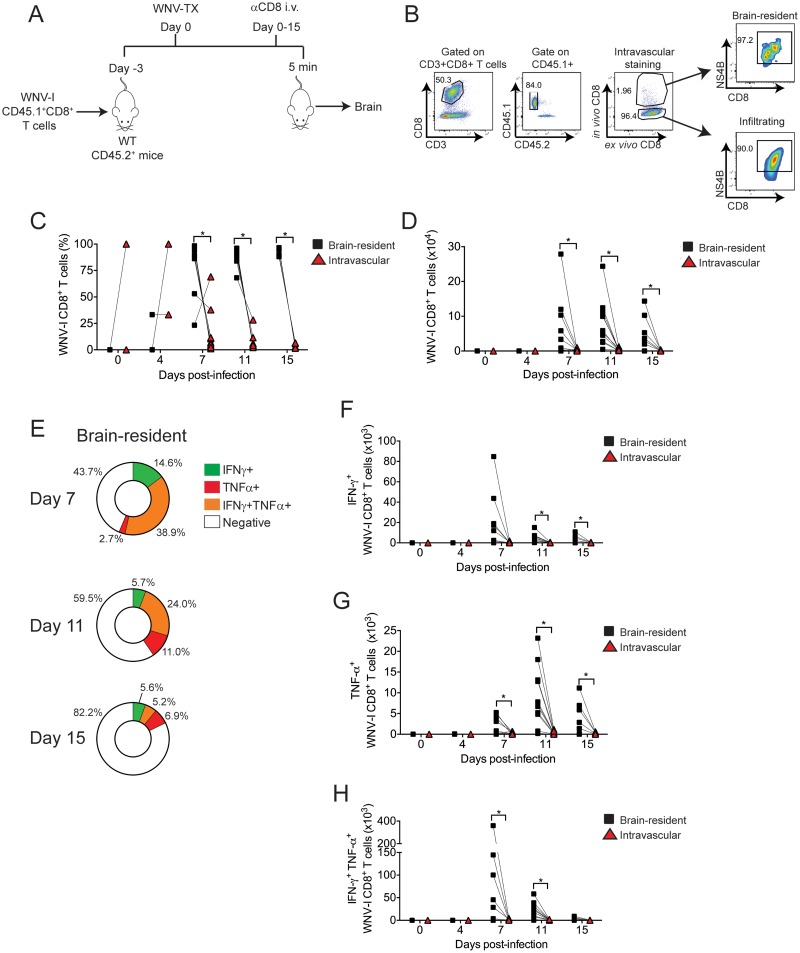
Next, we used in vivo intravascular immune cell staining to differentiate between intravascular and brain-resident (BR) CD8+ T cells during WNV infection (Fig. 5A and B). Similar to endogenous NS4B-specific CD8+ T cell responses during WNV infection (28, 29), WNV-I CD8+ T cells infiltrated the CNS beginning on day 7 p.i. and remained in the brain parenchyma through day 15 p.i. (Fig. 5C and D). Interestingly, from day 7 p.i. onward, a majority of the WNV-I CD8+ T cells were identified as brain resident rather than localized within the vasculature.
FIG 5.
WNV-I CD8+ T cells are polyfunctional in the brain parenchyma. (A) WNV-I CD8+ T cells (5 × 104) were transferred into congenically marked mice. Three days later, the recipients were infected with 100 PFU of WNV-TX s.c., and on the day of sacrifice, the mice received 10 μg of allophycocyanin-labeled anti-CD8 antibody for 5 min through the i.v. route. (B) Gating strategy to determine the anatomic location of WNV-I CD8+ T cells in the brain. (C and D) Frequency (C) and cell counts (D) of brain-resident and intravascular WNV-I CD8+ T cells during the course of WNV infection. (E) Percentages of IFN-γ-single-positive, TNF-α-single-positive, and IFN-γ- TNF-α-double-positive cells in the brain. (F to H) Cell counts of IFN-γ- and TNF-α-single- and double-positive WNV-I CD8+ T cells recovered from the brain. The data are representative of the results of two independent experiments with at least 4 mice per time point. *, P < 0.05; paired t test.
We next evaluated the functional properties of brain-resident WNV-I CD8+ T cells following ex vivo peptide restimulation. Similar to WNV-specific CD8+ T cells within the spleen, we observed both single- and double-positive IFN-γ- and TNF-α-secreting WNV-I CD8+ T cells through day 15 p.i. On days 7 and 11 p.i., IFN-γ- and TNF-α-double-positive cells were more prevalent and had higher cell counts than individual single-positive cells on brain-resident WNV-I CD8+ T cells (Fig. 5E to H). By day 15 p.i., brain-resident WNV-I CD8+ T cells showed markedly reduced cytokine responses. These findings demonstrate that early during CNS neuroinvasion, WNV-specific CD8+ T cell responses are multifunctional and the cells secrete both IFN-γ and TNF-α.
WNV-I CD8+ T cells in the brain parenchyma acquire a tissue-resident phenotype.
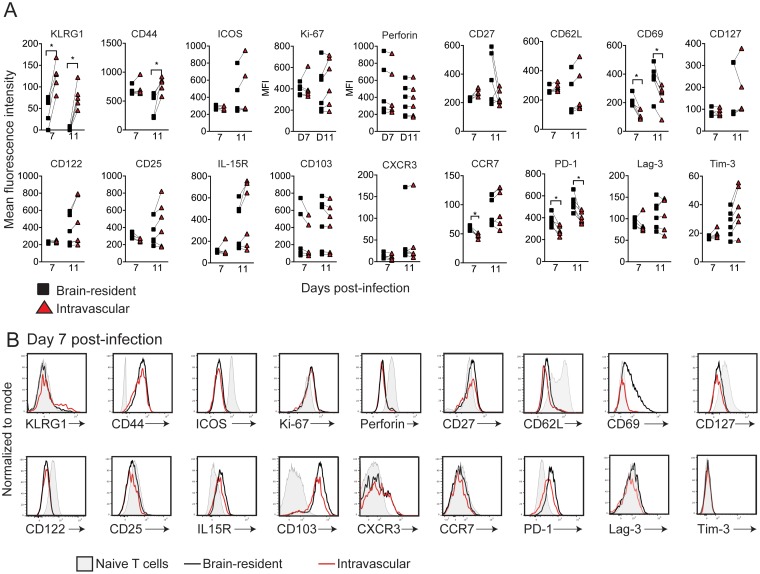
We next examined the expression of surface markers on brain-resident and intravascular WNV-I CD8+ T cells during WNV infection (Fig. 6A and B). We observed a significant decrease in KLRG1 expression on BR compared to i.v. WNV-I CD8+ T cells on days 7 (BR, MFI of 47.5 ± 32.7, versus i.v., 123.5 ± 32.3; P < 0.05) and 11 (BR, MFI of 1.7 ± 3.9, versus i.v., 77.38 ± 28.5; P < 0.05) p.i. Similarly, CD44 expression was significantly decreased on brain-resident compared to intravascular WNV-I CD8+ T cells on day 11 p.i. (BR, MFI of 447 ± 205.3, versus i.v., 782.8 ± 133.2; P < 0.05). We observed a significant reduction in ICOS expression in both vasculature and brain parenchyma WNV-I CD8+ T cells relative to naive CD8+ T cells. While slightly increased over naive CD8+ T cells, Ki-67 showed no difference between brain-resident and intravascular WNV-I CD8+ T cells. Intriguingly, perforin expression was profoundly decreased on brain-resident WNV-I CD8+ T cells compared to naive CD8+ T cells. Both CXCR3 and CCR7 showed little to no expression on either brain-resident or intravascular WNV-I CD8+ T cells compared to naive CD8+ T cells. Altogether, these findings strongly suggest that WNV-I CD8+ T cells undergo dynamic changes as they enter the brain region, with reduced activation and proliferation.
FIG 6.
WNV-I CD8+ T cells are tissue-resident T cells in the CNS. WNV-I CD8+ T cells (5 × 104) were transferred into congenically marked mice. Three days later, the recipients were infected with 100 PFU of WNV-TX s.c., and on the day of sacrifice, the mice received 10 μg of allophycocyanin-labeled anti-CD8 antibody for 5 min through the i.v. route. (A and B) MFI (A) and representative histograms (B) on day 7 p.i. The data are representative of the results of two independent experiments with at least 5 mice per condition. *, P < 0.05; paired t test.
In terms of cell surface expression of memory markers, we observed profound, yet similar, downregulation of CD62L, CD127, CD122, and CD25 expression on brain-resident and intravascular WNV-I CD8+ T cells compared to naive CD8+ T cell controls. Interestingly, CD27 and IL-15R showed increased expression on brain WNV-I CD8+ T cells compared to naive CD8+ T cell controls; however, no significant differences were observed between brain-resident and intravascular cells.
As CD8+ T cells enter a tissue during pathogen infection, they acquire a tissue-resident phenotype characterized by upregulation of CD69 (30) and CD103 (31, 32) expression. We observed increased expression of CD69 and CD103 on brain-resident WNV-I CD8+ T cells compared to naive CD8+ T cells on day 7 and day 11 p.i. Interestingly, CD69, but not CD103, displayed differential expression on brain-resident versus intravascular WNV-I CD8+ T cells, suggesting that CD69 is a marker of tissue-resident CD8+ T cells within the brain on day 7 (BR, MFI of 212.4 ± 40.6, versus i.v., 118.8 ± 33.3; P < 0.05) and on day 11 (BR, MFI of 367.5 ± 104.9, versus i.v., 243.5 ± 9; P < 0.05) p.i.
The inhibitory receptor PD-1, but not Lag-3 or Tim-3, showed significantly increased expression on brain-resident compared to intravascular WNV-I CD8+ T cells or naive CD8+ T cells on day 7 (BR, MFI of 377.8 ± 60.0, versus i.v., 284 ± 5; P < 0.05) and day 11 (BR, MFI of 531.5 ± 74.7, versus i.v., 402 ± 51.8; P < 0.05) p.i. Altogether, these data indicate that WNV-I CD8+ T cells undergo dynamic changes as they differentiate into brain-resident CD8+ T cells.
WNV-I CD8+ T cells reduce the viral burden.
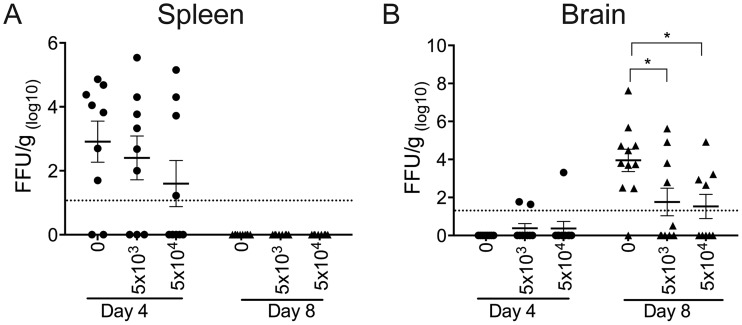
We next evaluated the antiviral potential of the WNV-I CD8+ T cells to reduce the WNV burden in the periphery and CNS. We adoptively transferred 5 × 103 or 5 × 104 WNV-I CD8+ T cells into C57BL/6J recipient mice and infected the mice with WNV-TX 3 days posttransfer. Spleen and brain tissues were harvested on days 4 and 8 p.i. On day 4 p.i., we observed a slight reduction in viral titers in the spleen with increasing amounts of WNV-I CD8+ T cells transferred into the recipient mice (Fig. 7A). On day 8 p.i., the viral titers were below the limit of detection in the spleens of all the experimental groups. In the brain, we detected WNV in the brains of mice that received 5 × 103 (2 out of 10 mice) or 5 × 104 (1 out of 9 mice) WNV-I CD8+ T cells on day 4 p.i., although these differences were not statistically significant. Most importantly, on day 8 p.i., we observed a statistically significant reduction in the WNV burden in mice that received 5 × 103 (76-fold reduction; P < 0.05) or 5 × 104 (400-fold reduction; P < 0.05) WNV-I CD8+ T cells compared to mice that received no WNV-I CD8+ T cells (Fig. 7B). Altogether, these findings demonstrate that WNV NS4B-specific CD8+ T cells are sufficient to reduce the WNV burden in the CNS.
FIG 7.
WNV-I CD8+ T cells reduce the viral burden in C57BL/6J mice. WNV-I CD8+ T cells (5 × 103 or 5 × 104) were transferred into C57BL/6J mice, and the mice were infected with WNV-TX via the s.c. route 3 days posttransfer. Tissues were harvested on days 4 and 8 p.i., and the viral burdens within the spleen (A) and brain (B) were determined by focus-forming assay. The data are represented as focus-forming units (FFU) per gram of tissue and are combined (n = 9 to 11 mice in total) from the results of two independent experiments with at least 5 mice per experimental condition. *, P < 0.05; one-way ANOVA.
DISCUSSION
In this study, we report the generation of TCR-transgenic mice to track and study virus-specific CD8+ T cell responses during WNV infection. We combined in vivo intravascular immune cell staining to characterize circulating and tissue-resident CD8+ T cell responses. Using an optimized number of transgenic WNV-I CD8+ T cells, we found that anatomic location within the spleen, brain, and vasculature influences the activation and effector functions of virus-specific CD8+ T cells during WNV infection. Most importantly, we found that the transgenic WNV-I CD8+ T cells were able to reduce the viral burden in the CNS during WNV infection.
Adoptive transfer of TCR-transgenic CD8+ T cells is a powerful way to track and study virus-specific CD8+ T cell responses during infection. During our initial characterization of WNV-I CD8+ T cells, we observed that increasing the number of WNV-I CD8+ T cells by 10- or 100-fold resulted in a dramatic reduction in the recovery of WNV-I CD8+ T cells on day 7 p.i. This phenomenon, also referred to as a “ceiling” of the effector T cell response, has been observed in other experimental model systems (33) and pathogen infection (34, 35). It is plausible that increasing the initial frequency of WNV-I CD8+ T cells results in altering viral pathogenesis (36) or the kinetics of T cell expansion or proliferative burst (37). Further studies are needed to better understand how the initial frequency of TCR-transgenic cells impacts the viral pathogenesis and kinetics of CD8+ T cell activation and expansion during WNV infection.
In the spleen, we observed that WNV-I CD8+ T cells are preferentially distributed within the RP compared to the WP of the spleen. In both the RP and WP, WNV-I CD8+ T cells differentiated into effector cells, as determined by cell surface expression of KLRG1, CD44, CD69, and CXCR3 and their ability to secrete inflammatory cytokines (TNF-α and IFN-γ). Similar to lymphocytic choriomeningitis virus (LCMV) infection, we also observed preferential expansion of SLECs and MPECs within the RP and WP of the spleen, respectively. It is possible that a combination of factors, such as antigen-presenting cell (APC) activation (38), viral replication, and regulatory T cells (23), may promote the differential expansion and polyfunctionality of WNV-I CD8+ T cells within the spleen during virus infection.
WNV-specific CD8+ T cells migrate to the CNS around 7 days p.i. and persist in the CNS through 6 months post-WNV infection despite the lack of detection of viral RNA within the brain (39, 40). In a similar manner, we found that CNS-resident WNV-I CD8+ T cells (CD69+ CD103+) persisted in the CNS through day 15 p.i. In other models, specifically LCMV and vesicular stomatitis virus (VSV), virus-specific CD8+ T cells have been reported to accumulate within the CNS following pathogen clearance (30, 31). However, it is not yet clear what role, if any, these tissue-resident CD8+ T cells play in mediating immune homeostasis within the CNS. A potential hypothesis is that virus-specific CD8+ T cells may persist in the CNS to block subsequent viral infections or virus reactivation. Furthermore, it will be interesting to determine the host factors that regulate maintenance of WNV-specific CD8+ T cells within the brain and whether antigen is required for CD8+ T cell persistence within the brain following WNV clearance.
The mechanisms by which CD8+ T cells mediate clearance within the CNS during WNV infection have been studied but are not yet well defined. Recent studies using global knockout mice and endogenous CD8+ T cells have identified potential host factors, including TNF-α, IFN-γ, TRAIL, and perforin, as important mediators in controlling WNV replication within the CNS (12, 13). Furthermore, dendritic cell (DC)-CD8+ T cell interactions within the CNS, as well as IL-1 signaling, have also been shown to play important roles in programming protective CD8+ T cells during WNV infection (41). These transgenic WNV-I CD8+ T cells can now be used to more accurately dissect mechanisms that mediate viral control within the CNS and protection against lethal WNV infection.
In summary, we have presented the generation and characterization of a novel WNV TCR-transgenic mouse that can be used to better understand CD8+ T cell responses during WNV infection. These mice are available to the scientific community and will allow future studies to uncover mechanisms underlying differentiation into memory CD8+ T cells, homing to the CNS, maintenance of tissue-resident CD8+ T cells within the CNS, and mechanisms of neuroprotection during WNV infection.
MATERIALS AND METHODS
Ethics statement.
This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Animal experiments were approved and performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at Emory University (protocol number YER-2002219-111016N).
Cells and viruses.
BHK-21 cells were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), HEPES, l-glutamine, sodium pyruvate, antibiotic-antimycotic solution, and nonessential amino acids. WNV isolate TX 2002-HC (WNV-TX) was described previously (42), and its titer was determined by a standard plaque assay on BHK-21 cells. Working stocks were generated by passaging WNV-TX twice on Vero cells (ATCC CCL81) and used for in in vivo experiments.
Mouse experiments.
Inbred C57BL/6J mice were commercially obtained (Jackson Laboratories, Bar Harbor, ME). WNV-I TCR-transgenic mice were obtained from the Bevan laboratory and were described previously (43). All the mice were genotyped and bred under specific-pathogen-free conditions in the rodent facility at Yerkes National Primate Research Center at Emory University. The methods for mouse use and care were in accordance with the Emory University Institutional Animal Care and Use Committee guidelines. Age-matched 8- to 12-week-old mice were inoculated subcutaneously in the left rear footpad with 100 PFU of WNV-TX in a 10-μl inoculum diluted in phosphate-buffered saline (PBS) supplemented with 2% heat-inactivated FBS. Mock-infected mice were inoculated in a similar manner with diluents alone. The mice were monitored daily for morbidity and mortality.
Generation of transgenic WNV-I CD8+ T cells.
To generate the TCR-transgenic mice with specificity for the WNV immunodominant epitope, a cytotoxic T cell specific for the SSVNWNATTA/Db epitope, the rearranged TCRα and -β chain cDNAs were cloned into a targeting construct and injected into C57BL/6 mouse eggs (Fig. 1A). Magnetically sorted T cells from infected C57BL/6 mice that were heterozygous for a TCRα locus deletion (to avoid any possibility of having two TCRα chains in the same cell) were cocultured with T cell-depleted, irradiated splenic antigen-presenting cells pulsed with 1 μM NS4B peptide. The frequency of NS4B/Db-specific cells was assayed at the initiation of culture and at day 2 or 3 of culture. T cell blasts were fused with the TCRαβ-negative BW5147 lymphoma cell line using polyethylene glycol (PEG). Hybridoma cells were screened for surface expression of CD3, CD4, and CD8 and for pan-TCRβ. Antigen specificity for NS4B was screened by the ability to produce IL-2, measured by enzyme-linked immunosorbent assay (ELISA), in response to stimulation with WNV peptide plus spleen antigen-presenting cells. The TCR specificity was identified by a combination of flow cytometry and PCR. Isolated RNA was used to make cDNA and amplified using the Cα and Cβ primers and V gene-specific primers designed to amplify V gene subfamilies for sequencing. After the chains were sequenced, the V region segment was amplified and inserted into a cassette vector that contained Cα or Cβ constant region cDNA sequences, as well as 100 to 200 bases of genomic sequence that contained a poly(A) sequence. The full-length cDNAs were cloned into the pcDNA3 human CD2 vector for injection into C57BL/6 eggs at the University of Washington Transgenic Facility. Following generation of the mice, we isolated peripheral blood mononuclear cells from the WNV-I transgenic mice and stained the cells with a WNV tetramer specific for the NS4B immunodominant epitope (43).
WNV-I CD8+ T cell adoptive transfers.
WNV-I CD8+ T cells were isolated from splenocyte suspensions using the CD8+ T cell negative-selection kit (BioLegend, San Diego, CA; catalog no. 480007) following the manufacturer's instructions. In most of the experiments, 5 × 104 WNV-I CD8+ T cells, unless otherwise specified, were adoptively transferred into congenically marked mismatched C57BL6/J mice in 200 μl of FBS-free DMEM via tail vein injection. Three days posttransfer, the mice were infected with 100 PFU WNV-TX through the s.c. route in the left rear footpad and monitored daily as described above.
In vivo intravascular staining.
In vivo intravascular staining of T cells was done as previously described (21, 23). At each time point, 10 μg of allophycocyanin-labeled anti-CD8α antibody (Tonbo Biosciences, San Diego, CA; catalog no. 20-1886) was administered intravenously in a volume of 200 μl. Five minutes post-antibody delivery, the mice were euthanized by isoflurane overdose. Whole blood, spleens, lymph nodes, and brains were collected immediately after euthanasia. Successful intravascular staining was defined as greater than 95% of in vivo and ex vivo double-stained CD8+ T cells in blood and less than 5% in lymph nodes.
Splenocyte isolation.
Spleens were collected, mechanically dispersed, and washed with 10 ml of ice-cold PBS. Red blood cells were lysed in 1 ml of ACK lysing buffer (Lonza, Basel, Switzerland) for 5 min on ice and washed with 10 ml of PBS. Single-cell suspensions were centrifuged for 5 min at 1,250 rpm at 4°C. The cell pellet was resuspended in 10 ml of PBS and passed through a 70-μm Falcon cell strainer (Corning Life Sciences, Corning, NY) and then centrifuged and resuspended in 1 ml fluorescence-activated cell sorting (FACS) buffer (PBS supplemented with 2% FBS and 10 mM EDTA). Single-cell suspensions were generated for antibody staining and CD8+ T cell ex vivo restimulation assays.
Intracerebral lymphocyte (ICL) isolation.
Brains were harvested, mechanically dispersed using a 10-ml syringe plunger, and resuspended in 5 ml of PBS supplemented with 50 μg/ml Liberase (Roche, Basel, Switzerland) and 50 μg/ml DNase I (Akron Biotech, Boca Raton, FL) and digested for 15 min at room temperature (RT). The homogenized brains were washed with 10 ml of cRPMI medium (10% FBS, nonessential amino acids, 50 μM β-mercaptoethanol, 10 mM HEPES, 50 IU/ml [each] penicillin and streptomycin). The digested brains were resuspended in 5 ml of cRPMI and passed through a 70-μm Falcon cell strainer. Immune cells were isolated after centrifugation at 2,000 rpm for 20 min at 4°C under a 30% Percoll cushion (GE, Uppsala, Sweden).
Flow cytometry and antibodies.
Immune cells from the brain and spleens were strained in FACS buffer with Fc blocking anti-mouse CD16/CD32 antibody (clone 2.4G2; Tonbo Bioscience), washed with FACS buffer, and stained for 30 min at room temperature with WNV Db-restricted NS4b tetramer conjugated to APCs (produced at the NIH Tetramer Core Facility at Emory University, Atlanta, GA). Following tetramer staining, the cells were stained with directly conjugated mouse-specific antibodies to CD3 (clone 2C11), CD8 (clone 53-6.7), CD62L (clone MEL14), CD44 (clone IM7), KLRG-1 (clone 2F1), CD45.1 (clone A20), and CD45.2 (clone 104) (Tonbo Bioscience); CD127 (clone SB/199), CD103 (clone OX-62), CXCR3 (clone CXCR3-173), PD-1 (clone J43), Tim-3 (clone SD12), CD69 (clone H1.2F3), CD122 (clone 5H4), Lag3 (clone T47-530), CD25 (clone 7D4), and IL-15R (clone DNT15Ra) (BD Bioscience); and CCR-7 (clone 4B12) (BioLegend). Following 30 min incubation at 4°C, the immune cells were washed with PBS and stained for dead cells in PBS with Ghost 510 dye (1:1,000 dilution; Tonbo Bioscience). The cells were washed with 200 μl of FACS buffer and fixed with Erilyse fixing buffer (BD Bioscience).
Ex vivo T cell assays.
Single-cell suspensions (1 × 106 cells) were seeded in 96-well U-bottom tissue culture-treated plates in 100 μl cRPMI. Lymphocytes were stimulated with 1 μg/ml of the WNV NS4b peptide (SSVWNATTAI) in the presence of Golgi Stop (BD Bioscience) for 5 h at 37°C. Following peptide stimulation, the cells were washed with FACS buffer and stained for cell surface markers and dead cells as described above. The lymphocytes were then incubated with 100 μl of permeabilization buffer (Tonbo Biosciences) for 60 min, washed twice with 200 μl of fixation buffer (Tonbo Biosciences), resuspended in 50 μl of FACS buffer, and stained with antibodies against IFN-γ (clone AN-18; BioLegend) and TNF-α (clone C9B7W; BD Bioscience) for 30 min at 4°C. The cells were washed with FACS buffer, and flow cytometry was performed on a BD LSRII machine using BD FACSDiva software. Flow cytometry data analysis was performed using FlowJo software version 10.1r5 (TreeStar, Ashland, OR).
Focus-forming assay.
WNV-infected mice were euthanized by overdose with isoflurane and perfused with 20 ml PBS. Spleens and brains were removed, weighed, placed in soft-tissue-homogenizing tubes (Omni International, Kennesaw, GA) containing 500 μl of PBS with 1% heat-inactivated FBS, and homogenized using a Precellys 24 (Bertin Technologies, France) at 1,500 rpm for 20 s. Focus-forming assays were performed as previously described with minor modifications (44). Briefly, tissue homogenates were diluted in DMEM supplemented with 1% FBS and used to infect Vero cells for 1 h at 37°C. The cells and inoculum were overlaid with methylcellulose (Opti-MEM [Corning Cellgro], 1% antibiotic/antimycotic [Corning Cellgro], 2% FBS, 2% methylcellulose [Sigma-Aldrich]) and incubated for 72 h at 37°C. The cells were washed with PBS, fixed with 200 μl of 4% paraformaldehyde (PFA), washed twice with PBS, and permeabilized (PBS supplemented with 2% saponin [Sigma-Aldrich] and 2% FBS or permeabilization solution). The cells were incubated with humanized WNV E16 antibody (kindly provided by Michael Diamond) in permeabilization solution for 2 h at RT. The cells were washed twice with PBS, followed by incubation with anti-human horseradish peroxidase (HRP)-conjugated secondary antibody (HRP-conjugated goat anti-mouse IgG) in permeabilization solution (1:1,200 dilution; Jackson ImmunoResearch, West Grove, PA) diluted 1:3,000 in permeabilization solution for 2 h at RT. Foci were developed with TrueBlue peroxidase substrate (SeraCare, Milford, MA). The plates were read on a CTL-ImmunoSpot S6 Micro Analyzer and counted with the help of ImageJ software version 1.51f. Counts were normalized to the weight of the tissue, and titers were expressed as focus-forming units per gram.
Statistical analyses.
Statistical analyses were performed in GraphPad 6 (Prism, La Jolla, CA, USA). Statistical significance was calculated using a paired t test for normally distributed data. One-way analysis of variance (ANOVA) was used for multiple comparisons. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank the Pediatrics Flow Cytometry Facility at Emory School of Medicine and the EVC Flow Cytometry Core. We thank Kendra Quicke and Justin O'Neal for critical readings of the manuscript. We acknowledge the NIH Tetramer Core Facility (contract HHSN272201300006C) for provision of tetramers.
This work was funded in part by National Institutes of Health grants U19AI083019 (M.S.S.), R56AI110516 (M.S.S.), R21AI113485 (M.S.S.), R01AI124680 (A.G.), R01AI070101 (A.G.), R01AI126890 (A.G.), and R01AI136533 (A.G.); by Children's Healthcare of Atlanta, Emory Vaccine Center, and the Georgia Research Alliance (M.S.S.); by P510D11132 to Yerkes National Primate Research Center; and by P30A050509 to the EVC Flow Cytometry Core. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.Grubaugh ND, Ebel GD. 2016. Dynamics of West Nile virus evolution in mosquito vectors. Curr Opin Virol 21:132–138. doi: 10.1016/j.coviro.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David S, Abraham AM. 2016. Epidemiological and clinical aspects on West Nile virus, a globally emerging pathogen. Infect Dis 48:571–586. doi: 10.3109/23744235.2016.1164890. [DOI] [PubMed] [Google Scholar]
- 3.Mostashari F, Bunning ML, Kitsutani PT, Singer DA, Nash D, Cooper MJ, Katz N, Liljebjelke KA, Biggerstaff BJ, Fine AD, Layton MC, Mullin SM, Johnson AJ, Martin DA, Hayes EB, Campbell GL. 2001. Epidemic West Nile encephalitis, New York, 1999: results of a household-based seroepidemiological survey. Lancet 358:261–264. doi: 10.1016/S0140-6736(01)05480-0. [DOI] [PubMed] [Google Scholar]
- 4.Petersen LR, Marfin AA. 2002. West Nile virus: a primer for the clinician. Ann Intern Med 137:173–179. doi: 10.7326/0003-4819-137-3-200208060-00009. [DOI] [PubMed] [Google Scholar]
- 5.Murray K, Baraniuk S, Resnick M, Arafat R, Kilborn C, Cain K, Shallenberger R, York TL, Martinez D, Hellums JS, Hellums D, Malkoff M, Elgawley N, McNeely W, Khuwaja SA, Tesh RB. 2006. Risk factors for encephalitis and death from West Nile virus infection. Epidemiol Infect 134:1325–1332. doi: 10.1017/S0950268806006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omalu BI, Shakir AA, Wang G, Lipkin WI, Wiley CA. 2003. Fatal fulminant pan-meningo-polioencephalitis due to West Nile virus. Brain Pathol 13:465–472. doi: 10.1111/j.1750-3639.2003.tb00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sampson BA, Ambrosi C, Charlot A, Reiber K, Veress JF, Armbrustmacher V. 2000. The pathology of human West Nile virus infection. Hum Pathol 31:527–531. doi: 10.1053/hp.2000.8047. [DOI] [PubMed] [Google Scholar]
- 8.James EA, Gates TJ, LaFond RE, Yamamoto S, Ni C, Mai D, Gersuk VH, O'Brien K, Nguyen QA, Zeitner B, Lanteri MC, Norris PJ, Chaussabel D, Malhotra U, Kwok WW. 2016. Neuroinvasive West Nile infection elicits elevated and atypically polarized T cell responses that promote a pathogenic outcome. PLoS Pathog 12:e1005375. doi: 10.1371/journal.ppat.1005375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanteri MC, Diamond MS, Law JP, Chew GM, Wu S, Inglis HC, Wong D, Busch MP, Norris PJ, Ndhlovu LC. 2014. Increased frequency of Tim-3 expressing T cells is associated with symptomatic West Nile virus infection. PLoS One 9:e92134. doi: 10.1371/journal.pone.0092134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perez-Ramirez E, Llorente F, Del Amo J, Fall G, Sall AA, Lubisi A, Lecollinet S, Vazquez A, Jimenez-Clavero MA. 2017. Pathogenicity evaluation of twelve West Nile virus strains belonging to four lineages from five continents in a mouse model: discrimination between three pathogenicity categories. J Gen Virol 98:662–670. doi: 10.1099/jgv.0.000743. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Chan YK, Lu B, Diamond MS, Klein RS. 2008. CXCR3 mediates region-specific antiviral T cell trafficking within the central nervous system during West Nile virus encephalitis. J Immunol 180:2641–2649. doi: 10.4049/jimmunol.180.4.2641. [DOI] [PubMed] [Google Scholar]
- 12.Shrestha B, Diamond MS. 2007. Fas ligand interactions contribute to CD8+ T-cell-mediated control of West Nile virus infection in the central nervous system. J Virol 81:11749–11757. doi: 10.1128/JVI.01136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha B, Samuel MA, Diamond MS. 2006. CD8+ T cells require perforin to clear West Nile virus from infected neurons. J Virol 80:119–129. doi: 10.1128/JVI.80.1.119-129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shrestha B, Diamond MS. 2004. Role of CD8+ T cells in control of West Nile virus infection. J Virol 78:8312–8321. doi: 10.1128/JVI.78.15.8312-8321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrestha B, Pinto AK, Green S, Bosch I, Diamond MS. 2012. CD8+ T cells use TRAIL to restrict West Nile virus pathogenesis by controlling infection in neurons. J Virol 86:8937–8948. doi: 10.1128/JVI.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brien JD, Uhrlaub JL, Nikolich-Zugich J. 2007. Protective capacity and epitope specificity of CD8(+) T cells responding to lethal West Nile virus infection. Eur J Immunol 37:1855–1863. doi: 10.1002/eji.200737196. [DOI] [PubMed] [Google Scholar]
- 17.Purtha WE, Myers N, Mitaksov V, Sitati E, Connolly J, Fremont DH, Hansen TH, Diamond MS. 2007. Antigen-specific cytotoxic T lymphocytes protect against lethal West Nile virus encephalitis. Eur J Immunol 37:1845–1854. doi: 10.1002/eji.200737192. [DOI] [PubMed] [Google Scholar]
- 18.Suthar MS, Ramos HJ, Brassil MM, Netland J, Chappell CP, Blahnik G, McMillan A, Diamond MS, Clark EA, Bevan MJ, Gale M Jr. 2012. The RIG-I-like receptor LGP2 controls CD8(+) T cell survival and fitness. Immunity 37:235–248. doi: 10.1016/j.immuni.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGavern DB, Christen U, Oldstone MB. 2002. Molecular anatomy of antigen-specific CD8(+) T cell engagement and synapse formation in vivo. Nat Immunol 3:918–925. doi: 10.1038/ni843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniels MA, Jameson SC. 2000. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J Exp Med 191:335–346. doi: 10.1084/jem.191.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson KG, Mayer-Barber K, Sung H, Beura L, James BR, Taylor JJ, Qunaj L, Griffith TS, Vezys V, Barber DL, Masopust D. 2014. Intravascular staining for discrimination of vascular and tissue leukocytes. Nat Protoc 9:209–222. doi: 10.1038/nprot.2014.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takamura S, Yagi H, Hakata Y, Motozono C, McMaster SR, Masumoto T, Fujisawa M, Chikaishi T, Komeda J, Itoh J, Umemura M, Kyusai A, Tomura M, Nakayama T, Woodland DL, Kohlmeier JE, Miyazawa M. 2016. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J Exp Med 213:3057–3073. doi: 10.1084/jem.20160938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo YJ, Jothikumar P, Suthar MS, Zhu C, Grakoui A. 2016. Local cellular and cytokine cues in the spleen regulate in situ T cell receptor affinity, function, and fate of CD8(+) T cells. Immunity 45:988–998. doi: 10.1016/j.immuni.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallin JJ, Liang L, Bakardjiev A, Sha WC. 2001. Enhancement of CD8+ T cell responses by ICOS/B7h costimulation. J Immunol 167:132–139. doi: 10.4049/jimmunol.167.1.132. [DOI] [PubMed] [Google Scholar]
- 25.Ahlers JD, Belyakov IM. 2010. Memories that last forever: strategies for optimizing vaccine T-cell memory. Blood 115:1678–1689. doi: 10.1182/blood-2009-06-227546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson JA, McDonald-Hyman C, Jameson SC, Hamilton SE. 2013. Effector-like CD8(+) T cells in the memory population mediate potent protective immunity. Immunity 38:1250–1260. doi: 10.1016/j.immuni.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abboud G, Desai P, Dastmalchi F, Stanfield J, Tahiliani V, Hutchinson TE, Salek-Ardakani S. 2016. Tissue-specific programming of memory CD8 T cell subsets impacts protection against lethal respiratory virus infection. J Exp Med 213:2897–2911. doi: 10.1084/jem.20160167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramos HJ, Lanteri MC, Blahnik G, Negash A, Suthar MS, Brassil MM, Sodhi K, Treuting PM, Busch MP, Norris PJ, Gale M Jr. 2012. IL-1beta signaling promotes CNS-intrinsic immune control of West Nile virus infection. PLoS Pathog 8:e1003039. doi: 10.1371/journal.ppat.1003039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suthar MS, Ma DY, Thomas S, Lund JM, Zhang N, Daffis S, Rudensky AY, Bevan MJ, Clark EA, Kaja MK, Diamond MS, Gale M Jr. 2010. IPS-1 is essential for the control of West Nile virus infection and immunity. PLoS Pathog 6:e1000757. doi: 10.1371/journal.ppat.1000757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinbach K, Vincenti I, Kreutzfeldt M, Page N, Muschaweckh A, Wagner I, Drexler I, Pinschewer D, Korn T, Merkler D. 2016. Brain-resident memory T cells represent an autonomous cytotoxic barrier to viral infection. J Exp Med 213:1571–1587. doi: 10.1084/jem.20151916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakim LM, Woodward-Davis A, Bevan MJ. 2010. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A 107:17872–17879. doi: 10.1073/pnas.1010201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wakim LM, Woodward-Davis A, Liu R, Hu Y, Villadangos J, Smyth G, Bevan MJ. 2012. The molecular signature of tissue resident memory CD8 T cells isolated from the brain. J Immunol 189:3462–3471. doi: 10.4049/jimmunol.1201305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemp RA, Powell TJ, Dwyer DW, Dutton RW. 2004. Cutting edge: regulation of CD8+ T cell effector population size. J Immunol 173:2923–2927. doi: 10.4049/jimmunol.173.5.2923. [DOI] [PubMed] [Google Scholar]
- 34.Badovinac VP, Haring JS, Harty JT. 2007. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity 26:827–841. doi: 10.1016/j.immuni.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butz EA, Bevan MJ. 1998. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity 8:167–175. doi: 10.1016/S1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.La Gruta NL, Doherty PC, Turner SJ. 2006. A correlation between function and selected measures of T cell avidity in influenza virus-specific CD8+ T cell responses. Eur J Immunol 36:2951–2959. doi: 10.1002/eji.200636390. [DOI] [PubMed] [Google Scholar]
- 37.Schildberg FA, Klein SR, Freeman GJ, Sharpe AH. 2016. Coinhibitory pathways in the B7-CD28 ligand-receptor family. Immunity 44:955–972. doi: 10.1016/j.immuni.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatum AM, Watson AM, Schell TD. 2010. Direct presentation regulates the magnitude of the CD8+ T cell response to cell-associated antigen through prolonged T cell proliferation. J Immunol 185:2763–2772. doi: 10.4049/jimmunol.0903920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart BS, Demarest VL, Wong SJ, Green S, Bernard KA. 2011. Persistence of virus-specific immune responses in the central nervous system of mice after West Nile virus infection. BMC Immunol 12:6. doi: 10.1186/1471-2172-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Appler KK, Brown AN, Stewart BS, Behr MJ, Demarest VL, Wong SJ, Bernard KA. 2010. Persistence of West Nile virus in the central nervous system and periphery of mice. PLoS One 5:e10649. doi: 10.1371/journal.pone.0010649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Durrant DM, Daniels BP, Klein RS. 2014. IL-1R1 signaling regulates CXCL12-mediated T cell localization and fate within the central nervous system during West Nile Virus encephalitis. J Immunol 193:4095–4106. doi: 10.4049/jimmunol.1401192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keller BC, Fredericksen BL, Samuel MA, Mock RE, Mason PW, Diamond MS, Gale M Jr. 2006. Resistance to alpha/beta interferon is a determinant of West Nile virus replication fitness and virulence. J Virol 80:9424–9434. doi: 10.1128/JVI.00768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim S, Pinto AK, Myers NB, Hawkins O, Doll K, Kaabinejadian S, Netland J, Bevan MJ, Weidanz JA, Hildebrand WH, Diamond MS, Hansen TH. 2014. A novel T-cell receptor mimic defines dendritic cells that present an immunodominant West Nile virus epitope in mice. Eur J Immunol 44:1936–1946. doi: 10.1002/eji.201444450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bowen JR, Quicke KM, Maddur MS, O'Neal JT, McDonald CE, Fedorova NB, Puri V, Shabman RS, Pulendran B, Suthar MS. 2017. Zika virus antagonizes type i interferon responses during infection of human dendritic cells. PLoS Pathog 13:e1006164. doi: 10.1371/journal.ppat.1006164. [DOI] [PMC free article] [PubMed] [Google Scholar]