ABSTRACT
The initial events after DNA virus infection involve a race between epigenetic silencing of the incoming viral DNA by host cell factors and expression of viral genes. Several host gene products, including the nuclear domain 10 (ND10) components PML (promyelocytic leukemia) and Daxx (death domain-associated protein 6), as well as IFI16 (interferon-inducible protein 16), have been shown to restrict herpes simplex virus 1 (HSV-1) replication. Whether IFI16 and ND10 components work together or separately to restrict HSV-1 replication is not known. To determine the combinatorial effects of IFI16 and ND10 proteins on viral infection, we depleted Daxx or PML in primary human foreskin fibroblasts (HFFs) in the presence or absence of IFI16. Daxx or IFI16 depletion resulted in higher ICP0 mutant viral yields, and the effects were additive. Surprisingly, small interfering RNA (siRNA) depletion of PML in the HFF cells led to decreased ICP0-null virus replication, while short hairpin RNA (shRNA) depletion led to increased ICP0-null virus replication, arguing that different PML isoforms or PML-related proteins may have restrictive or proviral functions. In normal human cells, viral DNA replication increases expression of all classes of HSV-1 genes. We observed that IFI16 repressed transcription from both parental and progeny DNA genomes. Taken together, our results show that the mechanisms of action of IFI16 and ND10 proteins are independent, at least in part, and that IFI16 exerts restrictive effects on both input and replicated viral genomes. These results raise the potential for distinct mechanisms of action of IFI16 on parental and progeny viral DNA molecules.
IMPORTANCE Many human DNA viruses transcribe their genomes and replicate in the nucleus of a host cell, where they exploit the host cell nuclear machinery for their own replication. Host factors attempt to restrict viral replication by blocking such events, and viruses have evolved mechanisms to neutralize the host restriction factors. In this study, we provide information about the mechanisms of action of three host cell factors that restrict replication of herpes simplex virus (HSV). We found that these factors function independently and that one acts to restrict viral transcription from parental and progeny viral DNA genomes. These results provide new information about how cells counter DNA virus replication in the nucleus and provide possible approaches to enhance the ability of human cells to resist HSV infection.
KEYWORDS: DNA virus, ND10, nuclear replication, restriction factor, IFI16
INTRODUCTION
Host factors control viral replication through positive and negative effects on viral replication at steps ranging from entry to replication and assembly of progeny viruses. Many DNA viruses transcribe and replicate their genomes in the host cell nucleus by using the cellular machinery and are therefore subject to regulation by host cell factors that can restrict viral processes. In turn, viruses have evolved strategies to evade host cell factor-mediated restriction. For example, herpes simplex virus 1 (HSV-1) infected cell protein 0 (ICP0) is not essential for viral replication but does stimulate viral replication, in particular at low multiplicities of infection (MOI) (1, 2). ICP0 is an E3 ubiquitin ligase that promotes the degradation of several host restriction factors, including the promyelocytic leukemia (PML) protein, Sp100, ATRX, and interferon-inducible protein 16 (IFI16) (3–6). Depletion of any of these factors increases the replication of ICP0-negative mutant viruses (7, 8).
Host restriction factors for nuclear DNA viruses include the components of nuclear domain 10 (ND10) bodies, or PML nuclear bodies (PML-NBs), which are nuclear punctate or ring-like structures consisting of approximately 150 permanently or transiently associated proteins (9). PML is a member of the tripartite (TRIM) family of proteins and contains a RING domain, two zinc-binding B-box domains, and a coiled-coil domain (10). According to current models, PML serves as a structural platform for recruitment of other proteins, depending on the specific function (11, 12). ND10 bodies can act in several key cellular pathways, including apoptosis (13), gene regulation (14, 15), oncogenesis (15), and the antiviral response (16). In the last capacity, three factors, PML, Sp100, and Daxx, have been found to inhibit HSV-1 infection in HepaRG cells and fibroblasts (7). This inhibitory effect was also shown for a panel of DNA and RNA viruses, including but not limited to other herpesviruses (17–19) and HIV (20). As described above, HSV-1 ICP0 targets certain of these restriction factors for degradation.
Two recent publications, however, raised the hypothesis that PML might also exhibit proviral activity for HSV-1 infection in Hep2 cells, in a multiplicity-of-infection (MOI)-dependent fashion (21, 22). Furthermore, Ishov et al. (23) reported that PML-NBs may provide a platform for human cytomegalovirus (HCMV) gene expression and replication. PML-NBs have been reported to localize near incoming and early-replicating viral DNA (vDNA) (24–26).
An additional ND10 protein of interest, Daxx, has been described as a histone H3.3 chaperone (27) which physically interacts with chromatin-associated proteins, such as histone deacetylase II (HDAC II) and histones H2A, H2B, H3, and H4 (28, 29). Thus, Daxx has been implicated in epigenetic regulation of gene expression (28). It is believed that Daxx cooperates with the alpha-thalassemia retardation syndrome X-linked (ATRX) protein to form an ATP-dependent SNF2-like chromatin remodeling complex. The combined action of ATRX and Daxx is believed to lead to HDAC II recruitment to the genome, whereby ATRX performs the ATPase activity and Daxx serves as the recruitment factor (30; reviewed in reference 31).
Interferon-inducible protein 16 (IFI16) has also emerged as an additional crucial restriction factor during lytic herpesviral infection (8, 32–34). IFI16 is a member of the PYHIN protein family (35) and is comprised of an N-terminal Pyrin domain, which mediates protein-protein interactions, and two C-terminal HIN domains, which confer a sequence-nonspecific double-stranded DNA (dsDNA)-binding ability (36). It was identified initially as a cytoplasmic DNA-binding protein (35), but it can be nuclear or cytoplasmic depending on the cell type (37). In the context of HSV-1 infection, IFI16 is targeted for proteasomal degradation by the viral ICP0 protein (6, 38). We and others have demonstrated that IFI16 can sense herpesviral DNA in the nuclei of infected cells, initiate innate signaling (6, 33, 39), and lead to silencing of the viral DNA via establishment of repressive heterochromatin (8, 32). Depletion of IFI16 leads to an increase in replication of ICP0-null viruses due to enhanced viral gene expression (8, 32). Moreover, IFI16 has been observed to associate with ND10 proteins (40).
PML-NB or ND10 components and IFI16 react dynamically to incoming viral DNA, with IFI16, Daxx, and PML being recruited to genome complexes within a scale of minutes after infection in diploid human fibroblasts (HFs) (8, 40, 41). In light of these observations, we asked if IFI16 acts independently of the ND10 proteins, Daxx and PML, through combinatorial depletion of these proteins. We found that the inhibitory effects of IFI16 and Daxx are additive but that the effects of PML are inhibitory or proviral, depending on whether PML depletion is via short hairpin RNAs (shRNAs) or small interfering RNAs (siRNAs). In addition, we present results arguing that IFI16 exerts its restrictive effect on input genomes as well as progeny genes in normal human cells.
RESULTS
KO of IFI16 in HFFs increases replication of an HSV-1 ICP0-null virus.
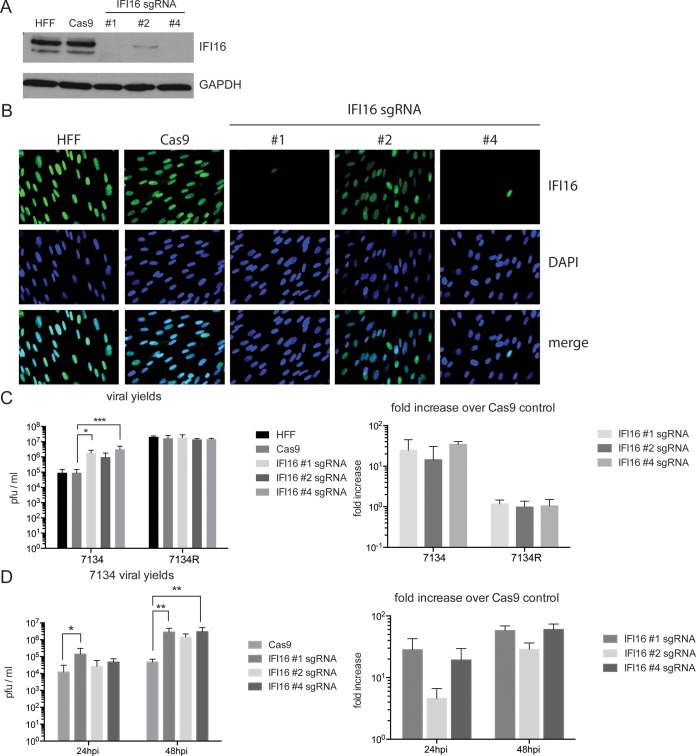
We showed previously that depletion of IFI16 in human foreskin fibroblast (HFF) cells by use of siRNAs increased replication of ICP0-null viruses (8). To confirm this effect by using gene knockout (KO) approaches, we established HFF IFI16 knockout cells by using the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 method. We used guide RNAs (gRNAs) complementary to three different regions of the IFI16 gene, mapping to the transcription start site (gRNA 2) or within the first 200 bp of the transcribed region (gRNAs 1 and 4). As a control, we used a cell line expressing only Cas9. Cell lines expressing Cas9 with gRNA 1 or 4 showed no detectable IFI16 protein by Western blotting, while expression of gRNA 2 led to an intermediate phenotype with a partial reduction of IFI16 (Fig. 1A). Levels of IFI16 expression were confirmed by immunofluorescence (Fig. 1B). We then tested the capacity of the three IFI16 knockout cell lines to support replication of the HSV-1 7134 ICP0-null virus or the HSV-1 7134R ICP0+ virus. Consistent with our previous siRNA results, we found that the IFI16 knockout cell lines showed increased replication of 7134 virus (Fig. 1C). Compared to those with either wild-type HFFs or HFFs expressing only Cas9, viral yields increased between 10- and 100-fold (Fig. 1C). This increase was statistically significant for gRNAs 1 (P ≤ 0.05 by t test) and 4 (P ≤ 0.001 by t test). Consistent with the extent of the knockout, cell lines 1 and 4 were affected the most, and cell line 2 exhibited an intermediate phenotype. No differences in viral yields were observed between the different cell lines infected with 7134R virus, likely due to degradation of IFI16 promoted by ICP0 encoded by 7134R virus. To analyze the kinetics of restriction, viral yields were determined at 24 h postinfection (hpi) and 48 hpi for 7134 virus (MOI = 0.1). We found that apart from the overall increase in viral titer from 24 to 48 h, failure of the IFI16 knockout cell lines to restrict 7134 virus was more pronounced after 48 h (P ≤ 0.01 by t test; both gRNAs) than at 24 hpi (P ≤ 0.05 by t test; only gRNA 1) (Fig. 1D). This observation was also reflected by a higher statistical significance of our results at 48 hpi than at 24 hpi.
FIG 1.
IFI16 knockout via CRISPR/Cas in HFF cells leads to a defect in restriction of an HSV-1 ICP0-null virus. (A) Immunoblot of whole-cell lysates probed with antibodies specific for IFI16 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in HFF, Cas9-expressing, and IFI16 knockout cells. (B) Immunofluorescence. HFF, Cas9-expressing, and IFI16 knockout (with gRNA 1, 2, or 4) cells were fixed, permeabilized, and incubated with DAPI (4′,6-diamidino-2-phenylindole; blue) and an antibody specific to IFI16 (green). Total magnification, ×400. (C) Wild-type HFF cells (HFF), HFF cells expressing Cas9 (Cas9), or HFF cells expressing Cas9 and one of three IFI16-specific synthetic guide RNAs (gRNA 1, 2, or 4) were infected with the HSV-1 ICP0-null virus 7134 or the ICP0+ virus 7134R at an MOI of 0.1. Viral yields were collected at 48 hpi and titrated on U2OS cells. Results were plotted as numbers of PFU per milliliter or normalized to the Cas9 control level. For statistical analysis, t tests were performed. (D) HFF, Cas9-expressing, or IFI16 knockout cells (with gRNA 1, 2, or 4) were infected with 7134 at an MOI of 0.1. Viral yields were harvested at 24 hpi or 48 hpi and titrated on U2OS cells. Results were plotted as numbers of PFU per milliliter or normalized to the Cas9 control level. Statistical analysis was performed as described above.
HSV-1 IE, E, and L proteins are expressed at higher levels and earlier in IFI16 knockout cell lines than in HFFs expressing only Cas9.
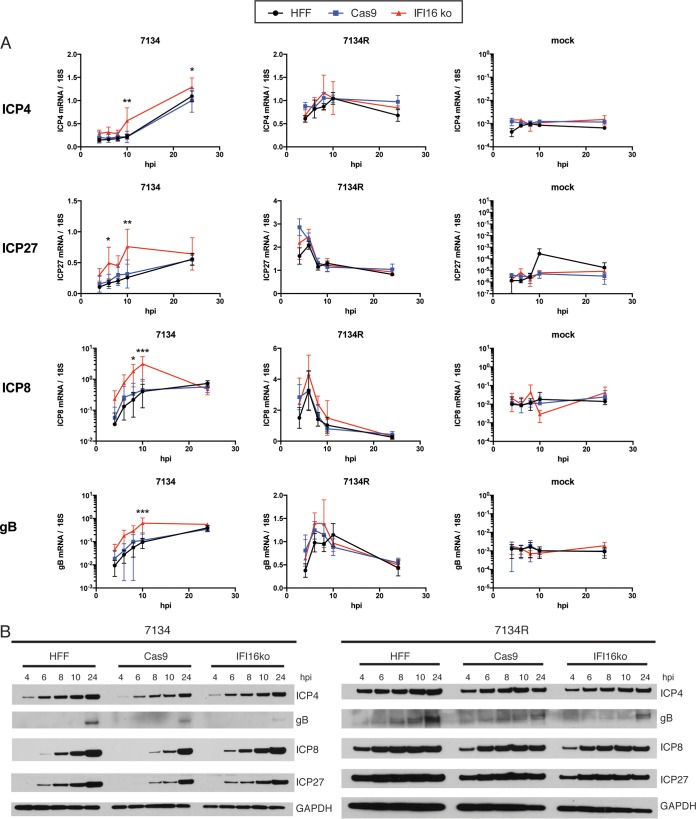
We further investigated the stage of infection at which IFI16 exerts its restriction effect. HFF, Cas9-expressing, or IFI16 knockout cells were infected with 7134 or 7134R virus (MOI = 5) to analyze immediate early (IE), early (E), and late (L) gene expression. We probed for representative IE (ICP4 and ICP27), E (ICP8), and L (gB) genes at 4 to 24 hpi. In cells infected with 7134R virus, we observed no differences in ICP4, ICP27, ICP8, and gB transcript and protein levels in HFF, Cas9-expressing, or IFI16 knockout cells (Fig. 2A and B). In contrast, after infection with the 7134 ICP0-null virus, ICP4, ICP27, ICP8, and gB transcript levels were all elevated compared to those in HFFs and Cas9-expressing cells (Fig. 2A). We detected the largest differences in viral transcript levels at 10 hpi for ICP4 (3-fold), ICP27 (2.5-fold), ICP8 (7-fold), and gB (6-fold), with levels increasing from IE to E to L genes. Compared to both HFF and Cas9-expressing control cells, differences in transcript levels were statistically significant by 2-way analysis of variance (ANOVA) with multiple comparisons for ICP4 at 10 hpi (P ≤ 0.01) and 24 hpi (P ≤ 0.05), for ICP27 at 6 hpi (P ≤ 0.05) and 10 hpi (P ≤ 0.01), for ICP8 at 8 hpi (P ≤ 0.05) and 10 hpi (P ≤ 0.001), and for gB at 10 hpi (P ≤ 0.001). At the protein level, it was evident that ICP27 and ICP8 were expressed earlier and to higher peak levels in IFI16 knockout cells than in the Cas9-expressing control cells (Fig. 2B). This effect was less pronounced with ICP4, in accordance with the observed smaller difference in mRNA levels between the cell lines. gB protein was detectable only at 24 hpi (in accordance with lower transcript levels than those of ICP4, ICP27, and ICP8 at earlier time points) but was increased in IFI16 KO cells compared to its level in the control cells. In total, these data argued that IFI16 acts negatively on IE, E, and L gene expression, with restriction of IE genes causing further reduction and delay in E and L gene expression. These results are consistent with published results arguing that IFI16 plays a role in epigenetic viral DNA silencing (8, 32, 34).
FIG 2.
Expression of IE, E, and L genes is earlier and increased in IFI16 knockout HFF cells. HFF, Cas9-expressing, and IFI16 knockout cells were infected with the 7134 and 7134R viruses at an MOI of 5 or mock infected. Cells were harvested for RNA and protein analysis at 4, 6, 8, 10, and 24 hpi. (A) ICP4, ICP27, ICP8, and gB transcript levels in HFF (black), Cas9-expressing (blue), and IFI16 knockout (red) cells were analyzed by qRT-PCR, and values were plotted relative to the 18S rRNA level. For statistical analysis, 2-way ANOVA including multiple comparisons was performed. Asterisks indicate significance compared to the values for HFF and Cas9-expressing cells. (B) Immunoblot showing ICP4, ICP27, ICP8, gB, cGAS, and GAPDH from 7134- or 7134R-infected cells.
Effects of IFI16 on gene expression from input and progeny genomes.
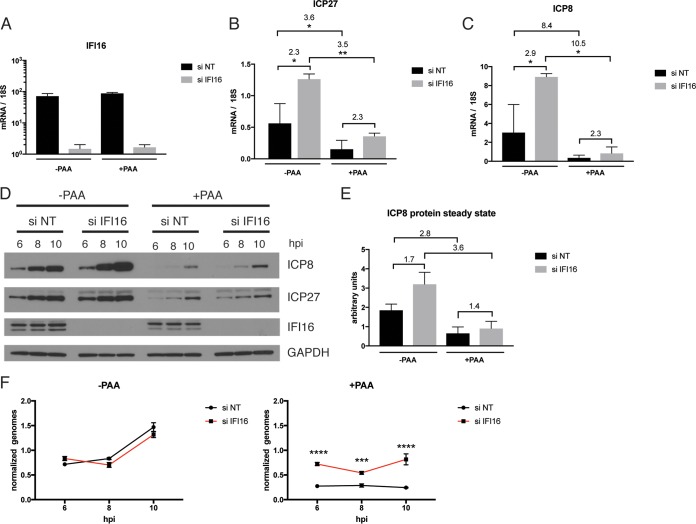
To determine if the restrictive effect of IFI16 was exerted on parental or progeny genomes, we examined gene expression at 10 hpi for the 7134 ICP0− virus with or without IFI16 under conditions where viral DNA replication occurred or was inhibited by phosphonoacetate (PAA). In this study, we utilized an siRNA-based strategy to deplete IFI16. The IFI16 knockdown was efficient (Fig. 3A and D), and IFI16 mRNA and protein levels were reduced by siRNA treatment but not affected by the addition of PAA (Fig. 3A and D). Both ICP27 and ICP8 transcript levels were reduced in a statistically significant fashion (2-way ANOVA) by addition of PAA (Fig. 3B and C). For ICP27 mRNA, this reduction was 3.6-fold (P ≤ 0.05) with siNT and 3.5-fold (P ≤ 0.01) with siIFI16. For ICP8 mRNA, the reduction was 8.4-fold with siNT and 10.5-fold (P ≤ 0.05) with siIFI16. Depletion of IFI16 increased ICP27 and ICP8 transcript levels (Fig. 3B and C), as observed above for IFI16 knockout cells. The magnitude of the RNA level increase due to IFI16 depletion, however, was independent of the presence of PAA (2.3- and 2.3-fold in the absence and presence of PAA, respectively, for ICP27 and 2.9- and 2.3-fold in the absence and presence of PAA, respectively, for ICP8) but dependent on the presence or absence of IFI16. These increases in both ICP27 and ICP8 mRNA levels were statistically significant in the absence of PAA (P ≤ 0.05 by 2-way ANOVA). The above observations were confirmed by Western blot analysis of protein levels (Fig. 3D). Depletion of IFI16 led to modest increases in ICP27 and ICP8 protein levels (1.7- and 1.4-fold for ICP8 at 10 hpi) (Fig. 3D and E), and addition of PAA led to decreases in ICP27 and ICP8 protein levels (2.8- and 3.6-fold for ICP8 at 10 hpi) (Fig. 3D and E). We further analyzed viral DNA replication (Fig. 3F). As expected, cells treated with PAA failed to replicate viral DNA, independent of the presence or absence of IFI16. In the presence of PAA, the viral genome copy number was slightly elevated at a constant level in the absence of IFI16. This difference in copy numbers was found to be highly statistically significant (P ≤ 0.001 at 8 hpi and P ≤ 0.0001 by 2-way ANOVA). In summary, the results for viral gene expression by ICP0-null viruses in HFF cells in the presence or absence of viral DNA replication argued that progeny vDNA is the major template for viral gene mRNA transcription for an ICP0-null mutant virus in HFF cells. Furthermore, the restrictive effect of IFI16 showed similar effects with and without viral DNA replication, so transcription from both input and progeny genes appeared to be affected by IFI16.
FIG 3.
IFI16 acts on gene expression from input and progeny genomes. HFF cells were treated with siRNAs specific for IFI16 or with nontargeting siRNA and infected with 7134 virus at an MOI of 5 in the presence or absence of PAA. Samples were harvested at 6, 8, and 10 hpi. (A to C) IFI16, ICP27, and ICP8 transcript levels at 10 hpi were analyzed by qRT-PCR and plotted relative to the 18S rRNA level. Numbers denote fold changes between the indicated columns. (D) Immunoblot probed with antibodies specific for ICP27, ICP8, IFI16, and GAPDH. (E) ICP8 protein levels at 10 hpi were quantified from the immunoblot, normalized to GAPDH, and plotted. Numbers indicate fold changes between columns. The diagram represents results for two biological replicates. (F) Total DNA was harvested at 1, 6, 8, and 10 hpi. Samples were analyzed via qPCR and plotted normalized to input DNA (1 hpi). For statistical analysis for panels B, C, and F, 2-way ANOVA including multiple comparisons was performed.
The effects on viral replication exerted by IFI16, PML, and Daxx are independent.
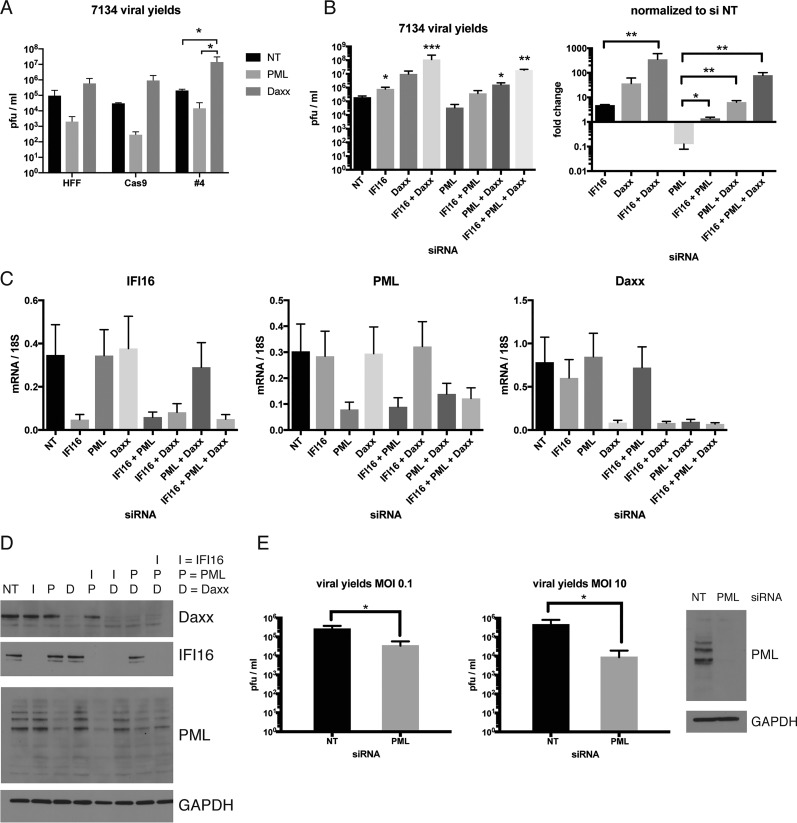
Antiviral effects of IFI16 and ND10 proteins against herpesviruses have been shown previously by us and others (4, 6, 7, 42). We focused on the ND10 proteins PML and Daxx to investigate if and how they might cooperate with IFI16 to restrict HSV in normal human cells. We used siRNAs to deplete PML or Daxx in HFFs, Cas9-expressing, or IFI16 KO cells and analyzed viral yields after infection with the 7134 virus. Consistent with previous results (7), Daxx depletion increased viral yields between 10- and 100-fold (Fig. 4A). Surprisingly, upon depletion of PML, we saw reductions in viral yields of between 10- and 100-fold (Fig. 4A). Interestingly, the number of PML foci per cell was slightly reduced, and foci appeared dimmer in IFI16 KO cells than in HFF and Cas9-expressing cells (see Fig. S1 in the supplemental material), suggesting that IFI16 affects the level or assembly of PML-NBs. This reduction in PML focus number was found to be highly statistically significant (P ≤ 0.0001 by t test) for comparisons of IFI16 KO cells to HFF and Cas9-expressing control cells. To ensure that our observations were not due to specific characteristics of the Cas/CRISPR-modified cells, we treated HFFs with siRNAs against IFI16, PML, and Daxx or combinations thereof and investigated viral yields of the 7134 virus. Knockdown efficiency was confirmed by mRNA and protein analyses for each factor (Fig. 4C and D). Consistent with our own and others' results (8, 43), we confirmed that depletion of IFI16 or Daxx increased viral yields by approximately 10- or 100-fold, respectively (Fig. 4B). This increase was statistically significant for depletion of IFI16 (P ≤ 0.05 by t test). IFI16 depletion increased viral yields less than IFI16 KO did (Fig. 1C versus Fig. 4B), possibly due to a more complete loss of IFI16 with gene knockout.
FIG 4.
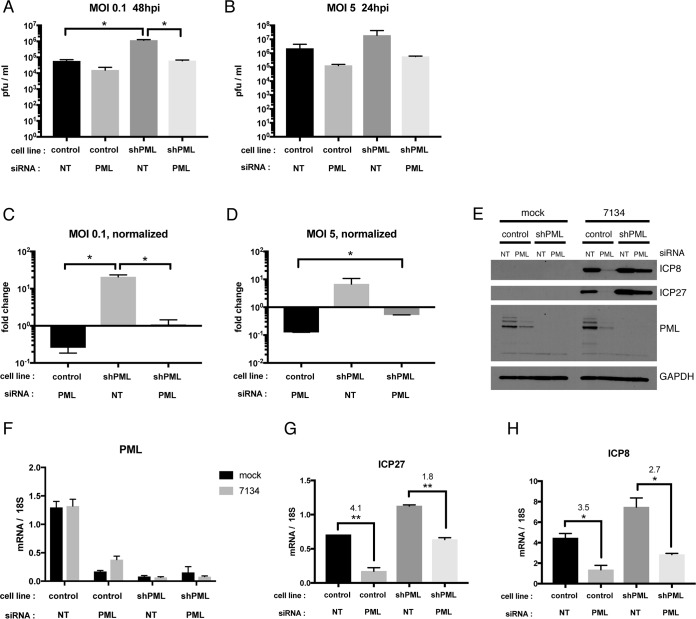
IFI16, PML, and Daxx exert their effects on replication of an ICP0-null virus in ways that are independent of each other. (A) HFF, Cas9-expressing, or IFI16 KO cells were treated with pooled siRNAs specific for PML or Daxx or with nontargeting siRNA and infected with 7134 virus at an MOI of 0.1. Viral yields were collected at 48 hpi and titrated on U2OS cells. (B) HFF cells were treated with pooled siRNAs specific for IFI16, PML, or Daxx or with nontargeting siRNA and infected with 7134 virus at an MOI of 0.1. Viral yields were collected at 48 hpi and titrated on U2OS cells. Viral yields were plotted as numbers of PFU per milliliter or normalized to nontargeting siRNA. For statistical analysis, t tests were performed. Asterisks located directly above columns indicate significance compared to the siNT value. (C) Relative IFI16, PML, and Daxx mRNA levels were analyzed via qRT-PCR prior to infection. (D) Immunoblot of protein samples harvested before infection and probed with antibodies specific for IFI16, PML, Daxx, and GAPDH. siRNA treatment is indicated by letters, as follows: I, IFI16; P, PML; and D, Daxx. (E) HFF cells were treated with siRNAs specific for PML or with nontargeting siRNA and infected with 7134 virus at an MOI of 0.1 or 10. Viral yields were collected at 48 or 24 hpi, respectively, and titrated on U2OS cells. The effectiveness of siRNA treatment was confirmed by immunoblot analysis of cell lysates prior to infection. Statistical analysis by the t test was performed as described above.
PML depletion led to a decrease in viral yields of approximately 10-fold. The reduction in viral yields upon PML depletion in both experiments was a surprising result, because previous studies had observed that PML is a restriction factor for HSV-1 and other viruses (17, 18, 20). Furthermore, our results showed that knockdown of two or more of the above proteins had additive or subtractive effects on 7134 viral yields. Depletion of IFI16 and Daxx increased viral yields by 2 to 3 orders of magnitude (Fig. 4B). When IFI16 or Daxx knockdown was combined with PML siRNA treatment, this resulted in a smaller increase in viral yields compared to that with treatment with IFI16 or Daxx siRNA alone (Fig. 4B). Depletion of all three proteins led to an intermediate phenotype. The additive or subtractive effects were statistically significant (t test) compared to the effects of siNT treatment for combined depletion of (i) IFI16 and Daxx (P ≤ 0.001), (ii) PML and Daxx (P ≤ 0.05), and (iii) IFI16, PML, and Daxx (P ≤ 0.01). Compared to depletion of IFI16 alone, additional siDaxx treatment enhanced viral yields in a statistically significant way (P ≤ 0.01 by t test). Compared to PML depletion alone, combined depletions of (i) IFI16 and PML, (ii) Daxx and PML, and (iii) IFI16, PML, and Daxx resulted in significantly enhanced viral yields (P ≤ 0.05, P ≤ 0.01, and P ≤ 0.01, respectively, by the t test). Thus, the anti- or proviral effects of IFI16, PML, and Daxx appeared to be independent of each other.
siRNA-mediated knockdown of PML decreases viral yields of an ICP0-null virus in an MOI-independent fashion.
Our initial experiments on the effect of PML on 7134 viral yields gave different results from those of most previously published studies (7, 43, 44). Because we had used an siRNA-based strategy, we sought to further validate our results. The initial experiments were done using a pool of 4 siRNAs specific for PML; therefore, to exclude the possibility that our observations were due to off-target effects, we tested individual siRNAs from the pool (Fig. S2). We found that treatments with the individual siRNAs affected PML to different extents. The combination of all four siRNAs was most effective at reducing PML protein levels as determined by Western blotting (Fig. S2B). Treatment with siRNA 06 reduced 7134 viral yields in HFF cells by 100-fold, making siRNA 06 the most effective of the single siRNAs (Fig. S2A). This reduction was statistically significant (P ≤ 0.05 by t test). Furthermore, a clear correlation between knockdown efficiencies determined by PML mRNA levels and reductions in viral yields could be established (Fig. S2D, pool marked by a triangle). In addition, we performed a human genome-wide reverse bioinformatic analysis to identify putative off-target sites of the siRNAs in the pool. We found no evidence that the transcript levels of the respective genes identified by the analysis changed upon treatment with any of the siRNAs in the pool (Table S1).
A previous study reported that PML might act antivirally or provirally depending on the MOI (21). To address this possibility, we infected HFF cells in which PML had been depleted or not with 7134 virus at an MOI of 0.1 or 10 and analyzed the viral yields. We found that depletion of PML in HFF cells by use of siRNAs led to statistically significant decreases in viral yields (P ≤ 0.05 by t test) that were independent of the MOI (Fig. 4E).
Effects of different PML knockdown strategies on ICP0-negative mutant virus replication.
To reconcile our results with published results (45), we obtained a previously used HFF-derived cell line in which a PML-specific shRNA is expressed, as well as its respective control cell line (43). For cells infected with 7134 virus, we confirmed that viral yields were increased upon shRNA-mediated PML knockdown, by about 10-fold, as reported before (Fig. 5A to D). Treatment of these cells with our PML-specific siRNA, however, resulted in a 10-fold reduction of 7134 viral yields, consistent with our results described above (Fig. 5A to D). These effects were observed at either a low or high MOI. At a low MOI, the increase in viral yields upon shRNA-mediated PML depletion in cells treated with siNT was statistically significant (P ≤ 0.05 by t test), as was the decrease in viral yields when cells expressing the PML-specific shRNA were additionally treated with PML-specific siRNA (P ≤ 0.05 by t test) (Fig. 5A and C). At a high MOI, the increase in viral yields in cells expressing the PML-specific shRNA was statistically significant (P ≤ 0.05 by t test) (Fig. 4D). At the protein level, immunoblot analysis showed the decrease of PML upon siRNA treatment of the control cell line (Fig. 5E). HFF cells expressing the shRNA did not show detectable PML protein. In control cells infected with 7134 virus, ICP27 and ICP8 protein expression was greatly reduced after siRNA treatment. This reduction was also observed, to a lesser extent, when the cells expressing the shRNA were treated with the siRNA (Fig. 5E). Both shRNA and siRNA treatments were effective at reducing PML mRNA levels (Fig. 5F). ICP27 and ICP8 mRNA levels were increased in PML shRNA-expressing cells and reduced upon PML siRNA treatment (Fig. 5G and H). The reduction upon PML siRNA treatment was found to be highly statistically significant for both ICP27 (P ≤ 0.01 by t test) and ICP8 (P ≤ 0.05 by t test). In total, these experiments confirmed previous results with PML as well as our own results.
FIG 5.
Different knockdown strategies affect HSV-1 replication differently. HFF cells expressing a PML-specific shRNA or control HFFs were treated with PML-specific siRNAs or nontargeting siRNA. Cells were infected with 7134 virus at an MOI of 0.1 or 5, and viral yields were harvested at 48 or 24 hpi, respectively, and titrated on U2OS cells. Total viral yields (A and B) or yields normalized to those for control HFFs treated with siNT (C and D) were plotted. (E) Immunoblot of lysates from mock- or 7134-infected cells at 6 hpi, probed with antibodies specific for PML, GAPDH, ICP8, and ICP27. (F) PML transcript levels at 6 hpi in mock- or 7134 virus-infected cells were assessed via qRT-PCR and plotted relative to 18S rRNA levels. (G and H) ICP8 and ICP27 transcript levels were measured by qRT-PCR and plotted relative to 18S rRNA levels. Numbers denote fold changes between columns. For all statistical analyses for this figure, t tests were performed.
PML knockdown reduces IE and E gene expression after infection with HSV-1 7134.
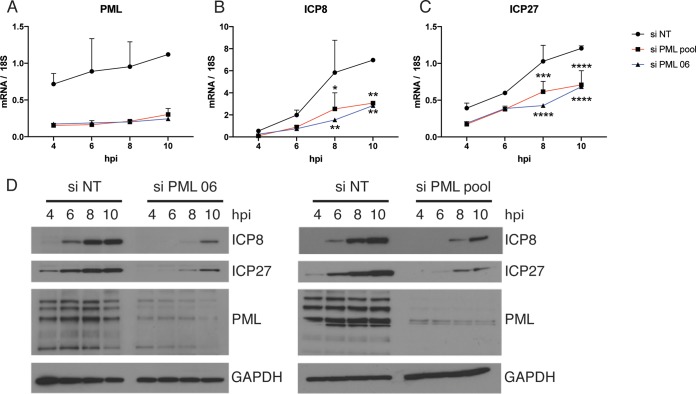
To gain further mechanistic insight into how the knockdown of PML affects viral replication in HFF cells, we measured the levels of ICP27 and ICP8 transcripts after knockdown with siRNA 06 or the siRNA pool (Fig. 6A) and subsequent infection with 7134 virus. Both ICP27 and ICP8 transcript levels were reduced upon PML knockdown (Fig. 6B and C). This effect was more pronounced for ICP8 than for ICP27 and was strongest at 10 hpi. The ICP27 transcript level reduction upon treatment with the siRNA pool or the single siRNA 06 versus treatment with siNT was statistically significant by 2-way ANOVA at 8 hpi (P ≤ 0.001 and P ≤ 0.0001, respectively) and 10 hpi (P ≤ 0.0001). The ICP8 transcript level reduction upon treatment with the siRNA pool or the single siRNA 06 versus treatment with siNT was statistically significant by 2-way ANOVA at 8 hpi (P ≤ 0.05 and P ≤ 0.01, respectively) and 10 hpi (P ≤ 0.01). At the protein level, it was evident that PML knockdown significantly reduced and delayed expression of both ICP27 and ICP8 (Fig. 6D). There was no difference in the effects observed when knockdown was done with the siRNA pool or the single siRNA 06. In total, these results argue that a form of PML or a PML-related function can exert a proviral effect on the expression of viral IE and E genes in HFF cells.
FIG 6.
A PML-related function promotes ICP0-null virus replication and viral IE and E gene expression. HFF cells were treated with siRNAs specific for PML, a single siRNA from that pool (siRNA 06), or nontargeting siRNA and infected with 7134 virus at an MOI of 5. Samples were harvested at 4 to 10 hpi, and transcript levels of PML (A), ICP8 (B), and ICP27 (C) were analyzed by qRT-PCR and plotted relative to 18S rRNA levels. (D) Immunoblots of cell lysates obtained at 4 to 10 hpi, probed with antibodies specific for ICP8, ICP27, PML, and GAPDH. For statistical analyses for panels B and C, 2-way ANOVA including multiple comparisons was performed.
A PML-related function acts at the stage of viral DNA replication.
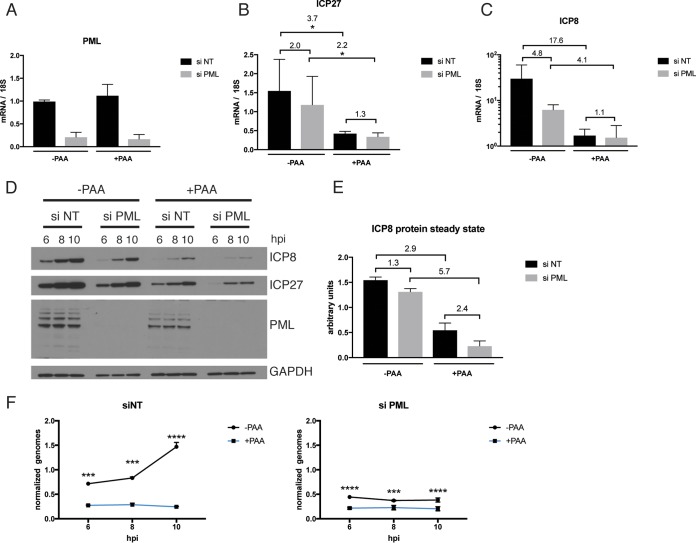
To determine whether the reduction in viral gene expression was independent of viral DNA replication, we treated HFF cells with either a nontargeting siRNA or the PML-specific siRNA pool and infected them with 7134 virus in the presence or absence of PAA for 10 h (Fig. 7). As expected, PML transcript and protein levels were not affected by the PAA treatment but were responsive only to the knockdown (Fig. 7A and D). In cells treated with the nontargeting siRNA, ICP27 and ICP8 gene transcript levels were significantly reduced in the presence of PAA, by factors of 3.7- and 18-fold, respectively (Fig. 7B and C). In contrast, upon PML knockdown, transcript levels were generally low, and the difference in the presence or absence of PAA was less pronounced (2.2-fold for ICP27 and 4.1-fold for ICP8) (Fig. 7B and C). These differences were found to be statistically significant for ICP27 (P ≤ 0.05 by 2-way ANOVA). We further confirmed our previous observations of a reduction in viral gene expression upon knockdown of PML (2.0-fold for ICP27 and 4.8-fold for ICP8) (Fig. 7B and C). On comparing ICP27 and ICP8 transcript levels in the presence and absence of PML in cells treated with PAA, no differences were observed (1.3-fold for ICP27 and 1.1-fold for ICP8) (Fig. 7B and C). At the protein level, effects similar to those at the mRNA level were seen (Fig. 7D and E).
FIG 7.
A PML-related function acts at the stage of viral DNA synthesis. HFF cells were treated with siRNAs specific for PML or with nontargeting siRNA and infected with 7134 virus at an MOI of 5 in the presence or absence of PAA. Samples were harvested at 6, 8, and 10 hpi. (A to C) IFI16, ICP27, and ICP8 transcript levels at 10 hpi were analyzed by qRT-PCR and plotted relative to 18S rRNA levels. Numbers denote fold changes between the indicated columns. (D) Immunoblot probed with antibodies specific for ICP27, ICP8, IFI16, and GAPDH. (E) ICP8 protein levels at 10 hpi were quantified from the immunoblot, normalized to GAPDH, and plotted. Numbers indicate fold changes between the respective columns. The diagram represents data for two biological replicates. (F) Total DNA was harvested at 1, 6, 8, and 10 hpi. Samples were analyzed via qPCR and plotted with normalization to input DNA (1 hpi). For statistical analyses for panels B and F, 2-way ANOVA including multiple comparisons was performed.
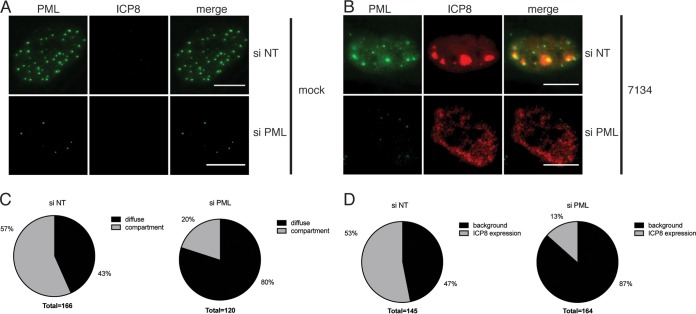
We also analyzed viral DNA replication and found that PML depletion largely blocked DNA replication. The differences in vDNA levels were highly statistically significant at each time point analyzed (P ≤ 0.001 at 8 hpi and P ≤ 0.0001 at 6 hpi and 10 hpi by 2-way ANOVA) (Fig. 7F). We speculated that the absence of DNA replication would affect formation of replication compartments or vice versa and carried out an immunofluorescence analysis of replication compartment size (Fig. 8). PML knockdown was efficient in HFFs and comparable to that described above (Fig. 8A). Upon infection with 7134 virus, PML-depleted cells seemed to show a rather diffuse distribution of ICP8 instead of formation of defined replication compartments (Fig. 8B). The percentage of cells exhibiting defined replication compartments as opposed to diffuse staining decreased from 57% to 20% upon depletion of PML (Fig. 8C). In addition, the overall percentage of cells that showed ICP8 protein expression beyond the background level was reduced from 53% to 13% (Fig. 8D).
FIG 8.
Absence of PML reduces replication compartment formation. HFF cells were cultivated on coverslips and treated with siRNAs specific for PML or with nontargeting siRNA. Cells were infected with 7134 virus at an MOI of 5 or mock infected and then were fixed at 6 hpi. Immunofluorescence staining was performed with antibodies specific for PML and ICP8. (A) Images showing nuclei of representative mock-infected cells in the presence or absence of PML. Channels specifically detected PML (green) or ICP8 (red), and merged images are also shown. (B) Same as panel A, but with 7134 virus infection. (C) Cells showing ICP8 expression above the background were sorted into two groups according to the morphology of staining (diffuse versus replication compartment). Percentages of cells with diffuse or replication compartment-like ICP8 staining are depicted for both siNT and siPML conditions. (D) Percentages of cells showing ICP8 staining above the background for either the siNT or siPML background.
Thus, in total, the PML siRNA knockdown phenotype was similar to that in the absence of DNA replication. Our results argue that a PML-related function acts in a proviral fashion at the stage of viral DNA replication and is required for efficient formation of replication compartments.
ICP0-null mutant viral gene expression is promoted by viral DNA replication to different extents in different human cell types.
In the course of these studies, we observed that viral DNA replication stimulated IE and E gene expression differently from that in the original cascade model of HSV gene expression (46). To examine the regulation of HSV-1 gene expression of an ICP0-null virus in other cell types, we studied HeLa cells, Vero E6 cells, primary HFF cells, and normal oral keratinocytes (NOKs). Equal numbers of cells were seeded and infected at the same MOI for each cell type in the presence or absence of PAA. In the absence of PAA, ICP27 transcript levels declined from 6 to 10 hpi in HeLa and Vero cells (Fig. S3A and C) but rose in HFFs and NOKs (Fig. S3E and G). ICP8 transcripts reached peak levels in HeLa, Vero, and NOK cells between 6 hpi and 10 hpi but were still rising in HFFs in the same time frame. For ICP27, the observed differences were statistically significant for all four cell types (for HeLa cells, P ≤ 0.001 at 6 hpi and P ≤ 0.01 at 8 hpi and 10 hpi; for Vero cells, P ≤ 0.0001 for all time points by 2-way ANOVA; for HFFs, P ≤ 0.01 at 6 hpi and P ≤ 0.0001 at 8 hpi and 10 hpi; and for NOKs, P ≤ 0.001 at 6 hpi, P ≤ 0.01 at 8 hpi, and P ≤ 0.0001 at 10 hpi). Also, for ICP8, the observed differences were statistically significant for all four cell types (for HeLa cells, P ≤ 0.05 for all time points; for Vero cells, P ≤ 0.01 at 8 hpi and 10 hpi; for HFFs, P ≤ 0.01 at 6 hpi and P ≤ 0.0001 at 8 hpi and 10 hpi; and for NOKs, P ≤ 0.05 at 6 hpi and 8 hpi and P ≤ 0.01 at 10 hpi by 2-way ANOVA). The respective protein levels followed the same trends but trailed behind the transcript kinetics (Fig. S3B, D, F, and H). In the absence of vDNA replication, both ICP27 and ICP8 expression levels were decreased, and respective mRNA and protein levels gradually rose over time. We analyzed viral protein expression at 10 hpi for the different cell types on one Western blot for better comparison (Fig. S3I), which confirmed that HFF and NOK cells depended more on newly synthesized DNA as the template for viral IE and E gene expression, as indicated by ICP27 and ICP8 protein expression. We found that the relative ICP8 protein signal intensity differences were statistically significant (P ≤ 0.05 by t test) in the absence of PAA. When vDNA replication was blocked, differences in relative steady-state ICP8 protein expression were statistically significant (P ≤ 0.05 by t test) for HeLa versus Vero, Vero versus HFF, and Vero versus NOK cells. We speculate that the observed differences are possibly due to a more restrictive environment toward incoming genomes in certain cell types.
DISCUSSION
Previous studies have shown that the ND10 proteins PML and Daxx (7), as well as the innate DNA sensor IFI16 (8, 32, 34), can restrict replication of various HSV-1 ICP0-negative mutant strains. Our initial experiments confirmed the role of IFI16 as a crucial restriction factor for HSV-1 ICP0-null strains, as reflected by 10- to 100-fold increases in viral yields upon knockout of the IFI16 gene in primary human cells. This confirmed previous results showing an increase in replication of ICP0-null mutant viruses and no effect on ICP0-positive viruses when IFI16 was depleted by use of siRNAs or shRNAs (8, 47). In contrast, one study showed that IFI16 knockdown also increased wild-type HSV-1 replication (32). The differences may be due to the latter study using microporation to introduce the siRNAs, as opposed to the transfection approach used in the other studies. We observed that viral DNA replication enhances expression of IE, E, and L genes by an ICP0-negative mutant virus and that depletion of IFI16 increases expression of transcripts from all kinetic classes of genes, with or without viral DNA synthesis. This argues that IFI16 can repress either input or progeny viral DNA molecules.
We observed that a PML-related function has a positive effect on replication of an ICP0-null virus in primary HFF cells. Under siRNA knockdown conditions, this PML-related function seems to be required for efficient replication compartment formation and thus for viral DNA replication. Different PML depletion strategies revealed that PML or separate PML-related functions might act in a dual fashion, having both antiviral and proviral activities in HSV-1 infection of primary human cells.
Expression of all kinetic classes of genes of an ICP0-null virus is increased by viral DNA synthesis.
Our observations indicate that viral IE and E gene expression in HFF cells in the absence of ICP0 is strongly stimulated by newly synthesized viral DNA. IE and E gene expression was less dependent on viral DNA synthesis in HeLa and Vero cell lines, whereas NOK cells showed a phenotype similar to that of HFF cells. Therefore, two normal human cell types depend largely on newly synthesized DNA for efficient viral gene expression. While the progression through IE, E, and L gene expression may initially occur from incoming genomes, the majority of IE and E gene expression in HFF and NOK cells seems to be accomplished after DNA replication, without a clear temporal resolution. This observation may reflect the restriction of parental genomes being more effective in normal human cells. While our experiments were not designed to give a detailed answer to the question of which stage of NOK and HFF cells is more restrictive toward HSV-1, we further emphasize that experimental results regarding restriction of HSV-1 might differ greatly depending on the cell types used in the respective assays.
IFI16 affects viral transcription from input and progeny viral genomes.
We observed that IFI16 reduces the levels of IE, E, and L transcripts with or without viral DNA synthesis. Because the levels of viral gene expression are significantly higher after viral DNA replication, these results argue that IFI16 acts on both parental and progeny viral DNA molecules. Our previous models involved IFI16 binding to input viral DNA, multimerizing, and recruiting epigenetic factors that load heterochromatin or add heterochromatic modifications on viral chromatin (48). IFI16 was previously shown to bind to viral DNA in infected cells (39), but that analysis was done at later times of infection. One paper showed colocalization of 5-ethynyl-2’-deoxyuridine (EdU)-labeled HSV DNA with IFI16 at 30 and 60 min postinfection (32), but the EdU-labeled foci appeared to be too numerous for input viral genomes and may have been caused by residual labeling of cellular DNA by free EdU. At early times postinfection, IFI16 colocalizes with viral genome complexes (8), consistent with the model of IFI16 recruiting epigenetic factors to the input DNA. In this light, it is interesting that significantly higher levels of input vDNA were observed in IFI16-depleted cells, suggesting that IFI16 can promote the turnover or loss of input vDNA. In contrast, in cells infected with an ICP0-null mutant so that IFI16 is not degraded, IFI16 localizes to or within replication compartments, but only to part of the compartments, not throughout the replication compartments (6; P. Merkl and D. M. Knipe, unpublished data). Therefore, IFI16 may function by different mechanisms to restrict transcription from input versus progeny HSV-1 genomes.
IFI16 and Daxx act independently in HSV-1 gene expression regulation.
Our results are consistent with the idea that the restrictive effects of IFI16 and Daxx are additive and thus independent of each other. We and others have shown that IFI16 is a primary viral DNA sensor and promotes the formation of heterochromatin on incoming viral genomes (8, 32). This in turn results in epigenetic silencing of viral gene expression. Because there is no evidence that IFI16 has enzymatic methyltransferase activity or functions as a histone chaperone, we proposed that it has the capability to recruit histone modifiers (48). IFI16 depletion leads to an increase of H3K4me3 signals on the ICP4 gene promoter, correlated with a decrease in H3K9me3 signals, and thus with increased gene expression (8, 32). In addition, overexpression of IFI16 in U2OS cells results in less IE gene expression from ICP0-null genomes (8). As described above, the additive effects of IFI16 and Daxx could be exerted on different forms of the viral DNA, such as input and parental viral DNAs.
As described above, there is evidence from other biological systems that Daxx acts through epigenetic mechanisms and can lead to a restrictive effect such as that observed for IFI16. The mechanism of action of Daxx is better understood in the context of HCMV infection than in that of HSV infection. HCMV infection of Daxx-depleted fibroblasts shows increased H3K9 acetylation on major IE gene promoters compared to that for fibroblasts containing Daxx (49). However, Daxx interacts with ATRX to form a SNF2-like, ATP-dependent chromatin remodeling complex, a function that has not been attributed to IFI16. IFI16 has direct DNA-binding capabilities via its two HIN domains, an additional feature not found in Daxx. A putative mechanism of IFI16 recruitment of histone-modifying factors would therefore involve IFI16 binding to naked incoming viral DNA, as supported by evidence that IFI16 localizes to incoming viral genomes (J. Cabral and D. M. Knipe, unpublished data).
We speculate that IFI16-mediated restriction is initiated by IFI16 binding to incoming naked viral DNA, forming a platform for recruitment of chromatin-remodeling factors and HDACs, which then introduce repressive chromatin marks, such as H3K9me3. We propose that the Daxx/ATRX SNF2-like complex does not require recruitment via IFI16 but has intrinsic capabilities to recognize target histones (Daxx) and to remodel the chromatin structure (ATRX). In the end, both IFI16 and Daxx/ATRX actions lead to silencing of incoming viral DNA, but they appear to be part of distinct mechanisms. This model differs from one recently presented (50) in which the PML nuclear bodies “entrap” the input viral DNA to “restrict the initiation” of ICP0-negative viral replication. Viral DNA is loaded with heterochromatin by as early as 1 to 2 hpi (51), so it seems more likely that early host restriction factors recruit host epigenetic silencing factors rather than physically excluding proviral host factors.
A PML-related function is required for replication compartment formation in primary HFF cells, thus exhibiting a novel proviral role.
As described above, we found that a PML-related function can exert a proviral effect on HSV-1 ICP0-null virus replication. This observation appears to be contradictory to the literature, but some evidence exists that PML may have a dual behavior in the context of DNA virus infection. There is a body of evidence showing that PML is a restriction factor for HSV-1 and HCMV infections (43, 44, 52, 53), but our findings obtained by use of PML siRNAs are more in accord with two recent publications (21, 22) supporting the notion that PML can act provirally under certain circumstances. The latter two papers reported that wild-type HSV-1 replication is decreased in PML-deficient Hep2 cells under low-MOI infection conditions. In our system, the proviral effect of PML is not dependent on the MOI. From a mechanistic point of view, the PML-related function seems to be required for vDNA replication, and depletion of PML apparently prohibits efficient formation of replication compartments. This is in line with previous studies that have shown that PML is recruited to or found in the proximity of replication compartments (23, 24, 26) and that it might provide a platform for replication and gene expression in HCMV infection (23).
The method of PML depletion and the multiplicity of PML isoforms might provide an explanation for the attribution of anti- or proviral properties.
Because siRNA-based strategies come with the risk of off-target effects, we deconvolved the siRNA pool and found a correlation between the PML knockdown efficiency of each single siRNA and the decrease in viral yields, arguing for specificity. Furthermore, our genome-wide bioinformatic analysis revealed only a few potential off-target genes, which we found to be unresponsive to PML siRNA treatment. The four siRNAs in the pool target sites in exons 2, 3, and 4, which are supposed to be common to all known PML isoforms.
In contrast, Everett et al. (43) used an shRNA strategy that targeted exon 4 to deplete PML in human fibroblasts, while Xu et al. (21) used the CRISPR/Cas system with a set of guide RNAs targeting exon 1 to knock out the PML gene in Hep2 cells. Depending on the source of information, 7, 11, or 14 PML isoforms exist (10, 54) (Uniprot and NCBI databases). However, there is no uniform nomenclature. Cuchet et al. (44) demonstrated that the two larger PML isoforms, PML I and II, were partially responsible for the restrictive effect of PML in HepaRG cells but could not fully restore the phenotype observed with expression of all PML isoforms. However, their study did not address the effect of the cytoplasmic major isoform VIIb (see reference 54 for details on the nomenclature) and acknowledged that other splice isoforms might exist and influence viral replication. Our own siRNA-mediated knockdown experiments with HFF cells expressing the PML-specific shRNA mentioned above revealed that we might be looking at two different functions of PML or functions correlated with PML expression. In this light, possible explanations for the apparently opposing results might be the existence of currently unaccounted PML isoforms, different isoform expression patterns in different cell types, or different parental virus strains. It is also conceivable that certain forms of PML are essential for cell growth and are not depleted under long-term knockdown conditions.
Summary.
Our results further define the role of IFI16 as an important restriction factor for HSV-1, and we are able to map the mechanism of action to the transcription of IE genes from both parental and progeny viral genomes. Our results raise the possibility that IFI16 may act by different mechanisms on parental and progeny genomes. In addition, we provide evidence that PML has both antiviral and proviral properties in HSV infection and give a mechanistic explanation for the proviral activity of PML, that is, the requirement of the protein for the formation of replication compartments and viral DNA replication. Finally, our results argue that IFI16, PML, and Daxx exert at least some of their respective effects by independent mechanisms during ICP0-null virus infection of normal human cells. Additional studies are needed to further define each of the restriction mechanisms.
MATERIALS AND METHODS
Cell culture, viruses, and infections.
HFF cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum (FBS). The ICP0-null virus 7134 and the respective rescued virus 7134R (55) were propagated and titrated in parallel on U2OS cells (56). For infections, viruses were diluted in phosphate-buffered saline (PBS) containing 0.1% (wt/vol) glucose and 1% (vol/vol) bovine calf serum (BCS). Infections were carried out at the respective MOI for 1 h at 37°C, the inoculum was replaced with DMEM containing 1% BCS, and the cells were kept at 37°C for the indicated times.
To discern between parental and progeny virus effects, HFF cells were infected with HSV-1 7134 in the presence of 400 μg/ml phosphonoacetic acid (PAA) and 10 mM HEPES, pH 7.4. Cells were incubated for 1 h at 37°C, overlaid with DMEM containing 0.1% BCS, 400 μg/ml PAA, and 10 mM HEPES, pH 7.4, and harvested at the indicated times.
IFI16 Cas9/CRISPR knockout cell lines.
Double-stranded DNA oligonucleotides encoding specific gRNAs were inserted into the pRRL-Cas9-Puro lentivirus vector (57) by homologous recombination (In-Fusion; Clontech). pRRL-Cas9-Puro plasmids expressing Cas9 alone or coexpressing the indicated gRNAs were cotransfected with the psPAX2 packaging vector and pVSV-G into 10-mm dishes of 50% confluent HEK293T cells. Medium containing transfection complexes was replaced at 24 h posttransfection. Supernatants from transfected cells were harvested at 48 h posttransfection and filtered through a 0.45-μm syringe filter (Millipore) to generate lentiviral preparations. Four of 5 ml of lentivirus-containing solution was added to 1 × 105 HFF cells in a 6-well plate and spin infected at 2,000 rpm for 30 min. Lentivirus-containing medium was replaced at 24 h posttransduction. At 3 days posttransduction, cells were selected for puromycin resistance (2.5 μg/ml) and expanded over a 2-week period. Guide RNA sequences are as follows: gRNA 1, GUUCCGAGGUGAUGCUGGUU; gRNA 2, UCACUUAUGUCUGUAAAGAU; and gRNA 4, UUGAUGGAAGAAAAGUUCCG.
Relative quantification of viral DNA by qPCR and relative quantification of RNA by quantitative reverse transcription-PCR (qRT-PCR).
mRNAs were prepared (Qiagen RNeasy kit), treated with DNase (DNAfree kit; Ambion), and reverse transcribed (High Capacity cDNA RT kit; Applied Biosystems). Total DNA was extracted by use of a Qiagen blood and tissue kit according to the manufacturer's standard operating procedure (SOP). A list of the primers used for subsequent quantitative PCR (qPCR) (Fast SYBR green reagents; Thermo Fisher) is provided in Table 1.
TABLE 1.
Primers for qRT-PCR and qPCR
| Name | Sequence or reference |
|---|---|
| ICP4 (mRNA) for | GCGTCGTCGAGGTCGT |
| ICP4 (mRNA) rev | CGCGGAGACGGAGGAG |
| ICP27 (mRNA) for | GCATCCTTCGTGTTTGTCATT |
| ICP27 (mRNA) rev | GCATCTTCTCTCCGACCCCG |
| ICP8 (mRNA) for | GGAGGTGCACCGGATACC |
| ICP8 (mRNA) rev | GGCTAACCGGCATGAAC |
| gB (mRNA) for | TGTGTACATGTCCCCGTTTTACG |
| gB (mRNA) rev | GCGTAGAAGCCGTCAACCT |
| ICP8 (genomic) for | CAGGCGCCCAATACGACCAAATC |
| ICP8 (genomic) rev | GAGACCGGGGTTGGGGAATGAATC |
| GAPDH (genomic) for | CAGGCGCCCAATACGACCAAATC |
| GAPDH (genomic) rev | TTCGACAGTCAGCCGCATCTTCTT |
| IFI16 (mRNA) for | Orzalli et al. (6) |
| IFI16 (mRNA) rev | Orzalli et al. (6) |
| PML (mRNA) for | GGACCCTATTGACGTTGACC |
| PML (mRNA) rev | TTGATGGAGAAGGCGTACAC |
| Daxx (mRNA) for | AGAAGCAAACAGGATCAGGG |
| Daxx (mRNA) rev | GCTGGGCAGGGTACATATC |
Immunofluorescence.
Cells were grown, fixed with formaldehyde, permeabilized, and incubated with antibodies (Table 2) as described before (6). Images were acquired with a Zeiss Axioplan microscope using a Hamamatsu charge-coupled device (CCD) camera and processed with AxioVision software.
TABLE 2.
Antibodies used for this studya
| Antibody specificity | Name | Source or reference |
|---|---|---|
| IFI16 | ab50004 | Abcam |
| PML (Western blotting) | A301-167A | Bethyl |
| PML (IFA) | ab96051 | Abcam |
| Daxx | D7810 | Sigma |
| GAPDH | ab8245 | Abcam |
| ICP8 | Knipe and Smith (59) | |
| ICP27 | ab31631 | Abcam |
| ICP4 | ab6514 | Abcam |
| gB | ab6506 | Abcam |
| Mouse IgG | Alexa 488-conjugated secondary Ab for IFA; ab150113 | Abcam |
| Rabbit IgG | Alexa 594-conjugated secondary Ab for IFA; 150080 | Abcam |
IFA, immunofluorescence assay.
Depletion of proteins.
siRNAs specific for IFI16, PML, and Daxx and a nontargeting siRNA were purchased from Dharmacon (Table 3). siRNA transfection into HFF cells was done using RNAi Max (Invitrogen) according to the manufacturer's SOP.
TABLE 3.
siRNAs used for this study
| siRNA | Sequence or reference |
|---|---|
| IFI16 | Orzalli et al. (6) |
| PML (pool) | Combination of the PML siRNAs below |
| PML 05 | GGAAAGAUGCAGCUGUAUC |
| PML 06 | GAGCUCAAGUGCGACAUCA |
| PML 07 | GGACAUGCACGGUUUCCUG |
| PML 08 | GCAACCAGUCGGUGCGUGA |
| Daxx | CAGCCAAGCUCUAUGUCUA |
| GAGGUUAACAGGCGCGCAUUG | |
| GCAAAACAAAGGACGCAUA | |
| GGAGUUGGAUCUCUCAGAA |
Immunoblotting.
Samples were run in 4 to 12% NuPage (Thermo Fisher) SDS-PAGE gradient gels and blotted onto polyvinylidene difluoride (PVDF) membranes as described previously (58). A list of antibodies used is provided in Table 2.
Statistical analysis.
All statistical analyses were performed with GraphPad Prism 7. For single-column comparisons, t tests were run. Statistical significance is indicated in the figures, as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. For grouped comparisons with two variables, 2-way ANOVA was performed, including multiple comparisons between variables of interest. Statistical significance indicators for 2-way ANOVA follow the same scheme as that for t tests.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Feodor Lynen Fellowship from the Alexander von Humboldt Foundation to P.E.M. and by NIH grant AI106934 to D.M.K.
We thank Thomas Stamminger for kindly providing us with the HFF control and HFF shPML cell lines. We thank Jeho Shin for technical assistance and Patrick T. Waters for assistance with the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00057-18.
REFERENCES
- 1.Sacks WR, Schaffer PA. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol 61:829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stow ND, Stow EC. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol 67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 3.Chelbi-Alix MK, de The H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 4.Lukashchuk V, Everett RD. 2010. Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx. J Virol 84:4026–4040. doi: 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurak I, Silverstein LB, Sharma M, Coen DM. 2012. Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity. J Virol 86:10093–10102. doi: 10.1128/JVI.00930-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orzalli MH, DeLuca NA, Knipe DM. 2012. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A 109:E3008–E3017. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glass M, Everett RD. 2013. Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J Virol 87:2174–2185. doi: 10.1128/JVI.02950-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orzalli MH, Conwell SE, Berrios C, Decaprio JA, Knipe DM. 2013. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci U S A 110:E4492–E4501. doi: 10.1073/pnas.1316194110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Damme E, Laukens K, Dang TH, Van Ostade X. 2010. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci 6:51–67. doi: 10.7150/ijbs.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jensen K, Shiels C, Freemont PS. 2001. PML protein isoforms and the RBCC/TRIM motif. Oncogene 20:7223–7233. doi: 10.1038/sj.onc.1204765. [DOI] [PubMed] [Google Scholar]
- 11.Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med 193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salomoni P, Pandolfi PP. 2002. The role of PML in tumor suppression. Cell 108:165–170. doi: 10.1016/S0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 13.Bernardi R, Pandolfi PP. 2003. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene 22:9048–9057. doi: 10.1038/sj.onc.1207106. [DOI] [PubMed] [Google Scholar]
- 14.Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. 2001. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J 20:4547–4559. doi: 10.1093/emboj/20.16.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Peres L, Honore N, Nasr R, Zhu J, de The H. 2006. Dimerization-induced corepressor binding and relaxed DNA-binding specificity are critical for PML/RARA-induced immortalization. Proc Natl Acad Sci U S A 103:9238–9243. doi: 10.1073/pnas.0603324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geoffroy MC, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res 31:145–158. doi: 10.1089/jir.2010.0111. [DOI] [PubMed] [Google Scholar]
- 17.Scherer M, Schilling EM, Stamminger T. 2017. The human CMV IE1 protein: an offender of PML nuclear bodies. Adv Anat Embryol Cell Biol 223:77–94. doi: 10.1007/978-3-319-53168-7_4. [DOI] [PubMed] [Google Scholar]
- 18.Schilling EM, Scherer M, Reuter N, Schweininger J, Muller YA, Stamminger T. 2017. The human cytomegalovirus IE1 protein antagonizes PML nuclear body-mediated intrinsic immunity via the inhibition of PML de novo SUMOylation. J Virol 91:e02049-16. doi: 10.1128/JVI.02049-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zerboni L, Che X, Reichelt M, Qiao Y, Gu H, Arvin A. 2013. Herpes simplex virus-1 tropism for human sensory ganglion neurons in the severe combined immunodeficiency mouse model of neuropathogenesis. J Virol 87:2791–2802. doi: 10.1128/JVI.01375-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kahle T, Volkmann B, Eissmann K, Herrmann A, Schmitt S, Wittmann S, Merkel L, Reuter N, Stamminger T, Gramberg T. 2015. TRIM19/PML restricts HIV infection in a cell type-dependent manner. Viruses 8:2. doi: 10.3390/v8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu P, Mallon S, Roizman B. 2016. PML plays both inimical and beneficial roles in HSV-1 replication. Proc Natl Acad Sci U S A 113:E3022–E3028. doi: 10.1073/pnas.1605513113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu P, Roizman B. 2017. The SP100 component of ND10 enhances accumulation of PML and suppresses replication and the assembly of HSV replication compartments. Proc Natl Acad Sci U S A 114:E3823–E3829. doi: 10.1073/pnas.1703395114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishov AM, Stenberg RM, Maul GG. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J Cell Biol 138:5–16. doi: 10.1083/jcb.138.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maul GG, Ishov AM, Everett RD. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type 1. Virology 217:67–75. doi: 10.1006/viro.1996.0094. [DOI] [PubMed] [Google Scholar]
- 25.Maul GG. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660–667. doi:. [DOI] [PubMed] [Google Scholar]
- 26.Uprichard SL, Knipe DM. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113–125. doi: 10.1006/viro.1996.8430. [DOI] [PubMed] [Google Scholar]
- 27.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. 2010. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A 107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J Cell Sci 115:3319–3330. [DOI] [PubMed] [Google Scholar]
- 29.Li J, O'Malley BW, Wong J. 2000. p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol Cell Biol 20:2031–2042. doi: 10.1128/MCB.20.6.2031-2042.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torii S, Egan DA, Evans RA, Reed JC. 1999. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J 18:6037–6049. doi: 10.1093/emboj/18.21.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dellaire G, Bazett-Jones DP. 2004. PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26:963–977. doi: 10.1002/bies.20089. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KE, Bottero V, Flaherty S, Dutta S, Singh VV, Chandran B. 2014. IFI16 restricts HSV-1 replication by accumulating on the HSV-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog 10:e1004503. doi: 10.1371/journal.ppat.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe 9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Diner BA, Lum KK, Toettcher JE, Cristea IM. 2016. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. mBio 7:e01553-16. doi: 10.1128/mBio.01553-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, Sharma S, Sirois CM, Jin T, Latz E, Xiao TS, Fitzgerald KA, Paludan SR, Bowie AG. 2010. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol 11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin T, Perry A, Jiang J, Smith P, Curry JA, Unterholzner L, Jiang Z, Horvath G, Rathinam VA, Johnstone RW, Hornung V, Latz E, Bowie AG, Fitzgerald KA, Xiao TS. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity 36:561–571. doi: 10.1016/j.immuni.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choubey D, Deka R, Ho SM. 2008. Interferon-inducible IFI16 protein in human cancers and autoimmune diseases. Front Biosci 13:598–608. doi: 10.2741/2705. [DOI] [PubMed] [Google Scholar]
- 38.Johnson KE, Chikoti L, Chandran B. 2013. Herpes simplex virus 1 infection induces activation and subsequent inhibition of the IFI16 and NLRP3 inflammasomes. J Virol 87:5005–5018. doi: 10.1128/JVI.00082-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li T, Diner BA, Chen J, Cristea IM. 2012. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc Natl Acad Sci U S A 109:10558–10563. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diner BA, Lum KK, Javitt A, Cristea IM. 2015. Interactions of the antiviral factor interferon gamma-inducible protein 16 (IFI16) mediate immune signaling and herpes simplex virus-1 immunosuppression. Mol Cell Proteomics 14:2341–2356. doi: 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Everett RD. 2016. Dynamic response of IFI16 and promyelocytic leukemia nuclear body components to herpes simplex virus 1 infection. J Virol 90:167–179. doi: 10.1128/JVI.02249-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J Gen Virol 75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 43.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuchet D, Sykes A, Nicolas A, Orr A, Murray J, Sirma H, Heeren J, Bartelt A, Everett RD. 2011. PML isoforms I and II participate in PML-dependent restriction of HSV-1 replication. J Cell Sci 124:280–291. doi: 10.1242/jcs.075390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boutell C, Everett RD. 2013. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol 94:465–481. doi: 10.1099/vir.0.048900-0. [DOI] [PubMed] [Google Scholar]
- 46.Honess RW, Roizman B. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc Natl Acad Sci U S A 72:1276–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuchet-Lourenco D, Anderson G, Sloan E, Orr A, Everett RD. 2013. The viral ubiquitin ligase ICP0 is neither sufficient nor necessary for degradation of the cellular DNA sensor IFI16 during herpes simplex virus 1 infection. J Virol 87:13422–13432. doi: 10.1128/JVI.02474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knipe DM. 2015. Nuclear sensing of viral DNA, epigenetic regulation of herpes simplex virus infection, and innate immunity. Virology 479–480:153–159. doi: 10.1016/j.virol.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J Biol Chem 281:37652–37660. doi: 10.1074/jbc.M604273200. [DOI] [PubMed] [Google Scholar]
- 50.Alandijany T, Roberts APE, Conn KL, Loney C, McFarlane S, Orr A, Boutell C. 2018. Distinct temporal roles for the promyelocytic leukaemia (PML) protein in the sequential regulation of intracellular host immunity to HSV-1 infection. PLoS Pathog 14:e1006769. doi: 10.1371/journal.ppat.1006769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee JS, Raja P, Knipe DM. 2016. Herpesviral ICP0 protein promotes two waves of heterochromatin removal on an early viral promoter during lytic infection. mBio 7:e02007-15. doi: 10.1128/mBio.02007-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Everett RD, Parada C, Gripon P, Sirma H, Orr A. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol 82:2661–2672. doi: 10.1128/JVI.02308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80:8006–8018. doi: 10.1128/JVI.00743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. 2013. Differential roles of PML isoforms. Front Oncol 3:125. doi: 10.3389/fonc.2013.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai WZ, Schaffer PA. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J Virol 63:4579–4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai W, Schaffer PA. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J Virol 66:2904–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eckard SC, Rice GI, Fabre A, Badens C, Gray EE, Hartley JL, Crow YJ, Stetson DB. 2014. The SKIV2L RNA exosome limits activation of the RIG-I-like receptors. Nat Immunol 15:839–845. doi: 10.1038/ni.2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Orzalli MH, Broekema NM, Knipe DM. 2016. Varying roles of herpes simplex virus 1 ICP0 and Vhs in loss of cellular IFI16 in different cell types. J Virol 90:8351–8359. doi: 10.1128/JVI.00939-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Knipe DM, Smith JL. 1986. A mutant herpesvirus protein leads to a block in nuclear localization of other viral proteins. Mol Cell Biol 6:2371–2381. doi: 10.1128/MCB.6.7.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.