ABSTRACT
Endogenous viral elements (paleoviruses) provide “molecular fossils” for studying the deep history and macroevolution of viruses. Endogenous plant pararetroviruses (EPRVs) are widespread in angiosperms, but little is known about EPRVs in earlier-branching plants. Here we use a large-scale phylogenomic approach to investigate the diversity and macroevolution of plant pararetroviruses (formally known as Caulimoviridae). We uncover an unprecedented and unappreciated diversity of EPRVs within the genomes of gymnosperms and ferns. The known angiosperm viruses constitute only a minor part of the Caulimoviridae diversity. By characterizing the distribution of EPRVs, we show that no major euphyllophyte lineages escape the activity of Caulimoviridae, raising the possibility that many exogenous Caulimoviridae remain to be discovered in euphyllophytes. We find that the copy numbers of EPRVs are generally high, suggesting that EPRVs might define a unique group of repetitive elements and represent important components of euphyllophyte genomes. Evolutionary analyses suggest an ancient origin of Caulimoviridae and at least three independent origins of Caulimoviridae in angiosperms. Our findings reveal the remarkable diversity of Caulimoviridae and have important implications for understanding the origin and macroevolution of plant pararetroviruses.
IMPORTANCE Few viruses have been documented in plants outside angiosperms. Viruses can occasionally integrate into host genomes, forming endogenous viral elements (EVEs). Endogenous plant pararetroviruses (EPRVs) are widespread in angiosperms. In this study, we performed comprehensive comparative and phylogenetic analyses of EPRVs and found that EPRVs are present in the genomes of gymnosperms and ferns. We identified numerous EPRVs in gymnosperm and fern genomes, revealing an unprecedented depth in the diversity of plant pararetroviruses. Plant pararetroviruses mainly underwent cross-species transmission, and angiosperm pararetroviruses arose at least three times. Our study provides novel insights into the diversity and macroevolution of plant pararetroviruses.
KEYWORDS: transposable elements, phylogenetics, paleovirology, Caulimoviridae
INTRODUCTION
Endogenous viral elements (EVEs), viral sequences integrated into their hosts' genomes, document past virus (paleovirus) infections and provide “molecular fossils” for studying the deep history of viruses (1). EVEs lay the foundation for an emerging field, paleovirology (1, 2). The best-characterized EVEs are endogenous retroviruses (ERVs) (3). The replication of retroviruses requires integration into their hosts' genomes. On occasion, retroviruses infect germ lines of their hosts, and the integrated retroviruses, namely, ERVs, become vertically inherited. ERVs are widespread and highly abundant in the genomes of vertebrates (3); for example, ERVs make up 5% to 8% of the human genome (4). Recently, endogenous nonretroviral elements have been increasingly identified by comparative genomic analyses, which provide many novel insights into the remarkable diversity, deep history, and macroevolution of related viruses (5, 6). EVEs (especially ERVs) were pervasively coopted for the hosts' biology, ranging from placentation to the inhibition of exogenous viral infection to the regulation of innate immunity (7–9).
Like retroviruses, two families of viruses with double-stranded DNA genomes replicate through RNA intermediates known as pararetroviruses or DNA reverse-transcribing viruses (10). Unlike retroviruses, these pararetroviruses lack integrase, and thus, integration into host genomes is not essential for their replication. Pararetroviruses infect vertebrates (Hepadnaviridae) and plants (Caulimoviridae). Evolutionary analyses suggest that Hepadnaviridae and Caulimoviridae originated independently from retrotransposons with long terminal repeats (LTRs) (11). Endogenous hepadnaviruses have been increasingly identified in the genomes of birds and reptiles (12–14). The copy number of endogenous hepadnaviruses in host genomes is very low (around 10 copies) (12–14). The identification of endogenous hepadnaviruses reveals the prevalent nature and deep history (more than 207 million years) of Hepadnaviridae in vertebrates (12–15).
The Caulimoviridae family, pararetroviruses infecting plants, is classified into eight genera, namely, Caulimovirus, Soymovirus, Tungrovirus, Badnavirus, Solendovirus, Cavemovirus, Rosadnavirus, and Petuvirus (16). The genome size of Caulimoviridae is usually between 6,000 and 8,000 bp, encoding one to eight open reading frames (ORFs). The proteins (or domains) common to Caulimoviridae include movement protein (MP), coat protein (CP), aspartic protease (AP or PR), reverse transcriptase (RT), and RNase H1 (RH) (16). While the replication of Caulimoviridae does not require integration into host genomes, endogenous plant pararetroviruses (EPRVs) were identified in many angiosperms in the pregenomic era (17), for example, banana (18) and tobacco (19). Genome-scale data provide important resources to explore the distribution and diversity of EPRVs within plant genomes, which would improve our understanding of the macroevolution of Caulimoviridae and the relationship between viruses and their hosts (20). By mining a variety of plant genomes, Geering et al. (21) identified a novel lineage of EPRVs in flowering plants (angiosperms), sometimes with a high copy number, which was designated “Florendovirus” and was thought to constitute a new genus within the Caulimoviridae. By analyzing the movement protein of plant viruses, Mushegian and Elena (22) provide some clues for the presence of EPRVs within the genomes of ferns and gymnosperms. However, EPRVs have not been systematically analyzed in the genomes of plants outside angiosperms, and much remains unknown about the diversity and macroevolutionary mode of Caulimoviridae.
In this study, we use a large-scale phylogenomic approach to investigate whether EPRVs are widespread in the genomes of plants outside angiosperms. By mining 10 gymnosperm and 6 fern genomes, we identified EPRVs in the genomes of nearly all these gymnosperms and ferns. Phylogenetic analyses using the newly identified EPRVs together with other angiosperm viruses reveal an unappreciated diversity of Caulimoviridae and show that the known angiosperm viruses constitute only a minor part of the Caulimoviridae diversity. The newly identified EPRVs in gymnosperms and ferns provide many important and novel insights into the diversity, distribution, and macroevolution of Caulimoviridae.
(This article was submitted to an online preprint archive [23].)
RESULTS
Identification of EPRVs in gymnosperms and ferns.
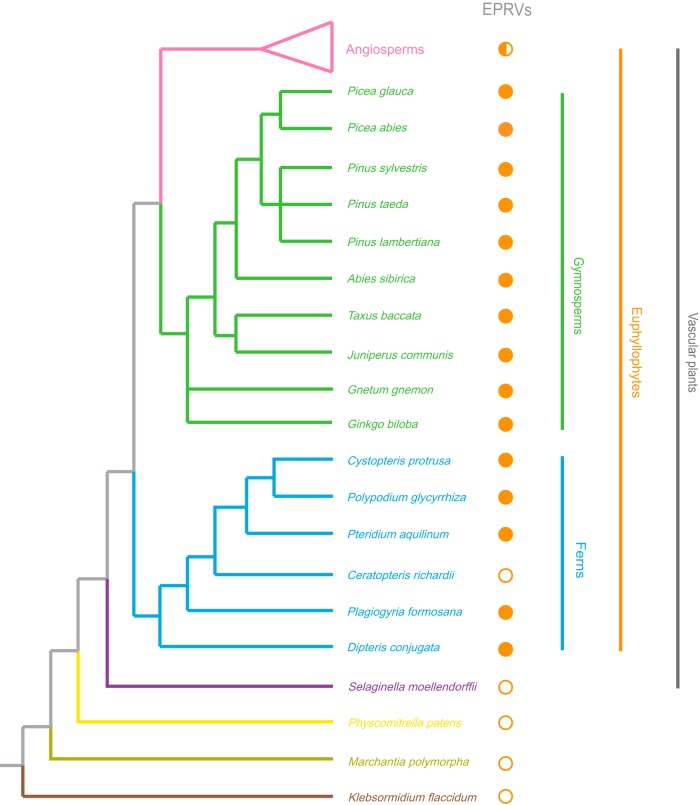
We used a combined similarity search and phylogenetic analysis approach to screen the genomes of 10 gymnosperms, 6 ferns, and 4 other earlier-branching plant species (Selaginella moellendorffii, Physcomitrella patens, Marchantia polymorpha, and Klebsormidium flaccidum) for the presence of EPRVs (Fig. 1 and Table 1). Briefly, a similarity search with the protein sequences of representative members of the Caulimoviridae was performed against these plant genomes (Fig. 1 and Table 1). Given that RT and RH of Caulimoviridae share significant similarity with retrotransposons and other reverse-transcribing viruses, EPRVs were further identified and confirmed by phylogenetic analyses (see Materials and Methods). We found that EPRVs are present in the genomes of nearly all the gymnosperms and ferns investigated in this study (Fig. 1), suggesting that EPRVs are prevalent and widespread in gymnosperms and ferns. EPRVs were not identified in the genome of the fern Ceratopteris richardii, which does not necessarily indicate the absence of EPRVs but is more likely due to the low-density coverage (1.082×) of its genome sequencing (only 3% of its genome is covered by the genome assembly). To test whether ERPVs are present in the genomes of Ceratopteris species, we obtained a sample of Ceratopteris thalictroides, a species closely related to C. richardii, and succeeded in amplifying EPRV insertions within the genome of C. thalictroides via PCR with degenerated primers designed for conserved regions of RT and RH. No EPRV was detected in the genomes of the lycophyte S. moellendorffii, the moss P. patens, the liverwort M. polymorpha, and the charophyte K. flaccidum (Fig. 1). Together with data from previous reports of EPRVs in angiosperms (17–22), we conclude that EPRVs are widespread in the genomes of euphyllophytes (ferns and seed plants).
FIG 1.
Distribution of EPRVs within plant genomes. The phylogenetic relationships of plant species are based on data reported previously (24, 39–41, 56–58). Different plant divisions are labeled in different colors. The presence and absence of EPRVs are marked with solid and open circles next to the related species, respectively. The half-filled circle indicates that EPRVs have been identified in some but not all the angiosperms.
TABLE 1.
Copy numbers of EPRVs identified in plant genomes
| Species | Division | NCBI accession no. or website | Assembly size (Gbp) | Genome size (Gbp) | No. of EPRVs identified |
|---|---|---|---|---|---|
| Pinus lambertiana | Pinophyta | GCA_001447015.2 | 27.603 | 31.0 | 4,556 |
| Pinus taeda | Pinophyta | GCA_000404065.3 | 20.148 | 21.9 | 6,194 |
| Picea glauca | Pinophyta | GCA_000411955.5 | 20.8 | 20 | 2,520 |
| Juniperus communis | Pinophyta | http://congenie.org/ | 1.861 | 11.6 | 18 |
| Taxus baccata | Pinophyta | http://congenie.org/ | 3.001 | 10.8 | 112 |
| Abies sibirica | Pinophyta | http://congenie.org/ | 2.183 | 15.5 | 130 |
| Pinus sylvestris | Pinophyta | http://congenie.org/ | 6.795 | 22.5 | 300 |
| Picea abies | Pinophyta | http://congenie.org/ | 12.019 | 19.6 | 1,238 |
| Ginkgo biloba | Ginkgophyta | http://gigadb.org/dataset/100209 | 10.61 | 10 | 1,156 |
| Gnetum gnemon | Gnetophyta | http://congenie.org/ | 1.837 | 3.3 | 1,350 |
| Dipteris conjugata | Pteridophyta | http://digitalcommons.usu.edu/fern_genome/2/ | 0.232 | 2.45 | 144 |
| Plagiogyria formosana | Pteridophyta | http://digitalcommons.usu.edu/fern_genome/2/ | 0.046 | 14.81 | 64 |
| Ceratopteris richardii | Pteridophyta | http://digitalcommons.usu.edu/fern_genome/2/ | 0.350 | 11.25 | 0 |
| Pteridium aquilinum | Pteridophyta | http://digitalcommons.usu.edu/fern_genome/2/ | 0.620 | 15.65 | 48 |
| Polypodium glycyrrhiza | Pteridophyta | http://digitalcommons.usu.edu/fern_genome/2/ | 0.053 | 10.02 | 21 |
| Cystopteris protrusa | Pteridophyta | http://digitalcommons.usu.edu/fern_genome/2/ | 0.043 | 4.23 | 12 |
| Physcomitrella patens | Bryophyta | GCA_000002425.1 | 0.478 | 0 | |
| Selaginella moellendorffii | Lycopodiophyta | GCA_000143415.2 | 0.213 | 0 | |
| Marchantia polymorpha | Marchantiophyta | GCA_001641455.1 | 0.206 | 0 | |
| Klebsormidium flaccidum | Charophyta | GCA_000708835.1 | 0.104 | 0 |
The copy numbers of EPRVs within the genomes of gymnosperms and ferns appear to be generally high (Table 1). It should be noted that the copy numbers of the EPRVs identified might not represent the actual copy numbers of EPRVs, because (i) the genomes of some gymnosperms and ferns are of low coverage, (ii) assembly is challenging for genomes that are highly abundant in repetitive sequences, and (iii) EPRVs might not be evenly distributed. However, some genomes seem to be of high quality; for example, 98.63% of the total length of the contigs assembled from a large pool of approximately 4,600 fosmid clones was covered by the genome assembly of loblolly pine (Pinus taeda) (24). Nevertheless, our results suggest that EPRVs might represent important components of plant genomes.
Diversity and classification of Caulimoviridae.
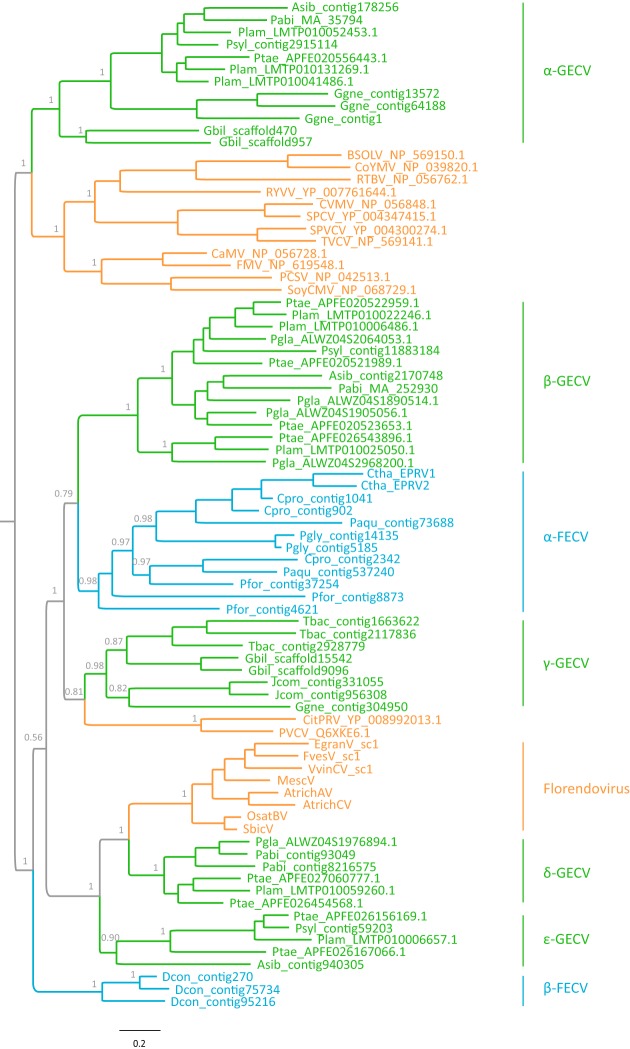
To explore the relationship between the newly identified gymnosperm and fern EPRVs and the known angiosperm Caulimoviridae, we inferred a phylogeny of representative exogenous and endogenous viruses of Caulimoviridae using the highly conserved RT-RH proteins. The root of the Caulimoviridae phylogeny was identified by using a state-of-the-art rooting approach, the minimal ancestor deviation (MAD) method (25). Our phylogenetic analysis reveals an extraordinarily large diversity of the Caulimoviridae family, which has never been appreciated previously (Fig. 2). The eight known viral genera and florendoviruses fall well within the diversity of EPRVs of gymnosperms and ferns. It follows that the previously known angiosperm viruses constitute only a minor part of the diversity of Caulimoviridae.
FIG 2.
Phylogenetic relationship of representative exogenous and endogenous Caulimoviridae. The phylogenetic tree was inferred based on the RT-RH protein sequences using a Bayesian method. The tree was rooted by using the MAD approach. Bayesian posterior probabilities are shown on the selected nodes. Caulimoviridae of angiosperms, gymnosperms, and ferns are highlighted in orange, green, and blue, respectively. Abbreviations: CVMV, cassava vein mosaic virus; SPCV, sweet potato caulimo-like virus; SPVCV, sweet potato vein clearing virus; TVCV, tobacco vein clearing virus; BSOLV, banana streak OL virus; CoYMV, Commelina yellow mottle virus; RTBV, rice tungro bacilliform virus; CaMV, cauliflower mosaic virus; FMV, figwort mosaic virus; RYVV, rose yellow vein virus; PCSV, peanut chlorotic streak virus; SoyCMV, soybean chlorotic mottle virus; CitPRV, citrus endogenous pararetrovirus; PVCV, petunia vein clearing virus; VvinCV_sc1, Vitis vinifera C virus sequence cluster 1; MescV, Manihot esculenta virus; FvesV_sc1, Fragaria vesca virus sequence cluster 1; AtrichAV, Amborella trichopoda A virus; AtrichCV, Amborella trichopoda C virus; EgranV_sc1, Eucalyptus grandis virus sequence cluster 1; OsatBV, Oryza sativa B virus; SbicV, Sorghum bicolor virus; Pabi, Picea abies; Pgla, Picea glauca; Ptae, Pinus taeda; Plam, Pinus lambertiana; Psyl, Pinus sylvestris; Asib, Abies sibirica; Gbil, Ginkgo biloba; Jcom, Juniperus communis; Ggne, Gnetum gnemon; Tbac, Taxus baccata; Pfor, Plagiogyria formosana; Pgly, Polypodium glycyrrhiza; Ctha, Ceratopteris thalictroides; Cpro, Cystopteris protrusa; Paqu, Pteridium aquilinum; GECV, gymnosperm endogenous caulimovirus-like virus; FECV, fern endogenous caulimovirus-like virus.
Our phylogenetic analysis identified at least seven monophyletic groups of EPRVs with high levels of support (Bayesian posterior probability of >0.90) in gymnosperms and ferns. These clades were designated α-type gymnosperm endogenous caulimovirus-like virus (α-GECV), β-GECV, γ-GECV, δ-GECV, ε-GECV, α-type fern endogenous caulimovirus-like virus (α-FECV), and β-FECV. The host of each clade is restricted to one plant division (except for α-GECV and γ-GECV) (Fig. 2). The divergence within one of these clades is comparable to and even greater than those of some known Caulimoviridae genera.
Macroevolutionary mode of Caulimoviridae.
To study the relative importance of cospeciation and host switching in the macroevolution of Caulimoviridae, we performed a global assessment of the correspondence between Caulimoviridae and host phylogenetic trees using an event-based approach. We did not detect significant congruence between host and virus trees (P values of >0.05) (Table 2), suggesting that cospeciation might not play a predominant role in the diversification of Caulimoviridae (see Fig. S3 at https://figshare.com/articles/Supplemental_figures_pdf/5895757).
TABLE 2.
Numbers of events experienced by virus lineages
| Event costsa | Total cost | No. of eventsb |
P valuec | ||||
|---|---|---|---|---|---|---|---|
| Cospeciation | Duplication | Duplication and host switching | Loss | Failure to diverge | |||
| −1, 0, 0, 0, 0 | −10 | 10–10 | 1–1 | 8–8 | 6–8 | 0–0 | >0.05 |
| 0, 1, 1, 2, 0 | 13 | 6 | 0 | 13 | 0 | 0 | >0.05 |
| 0, 1, 2, 1, 1 | 23 | 6–8 | 0–4 | 9–11 | 1–1 | 0–0 | >0.05 |
Event costs are for cospeciation, duplication, duplication and host switching, loss, and failure to diverge, respectively.
The numbers of events are expressed as ranges that result in the same cost.
Random-parasite-tree method with a sample size of 500.
Our phylogenetic analysis also shows that the angiosperm viruses form three independent monophyletic groups: two consist of the eight known genera of exogenous viruses, and one consists of florendoviruses (Fig. 2). The three angiosperm virus groups are only distantly related to each other. The phylogenetic relationship among euphyllophyte viruses indicates that the angiosperm viruses originated multiple times, probably through cross-division transmissions from gymnosperms (Fig. 2). Ancestral-state reconstruction reveals that the Caulimoviridae family originated in gymnosperms (see Fig. S1 at https://figshare.com/articles/Supplemental_figures_pdf/5895757); however, this conclusion should be taken with caution, given that all the fern genomes used in this study are of low coverage and there might be more novel EPRVs in ferns.
Genome structure evolution of Caulimoviridae.
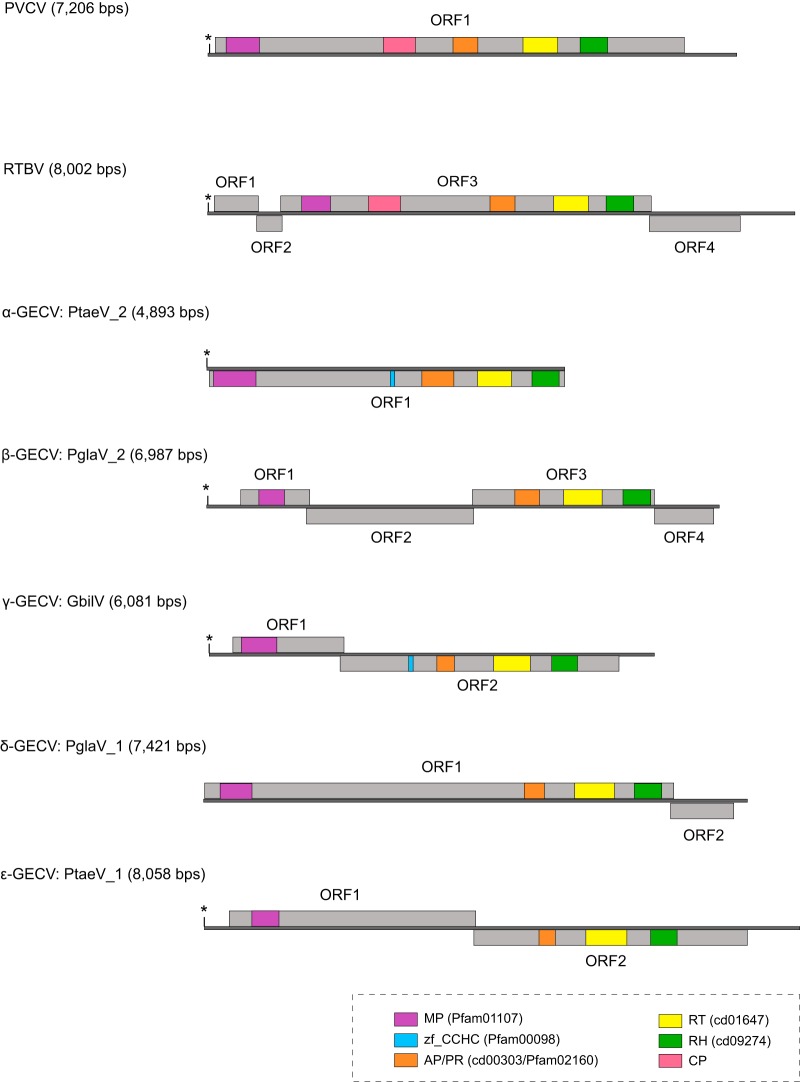
To explore the genome structure evolution within the Caulimoviridae family, we reconstructed consensus genome sequences of EPRVs (see Data set S1 in the supplemental material). Given that the fern genomes are of low coverage, we reconstructed only the genomes of gymnosperm EPRVs, and one representative of each of the five gymnosperm EPRV clades was inferred. These gymnosperm EPRV genomes vary wildly in size and ORF organization (Fig. 3). Conserved Domain (CD) searches show that protein domains common to all the EPRVs include MP, AP, RT, and RH, suggesting that the gymnosperm EPRVs exhibit a protein architecture similar to that of angiosperm Caulimoviridae (Fig. 3). No homologs of CP were identified in the consensus genome sequences, possibly due to the rapid nature of its evolution. However, we identified the zinc finger CCHC motif, a hallmark of CP, in Pinus taeda virus 2 (PtaeV_2) and Ginkgo biloba virus (GbilV) (Fig. 3). CD searches did not find any integrase-like domain, a pattern similar to that of angiosperm Caulimoviridae, indicating that integration might not be necessary for the replication of gymnosperm EPRVs either.
FIG 3.
Genome structures of representative gymnosperm EPRVs and exogenous Caulimoviridae. tRNAMet, which represents the beginning of viral replication, is indicated by an asterisk. The gray lines represent the genomes, and the rectangles represent the putative open reading frames (ORFs). The conserved protein domains are labeled in different colors. Abbreviations: RTBV, rice tungro bacilliform virus; PVCV, petunia vein clearing virus; PglaV_1, Picea glauca virus 1; PglaV_2, Picea glauca virus 2; PtaeV_1, Pinus taeda virus 1; PtaeV_2, Pinus taeda virus 2; GbilV, Ginkgo biloba virus; MP, movement protein; PR/AP, protease/pepsin-like aspartate protease; RT, reverse transcriptase; RH, RNase H1; zf_CCHC, zinc finger CCHC motif.
Age estimate of EPRV integrations and bursts.
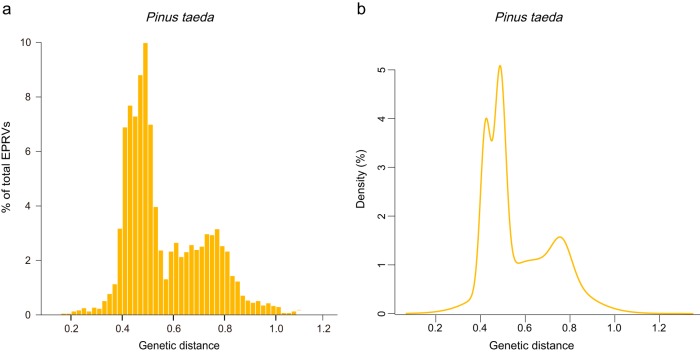
Because the genome of loblolly pine (P. taeda) is of relatively high quality (24), it was used to infer the evolutionary dynamics of EPRVs within the host genome. Mixture model analysis of the genetic divergence between EPRV copies and their consensus nucleotide sequence shows that there are four peaks in P. taeda (Bayesian information criterion [BIC] value of 6,415.5), with mean genetic distances (standard deviations) of 0.422 (0.022), 0.488 (0.024), 0.633 (0.168), and 0.763 (0.044), respectively (Fig. 4). This result suggests that there are at least four independent EPRV integration events occurring along the lineages leading to P. taeda. Based on the mixture analyses and phylogenetic analyses (see Fig. S2 at https://figshare.com/articles/Supplemental_figures_pdf/5895757), the ERPVs within the genome of P. taeda were classified into four families.
FIG 4.
Proliferation dynamics of EPRVs within the P. taeda genome. (a) Distribution of the genetic distances between EPRV copies and their consensus sequences (yellow bars). (b) Fitted Gaussian mixture models.
We calculated the median pairwise genetic distance within each family (0.525, 0.537, 0.475, and 0.666), which corresponds to the age of burst for each EPRV family (119.3 million years ago [MYA] to 183.6 MYA, 122 MYA to 187.8 MYA, 108 MYA to 166.1 MYA, and 151.4 MYA to 232.9 MYA) (26, 27). We found that all of the EPRV families investigated here experienced proliferation peaks tens or hundreds of million years ago. Consistently, the EPRV copies contain many frameshift mutations and premature stop codons (see Fig. S4 at https://figshare.com/articles/Supplemental_figures_pdf/5895757). However, these analyses come with two caveats: (i) it is uncertain whether the EPRV proliferation activity within the host genome follows a Gaussian distribution, and (ii) the actual evolutionary rate of EPRVs remains unclear, and the proliferation of EPRVs might undergo an error-prone reverse transcription process. Thus, the host rate range that we used can be used only as the lower bounds.
We identified an orthologous integration event of EPRVs in the genomes of Picea glauca and Picea abies, which diverged from each other ∼16.9 million years ago (26) (see Fig. S5 at https://figshare.com/articles/Supplemental_figures_pdf/5895757). Moreover, we identified an orthologous integration event of EPRVs in the genomes of P. taeda and Pinus lambertiana, which diverged from each other ∼75 million years ago (26) (see Fig. S5 at https://figshare.com/articles/Supplemental_figures_pdf/5895757). These results suggest that pararetroviruses invaded the common ancestor of P. glauca and P. abies and the common ancestor of P. taeda and P. lambertiana at least 16.9 million years ago and 75 million years ago, respectively. Taken together, our results suggest that EPRVs evolved within their host genomes for hundreds of millions of years and reveal an ancient origin of Caulimoviridae.
DISCUSSION
In this study, we report the identification of EPRVs within the genomes of gymnosperms and ferns. Together with data from previous reports of exogenous and endogenous Caulimoviridae in angiosperms, our results demonstrate that all the major lineages of euphyllophytes (ferns and seed plants) are/were infected by the Caulimoviridae family. Few viruses in plant species, outside angiosperms, have been documented (28). The identification of EPRVs in gymnosperms and ferns makes Caulimoviridae the only known virus family that infects all major lineages of euphyllophytes.
Our findings show that the newly identified EPRVs exhibit an unprecedented diversity, and the known angiosperm virus diversity accounts for only a minority of the Caulimoviridae diversity. The current Caulimoviridae classification system (16) cannot readily account for the diversity of EPRVs in gymnosperms and ferns. Indeed, the divergence within one clade of gymnosperm or fern EPRVs is comparable to the divergence of one exogenous virus genus or florendoviruses. Therefore, an updated classification incorporating gymnosperm and fern EPRVs should be developed. Most of the EPRV clades lack exogenous counterparts, either because the ancient virus lineages completely died out or because many exogenous viruses remain to be discovered. Similar patterns are also observed for retroviruses; for example, exogenous epsilonretroviruses infect only fish, but endogenous epsilonretroviruses were found in the genomes of amphibians and primates (29).
Two possible macroevolutionary modes of Caulimoviridae could be conceived: (i) a cospeciation model, where the viruses have coevolved with their euphyllophyte hosts for >400 million years and undergone sporadic cross-species transmission (Fig. 5a), and (ii) a cross-species transmission model, where frequent cross-species transmissions predominated in the evolution of Caulimoviridae (Fig. 5b). In this study, we failed to find a cospeciation signal between Caulimoviridae and their hosts, suggesting that cospeciation might not be predominant in the macroevolution of Caulimoviridae (Table 2; see also Fig. S3 at https://figshare.com/articles/Supplemental_figures_pdf/5895757). However, taxon sampling and event costs might have certain impacts on cospeciation analyses. On the other hand, the pattern of cospeciation is more sensitive to taxon sampling. In the literature, there are many cases in which some pathogens were first reported to codiverge with their hosts but were subsequently demonstrated not to codiverge with their hosts with increasing taxon sampling, for example, simian immunodeficiency viruses and their primate hosts (30). As for event costs, we used three different settings and detected no significant host-virus congruence. Nevertheless, our results suggest that Caulimoviridae might not mainly codiverge with their plant hosts. Indeed, it appears that the angiosperm viruses originated multiple times via independent cross-division transmission events (gymnosperms to angiosperms). For plant viruses, cross-division transmission events were rarely documented, partially because much remains unknown about the virosphere in plants outside angiosperms.
FIG 5.
Models of Caulimoviridae macroevolution. The evolution of plant hosts and viruses are indicated by blue lines and yellow dashed lines, respectively. (a) Cospeciation model where the viruses have coevolved with their euphyllophyte hosts and undergone sporadic cross-species transmission. (b) Cross-species transmission model where frequent cross-species transmission predominated in the evolution of Caulimoviridae.
We did not find any EPRV in earlier-branching plants (lycophytes and nonvascular plants). The absence of EPRVs in earlier-branching plants might be due to either (i) no viral infection, (ii) no viral integration occurring, or (iii) no fixation of endogenous viruses occurring when integration occurred. The paleoviruses provide molecular fossils for estimating the age of related viruses. Previously, the integration of banana streak virus into the Musa balbisiana genome was estimated to have occurred 0.63 million years ago (31). The endogenization of florendoviruses in Oryza species was estimated to take place at least 1.8 million years ago (21). Our analyses of EPRV activities indicate that they might have been activated within the host genomes for tens of millions or even hundreds of millions of years. A previous analysis of cauliflower mosaic virus (CMV), the type member of the genus Caulimovirus, using a tip-dating method suggests that CMV has a very recent origin (around several hundred years) (32). The discrepancy between short-term evolution and long-term evolution might be explained by a lack of temporal structure in serially sampled virus data sets (33) or the death of old virus lineages (34). Nevertheless, our findings pinpoint a possible ancient origin of Caulimoviridae.
Unlike EVEs of nonretroviral sources, the copy numbers of EPRVs are generally high, suggesting that EPRVs contribute significantly to the complexity of host genomes. Caulimoviridae are closely related to LTR retrotransposons (11). On the other hand, EPRVs lack LTRs, which makes it inappropriate for them to be classified as an LTR retrotransposon. It seems to be more appropriate to define EPRVs as a unique group of transposable elements (19).
Our findings suggest that Caulimoviridae integrated into and were amplified within host genomes multiple times. However, the integration and amplification mechanisms of EPRVs remain unclear, as the Caulimoviridae genomes lack integrase-like proteins and integration is not essential for their replication. Several potential mechanisms might be involved. (i) EPRVs encode a “cryptic” integrase without significant similarity to known proteins that function in integration. No integrase domain was found in the Petunia vein clearing virus (PVCV) genome, but one of its proteins encodes two distinctive motifs [HHCC and DD(35)E] that are shared by the integrase domains of retroviruses and LTR retrotransposons (35). However, it remains unknown whether this protein performs a function similar to that of integrase. (ii) Microhomology-mediated recombination between EPRV sequences and host sequences during the host gap repair process could result in the integration of viral sequences into the host genomes (17). This mechanism requires the free ends of open circular viral sequences produced during virus replication (19, 36). We did not find any conserved motif around EPRV insertion sites. (iii) Like short interspersed elements (SINEs), EPRVs might integrate and amplify themselves within the host genomes via hijacking the integrases of other retrotransposons (37, 38).
MATERIALS AND METHODS
Identification of EPRVs in plant genomes.
The genome sequences of 20 plant species were used to screen for the presence of EPRVs, including 10 gymnosperms (P. taeda, Pinus lambertiana, Pinus sylvestris, Picea abies, Picea glauca, Ginkgo biloba, Gnetum gnemon, Juniperus communis, Taxus baccata, and Abies sibirica), 6 ferns (C. richardii, Dipteris conjugata, Plagiogyria formosana, Pteridium aquilinum, Polypodium glycyrrhiza, and Cystopteris protrusa), 1 moss (P. patens), 1 liverwort (M. polymorpha), 1 lycophyte (S. moellendorffii), and 1 charophyte (K. flaccidum) (24, 39–41) (Table 1). To identify putative EPRVs within these genomes, we employed a two-step phylogenomic approach. First, the tBLASTn algorithm was employed for searches against the plant genomes using the RT-RH domain sequences of Rice tungro bacilliform virus (RTBV) (GenBank accession no. NP_056762) (amino acids [aa] 1175 to 1675) and PVCV (GenBank accession no. Q6XKE6) (aa 1351 to 1849) as queries, with an E cutoff value of 10−10. Next, all the significant hits obtained were aligned with RT-RH sequences of representative LTR retrotransposons, retroviruses, Hepadnaviridae, and Caulimoviridae (42) by using MAFFT with default parameters (43). The representative LTR retrotransposon sequences cover major diverse populations of currently known LTR retroelements of eukaryotes (42). Putative EPRVs, which form a monophyletic group with other Caulimoviridae with high support values, were identified based on phylogenetic analyses (see Fig. S6 at https://figshare.com/articles/Supplemental_figures_pdf/5895757 for an example). EPRVs were confirmed by further rounds of phylogenetic analyses with putative EPRVs and representative LTR retrotransposons, retroviruses, Hepadnaviridae, and Caulimoviridae. Phylogenetic analyses were performed by using an approximate maximum likelihood method implemented in FastTree 2.1.9 with default parameters (44). The copy numbers of EPRVs within each species were then counted. If the length between hits was <5,000 bp and the hits were in the same order as the query, the hits were treated as a single copy.
PCR amplification and EPRV cloning.
A sample of C. thalictroides was purchased from a local market in Guangxi Province, China. Genomic DNA was extracted by using a modified cetyltrimethylammonium bromide (CTAB) method (45). Amplification of a conserved RT-RH fragment was performed with degenerated primers FECVf (5′-TGGTAATCAATTATAAACCTCTTAAC-3′) and FECVr (5′-GGAACAATGAAGGCTGTTTT-3′). PCR was performed with 25-μl (final volume) reaction mixtures containing 0.5 μl EasyTaq (Transgen, Beijing), 2.5 μl 10× buffer, 2 μl deoxynucleoside triphosphate (dNTP) (2.5 mM), 2 μl of each primer (10 μM), 2 μl of template DNA, and 14 μl of water. The PCRs were cycled under the following conditions: an initial denaturation step at 94°C for 3 min; 32 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 40 s; and a final elongation step at 72°C for 5 min. The PCR products were purified by using DNA fragment purification kit version 4.0 (TaKaRa, Japan). Purified PCR products were cloned into the pMD19-T vector by using the pMD19-T vector cloning kit (TaKaRa, Japan). The cloned products were sequenced by TsingKe Biotech, Beijing, China.
Phylogenetic analysis.
To further analyze the relationship among members of the Caulimoviridae, phylogenetic analysis was performed by using the RT-RH protein sequences from representative EPRV sequences of each gymnosperm and fern species, exogenous viruses, and florendoviruses. These protein sequences were aligned by using the MAFFT algorithm with an accurate method with the L-INS-i strategy (43). Phylogenetic analysis was performed by using a Bayesian method implemented in MrBayes 3.2.6 (46). Because retroelements might undergo selective pressure different from that of other proteins represented in standard models, we used an empirical model of amino acid substitution, namely, RtRev, which is specific for retroviral genes and other elements containing RT (47). A total of 912,000 generations in four chains were run, with sampling of posterior trees every 100 generations. The first 25% of the posterior trees were discarded for further analysis. The root of the Caulimoviridae phylogeny was identified by using the MAD method, which is superior to other rooting approaches, such as outgroup, midpoint, and relaxed-molecular-clock rooting methods (25).
Reconstruction of consensus genome sequences.
Relatively complete EPRV sequences with MP-AP-RT-RH domains with extended flanking regions (∼5,000 bp for each end) were extracted. These sequences were then used as queries to search sequences with high similarity within their own host genome by using the BLASTn algorithm with an E cutoff value of 10−25. The significant hits were aligned by using MAFFT (43), and consensus sequences were generated by using Geneious 10 (48) and manually edited. ORFs with nucleotide sequences of 500 bp or longer were found by using Geneious 10 (48). Protein domains within these reconstructed genomes were detected by using a CD search (49).
Analysis of EPRV activity within host genomes.
Because the genome of P. taeda was of relatively high quality, we used its genome to infer the evolutionary dynamics of EPRVs within the host genome. AP-RT-RH nucleotide sequences with lengths of >500 bp for all the EPRVs within the P. taeda genome were extracted independently. These sequences were aligned by using MAFFT, and consensus sequences were inferred by using Geneious 10 (48). The genetic distance between the consensus sequences and EPRVs was calculated based on the Kimura two-parameter model. To identify significant peaks in the genetic distance data sets, Gaussian mixture models were fitted by using the R package mclust. The number of components (each component is modeled by the Gaussian distribution) was estimated by fitting models. The BIC was used as the model selection criterion.
The phylogenetic tree of the AP-RT-RH nucleotide sequences extracted as described above was reconstructed by using FastTree 2.1.9 with a GTR+CAT model (44). The different EPRV families within the phylogenetic tree were allocated based on the results of mixture model analysis. The nucleotide sequences of each EPRV family were extracted and aligned by using MAFFT (43). Pairwise genetic distances were calculated based on the Kimura two-parameter model. The age of the burst (T) for each EPRV family was estimated by the formula T = D/2μ, where D represents the median pairwise distance and μ represents the evolutionary rate of the host (∼1.43 × 10−9 to 2.2 × 10−9 substitutions per site per year) (39).
Cospeciation analysis.
We explore the host-virus cospeciation signal at the level of class, because the complex evolutionary history of EPRVs after integration might complicate cospeciation analyses. The relationships between virus and host phylogenetic trees were assessed by using an event-based method implemented in Jane 4 (50). Briefly, five events (cospeciation, duplication, duplication and host switch, loss, and failure to diverge) were assigned a cost. The numbers of each event were estimated by finding the solution with the minimum total cost. The event cost schemes (cospeciation-duplication-duplication and host switch-loss-failure to diverge) were set as follows: −1-0-0-0-0 (51, 52), 0-1-1-2-0 (known as Charleston's cost scheme) (50, 53), and 0-1-2-1-1 (Jane's default setting). Host-virus phylogeny congruence was assessed by statistical tests with the random-parasite-tree method, with a sample size of 500, which generates samples of random parasite trees and solves these samples to obtain their best costs (50). These costs are then compared to the cost of the original instance to quantify the statistical significance of cospeciation evidence (50). If the original cost is significantly different from the large proportion of costs of these randomly generated trees, this indicates a global fit between host and parasite trees (50, 54).
Reconstruction of ancestral states.
To detect the macroevolutionary pattern among members of the Caulimoviridae, we performed ancestral-state reconstruction with Mesquite 3.10 (55). We assigned the 82 virus taxa (Fig. 2) using their hosts (gymnosperm, angiosperm, and fern) as characters. The parsimony model was used to trace character evolution over the posterior trees sampled in the above-mentioned Bayesian analysis.
Accession number(s).
The sequences reported here have been deposited in GenBank (accession no. MF661773 to MF661774).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (31701091), the Natural Science Foundation of Jiangsu Province (BK20161016), the Program for Jiangsu Excellent Scientific and Technological Innovation Team (17CXTD00014), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02043-17.
REFERENCES
- 1.Katzourakis A, Gifford RJ. 2010. Endogenous viral elements in animal genomes. PLoS Genet 6:e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Emerman M, Malik HS. 2010. Paleovirology—modern consequences of ancient viruses. PLoS Biol 8:e1000301. doi: 10.1371/journal.pbio.1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayward A, Grabherr M, Jern P. 2013. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc Natl Acad Sci U S A 110:20146–20151. doi: 10.1073/pnas.1315419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A 101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feschotte C, Gilbert C. 2012. Endogenous viruses: insight into viral evolution and impact on host biology. Nat Rev Genet 13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 6.Aiewsakun P, Katzourakis A. 2015. Endogenous viruses: connecting recent and ancient viral evolution. Virology 479–480:26–37. doi: 10.1016/j.virol.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Aswad A, Katzourakis A. 2012. Paleovirology and virally derived immunity. Trends Ecol Evol 27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esnault C, Cornelis G, Heidmann O, Heidmann T. 2013. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet 9:e1003400. doi: 10.1371/journal.pgen.1003400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Temin HM. 1985. Reverse transcription in the eukaryotic genome: retroviruses, pararetroviruses, retrotransposons, and retrotranscripts. Mol Biol Evol 2:455–468. [DOI] [PubMed] [Google Scholar]
- 11.Xiong Y, Eickbush TH. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J 9:3353–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert C, Feschotte C. 2010. Genomic fossils calibrate the long-term evolution of hepadnaviruses. PLoS Biol 8:e1000495. doi: 10.1371/journal.pbio.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert C, Meik JM, Dashevsky D, Card DC, Castoe TA, Schaack S. 2014. Endogenous hepadnaviruses, bornaviruses and circoviruses in snakes. Proc Biol Sci 281:20141122. doi: 10.1098/rspb.2014.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suh A, Weber CC, Kehlmaier C, Braun EL, Green RE, Fritz U, Ray DA, Ellegren H. 2014. Early mesozoic coexistence of amniotes and hepadnaviridae. PLoS Genet 10:e1004559. doi: 10.1371/journal.pgen.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dill JA, Camus AC, Leary JH, Di Giallonardo F, Holmes EC, Ng TF. 2016. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J Virol 90:7920–7933. doi: 10.1128/JVI.00832-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2012. Virus taxonomy. Classification and nomenclature of viruses. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 17.Staginnus C, Richert-Pöggeler KR. 2006. Endogenous pararetroviruses: two-faced travelers in the plant genome. Trends Plant Sci 11:485–491. doi: 10.1016/j.tplants.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Harper G, Osuji JO, Heslop-Harrison JS, Hull R. 1999. Integration of banana streak badnavirus into the Musa genome: molecular and cytogenetic evidence. Virology 255:207–213. doi: 10.1006/viro.1998.9581. [DOI] [PubMed] [Google Scholar]
- 19.Jakowitsch J, Mette MF, van der Winden J, Matzke MA, Matzke AJM. 1999. Integrated pararetroviral sequences define a unique class of dispersed repetitive DNA in plants. Proc Natl Acad Sci U S A 96:13241–13246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Zheng H, Kishima Y. 2017. Genomic fossils reveal adaptation of non-autonomous pararetroviruses driven by concerted evolution of noncoding regulatory sequences. PLoS Pathog 13:e1006413. doi: 10.1371/journal.ppat.1006413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geering AD, Maumus F, Copetti D, Choisne N, Zwickl DJ, Zytnicki M, McTaggart AR, Scalabrin S, Vezzulli S, Wing RA, Quesneville H, Teycheney PY. 2014. Endogenous florendoviruses are major components of plant genomes and hallmarks of virus evolution. Nat Commun 5:5269. doi: 10.1038/ncomms6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mushegian AR, Elena SF. 2015. Evolution of plant virus movement proteins from the 30K superfamily and of their homologs integrated in plant genomes. Virology 476:304–315. doi: 10.1016/j.virol.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 23.Gong Z, Han G-Z. 2017. Hidden diversity and macroevolutionary mode of Caulimoviridae uncovered by euphyllophyte paleoviruses. bioRxiv doi: 10.1101/170415. [DOI] [PMC free article] [PubMed]
- 24.Neale DB, Wegrzyn JL, Stevens KA, Zimin AV, Puiu D, Crepeau MW, Cardeno C, Koriabine M, Holtz-Morris AE, Liechty JD, Martínez-García PJ, Vasquez-Gross HA, Lin BY, Zieve JJ, Dougherty WM, Fuentes-Soriano S, Wu LS, Gilbert D, Marçais G, Roberts M, Holt C, Yandell M, Davis JM, Smith KE, Dean JF, Lorenz WW, Whetten RW, Sederoff R, Wheeler N, McGuire PE, Main D, Loopstra CA, Mockaitis K, deJong PJ, Yorke JA, Salzberg SL, Langley CH. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15:R59. doi: 10.1186/gb-2014-15-3-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tria FDK, Landan G, Dagan T. 2017. Phylogenetic rooting using minimal ancestor deviation. Nat Ecol Evol 1:193. doi: 10.1038/s41559-017-0193. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AB, Beaulieu JM, Rai HS, Crane PR, Donoghue MJ, Mathews S. 2012. Hemisphere-scale differences in conifer evolutionary dynamics. Proc Natl Acad Sci U S A 109:16217–16221. doi: 10.1073/pnas.1213621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ray DA, Feschotte C, Pagan HJ, Smith JD, Pritham EJ, Arensburger P, Atkinson PW, Craig NL. 2008. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res 18:717–728. doi: 10.1101/gr.071886.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hull R. 2014. Plant virology, 5th ed Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 29.Brown K, Emes RD, Tarlinton RE. 2014. Multiple groups of endogenous epsilon-like endogenous retroviruses conserved across primates. J Virol 88:12464–12471. doi: 10.1128/JVI.00966-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wertheim JO, Worobey M. 2007. A challenge to the ancient origin of SIVagm based on African green monkey mitochondrial genomes. PLoS Pathog 3:e95. doi: 10.1371/journal.ppat.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gayral P, Noa-Carrazana JC, Lescot M, Lheureux F, Lockhart BE, Matsumoto T, Piffanelli P, Iskra-Caruana ML. 2008. A single Banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. J Virol 82:6697–6710. doi: 10.1128/JVI.00212-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yasaka R, Nguyen HD, Ho SY, Duchêne S, Korkmaz S, Katis N, Takahashi H, Gibbs AJ, Ohshima K. 2014. The temporal evolution and global spread of Cauliflower mosaic virus, a plant pararetrovirus. PLoS One 9:e85641. doi: 10.1371/journal.pone.0085641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patterson Ross Z, Klunk J, Fornaciari G, Giuffra V, Duchêne S, Duggan AT, Poinar D, Douglas MW, Eden JS, Holmes EC, Poinar HN. 2018. The paradox of HBV evolution as revealed from a 16th century mummy. PLoS Pathog 14:e1006750. doi: 10.1371/journal.ppat.1006750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holmes EC. 2003. Molecular clocks and the puzzle of RNA virus origins. J Virol 77:3893–3897. doi: 10.1128/JVI.77.7.3893-3897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richert-Pöggeler KR, Shepherd RJ. 1997. Petunia vein-clearing virus: a plant pararetrovirus with the core sequences for an integrase function. Virology 236:137–146. doi: 10.1006/viro.1997.8712. [DOI] [PubMed] [Google Scholar]
- 36.Kunii M, Kanda M, Nagano H, Uyeda I, Kishima Y, Sano Y. 2004. Reconstruction of putative DNA virus from endogenous rice tungro bacilliform virus-like sequences in the rice genome: implications for integration and evolution. BMC Genomics 5:80. doi: 10.1186/1471-2164-5-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. 2011. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet 12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. 2015. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr 3:MDNA3-0061-2014. doi: 10.1128/microbiolspec.MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, Vicedomini R, Sahlin K, Sherwood E, Elfstrand M, Gramzow L, Holmberg K, Hällman J, Keech O, Klasson L, Koriabine M, Kucukoglu M, Käller M, Luthman J, Lysholm F, Niittylä T, Olson A, Rilakovic N, Ritland C, Rosselló JA, Sena J, Svensson T, Talavera-López C, Theißen G, Tuominen H, Vanneste K, Wu ZQ, Zhang B, Zerbe P, Arvestad L, Bhalerao R, Bohlmann J, Bousquet J, Garcia Gil R, Hvidsten TR, de Jong P, MacKay J, Morgante M, Ritland K, Sundberg B, Thompson SL, et al. . 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- 40.Wolf PG, Sessa EB, Marchant DB, Li FW, Rothfels CJ, Sigel EM, Gitzendanner MA, Visger CJ, Banks JA, Soltis DE, Soltis PS, Pryer KM, Der JP. 2015. An exploration into fern genome space. Genome Biol Evol 7:2533–2544. doi: 10.1093/gbe/evv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan R, Zhao Y, Zhang H, Fan G, Liu X, Zhou W, Shi C, Wang J, Liu W, Liang X, Fu Y, Ma K, Zhao L, Zhang F, Lu Z, Lee SM, Xu X, Wang J, Yang H, Fu C, Ge S, Chen W. 2016. Draft genome of the living fossil Ginkgo biloba. Gigascience 5:49. doi: 10.1186/s13742-016-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Llorens C, Muñoz-Pomer A, Bernad L, Botella H, Moya A. 2009. Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol Direct 4:41. doi: 10.1186/1745-6150-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Report 15:8–15. doi: 10.1007/BF02772108. [DOI] [Google Scholar]
- 46.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 1:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dimmic MW, Rest JS, Mindell DP, Goldstein RA. 2002. rtREV: an amino acid substitution matrix for inference of retrovirus and reverse transcriptase phylogeny. J Mol Evol 55:65–73. doi: 10.1007/s00239-001-2304-y. [DOI] [PubMed] [Google Scholar]
- 48.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conow C, Fielder D, Ovadia Y, Libeskind-Hadas R. 2010. Jane: a new tool for the cophylogeny reconstruction problem. Algorithms Mol Biol 5:16. doi: 10.1186/1748-7188-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ronquist F. 1997. Phylogenetic approaches in coevolution and biogeography. Zool Scr 26:313–322. doi: 10.1111/j.1463-6409.1997.tb00421.x. [DOI] [Google Scholar]
- 52.Aiewsakun P, Katzourakis A. 2017. Marine origin of retroviruses in the early Palaeozoic Era. Nat Commun 8:13954. doi: 10.1038/ncomms13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Charleston MA. 1998. Jungles: a new solution to the host/parasite phylogeny reconciliation problem. Math Biosci 149:191–223. doi: 10.1016/S0025-5564(97)10012-8. [DOI] [PubMed] [Google Scholar]
- 54.Cruaud A, Rønsted N, Chantarasuwan B, Chou LS, Clement WL, Couloux A, Cousins B, Genson G, Harrison RD, Hanson PE, Hossaert-McKey M, Jabbour-Zahab R, Jousselin E, Kerdelhué C, Kjellberg F, Lopez-Vaamonde C, Peebles J, Peng YQ, Pereira RA, Schramm T, Ubaidillah R, van Noort S, Weiblen GD, Yang DR, Yodpinyanee A, Libeskind-Hadas R, Cook JM, Rasplus JY, Savolainen V. 2012. An extreme case of plant-insect codiversification: figs and fig-pollinating wasps. Syst Biol 61:1029–1047. doi: 10.1093/sysbio/sys068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maddison WP, Maddison DR. 2016. Mesquite: a modular system for evolutionary analysis, version 3.10 http://mesquiteproject.org/.
- 56.Bowe LM, Coat G, dePamphilis CW. 2000. Phylogeny of seed plants based on all three genomic compartments: extant gymnosperms are monophyletic and Gnetales' closest relatives are conifers. Proc Natl Acad Sci U S A 97:4092–4097. doi: 10.1073/pnas.97.8.4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang C, Bowman JL, Meyerowitz EM. 2016. Field guide to plant model systems. Cell 167:325–339. doi: 10.1016/j.cell.2016.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chaw SM, Zharkikh A, Sung HM, Lau TC, Li WH. 1997. Molecular phylogeny of extant gymnosperms and seed plant evolution: analysis of nuclear 18S rRNA sequences. Mol Biol Evol 14:56–68. doi: 10.1093/oxfordjournals.molbev.a025702. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.