ABSTRACT
Several viruses induce intestinal epithelial cell death during enteric infection. However, it is unclear whether proapoptotic capacity promotes or inhibits replication in this tissue. We infected mice with two reovirus strains that infect the intestine but differ in the capacity to alter immunological tolerance to new food antigen. Infection with reovirus strain T1L, which induces an inflammatory immune response to fed antigen, is prolonged in the intestine, whereas T3D-RV, which does not induce this response, is rapidly cleared from the intestine. Compared with T1L, T3D-RV infection triggered apoptosis of intestinal epithelial cells and subsequent sloughing of dead cells into the intestinal lumen. We conclude that the infection advantage of T1L derives from its capacity to subvert host restriction by epithelial cell apoptosis, providing a possible mechanism by which T1L enhances inflammatory signals during antigen feeding. Using a panel of T1L × T3D-RV reassortant viruses, we identified the viral M1 and M2 gene segments as determinants of reovirus-induced apoptosis in the intestine. Expression of the T1L M1 and M2 genes in a T3D-RV background was sufficient to limit epithelial cell apoptosis and enhance viral infection to levels displayed by T1L. These findings define additional reovirus gene segments required for enteric infection of mice and illuminate the antiviral effect of intestinal epithelial cell apoptosis in limiting enteric viral infection. Viral strain-specific differences in the capacity to infect the intestine may be useful in identifying viruses capable of ameliorating tolerance to fed antigen in autoimmune conditions like celiac disease.
IMPORTANCE Acute viral infections are thought to be cleared by the host with few lasting consequences. However, there may be much broader and long-lasting effects of viruses on immune homeostasis. Infection with reovirus, a common, nonpathogenic virus, triggers inflammation against innocuous food antigens, implicating this virus in the development of celiac disease, an autoimmune intestinal disorder triggered by exposure to dietary gluten. Using two reovirus strains that differ in the capacity to abrogate oral tolerance, we found that strain-specific differences in the capacity to replicate in the intestine inversely correlate with the capacity to induce apoptotic death of intestinal epithelial cells, providing a host-mediated process to restrict intestinal infection. This work contributes new knowledge about virus-host interactions in the intestine and establishes a foundation for future studies to define mechanisms by which viruses break oral tolerance in celiac disease.
KEYWORDS: apoptosis, cell death, enteroid, gastrointestinal infection, mucosal immunity, reovirus
INTRODUCTION
It is thought that following viral infection, cells undergo apoptosis to restrict viral dissemination and protect adjacent cells. However, some viruses have established mechanisms to inhibit cell death mediators to promote replication (1, 2). The factors that control such outcomes are not well understood but likely to be tissue and pathogen specific. Some enteric viruses can disrupt host immune responses against newly introduced dietary antigens (3), but the virus-host interplay that dictates this outcome is not clear.
Apoptosis, or programmed cell death, is characterized by distinct morphological changes that result from a tightly regulated proteolytic pathway. Prior to apoptosis, cysteine-aspartyl proteases, or caspases, are sequestered in the cell as proenzymes. There are two distinct apoptotic pathways that initiate the caspase cascade: the extrinsic or death receptor pathway and the intrinsic or mitochondrial pathway. The extrinsic and intrinsic pathways converge at the execution phase with activation of caspase-3 to mediate the biochemical changes characteristic of apoptosis. These include cytoskeletal and nuclear protein cleavage, protein cross-linking, DNA fragmentation, formation of apoptotic bodies, and expression of ligands for recognition by phagocytic cells (4, 5).
The intestinal epithelium uniquely functions to passively transport nutrients and water while remaining impermeable to the external environment. Any insult, whether microbial, toxic, or traumatic, can lead to cell death, loss of epithelial contiguity, and breakdown in gut barrier function. The intestinal epithelium of most mammals is capable of regenerating to alleviate disruptions in barrier function and maintain homeostasis. Newly generated intestinal epithelial cells (IECs), or enterocytes, migrate from the proliferation zone at the crypt base toward the villus tip, where they are extruded or sloughed into the gut lumen. The rapid renewal of the villus epithelium by stem cells occurs every 2 to 6 days in healthy adult mammals (6).
Spontaneous apoptosis rarely occurs in healthy intestinal tissue, with only 0.4% of villus cells expressing activated caspase-3 in mice (7). However, pathogenic insults can cause premature apoptosis of enterocytes. For example, intraperitoneal injections of mice with poly(I·C), a double-stranded RNA (dsRNA) analog, or rotavirus genomic dsRNA are sufficient to stimulate enterocyte death in mice as observed by activated caspase-3 immunostaining, villus shortening, and diarrhea (8, 9). Several viruses, including human norovirus (10), rotavirus (11), and simian immunodeficiency virus (12), cause enterocyte death in animals, but little is known about virus-induced cell death in the intestine in the absence of pathology.
Mammalian reoviruses are nonenveloped viruses with a genome consisting of 10 dsRNA segments. Reovirus infects the intestine following oral inoculation. In humans, infections are common during early childhood (13), and seroprevalence remains intact throughout life (14). Reovirus infections of the intestine are commonly nonpathogenic in humans and adult mice (3, 15). However, reovirus infection can disrupt the host immune response to food antigen during the development of celiac disease (3). Furthermore, the abrogation of oral tolerance during reovirus infection is strain specific, indicating that reovirus strains differ during intestinal infection to promote loss of oral tolerance.
Two human reovirus isolates, type 1 Lang (T1L) and type 3 Dearing (T3D), were isolated from children (16–18) and are studied to understand mechanisms of reovirus-host interactions. Following peroral (p.o.) inoculation, reovirus T1L transcytoses across M cells in the ileum and disseminates to the underlying intestinal tissue (19, 20). The virus is then detected in Peyer's patches (PPs), mesenteric lymph nodes (MLNs), and spleen, indicative of hematogenous dissemination (21). Reovirus also can be detected in the duodenum, jejunum, ileum, and colon. Reovirus strain T3D, however, does not infect the intestine due to polymorphisms in the S1 and L2 gene segments (22). Thus, T3D-RV, a reassortant virus containing the S1 and L2 gene segments of T1L in a T3D genetic background, was engineered for studies of intestinal infection by an additional reovirus strain. T1L intestinal infection promotes inflammatory responses to dietary antigen, while T3D-RV does not (3). The goal of this study was to define pathobiological properties that differ between these two strains during intestinal infection.
Reovirus infection causes cell death in many types of cell lines and primary cell cultures through activation of both extrinsic and intrinsic pathways of apoptosis (23, 24). Reovirus-induced apoptosis is triggered during recognition of the virus upon entry by cellular pattern recognition receptors (25). Strain-specific differences in reovirus-induced cell death suggest the existence of viral determinants of apoptosis. T3D induces apoptosis more efficiently than T1L, a property that segregates with the S1 and M2 gene segments (26–28). However, little is known about the viral determinants of apoptosis in the intestine and the effects of apoptosis on viral replication in this tissue.
Following p.o. inoculation with T1L, apoptotic cellular debris from infected enterocytes is observed in phagocytic vesicles of dendritic cells found in PPs (29). Newborn mice inoculated perorally with an apoptosis-deficient strain of reovirus develop high viral titers in the intestine compared with those produced by an apoptosis-proficient strain (30). Therefore, we hypothesized that infected enterocytes activate apoptosis and sloughing to restrict reovirus replication in the intestine.
To test this hypothesis, we quantified levels of apoptosis induced by T1L and T3D-RV during infection of cultured cells, intestinal organoids, and mice. We found that T3D-RV induces higher levels of apoptosis than T1L in all three assay systems. Additionally, T3D-RV intestinal infection was cleared more rapidly than infection by T1L. To examine the relationship between apoptosis and infection in the intestine, a panel of T1L × T3D-RV reassortant viruses was recovered using plasmid-based rescue. The reassortant viruses capable of suppressing apoptosis in the intestine also displayed enhanced infection capacity, similar to T1L. These results define apoptosis as a cellular process that restricts enteric viral infection and highlights strain-specific determinants that may be required for reovirus-induced oral tolerance blockade.
RESULTS
T1L-infected mice maintain higher titers in the intestine during acute infection.
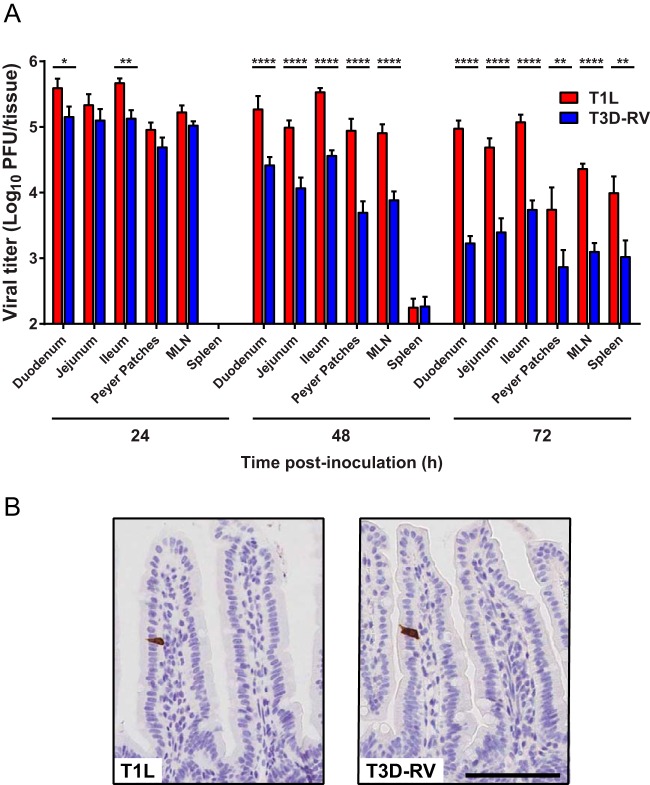
Inoculation of mice p.o. with T1L abrogates immunological tolerance to food antigen, whereas inoculation with strain T3D-RV does not (3). Although reovirus T1L intestinal infection is well characterized, little is known about T3D-RV in the intestine or how these viruses differ in interactions with intestinal tissue to induce tolerance blockade. To determine whether T1L and T3D-RV produce comparable viral loads in intestinal tissue, mice were infected perorally with each strain, and viral titers were determined at 24, 48, and 72 h postinoculation (hpi). T1L produced higher titers in mice than did T3D-RV at 48 and 72 hpi (Fig. 1A). Therefore, although the two virus strains produced comparable titers in the intestine, PPs, MLNs, and spleen 24 h after p.o. inoculation, T1L titers remain elevated throughout infection relative to titers of T3D-RV.
FIG 1.
Viral infection in murine tissues following inoculation with reoviruses T1L and T3D-RV. Mice were inoculated perorally with 1010 PFU of T1L or T3D-RV (n = 7 to 10 mice per virus strain). (A) Titers of T1L and T3D-RV in different regions of the intestine and secondary lymphoid organs were determined at the times shown by plaque assay. The small intestine was sectioned into thirds, approximating the duodenum, jejunum, and ileum. Viral titers are expressed as PFU per tissue. The 24-hpi titer values were previously published in reference 3; data are used with permission of the publisher. (B) One day after inoculation, intestines were resected, and the distal half was flushed, Swiss rolled, and processed for histology. Sections were stained with a polyclonal antiserum specific for reovirus. Representative sections of jejunum are shown (scale bar, 100 μm). Error bars indicate SEMs. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; one-way ANOVA and Sidak's multiple-comparison test.
To determine cell types in the intestine targeted by T1L and T3D-RV, mice were inoculated perorally and euthanized at 1 day postinoculation (dpi). Intestines were dissected, Swiss rolled, and processed for immunohistochemistry. In intestines from both T1L- and T3D-RV-infected mice, cells displaying morphological characteristics of mature IECs stained positive for reovirus antigen (Fig. 1B). Consistent with previous observations (19), the incidence of reovirus-positive cells was low. Thus, both T1L and T3D-RV infect mature enterocytes in intestines of adult mice.
T3D-RV infection induces caspase-3 activation and villus shedding in the gut.
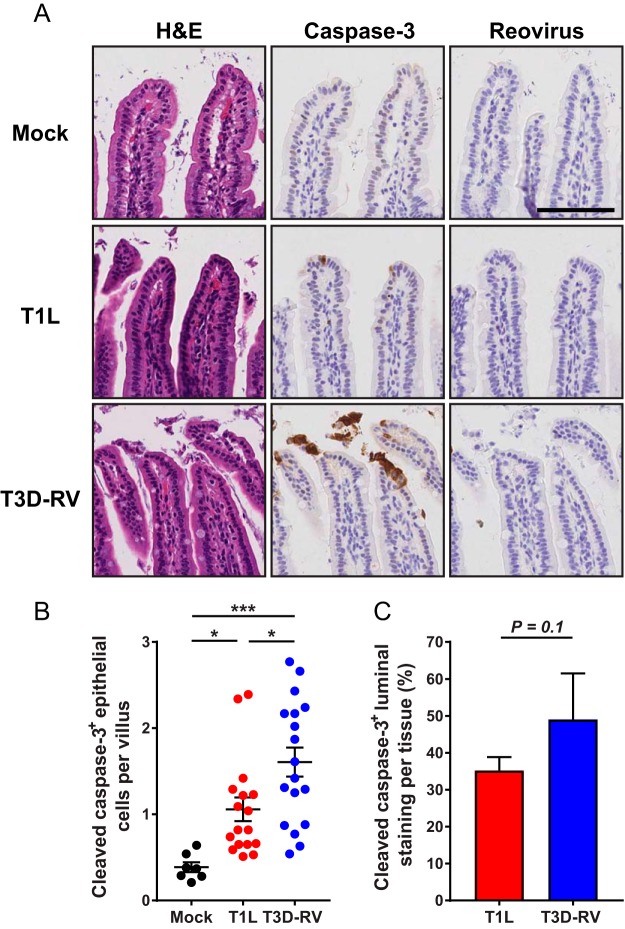
To determine whether T1L and T3D-RV induce cell death and cause tissue damage in vivo, mice were inoculated perorally and euthanized 1 and 8 dpi. Sections of intestinal tissue were stained for activated caspase-3 to identify cells undergoing apoptosis. Overall, viral infection led to an increase in caspase-3 activation relative to phosphate-buffered saline (PBS)-inoculated controls (Fig. 2A). Although activated caspase-3 levels increased during viral infection, no tissue injury was observed following hematoxylin and eosin (H&E) staining (Fig. 2A), and both strains were cleared by 8 dpi (data not shown).
FIG 2.
Cleaved caspase-3 in the intestines of mice following infection with reovirus T1L or T3D-RV. Mice were inoculated perorally with 108 PFU of T1L or T3D-RV or PBS (mock). One day after inoculation, intestines were resected. The distal half was flushed, Swiss rolled, and processed for histology. (A) Sections were stained with H&E, reovirus polyclonal antiserum, or antibody against cleaved caspase-3. Representative sections of jejunum are shown (scale bar, 100 μm). (B) Cells positive for cleaved caspase-3 were enumerated manually and normalized per villus. Each symbol represents an individual mouse (n = 5 to 18 mice per group). (C) Cleaved-caspase-3 staining in the lumen was quantified by outlining the luminal region using the Digital Histology Shared Resource tool (n = 3 mice per virus). The percent luminal staining was determined as follows: (area in the lumen positive for cleaved-caspase-3 staining/area in the whole tissue positive for cleaved-caspase-3 staining) × 100. (B) Error bars indicate SEMs. (C) Error bars indicate SDs. *, P < 0.05; ***, P < 0.001. P values were determined by one-way ANOVA and Tukey's multiple-comparison test (B) and Mann-Whitney test (C).
Since T1L and T3D differ in the capacity to induce apoptosis, we hypothesized that T3D-RV induces more apoptosis in the gut, which could stimulate sloughing of infected enterocytes to mediate the rapid viral clearance observed in Fig. 1A. To determine whether T1L and T3D-RV differ in the capacity to trigger apoptosis, epithelial cells positive for cleaved caspase-3 were enumerated and normalized to the total number of villi examined. T3D-RV-infected mice had significantly more epithelial cells positive for cleaved caspase-3 per villus than did those infected with T1L (Fig. 2B). To test whether T1L and T3D-RV differ in the shedding of apoptotic enterocytes into the intestinal lumen, the luminal region was outlined using Ariol Review software, the area positive for cleaved caspase-3 was demarcated, and the percentage of positive staining in the lumen was quantified relative to the positive staining in the entire tissue section. Compared with mice infected with T1L, cleaved-caspase-3 staining was increased in the lumens of mice infected with T3D-RV (Fig. 2C). The distribution of detectable reovirus antigen and apoptotic cells did not overlap, suggesting either that reovirus replication is not required for activation of caspase-3 in IECs (Fig. 2A) or that reovirus replication occurs in IECs but reovirus antigen is not detected due to the low sensitivity of the immunostaining technique. Therefore, T1L and T3D-RV infect the intestine comparably at 24 hpi, but T3D-RV promotes rapid enterocyte death and sloughing of apoptotic cells into the intestinal lumen, possibly mediating a strain-specific antiviral response.
T3D-RV provokes apoptosis and cell shedding in infected enteroids.
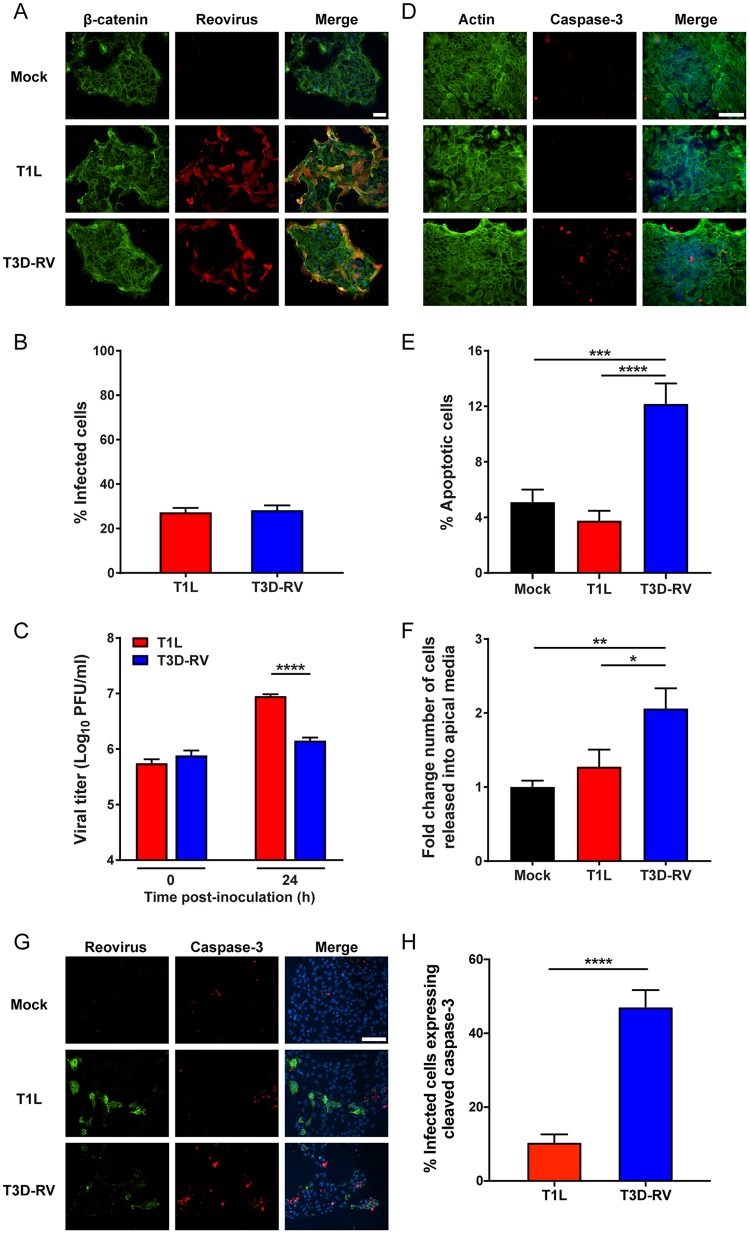
Techniques to cultivate intestinal organoids or “enteroids” in culture have advanced opportunities to study virus-host interactions at intestinal sites (31–33). To determine whether reovirus infects tissue-derived organoids from mice and promotes apoptotic cell death in these cultures, we dissected intestines from adult mice, isolated intestinal crypts, and propagated enteroid cultures in three-dimensional (3D) spheroids in Matrigel. These primary cultures contain crypt- and villus-like structures and all the differentiated cell types found in the healthy intestine. Following p.o. inoculation, IECs are exposed to reovirus from the luminal (apical) surface of the epithelium (34, 35). To examine the susceptibility of murine enterocytes to reovirus infection and subsequent virus-induced apoptosis, dissociated enteroids were seeded onto Transwell inserts, adsorbed with T1L or T3D-RV, and scored for reovirus infection and apoptosis. At 24 hpi, infectivities were comparable between the strains, as visualized by immunofluorescence staining using reovirus polyclonal antiserum (Fig. 3A and B). Murine enteroids infected with T3D-RV displayed a greater percentage of apoptotic cells, as determined by staining with an antibody specific for cleaved caspase-3 (Fig. 3D and E). At 24 hpi, the apical medium was collected, and cells in the medium were enumerated using an automated cell counter. T3D-RV-infected enteroids had a significantly larger number of cells in the apical medium, an indicator of cell sloughing (Fig. 3F). T1L titers in enteroids were significantly higher than T3D-RV titers (Fig. 3C), which correlates with increased numbers of caspase-3-positive cells and a slight decrease in the number of cells in the T3D-RV-infected cultures (data not shown). Furthermore, infection with T3D-RV significantly increased levels of cleaved caspase-3 within a given cell, as determined by costaining with an antibody specific for cleaved caspase-3 and a polyclonal antiserum specific for reovirus nonstructural protein μNS (Fig. 3G and H). Therefore, T1L and T3D-RV infect enteroids comparably but differ in the capacity to promote apoptosis and cell sloughing, suggesting that enteroids, like the intestine, undergo apoptotic cell death to inhibit viral replication.
FIG 3.
Viral infectivity and apoptosis in murine-derived enteroids following reovirus T1L and T3D-RV infection. Intestinal crypts were harvested from mice and established as enteroids in Matrigel. The enteroids were dissociated, seeded onto Transwell plates, and incubated at 37°C for 4 days. Enteroids were adsorbed with T1L or T3D-RV at an MOI of 100 PFU/cell or with PBS as a mock control. At 24 hpi, cells were fixed and stained with antibodies specific for β-catenin and reovirus polyclonal antiserum (A and B) or actin and cleaved caspase-3 (D and E). Nuclei were labeled with ProLong Gold antifade mountant containing DAPI (scale bar, 50 μm). (B) The percentage of infected cells was determined by enumeration of reovirus-positive cells from immunofluorescence images. (C) Viral titers were determined at the intervals shown by plaque assay and expressed as PFU per milliliter of cell homogenate. (E) The percentage of apoptotic cells was determined using the Nikon Elements Basic Research analysis software and represents the number of cleaved caspase-3-positive cells per total number of cells in the selected field. (F) The number of cells released into the apical medium was quantified using an automated cell counter. (G and H) At 24 hpi, cells were fixed and stained with an antibody specific for cleaved caspase-3 and a polyclonal antiserum specific for reovirus nonstructural protein μNS. Nuclei were labeled with ProLong Gold antifade mountant containing DAPI (scale bar, 50 μm). (H) The percentage of infected cells expressing cleaved caspase-3 was determined by dividing the number of costained reovirus-positive and cleaved-caspase-3-positive cells by the number of reovirus-positive cells and then multiplying the quotient by 100. Data represent results from two to four independent experiments performed in triplicate. Error bars indicate SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. P values were determined by Mann-Whitney test (C and H) and one-way ANOVA and Tukey's multiple-comparison test (E and F).
T3D-RV replicates comparably to T1L in cultured cells but stimulates enhanced levels of apoptosis.
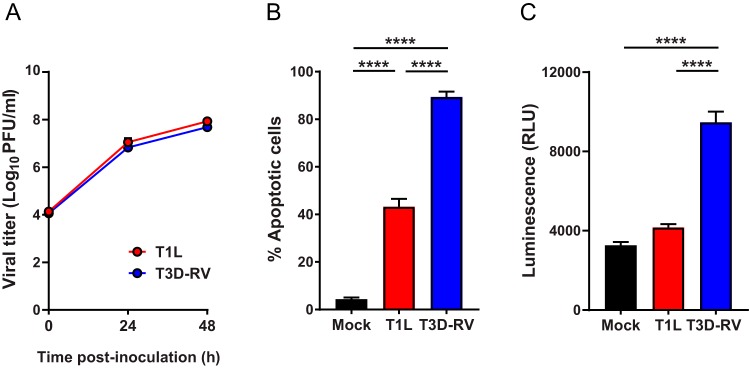
Viral reassortants occur naturally and are useful tools for identifying viral genes required for strain-specific polymorphisms. Before testing T1L × T3D-RV reassortants in mice, we assessed whether differences in apoptosis displayed by T1L and T3D-RV could be studied using cultured cells. By first testing the reassortants in cultured cells, we could define a subset of viral genes required for apoptosis and select viruses that express these genes to test using mice. To define the replication efficiency of T1L and T3D-RV, L929 (L) cells, a mouse fibroblast cell line highly susceptible to reovirus infection (36), were adsorbed with each virus strain, incubated for 0, 24, or 48 h, and lysed by freeze-thawing. Viral titers in cell lysates were determined by plaque assay. T1L and T3D-RV replicated comparably in these cells, producing nearly identical yields at all intervals tested (Fig. 4A). To determine whether T1L and T3D-RV differ in the capacity to induce apoptosis in cultured cells, L cells were adsorbed with each strain, and apoptosis was quantified at 38 hpi using an acridine orange (AO)-ethidium bromide staining assay. T3D-RV infection led to a significantly higher percentage of apoptotic cells than did T1L infection (Fig. 4B). To complement the AO staining assay, L cells were adsorbed with T1L and T3D-RV, and caspase-3/7 activity in cell lysates was quantified at 24 hpi using a fluorogenic substrate. T3D-RV-infected samples expressed higher levels of activated caspase-3/7 than did T1L- or mock-infected cells (Fig. 4C). Thus, T1L and T3D-RV replicate comparably in cultured cells, but T3D-RV induces the morphological and biochemical hallmarks of apoptosis more efficiently.
FIG 4.
Viral titers and apoptosis in L cells following reovirus T1L and T3D-RV infection. (A) L cells were adsorbed with T1L or T3D-RV at an MOI of 1 PFU/cell, and viral titers were determined at the intervals shown by plaque assay. Viral titers are expressed as PFU per milliliter of cell homogenate. (B and C) L cells were adsorbed with T1L or T3D-RV at an MOI of 100 PFU/cell. (B) Cells were evaluated by AO assay at 38 hpi. The results are expressed as percentage of apoptotic cells per field of view. (C) Cell lysates were subjected to a Caspase-Glo 3/7 assay at 24 hpi. Caspase-3 activity is expressed in relative luminescence units. Data represent results from three independent experiments performed in triplicate. Error bars indicate SEMs. ****, P < 0.0001; one-way ANOVA and Tukey's multiple-comparison test.
Viral gene segments M1 and M2 dictate reovirus apoptosis induction in cultured cells.
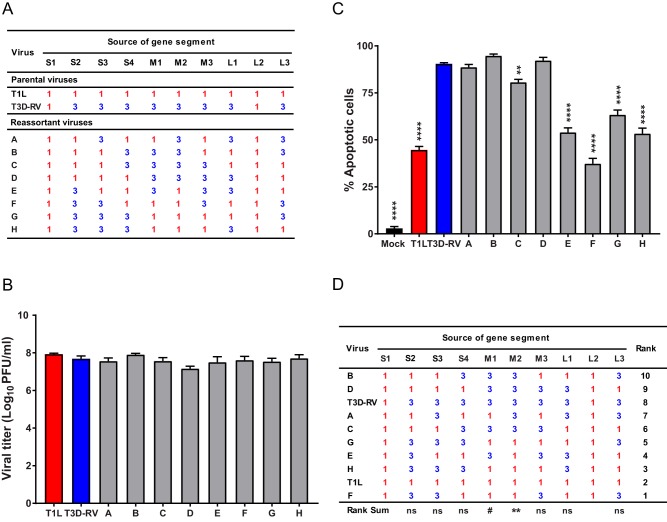
To identify viral genes that segregate with differences in apoptosis efficiency displayed by T1L and T3D-RV, eight T1L × T3D-RV reovirus reassortants (Fig. 5A) were recovered using reverse genetics (37). All reassortants contain the T1L S1 and L2 gene segments, which allow these strains to infect the murine intestine (38). To quantify the viral replication efficiency of the newly engineered reassortants, L cells were adsorbed with each virus strain and incubated for 24 h, and viral titers in cell lysates were determined by plaque assay. All reassortant viruses replicated comparably (Fig. 5B). To identify viral gene segments that segregate with apoptosis, L cells were adsorbed with T1L, T3D-RV, or one of eight T1L × T3D-RV reassortants and scored for apoptosis following AO staining. Reassortant viruses containing a T3D M2 gene induced more apoptosis by AO assay than those containing a T1L M2 gene (Fig. 5C and D). Additionally, the T3D M1 gene appeared to enhance apoptosis efficiency, albeit not as clearly as the T3D M2 gene (Fig. 5D). Therefore, the T3D M2 gene segment is required for increased levels of apoptosis in cultured cells, and the M1 gene also may influence apoptosis efficiency in this context.
FIG 5.
Viral replication and apoptosis in L cells following infection with reovirus reassortant viruses. (A) T1L × T3D-RV reassortant viruses used to identify genes that segregate with strain-specific differences in the capacity of reovirus to induce apoptosis in the intestine. Gene segments labeled “1” in red represent those derived from T1L; gene segments labeled “3” in blue represent those derived from T3D. (B) L cells were adsorbed with T1L, T3D-RV, or one of eight T1L × T3D-RV reassortants at an MOI of 1 PFU/cell, and viral titers were determined at 48 hpi by plaque assay. Viral titers are expressed as PFU per milliliter of cell homogenate. (C) L cells were adsorbed with T1L, T3D-RV, or one of eight T1L × T3D-RV reassortants at an MOI of 100 PFU/cell. PBS-inoculated samples (mock) were used as controls. Cells were evaluated by AO assay at 38 hpi. The results are expressed as the percentage of apoptotic cells per field of view. (D) Rank sum analysis of reassortants used to identify genes that segregate with the capacity to induce apoptosis. The rank number of each reassortant and the significance levels of each gene segment are shown. Data represent results from two or three experiments performed in triplicate. Error bars indicate SEMs. ns, nonsignificant. #, P = 0.06; **, P < 0.01; ****, P < 0.0001. P values were determined by one-way ANOVA and Dunnett's multiple-comparison test (compared with T3D-RV) (C) and Wilcoxon rank sum distribution test (D).
Viral gene segments M1 and M2 dictate reovirus-host interactions by altering apoptosis induction in the gut.
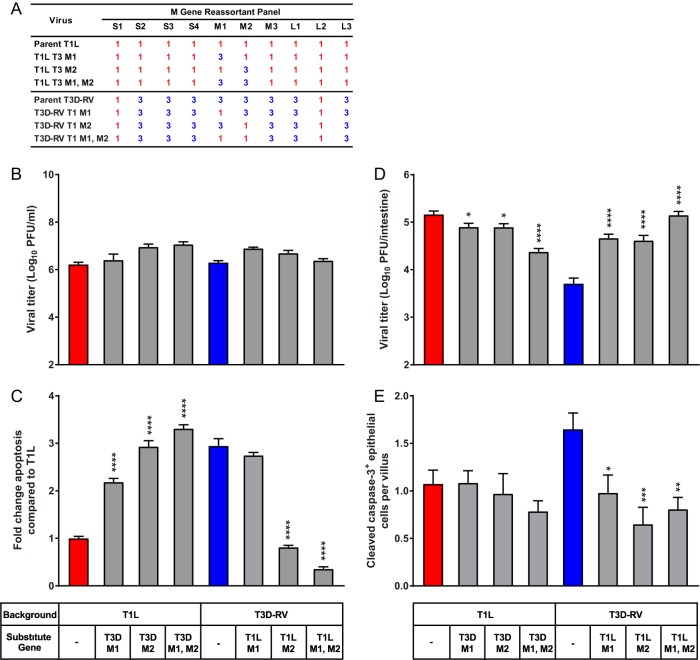
To examine the requirement of the M1 and M2 genes for apoptosis induction in the murine intestine, six T1L × T3D-RV M gene reciprocal reassortants were recovered. The reciprocal reassortants maintain the parental strain background but were constructed to exchange the M1 and M2 genes individually or together (Fig. 6A). To examine the replication capacity and subsequent virus-induced apoptosis of these reassortant viruses, L cells were adsorbed with each virus strain, incubated for various intervals, and scored for reovirus infection and apoptosis. The M gene reassortant viruses replicated comparably (Fig. 6B). At 38 hpi, the percentage of apoptotic cells was increased 3-fold following T3D-RV infection compared with the value obtained with T1L (Fig. 6C). The presence of a T3D M2 gene in a T1L background was sufficient to alleviate this difference, as the percentage of apoptotic cells was increased 2- to 3-fold following infection with these reassortant viruses compared to the value obtained with parental strain T1L (Fig. 6C). Additionally, reciprocal reassortant viruses containing the T1L M2 gene in a T3D-RV background displayed a significant reduction in apoptosis efficiency relative to that with parental strain T3D-RV (Fig. 6C). Surprisingly, the reassortant virus that contains the T3D M1 gene in a T1L background induced a 2-fold increase in the percentage of apoptotic cells compared with the value obtained with parental strain T1L (Fig. 6C). However, the reciprocal virus (T1L M1 gene in a T3D-RV background) produced a high percentage of apoptotic cells, likely due to the T3D M2 gene, which appears to override any effects of the T1L M1 gene (Fig. 6C). Furthermore, the reassortant virus that contains the T3D M1 and M2 genes in the T1L background slightly enhanced apoptosis levels compared with the levels obtained with viruses expressing the T3D M1 or M2 gene alone (Fig. 6C). Taken together, the results show that a T3D M2 gene in a T1L background is necessary and sufficient to stimulate apoptosis in cultured cells and, in conjunction with the T3D M1 gene, promotes maximum levels of apoptosis.
FIG 6.
Viral replication and apoptosis in cultured cells and murine intestine following infection with M gene reassortant viruses. (A) T1L × T3D-RV M gene reassortant viruses used to define genes that segregate with the apoptosis-inducing capacity of reovirus. Gene segments labeled “1” in red represent those derived from T1L; gene segments labeled “3” in blue represent those derived from T3D. (B) L cells were adsorbed with T1L, T3D-RV, or one of six T1L × T3D-RV M gene reassortants at an MOI of 1 PFU/cell, and viral titers were determined at 24 hpi by plaque assay. Viral titers are expressed as PFU per milliliter of cell homogenate. (C) L cells were adsorbed with T1L, T3D-RV, or one of six T1L × T3D-RV M gene reassortants at an MOI of 100 PFU/cell. Cells were evaluated by AO assay at 38 hpi. The results are expressed as fold change in apoptosis compared with the value for T1L. Data from two experiments performed in triplicate are presented. (D and E) Mice were inoculated perorally with 108 PFU of T1L, T3D-RV, or one of six T1L × T3D-RV M gene reassortants. The proximal half of the intestine was processed for viral titer, and the distal half was flushed, Swiss rolled, and processed for histology. (D) Titers of reovirus in the intestine were determined at 72 hpi by plaque assay (n ≥ 10 mice per virus). (E) At 24 hpi, histological sections were stained with an antibody against cleaved caspase-3. Cells positive for cleaved caspase-3 were enumerated manually and normalized per villus (n ≥ 8 mice per virus). Error bars indicate SEMs. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. P values in panels C to E were determined by one-way ANOVA and Dunnett's multiple-comparison test (compared with the parental strain).
Since T3D-RV activates apoptosis and subsequent villous sloughing and is cleared from the intestine more efficiently than T1L, we hypothesized that genes associated with enhanced apoptosis of T3D-RV also are associated with diminished infection capacity in the gut. To test this hypothesis, mice were inoculated perorally with T1L, T3D-RV, or each of the six T1L × T3D-RV M gene reassortant viruses and euthanized 24 hpi for histological analysis and 72 hpi for determination of viral titer. Tissue sections were stained for activated caspase-3 to identify cells undergoing apoptosis within the intestine. At 72 hpi, viral titers in the intestine of mice infected with T1L were approximately 30-fold greater than those in animals infected with T3D-RV. Reassortant viruses that express either or both of the T1L M1 and M2 genes in a T3D-RV background replicate comparably to T1L, indicating that these genes are sufficient to increase T3D-RV titers in the intestine (Fig. 6D). Furthermore, reduced numbers of cleaved caspase-3-positive enterocytes were observed in mice infected with these viruses (Fig. 6E). These findings suggest that enterocyte apoptosis serves as an antiviral response to inhibit viral infection in the intestine. Of note, the T3D M1 and M2 genes are not sufficient to limit T1L infection to levels comparable to those of T3D-RV or enhance apoptosis efficiency in the intestines of infected mice (Fig. 6D and E), suggesting that additional viral or host factors contribute to T1L infection capacity in the intestine.
DISCUSSION
In this study, we evaluated the function of apoptosis in T1L and T3D-RV intestinal infection and found that compared to T3D-RV, T1L induces less caspase-3 activation, less sloughing of intestinal villi, and prolonged infection. Using T1L × T3D-RV reassortant viruses, we found that the capacity to promote apoptosis in the intestine inversely correlates with infection, suggesting that enterocyte apoptosis and sloughing mediate an antiviral response to protect the host.
Enterocytes have the highest turnover rate of any cell population in the body (39, 40), most likely due to environmental insults and the necessity to maintain a healthy gut barrier. Despite the high rate of turnover, shedding events are rarely observed in healthy individuals and seldom visualized by biochemical markers such as cleaved caspase-3 (7, 40). However, a variety of stimuli and disease states can promote apoptosis during pathological cell shedding. Of note, some enteric viruses stimulate apoptotic death of IECs, which is usually associated with gastrointestinal pathology. In our study, infection with reovirus T3D-RV led to extensive caspase-3 activation in IECs and sloughing of caspase-3-positive cells into the intestinal lumen. Interestingly, T3D-RV-induced apoptosis was not associated with shortening of intestinal villi or other pathological markers, and barrier function was maintained. Activated caspase-3 was found predominantly at the villus tips, often with multiple cells undergoing apoptosis in a localized region of the villus. During infection with either T1L or T3D-RV, cleaved-caspase-3 staining was patchy but could be observed throughout the distal half of the small intestine. Virus-induced apoptosis was observed within the first 24 h following infection and declined at later time points.
The distribution of reovirus- and caspase-3-postive antigen staining in enterocytes did not overlap. Although this observation might be attributable to insensitivity of the immunohistochemical staining techniques, there are additional possibilities. Reovirus is capable of inducing apoptosis in the absence of viral replication (27, 41). During reovirus infection, pattern recognition receptors detect virus-associated molecular patterns (25, 42, 43) and trigger a signaling cascade that leads to activation of nuclear factor kappa light chain enhancer of activated B cells (NF-κB) (44) and interferon regulatory factor 3 (IRF-3) (25). Activated NF-κB and IRF-3 translocate to the nucleus and induce expression of many genes, including those encoding type I interferons (interferon alpha and interferon beta). Within reovirus-infected cells, NF-κB and IRF-3 are required for maximum levels of apoptosis (25, 44–46). It is possible that cytokines induced by NF-κB and IRF-3, including type I interferons, activate death pathways in adjacent, uninfected cells (47–50). In addition, triggering enterocyte death would restrict viral replication and limit the amount of antigen observed by immunohistochemical staining. Therefore, reovirus may induce apoptosis of either uninfected IECs by paracrine signaling or infected cells that cannot be detected.
Enteroids are useful for studying virus-host interactions ex vivo. There are several methods for enteroid isolation, which differ in the species types, starting material, expandability, and developmental stage. For our studies, we used murine tissue-derived organoids due to phenotypic similarities to the mouse intestine. We found that enteroids are susceptible to reovirus infection and observed strain-specific differences in apoptosis efficiency. The research described here will provide the basis for studies examining the capacity of other enteric viruses to promote apoptosis. Furthermore, enteroids harvested from mice deficient in antiviral response pathways may lead to discovery of host factors required for virus-induced apoptosis.
Differences in apoptosis induction segregate with the viral M2 gene (28), which differs between T1L and T3D-RV. It is not surprising that in our study the M2 gene segment segregated with differences in apoptosis efficiency following infection of cultured cells, as this association has been described previously (26–28). More interesting was the observation that the T1L M2 gene in a T3D-RV background was sufficient to increase viral titers in the intestine to levels comparable to those of T1L. The T1L M2 gene was identified in a previous study as a virulence factor required for infection of the intestine of neonatal mice (51). A subsequent study suggested that only the S1 and L2 genes are required for intestinal infection (22). Our findings resolve these discrepancies by using reassortant viruses with common S1 and L2 genes from T1L. Following p.o. inoculation with these strains, the M2 gene emerges as a determinant of reovirus intestinal infection. Viruses with a T1L M2 gene promoted less apoptosis in the intestine than those with a T3D M2 gene, providing additional evidence that epithelial cell death fosters an antiviral state and suggesting a mechanism by which the M2 gene dictates intestinal pathogenesis.
The M2 gene encodes outer-capsid protein μ1, which functions to penetrate endosomal membranes during viral entry, a property required for apoptosis induction (52). Following infection of IECs, it is possible that the T3D M2-encoded μ1 protein mediates penetration of endosomal membranes, exposing viral particles and dsRNA to pattern recognition receptors. Activation of these receptors stimulates proapoptotic signaling by NF-κB and IRF-3 and upregulation of interferons and other cytokines, which may evoke cell death. Such a scenario provides a mechanism by which T3D-RV could induce enterocyte apoptosis and extrusion from the villus to protect adjacent cells from subsequent infection.
The Φ domain of μ1 is necessary and sufficient to promote apoptosis in cell culture (53, 54), and polymorphisms in this domain influence the apoptotic efficiency of T3D (53). A virus containing an M2 gene with the apoptosis-attenuating polymorphisms in a T3D-RV background would likely lose apoptotic capacity in cultured cells, enteroids, and mice. Furthermore, other evidence suggests that reovirus strains that have diminished apoptotic capacity replicate to higher titers in the intestine (30).
Although the M2 gene is the primary determinant of differences in apoptosis efficiency exhibited by T1L and T3D-RV, our findings suggest that the M1 gene exacerbates cell death when combined with the M2 gene. The reovirus M1 gene-encoded μ2 protein is a strain-specific interferon antagonist that dictates pathogenesis in neonatal mice (55). A single amino acid polymorphism in μ2 modulates cytopathic effects by regulating the host interferon response (55). The M1 gene has not been identified in previous studies of viral determinants required for intestinal infection. The absence of such a linkage might be attributable to the use of newborn mice, which may lack mature interferon signaling. The T1L μ2-encoded interferon antagonist may subvert host antiviral responses in the gut, including the induction of apoptosis, allowing this strain to replicate more efficiently in this tissue. T1L infection induces greater levels of type I interferons and interferon-stimulated genes after 24 hpi than does infection with T3D-RV (3), which could be a by-product of prolonged T1L infection.
Inoculation perorally with reovirus strain T1L stimulates inflammatory responses to fed antigen, whereas T3D-RV does not produce this effect. In this study, we found that T3D-RV titers are diminished at later time points of infection, which is linked to its capacity to promote apoptosis in IECs. We predict that prolonged infection by T1L during the introduction of new food proteins stimulates inflammatory signals, such as type I interferons, to promote food antigen-specific T cell responses. From studies of reassortant viruses, we found that the T1L M1 and M2 genes in a T3D-RV background are sufficient to increase viral titers in the intestine to levels comparable to those of T1L. Using these reassortant viruses, future studies will determine whether decreased intestinal apoptosis and prolonged viral infection are associated with tolerance blockade. Consequently, this work provides a possible mechanism by which certain enteric, nonpathogenic viruses establish prolonged infection by subverting the host cell death response to stimulate the inflammatory signals required to break immunological tolerance to food antigens.
MATERIALS AND METHODS
Cells and viruses.
Spinner-adapted murine L cells were maintained in either suspension or monolayer cultures in Joklik's modified Eagle's minimal essential medium (SMEM; Lonza) supplemented to contain 5% fetal bovine serum (FBS; Gibco), 2 mM l-glutamine, 100 U/ml of penicillin, 100 μg/ml of streptomycin (Gibco), and 25 ng/ml of amphotericin B (Sigma). Baby hamster kidney (BHK) cells that constitutively express the bacteriophage T7 RNA polymerase (BHK-T7 cells) were maintained in Dulbecco's modified Eagle's minimal essential medium (DMEM; Gibco) supplemented to contain 5% FBS, 2 mM l-glutamine, 1 mg/ml of Geneticin (Gibco), and nonessential amino acids (Sigma).
Recombinant reoviruses were engineered using plasmid-based reverse genetics (37). Recombinant strain T1L is a stock recovered by plasmid-based rescue from cloned T1L cDNAs (56). The engineered reassortant virus strain, T3D-RV, was recovered following transfection of BHK-T7 cells with plasmid constructs encoding the S1 and L2 gene segments of strain T1L and the remaining eight gene segments of strain T3D. T1L × T3D-RV reassortant viruses were isolated using cloned T1L or T3D cDNAs. After 3 to 5 days of incubation, cells were frozen and thawed three times, and virus was isolated by plaque purification using monolayers of L cells (57). Reovirus virions were purified from second- or third-passage L-cell lysate stocks (58). Viral particles were extracted from infected cell lysates using Vertrel XF (DuPont), layered onto 1.2- to 1.4-g/cm3 CsCl gradients, and centrifuged at 62,000 × g for 16 h. Bands corresponding to virions (1.36 g/cm3) were collected and dialyzed in virion storage buffer (150 mM NaCl, 15 mM MgCl2, and 10 mM Tris-HCl [pH 7.4]) (59). Viral titer was determined by plaque assay using L cells (57). Purified viral particles were electrophoresed in SDS-polyacrylamide gels, which were stained with ethidium bromide to visualize viral gene segments.
Mice.
C57BL/6 mice were purchased from Jackson Laboratories. For all experiments, mice were inoculated at 6 to 8 weeks of age. All mice were maintained in a specific-pathogen-free (SPF) environment at the University of Pittsburgh or Vanderbilt University. Animal husbandry and experimental procedures were performed in accordance with Public Health Service policy and approved by the University of Pittsburgh School of Medicine and Vanderbilt University School of Medicine Institutional Animal Care and Use Committees.
Infection of mice.
Mice were inoculated perorally with 108 or 1010 PFU/mouse of purified reovirus diluted in PBS using a 22-gauge round-tipped needle (Cadence Science) (19). Titers of virus in the inocula were determined to confirm the number of infectious particles in the administered dose. For analysis of viral loads, mice were euthanized at various intervals postinoculation, and organs were harvested into 1 ml of PBS and stored at −20°C prior to assay. Samples were thawed (37°C), homogenized with a TissueLyser LT (Qiagen) for 8 min, frozen (−20°C), and homogenized again for 5 min prior to dilution for plaque assay. Viral titers in organ homogenates were determined as the number of PFU per tissue or intestine (57).
Antibodies.
Immunoglobulin G (IgG) fractions of rabbit antisera raised against reovirus strains T1L and T3D (60) were purified by protein A Sepharose chromatography (61). Rabbit anti-cleaved-caspase-3 monoclonal antibody was purchased from Cell Signaling Technologies. Anti-β-catenin antibody was purchased from BD Biosciences. Chicken polyclonal antiserum specific for μNS (62) was provided by John Parker. Primary antibodies were detected using Alexa Fluor-conjugated secondary antibodies from Thermo Fisher Scientific, while actin was visualized using ActinGreen ReadyProbe (Thermo Fisher Scientific).
Histology and immunohistochemistry.
Mice were inoculated perorally with purified reovirus diluted in PBS. At 1 dpi, mice were euthanized, and intestines were resected. The proximal half was prepared for viral titer determination by plaque assay, and the distal half was flushed with 10% formalin and Swiss rolled. Samples were submerged in 10% formalin at room temperature for 24 or 48 h and embedded in paraffin. Consecutive 6-μm sections were stained with H&E for evaluation of histopathologic changes or processed for immunohistochemical detection of reovirus antigen and cleaved (active) caspase-3. Images were captured at a magnification of ×20 and a resolution of 0.5 μm/pixel using a high-throughput Leica SCN400 slide scanner automated digital imaging system.
Quantification of histology.
For the caspase-3-positive staining area, tissue regions were differentiated from the glass slide using Ariol Review software. Upper and lower thresholds for color, saturation, and intensity were set for both blue, hematoxylin staining of nuclei and for brown, 3′diaminobenzidine (DAB) reaction products. Thus, brown positive areas were distinguished from blue-only negative areas. The area of positive staining per whole tissue region was calculated as a percent area of brown (DAB-positive) pixels divided by the total area analyzed.
Cleaved-caspase-3-staining epithelial cells were quantified by an observer blinded to the conditions of the experiment by enumerating all positive cells (brown) throughout the distal half of the small intestine and normalizing to the number of villi counted. Only epithelial cells and villi from properly oriented villus structures were included in the analysis. Proper orientation was defined in villi containing a clear exterior epithelial cell column, a crypt-to-villus ratio of at least 2:1, and an unfragmented lamina propria. Intestinal sections with fewer than 70 properly oriented villi were excluded.
Establishing small intestinal enteroids.
Crypt enteroid cultures were established as described previously (63). Eight centimeters of the proximal small intestine was dissected, flushed with PBS, and minced. Following a PBS wash, tissue was transferred to 5 ml of chelation buffer (2 mM EDTA in PBS) and incubated at 4°C for 30 min. After removal of chelation buffer, tissue fragments were resuspended in 5 ml of shaking buffer (PBS with 43.3 mM sucrose and 54.9 mM sorbitol) and agitated gently for 2 min to release intestinal crypts. Crypt-containing supernatants were filtered through a 100-μm filter, enumerated, and transferred into round-bottomed polystyrene tubes. Following centrifugation, crypts were resuspended in growth factor-reduced Matrigel (BD Bioscience) at a concentration of 300 crypts per 50 μl of Matrigel. Following polymerization, the crypt/Matrigel mixture was overlaid with 500 μl of mini-gut culture medium (Advanced Dulbecco's modified Eagle's medium–F-12 [Invitrogen] supplemented to contain 1× GlutaMAX [Thermo Fisher], 100 U/ml of penicillin [Invitrogen], 100 μg/ml of streptomycin [Invitrogen], 1 mM HEPES [Gibco], N-2 supplement [R&D Systems], B27 supplement [Invitrogen], 50 ng/ml of epidermal growth factor [EGF; R&D Systems], 500 ng/ml of R-spondin [Vanderbilt Antibody and Protein Resource], 100 ng/ml of noggin [R&D Systems], and 50% Wnt3a conditioned medium). The medium was replaced every 2 to 4 days. Enteroids were passaged as needed by collection and shearing through a 25-gauge needle prior to plating in Matrigel.
Enteroid Transwell plating.
Enteroids were collected 2 days after passage and incubated with TrypLE (Thermo Fisher Scientific) solution at 37°C for 90 min to dissociate cells into single-cell suspensions. Cells (2 × 105) were plated onto the top of a 24-well, 0.4-μm-pore Transwell filter coated with collagen 1 (Gibco) in medium supplemented to contain 10 μM Y27632 (Sigma). After 24 h, medium on the top and bottom of the Transwell filters was replaced with differentiation medium (Advanced DMEM–F-12 supplemented to contain 1× GlutaMAX, 100 U/ml of penicillin, 100 μg/ml of streptomycin, and 20% FBS).
Assays for reovirus replication, infectivity, and cell death in enteroids.
On day 4 post-Transwell plating, enteroids were adsorbed with virus at a multiplicity of infection (MOI) of 100 PFU/cell, assuming 2.5 × 105 cells/well, by adding 30 μl of virus inoculum to the apical compartment. After adsorption of virus at room temperature for 1 h, cells were washed twice with PBS, 200 μl of medium was added to the apical compartment, and 1 ml of medium was added to the basolateral compartment. Enteroids were incubated at 37°C for 24 h. For quantification of released cells, 100 μl of apical medium was collected, and cells in the medium were enumerated using a Scepter handheld automated cell counter. For viral replication assays, Transwell membrane inserts were removed from Transwells with a scalpel, submerged in 500 μl of medium, and frozen and thawed twice. Viral titers in cell lysates were determined by plaque assay using L cells (57). For enteroid staining, Transwell inserts were gently washed with PBS and fixed at room temperature for 30 min in 3% paraformaldehyde before continuing with the Transwell staining protocol.
Transwell insert staining and quantification.
After being washed and fixed, cells were permeabilized with 0.2% Triton-X (Sigma) followed by blocking in 3% milk solution. The following primary antibodies were used: reovirus polyclonal antiserum (60) at 1:1,000, anti-β-catenin (BD Biosciences) at 1:500, anti-cleaved-caspase-3 (Cell Signaling Technology) at 1:400, and μNS polyclonal antiserum at 1:1,000. Appropriate species-specific Alexa Fluor secondary antibodies (Thermo Fisher Scientific) were used at a dilution of 1:1,000. Enteroids were stained with ActinGreen ReadyProbe (Thermo Fisher Scientific). Following antibody incubation, inserts were excised from the Transwell and mounted on glass slides with 4′,6-diamidino-2-phenylindole (DAPI)-containing ProLong Gold mounting medium (Thermo Fisher Scientific). Images were collected using a Nikon Eclipse E800 microscope, and cells positive for cleaved caspase-3 were enumerated using Nikon Elements Basic Research software. For viral infectivity, the percentage of reovirus antigen-positive cells was determined. No background staining of uninfected control cells was detected.
Assays of reovirus replication and gene expression in cell culture.
L cells (2 × 105) in 24-well plates (Costar) were adsorbed with reovirus at an MOI of 1 PFU/cell, washed with PBS, and incubated at 37°C for various intervals. Cells were frozen and thawed twice prior to determination of viral titer by plaque assay using L cells (57). Viral titers are expressed as PFU per milliliter of cell homogenate.
Quantification of apoptosis by AO staining.
L cells (2 × 105) in 24-well plates were adsorbed with reovirus at an MOI of 100 PFU/cell, washed with PBS, and incubated at 37°C for 38 h. The percentage of apoptotic cells was determined using AO staining (27). Images were collected from three fields of view per well by epi-illumination fluorescence microscopy using a Lionheart FX automated microscope (Biotek). The percentage of apoptotic cells exhibiting orange nuclei with condensed and fragmented chromatin versus live cells exhibiting structurally normal green nuclei was quantified using an imaging algorithm developed in Gen5 software (Biotek). The average nucleus size and a fluorescence threshold were established by the user. Color values from each individual-pixel RGB (red, green, blue) triplet were obtained and compared to the user-defined threshold. To exclude false-positive signals, an exclusion criterion was established based on the average pixels of false-positive cells and included in the imaging algorithm. Therefore, green nuclei with condensed chromatin (early apoptotic) and red, structurally normal nuclei were subtracted from the final count.
Detection of caspase-3/7 activity.
L cells (5 × 104) in black, clear-bottom 96-well plates (Costar) were adsorbed with reovirus at an MOI of 100 PFU/cell at room temperature for 1 h. Following incubation of cells at 37°C for 24 h, caspase-3/7 activity was quantified using the Caspase-Glo 3/7 assay (Promega).
Statistical analysis.
Experiments were performed in triplicate and repeated at least twice. Representative results of single experiments are shown in figures as indicated. Mean values were compared using the Mann-Whitney test, Wilcoxon rank sum distribution test, or one-way analysis of variance (ANOVA) for multiple comparisons. ANOVA was followed by Dunnett's post hoc test for multiple comparisons to a control group, Sidak's post hoc test for multiple comparisons between sets of data, or Tukey's post hoc test for multiple comparisons between all groups. Grubb's test was used for exclusion of outliers. The statistical test used and P values are indicated in each figure legend. P values of <0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank members of the Dermody laboratory for helpful discussions and critical review of the manuscript and Brian Leibowitz for experimental expertise in intestinal apoptosis. We thank the Vanderbilt University and University of Pittsburgh Divisions of Animal Care for animal husbandry and Kay Washington for assistance with histological analyses. Histological slides were prepared by the Vanderbilt Translational Pathology Shared Resource, supported by Cancer Center support grant P30 CA068485 and the Vanderbilt Mouse Metabolic Phenotyping Center grant U24 DK059637. Whole-slide imaging and quantification of immunostaining were conducted in the Digital Histology Shared Resource at Vanderbilt University Medical Center (www.mc.vanderbilt.edu/dhsr).
This work was supported by United States Public Health Service awards T32 HL007751 and F31 DK108562 (J.J.B.) and R01 DK098435 (B.J. and T.S.D.). Additional support was provided by the Elizabeth B. Lamb Center for Pediatric Research.
REFERENCES
- 1.Cuconati A, Degenhardt K, Sundararajan R, Anschel A, White E. 2002. Bak and Bax function to limit adenovirus replication through apoptosis induction. J Virol 76:4547–4558. doi: 10.1128/JVI.76.9.4547-4558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Handke W, Luig C, Popovic B, Krmpotic A, Jonjic S, Brune W. 2013. Viral inhibition of BAK promotes murine cytomegalovirus dissemination to salivary glands. J Virol 87:3592–3596. doi: 10.1128/JVI.02657-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouziat R, Hinterleitner R, Brown JJ, Stencel-Baerenwald JE, Ikizler M, Mayassi T, Meisel M, Kim SM, Discepolo V, Pruijssers AJ, Ernest JD, Iskarpatyoti JA, Costes LM, Lawrence I, Palanski BA, Varma M, Zurenski MA, Khomandiak S, McAllister N, Aravamudhan P, Boehme KW, Hu F, Samsom JN, Reinecker HC, Kupfer SS, Guandalini S, Semrad CE, Abadie V, Khosla C, Barreiro LB, Xavier RJ, Ng A, Dermody TS, Jabri B. 2017. Reovirus infection triggers inflammatory responses to dietary antigens and development of celiac disease. Science 356:44–50. doi: 10.1126/science.aah5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengartner MO. 2000. The biochemistry of apoptosis. Nature 407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 5.Elmore S. 2007. Apoptosis: a review of programmed cell death. Toxicol Pathol 35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayhew TM, Myklebust R, Whybrow A, Jenkins R. 1999. Epithelial integrity, cell death and cell loss in mammalian small intestine. Histol Histopathol 14:257–267. [DOI] [PubMed] [Google Scholar]
- 7.Marshman E, Ottewell PD, Potten CS, Watson AJ. 2001. Caspase activation during spontaneous and radiation-induced apoptosis in the murine intestine. J Pathol 195:285–292. doi: 10.1002/path.967. [DOI] [PubMed] [Google Scholar]
- 8.McAllister CS, Lakhdari O, Pineton de Chambrun G, Gareau MG, Broquet A, Lee GH, Shenouda S, Eckmann L, Kagnoff MF. 2013. TLR3, TRIF, and caspase 8 determine double-stranded RNA-induced epithelial cell death and survival in vivo. J Immunol 190:418–427. doi: 10.4049/jimmunol.1202756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou R, Wei H, Sun R, Tian Z. 2007. Recognition of double-stranded RNA by TLR3 induces severe small intestinal injury in mice. J Immunol 178:4548–4556. doi: 10.4049/jimmunol.178.7.4548. [DOI] [PubMed] [Google Scholar]
- 10.Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol 80:10372–10381. doi: 10.1128/JVI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boshuizen JA, Reimerink JH, Korteland-van Male AM, van Ham VJ, Koopmans MP, Buller HA, Dekker J, Einerhand AW. 2003. Changes in small intestinal homeostasis, morphology, and gene expression during rotavirus infection of infant mice. J Virol 77:13005–13016. doi: 10.1128/JVI.77.24.13005-13016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, Estes JD, Duan L, Jessurun J, Pambuccian S, Forster C, Wietgrefe S, Zupancic M, Schacker T, Reilly C, Carlis JV, Haase AT. 2008. Simian immunodeficiency virus-induced intestinal cell apoptosis is the underlying mechanism of the regenerative enteropathy of early infection. J Infect Dis 197:420–429. doi: 10.1086/525046. [DOI] [PubMed] [Google Scholar]
- 13.Tai JH, Williams JV, Edwards KM, Wright PF, Crowe JE Jr, Dermody TS. 2005. Prevalence of reovirus-specific antibodies in young children in Nashville, Tennessee. J Infect Dis 191:1221–1224. doi: 10.1086/428911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Selb B, Weber B. 1994. A study of human reovirus IgG and IgA antibodies by ELISA and Western blot. J Virol Methods 47:15–25. doi: 10.1016/0166-0934(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 15.Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, Kelsall BL. 2007. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J Exp Med 204:1349–1358. doi: 10.1084/jem.20061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos-Alvarez M, Sabin AB. 1954. Characteristics of poliomyelitis and other enteric viruses recovered in tissue culture from healthy American children. Proc Soc Exp Biol Med 87:655–661. doi: 10.3181/00379727-87-21474. [DOI] [PubMed] [Google Scholar]
- 17.Ramos-Alvarez M, Sabin AB. 1958. Enteropathogenic viruses and bacteria; role in summer diarrheal diseases of infancy and early childhood. JAMA 167:147–156. doi: 10.1001/jama.1958.02990190001001. [DOI] [PubMed] [Google Scholar]
- 18.Sabin AB. 1956. The significance of viruses recovered from the intestinal tracts of healthy infants and children. Ann N Y Acad Sci 66:226–230. doi: 10.1111/j.1749-6632.1956.tb40126.x. [DOI] [PubMed] [Google Scholar]
- 19.Rubin DH, Kornstein MJ, Anderson AO. 1985. Reovirus serotype 1 intestinal infection: a novel replicative cycle with ileal disease. J Virol 53:391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubin DH. 1987. Reovirus serotype 1 binds to the basolateral membrane of intestinal epithelial cells. Microb Pathog 3:215–219. doi: 10.1016/0882-4010(87)90098-2. [DOI] [PubMed] [Google Scholar]
- 21.Kauffman RS, Wolf JL, Finberg R, Trier JS, Fields BN. 1983. The s1 protein determines the extent of spread of reovirus from the gastrointestinal tract of mice. Virology 124:403–410. doi: 10.1016/0042-6822(83)90356-2. [DOI] [PubMed] [Google Scholar]
- 22.Bodkin DK, Fields BN. 1989. Growth and survival of reovirus in intestinal tissue: role of the L2 and S1 genes. J Virol 63:1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dermody TS, Parker JS, Sherry B. 2013. Orthoreoviruses, p 1304–1346. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 24.Kominsky DJ, Bickel RJ, Tyler KL. 2002. Reovirus-induced apoptosis requires both death receptor- and mitochondrial-mediated caspase-dependent pathways of cell death. Cell Death Differ 9:926–933. doi: 10.1038/sj.cdd.4401045. [DOI] [PubMed] [Google Scholar]
- 25.Holm GH, Zurney J, Tumilasci V, Danthi P, Hiscott J, Sherry B, Dermody TS. 2007. Retinoic acid-inducible gene-I and interferon-b promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J Biol Chem 282:21953–21961. doi: 10.1074/jbc.M702112200. [DOI] [PubMed] [Google Scholar]
- 26.Rodgers SE, Barton ES, Oberhaus SM, Pike B, Gibson CA, Tyler KL, Dermody TS. 1997. Reovirus-induced apoptosis of MDCK cells is not linked to viral yield and is blocked by Bcl-2. J Virol 71:2540–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tyler KL, Squier MK, Rodgers SE, Schneider SE, Oberhaus SM, Grdina TA, Cohen JJ, Dermody TS. 1995. Differences in the capacity of reovirus strains to induce apoptosis are determined by the viral attachment protein s1. J Virol 69:6972–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyler KL, Squier MKT, Brown AL, Pike B, Willis D, Oberhaus SM, Dermody TS, Cohen JJ. 1996. Linkage between reovirus-induced apoptosis and inhibition of cellular DNA synthesis: role of the S1 and M2 genes. J Virol 70:7984–7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fleeton MN, Contractor N, Leon F, Wetzel JD, Dermody TS, Kelsall BL. 2004. Peyer's patch dendritic cells process viral antigen from apoptotic epithelial cells in the intestine of reovirus-infected mice. J Exp Med 200:235–245. doi: 10.1084/jem.20041132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pruijssers AJ, Hengel H, Abel TW, Dermody TS. 2013. Apoptosis induction influences reovirus replication and virulence in newborn mice. J Virol 87:12980–12989. doi: 10.1128/JVI.01931-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou WY, Blutt SE, Crawford SE, Ettayebi K, Zeng XL, Saxena K, Ramani S, Karandikar UC, Zachos NC, Estes MK. 31 March 2017. Human intestinal enteroids: new models to study gastrointestinal virus infections. Methods Mol Biol doi: 10.1007/7651_2017_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drummond CG, Bolock AM, Ma C, Luke CJ, Good M, Coyne CB. 2017. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc Natl Acad Sci U S A 114:1672–1677. doi: 10.1073/pnas.1617363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antar AAR, Konopka JL, Campbell JA, Henry RA, Perdigoto AL, Carter BD, Pozzi A, Abel TW, Dermody TS. 2009. Junctional adhesion molecule-A is required for hematogenous dissemination of reovirus. Cell Host Microbe 5:59–71. doi: 10.1016/j.chom.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boehme KW, Guglielmi KM, Dermody TS. 2009. Reovirus nonstructural protein sigma1s is required for establishment of viremia and systemic dissemination. Proc Natl Acad Sci U S A 106:19986–19991. doi: 10.1073/pnas.0907412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark KM, Wetzel JD, Gu Y, Ebert DH, McAbee SA, Stoneman EK, Baer GS, Zhu Y, Wilson GJ, Prasad BVV, Dermody TS. 2006. Reovirus variants selected for resistance to ammonium chloride have mutations in viral outer-capsid protein s3. J Virol 80:671–681. doi: 10.1128/JVI.80.2.671-681.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kobayashi T, Antar AA, Boehme KW, Danthi P, Eby EA, Guglielmi KM, Holm GH, Johnson EM, Maginnis MS, Naik S, Skelton WB, Wetzel JD, Wilson GJ, Chappell JD, Dermody TS. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bodkin DK, Nibert ML, Fields BN. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol 63:4676–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelberg HB. 2007. Alimentary system, p 342–360. In McGavin MD, Zachary JF (ed), Pathologic basis of veterinary disease, 4th ed Elsevier Mosby, St. Louis, MO. [Google Scholar]
- 40.Williams JM, Duckworth CA, Burkitt MD, Watson AJ, Campbell BJ, Pritchard DM. 2015. Epithelial cell shedding and barrier function: a matter of life and death at the small intestinal villus tip. Vet Pathol 52:445–455. doi: 10.1177/0300985814559404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly JL, Dermody TS. 2002. Virion disassembly is required for apoptosis induced by reovirus. J Virol 76:1632–1641. doi: 10.1128/JVI.76.4.1632-1641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M Jr. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Connolly JL, Rodgers SE, Clarke P, Ballard DW, Kerr LD, Tyler KL, Dermody TS. 2000. Reovirus-induced apoptosis requires activation of transcription factor NF-kB. J Virol 74:2981–2989. doi: 10.1128/JVI.74.7.2981-2989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Connolly JL, Barton ES, Dermody TS. 2001. Reovirus binding to cell surface sialic acid potentiates virus-induced apoptosis. J Virol 75:4029–4039. doi: 10.1128/JVI.75.9.4029-4039.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Donnell SM, Hansberger MW, Connolly JL, Chappell JD, Watson MJ, Pierce JM, Wetzel JD, Han W, Barton ES, Forrest JC, Valyi-Nagy T, Yull FE, Blackwell TS, Rottman JN, Sherry B, Dermody TS. 2005. Organ-specific roles for transcription factor NF-kappaB in reovirus-induced apoptosis and disease. J Clin Invest 115:2341–2350. doi: 10.1172/JCI22428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apelbaum A, Yarden G, Warszawski S, Harari D, Schreiber G. 2013. Type I interferons induce apoptosis by balancing cFLIP and caspase-8 independent of death ligands. Mol Cell Biol 33:800–814. doi: 10.1128/MCB.01430-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kotredes KP, Gamero AM. 2013. Interferons as inducers of apoptosis in malignant cells. J Interferon Cytokine Res 33:162–170. doi: 10.1089/jir.2012.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stawowczyk M, Van Scoy S, Kumar KP, Reich NC. 2011. The interferon stimulated gene 54 promotes apoptosis. J Biol Chem 286:7257–7266. doi: 10.1074/jbc.M110.207068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jarry A, Florent M, Bou-Hanna C, Meurette G, Mohty M, Mosnier F, Laboisse CL, Bossard C. 2017. Interferon-alpha promotes Th1 response and epithelial apoptosis via inflammasome activation in human intestinal mucosa. Cell Mol Gastroenterol Hepatol 3:72–81. doi: 10.1016/j.jcmgh.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubin DH, Fields BN. 1980. Molecular basis of reovirus virulence. Role of the M2 gene. J Exp Med 152:853–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danthi P, Kobayashi T, Holm GH, Hansberger MW, Abel TW, Dermody TS. 2008. Reovirus apoptosis and virulence are regulated by host cell membrane penetration efficiency. J Virol 82:161–172. doi: 10.1128/JVI.01739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danthi P, Coffey CM, Parker JS, Abel TW, Dermody TS. 2008. Independent regulation of reovirus membrane penetration and apoptosis by the mu1 phi domain. PLoS Pathog 4:e1000248. doi: 10.1371/journal.ppat.1000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Coffey CM, Sheh A, Kim IS, Chandran K, Nibert ML, Parker JS. 2006. Reovirus outer capsid protein m1 induces apoptosis and associates with lipid droplets, endoplasmic reticulum, and mitochondria. J Virol 80:8422–8438. doi: 10.1128/JVI.02601-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irvin SC, Zurney J, Ooms LS, Chappell JD, Dermody TS, Sherry B. 2012. A single-amino-acid polymorphism in reovirus protein mu2 determines repression of interferon signaling and modulates myocarditis. J Virol 86:2302–2311. doi: 10.1128/JVI.06236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi T, Ooms LS, Ikizler M, Chappell JD, Dermody TS. 2010. An improved reverse genetics system for mammalian orthoreoviruses. Virology 398:194–200. doi: 10.1016/j.virol.2009.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Virgin HWt Bassel-Duby R, Fields BN, Tyler KL. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J Virol 62:4594–4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Furlong DB, Nibert ML, Fields BN. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol 62:246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith RE, Zweerink HJ, Joklik WK. 1969. Polypeptide components of virions, top component and cores of reovirus type 3. Virology 39:791–810. doi: 10.1016/0042-6822(69)90017-8. [DOI] [PubMed] [Google Scholar]
- 60.Wetzel JD, Chappell JD, Fogo AB, Dermody TS. 1997. Efficiency of viral entry determines the capacity of murine erythroleukemia cells to support persistent infections by mammalian reoviruses. J Virol 71:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barton ES, Connolly JL, Forrest JC, Chappell JD, Dermody TS. 2001. Utilization of sialic acid as a coreceptor enhances reovirus attachment by multistep adhesion strengthening. J Biol Chem 276:2200–2211. doi: 10.1074/jbc.M004680200. [DOI] [PubMed] [Google Scholar]
- 62.Desmet EA, Anguish LJ, Parker JS. 2014. Virus-mediated compartmentalization of the host translational machinery. mBio 5:e01463-14. doi: 10.1128/mBio.01463-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. 2009. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]