ABSTRACT
Hepatitis C virus (HCV) infection causes 500,000 deaths annually, in association with end-stage liver diseases. Investigations of the HCV life cycle have widened the knowledge of virology, and here we discovered that two piperazinylbenzenesulfonamides inhibit HCV entry into liver cells. The entry of HCV into host cells is a complex process that is not fully understood but is characterized by multiple spatially and temporally regulated steps involving several known host factors. Through a high-content virus infection screening analysis with a library of 1,120 biologically active chemical compounds, we identified SB258585, an antagonist of serotonin receptor 6 (5-HT6), as a new inhibitor of HCV entry in liver-derived cell lines as well as primary hepatocytes. A functional characterization suggested a role for this compound and the compound SB399885, which share similar structures, as inhibitors of a late HCV entry step, modulating the localization of the coreceptor tight junction protein claudin-1 (CLDN1) in a 5-HT6-independent manner. Both chemical compounds induced an intracellular accumulation of CLDN1, reflecting export impairment. This regulation correlated with the modulation of protein kinase A (PKA) activity. The PKA inhibitor H89 fully reproduced these phenotypes. Furthermore, PKA activation resulted in increased CLDN1 accumulation at the cell surface. Interestingly, an increase of CLDN1 recycling did not correlate with an increased interaction with CD81 or HCV entry. These findings reinforce the hypothesis of a common pathway, shared by several viruses, which involves G-protein-coupled receptor-dependent signaling in late steps of viral entry.
IMPORTANCE The HCV entry process is highly complex, and important details of this structured event are poorly understood. By screening a library of biologically active chemical compounds, we identified two piperazinylbenzenesulfonamides as inhibitors of HCV entry. The mechanism of inhibition was not through the previously described activity of these inhibitors as antagonists of serotonin receptor 6 but instead through modulation of PKA activity in a 5-HT6-independent manner, as proven by the lack of 5-HT6 in the liver. We thus highlighted the involvement of the PKA pathway in modulating HCV entry at a postbinding step and in the recycling of the tight junction protein claudin-1 (CLDN1) toward the cell surface. Our work underscores once more the complexity of HCV entry steps and suggests a role for the PKA pathway as a regulator of CLDN1 recycling, with impacts on both cell biology and virology.
KEYWORDS: hepatitis C virus, virus entry, virus-host interaction, protein kinase A, piperazinylbenzenesulfonamide, tight junction protein
INTRODUCTION
Virus entry into specific host cells is a tightly regulated multistep process. Among known viruses, the liver pathogen hepatitis C virus (HCV), which causes 500,000 deaths annually, has probably one of the most complex multistep entry processes, involving a growing number of cellular entry factors (1). Furthermore, the regulation of trafficking of these proteins provides an additional level of complexity. Although the involvement of a large number of host factors has been demonstrated for HCV entry, several gaps remain in our comprehension of this process. Further functional studies are therefore needed to better understand the regulation of the early steps of the HCV life cycle.
After binding to attachment factors, such as heparan sulfate proteoglycans and the low-density lipoprotein (LDL) receptor, HCV particles bind specifically to the entry receptors scavenger receptor BI (SRB1) and CD81 tetraspanin (2). HCV binding to CD81 induces diffusion of this virion-tetraspanin complex on the plasma membrane, toward the sites of viral internalization (3). Only a specific fraction of CD81, located outside tetraspanin-enriched areas, is involved in HCV entry (4), through its interaction with the tight junction protein claudin-1 (CLDN1) (5–7). Importantly, CD81-CLDN1 interaction is a major event driving HCV endocytosis (8). Among other identified host factors involved in HCV entry (2), occludin (OCLN), another tight junction protein, also plays a major role in a late step of HCV entry, through an unknown mechanism (9). Together these observations indicate that tight junction proteins play a central role in HCV entry. However, the regulation of these proteins in the liver remains poorly understood, and little information is available on CLDN1 trafficking and its interplay with CD81 and OCLN in both infected and noninfected cells.
In this study, we implemented a high-content screening (HCS) approach using a library of chemical compounds to search for new regulators of HCV entry. We identified the serotonin receptor 6 (5-HT6) antagonists SB258585 and SB399885 as inhibitors of a late step of HCV entry. More specifically, we showed a reduction of protein kinase A (PKA) activation upon treatment, in a 5-HT6-independent manner. We confirmed the involvement of the PKA pathway in HCV entry and a role for its activation in CLDN1 export. Taken together, these data identify new inhibitors of the HCV postbinding step and provide new findings on the regulation of CLDN1 transport and function. This work highlights once more the complexity of the HCV entry process, proving that the presence of CLDN1 at the cell surface is not sufficient to mediate its interaction with CD81. This confirms the importance of the membrane environment and the need for the synergistic action of several receptors in HCV entry.
RESULTS
Identification of the 5-HT6 antagonist SB258585 as a new inhibitor of the HCV life cycle.
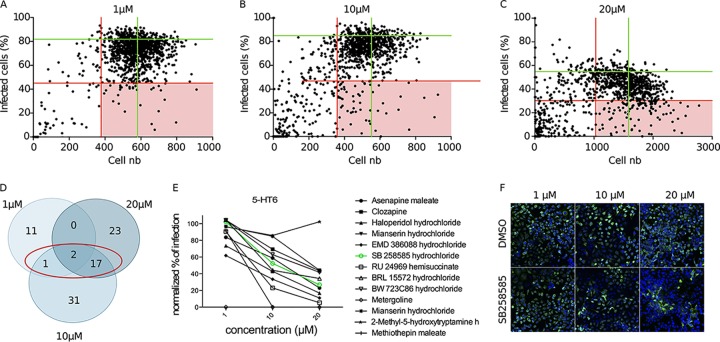
To identify new cellular factors involved in the early steps of the HCV infectious cycle, we performed HCS by using a library of 1,120 biologically active chemical compounds. The screen was performed at three different concentrations (1 μM, 10 μM, and 20 μM) with Huh-7 cells infected with cell culture-derived HCV (HCVcc) strain JFH1 (genotype 2a) (10, 11). A reinfection experiment was performed as an additional filter to eliminate false-positive compounds (see Data Set S1 in the supplemental material). Compounds reducing the infection by at least 45% compared to the average for dimethyl sulfoxide (DMSO) control wells and affecting cell viability by less than 35% were considered positive hits (Fig. 1A to C). Finally, we retained only the compounds confirmed in the reinfection experiment and inhibiting HCV infection for at least two of the three tested concentrations (Fig. 1D and Table 1). According to these criteria, 20 compounds were selected. Nine of them had previously been described as having an inhibitory effect on HCV or targeting cellular pathways identified as important for HCV infection, thus validating our screen (12–16). Among the 11 remaining hits, we identified 5 compounds targeting serotonin (5-HT) receptors or transporters. Serotonin receptors constitute a class of 16 human receptors, none of which have ever been identified as host factors involved in the HCV infectious cycle. To identify the 5-HT receptor(s) playing a role in HCV infection, we searched the Tocriscreen Total library for all compounds targeting each of these receptors in a more or less specific manner, in accordance with data available at http://www.guidetopharmacology.org/ and http://stitch.embl.de (17). Interestingly, with only one exception, all the compounds targeting serotonin receptor 6 (5-HT6) showed a dose-response inhibitory effect on HCV JFH1 infection (Fig. 1E), in contrast to the compounds targeting other 5-HT receptors, in which case, for most of them, only the highest concentration showed an inhibitory activity. Among the inhibitors of 5-HT6, SB258585 was identified as a hit according to the criteria of lack of cell toxicity and inhibition of infection chosen for the screen (Fig. 1F and Table 1).
FIG 1.
Identification of the 5-HT6 antagonist SB258585 as a new inhibitor of the HCV life cycle. (A to C) An HCS screen with a library of 1,120 chemical compounds was performed at three different concentrations (1 μM, 10 μM, and 20 μM). Dot plots show the distributions of the populations on the bases of cell number and percentage of HCVcc JFH1 (genotype 2a)-infected cells at the indicated concentrations. Each dot represents one compound. Green lines represent the average values corresponding to DMSO control wells. Red lines represent the selected cutoffs determining the positive hits (light red squares). (D) Summary of the positive hits selected according to the cutoffs for each concentration. The red circle indicates compounds confirmed as positive hits for at least two concentrations. (E) Graph showing percentages of HCVcc infection corresponding to the wells treated with chemical compounds targeting 5-HT6 in a more or less specific way, normalized to the mean for the DMSO control wells at each concentration. The green line corresponds to the compound SB258585 hydrochloride. (F) Wells corresponding to DMSO and SB258585 for each concentration of the screen. Nuclei are shown in blue, and HCV E1 staining is shown in green.
TABLE 1.
Confirmed hits for at least two of three concentrations in HCS screen with a library of 1,120 chemical compounds on Huh-7 cells infected with HCVcc JFH1 (genotype 2a)a
| Concentrations | CAS no. | Product name | Description | Reference(s) for described HCV inhibitory mechanism |
|---|---|---|---|---|
| 1, 10, and 20 μM | 35943-35-2 | API-2 | Selective inhibitor of Akt/PKB signaling | 15, 61, 62 |
| 40796-97-2 | MDL 72222 | 5-HT3 antagonist | ||
| 1 and 10 μM | 30484-77-6 | Flunarizine dihydrochloride | Dual Na+/Ca2+ channel (T-type) blocker | 16 |
| 10 and 20 μM | 627536-09-8 | SD 208 | Potent ATP-competitive transforming growth factor beta receptor I (TGF-βRI) inhibitor | 63 |
| 152121-47-6 | SB203580 | Selective inhibitor of p38 mitogen-activated protein kinase (MAPK) | 64, 65 | |
| 152121-30-7 | SB202190 | Potent, selective inhibitor of p38 MAPK | 14, 66 | |
| 869185-85-3 | SB203580 hydrochloride | Selective inhibitor of p38 MAPK | 64, 65 | |
| 129453-61-8 | ICI 182,780 | Estrogen receptor antagonist | 13 | |
| 138356-20-4 | BD 1047 dihydrobromide | σ1 selective antagonist | 12 | |
| 138356-09-9 | BD 1008 dihydrobromide | Potent, selective σ ligand | 12 | |
| 35838-58-5 | Etazolate hydrochloride | PDE4 inhibitor | ||
| 67197-96-0 | (−)-U-50488 hydrochloride | Standard selective κ agonist | ||
| 114528-79-9 | (±)-U-50488 hydrochloride | Standard selective κ agonist | ||
| 207403-36-9 | MR 16728 hydrochloride | Stimulates ACh release | ||
| 155059-42-0 | SM-21 maleate | Presynaptic cholinergic modulator | ||
| 155649-00-6 | PG-9 maleate | Presynaptic cholinergic modulator | ||
| 209480-63-7 | SB258585 hydrochloride | Potent, selective 5-HT6 antagonist | ||
| 66611-27-6 | RU 24969 hemisuccinate | 5-HT1B/1A agonist | ||
| 61718-82-9 | Fluvoxamine maleate | 5-HT reuptake inhibitor | ||
| 59729-32-7 | Citalopram hydrobromide | Highly potent and selective 5-HT uptake inhibitor |
CAS numbers of the compounds, as well as their main known targets, are indicated. Compounds described as having an inhibitory effect on HCV or targeting a pathway described to be involved in HCV infection are indicated.
SB258585 inhibits HCV infection, downregulating PKA activity in a manner independent of the 5-HT6 receptor.
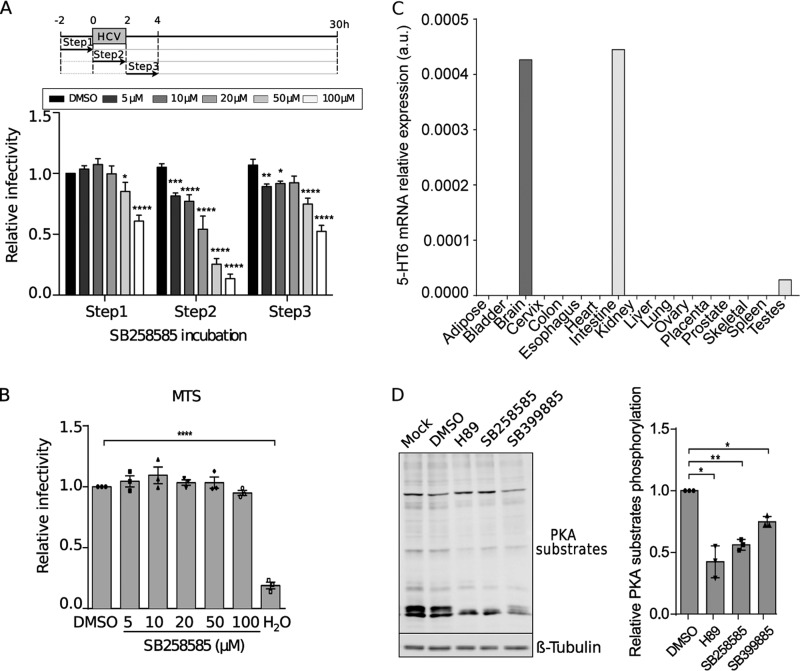
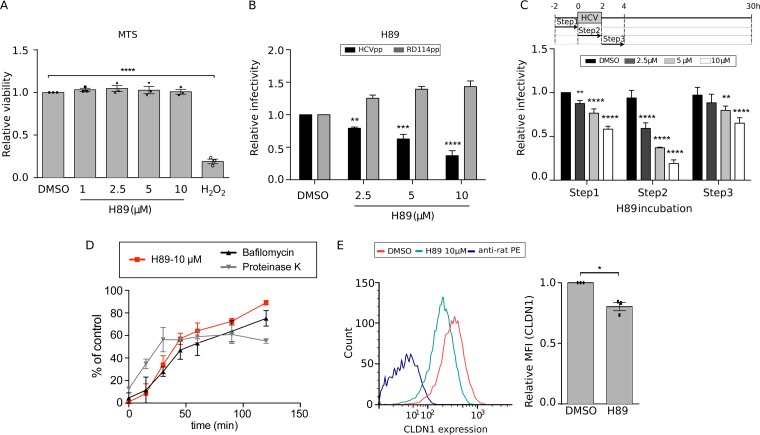
In order to confirm the inhibitory effect of the 5-HT6 antagonist SB258585 on the HCV viral cycle, kinetics of JFH1 infection were determined in the presence of this compound. The maximum inhibitory effect of SB258585 was observed when the chemical compound was added during the inoculation of the virus (Fig. 2A), suggesting an effect on virus entry. As shown in Fig. 2B, the inhibition was not due to drug toxicity.
FIG 2.
SB258585 inhibits HCV infection, modulating PKA in a 5-HT6-independent manner. (A) Huh-7 cells were treated with SB258585 at different concentrations for 2 h prior to, during, or after HCVcc JFH1 incubation, following the schematized kinetics. Infection at 30 h postinfection was quantified by immunofluorescence assay. (B) Huh-7 cells were treated with the drug for 2 h, followed by 28 h of rest. An MTS assay was performed in order to evaluate cell toxicity. (C) Quantification of 5-HT6 mRNA levels in 17 tissues from human biopsy specimens by qRT-PCR. a.u., arbitrary units. (D) Huh-7 cells were treated for 2 h with DMSO, H89 (10 μM), SB258585 (100 μM), or SB399885 (100 μM). A representative Western blot (n = 3) and relative quantification of the total phosphorylation of PKA substrates normalized to the loading control (β-tubulin) are presented. Results are presented as means ± SEM (n = 3) in panels A, B, and D. One-way (B and D) or two-way (A) analysis of variance (ANOVA) followed by the Dunnett or Bonferroni posttest was performed for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001.
The 5-HT6 receptor, mainly studied in the central nervous system, has not been characterized for the liver or hepatocytes. In order to determine its real involvement in HCV infection as a target of SB258585, we quantified its expression level in the liver. To do so, we compared its distribution in 17 different human tissues by quantitative reverse transcription-PCR (qRT-PCR). This analysis showed that 5-HT6 was highly expressed in brain tissues and the intestine (Fig. 2C). It was also expressed in testes, while it was not detected in all the other tissues, including the liver (Fig. 2C). Quantification of 5-HT6 mRNA in Huh-7 cells by qRT-PCR showed a ΔCT value of around 18 for comparison to the housekeeping gene RPLP0, confirming an almost complete absence of detection of 5-HT6 in this hepatic cell line. Not surprisingly, we were unable to detect the 5-HT6 protein by Western blotting and flow cytometry by using different antibodies (data not shown). This observation implies that the effect observed on HCV infection is probably not connected to 5-HT6.
5-HT6 is a G-protein-coupled receptor (GPCR) associated with a G alpha stimulatory protein (Gαs). This Gαs activates the adenylyl cyclase to produce cAMP, which in turn activates PKA (18). However, GPCR agonists and antagonists often show affinity for other GPCRs in addition to the one specifically targeted. Therefore, we evaluated whether the presence of the 5-HT6 antagonist leads to a regulation of the PKA pathway, likely through modulation of other GPCRs. We thus performed Western blotting with an antibody specific for PKA-phosphorylated substrates. A cell-permeating inhibitor of cAMP-dependent PKA, H89, was used as a positive control. This compound was described to inhibit PKA by competitive binding to the ATP site on the PKA catalytic subunit (19). As shown in Fig. 2D, SB258585 reduced the phosphorylation level of PKA substrates in a manner similar to that of the PKA inhibitor H89.
Therefore, our observations about the phosphorylation levels of PKA substrates suggest that the off-target effect of SB258585 targets a factor involved in PKA activation, likely another GPCR coupled to a Gαs protein.
SB258585 and SB399885 inhibit a late step of HCV entry, altering cell surface localization of CLDN1.
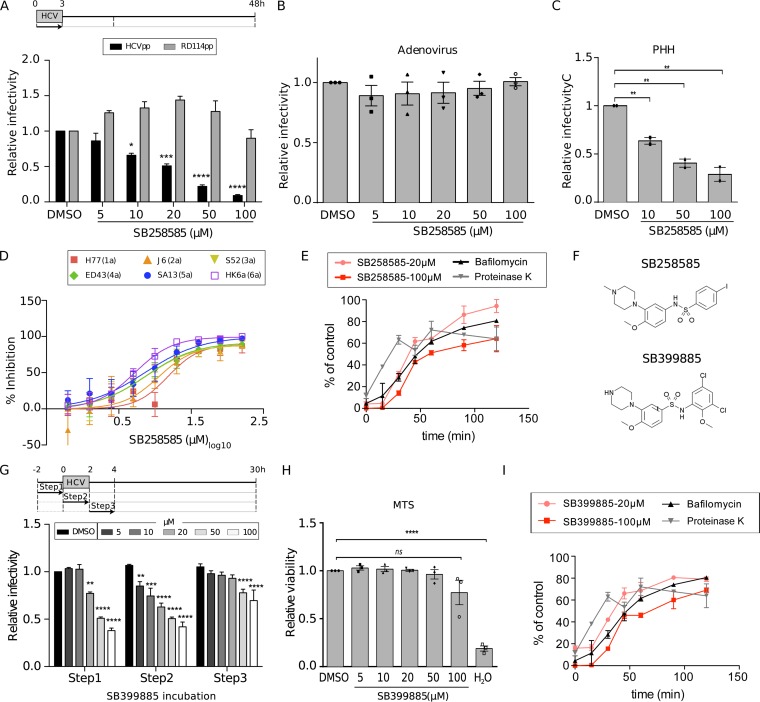
According to the kinetics shown in Fig. 2A, SB258585 seems to inhibit HCV entry. The inhibitory effect of SB258585 on HCV entry was then validated with the help of retroviral pseudoparticles harboring HCV envelope glycoproteins (HCVpp) from strain JFH1 (genotype 2a) (Fig. 3A). SB258585 had no effect on adenovirus infection, indicating that this compound does not have a global effect on viral entry (Fig. 3B). We also confirmed its inhibitory effect on primary human hepatocytes (PHHs) infected by HCV strain JFH1 (genotype 2a) (Fig. 3C) (20), as well as on 18 different JFH1-based core-NS2 strain recombinants representing the six major HCV genotypes and important subtypes (Fig. 3D and Table 2) (21–27).
FIG 3.
Piperazinylbenzenesulfonamides inhibit a late step of HCV entry for all the major HCV genotypes. (A) Huh-7 cells were treated for 3 h with SB258585 at different concentrations, concomitant with HCVpp JFH1 or RD114pp incubation. At 48 h postinfection, cells were lysed and luciferase activity measured in order to quantify the infection. (B) Huh-7 cells were incubated for 2 h with SB258585 at different concentrations, along with a GFP-expressing adenovirus. The percentage of infection was determined by quantifying GFP-positive cells at 24 h postinfection. (C) Primary human hepatocytes (PHHs) were treated for 2 h with SB258585 at different concentrations, in addition to infection with HCVcc JFH1. Infection was quantified by qRT-PCR at 24 h postinfection. (D) Huh-7.5 cells were treated for 3 h with SB258585, in addition to infection with JFH1-based HCV genotype 1 to 6 recombinant viruses with strain-specific core-NS2. Infection was quantified by immunofluorescence assay at 48 h postinoculation. Error bars represent standard deviations (SD). (E) After 1 h of viral attachment at 4°C, Huh-7 cells were shifted to 37°C and treated with SB258585 at the indicated concentrations, following 15-min kinetics for 2 h. Proteinase K (50 μg/ml) and bafilomycin A (25 nM) were used as controls for early and late entry steps, respectively. Infection was quantified at 30 h postinfection by immunofluorescence assay, and the value for each time point was normalized to that for the corresponding DMSO condition. (F) Chemical structures of SB258585 and SB399885. (G) Huh-7 cells were treated with SB399885 at different concentrations for 2 h prior to, during, or after HCVcc JFH1 incubation, following the schematized kinetics. Infection was quantified at 30 h postinfection by immunofluorescence assay. (H) Huh-7 cells were treated with SB399885 for 2 h, followed by 28 h of rest. An MTS assay was performed in order to evaluate cell toxicity. (I) After 1 h of viral attachment at 4°C, Huh-7 cells were shifted to 37°C and treated with SB399885 at the indicated concentrations, following 15-min kinetics for 2 h. Proteinase K (50 μg/ml) and bafilomycin A (25 nM) were used as controls for early and late entry steps, respectively. Infection was quantified at 30 h postinfection by immunofluorescence assay, and the value for each time point was normalized to that for the corresponding DMSO condition. Results are presented as means ± SEM (n = 3 [A to D, G, and H] and n = 2 [E and I]). PHH results are presented as means for triplicates ± SEM for two independent experiments. One-way (B, C, and H) or two-way (A and G) ANOVA followed by the Dunnett or Bonferroni posttest was performed for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001; ns, nonsignificant.
TABLE 2.
IC50s of SB258585 calculated for Huh-7.5 cells infected with the indicated JFH1-based HCV genotype 1 to 6 recombinant viruses with strain-specific core-NS2a
| Virus (genotype) | IC50 (μM) |
|---|---|
| H77 (1a) | 15.03 |
| TN (1a) | 12.44 |
| DH6 (1a) | 6.971 |
| J4 (1b) | 8.468 |
| DH1 (1b) | 5.585 |
| DH5 (1b) | 11.65 |
| J6 (2a) | 11.15 |
| T9 (2a) | 17.79 |
| J8 (2b) | 12.54 |
| DH8 (2b) | 13.93 |
| DH10 (2b) | 7.108 |
| S83 (2c) | 7.919 |
| S52 (3a) | 7.436 |
| DBN (3a) | 17.06 |
| DH11 (3a) | 14.01 |
| ED43 (4a) | 7.591 |
| SA13 (5a) | 7.237 |
| HK6a (6a) | 5.234 |
| J6ΔHVR1 (2a) | 8.235 |
IC50, 50% inhibitory concentration.
In order to better understand the mechanism of action of SB258585 as an inhibitor of HCV infection, we decided to test another characterized 5-HT6 antagonist, SB399885, which shares a very similar structure (Fig. 3F). The fact that both antagonists inhibit a common step of HCV infection (Fig. 2A and 3G and H) in the absence of their main targeted receptor, modulating PKA activity (Fig. 2D), suggests that their common piperazinylbenzenesulfonamide structure is likely responsible for a common off-target effect.
To better identify the entry step targeted by these compounds, we performed a time-of-addition assay, using proteinase K and bafilomycin A as controls of early and late steps of viral entry, respectively. Proteinase K blocks viral internalization through proteolysis of viral particles exposed to the extracellular compartment, while bafilomycin A blocks endosomal acidification, resulting in the inhibition of viral fusion (28). A comparison of SB258585 and SB399885 kinetics to the kinetics of proteinase K and bafilomycin A treatment revealed an inhibition of a late step of the entry process (Fig. 3E and I). It is worth noting that the slight difference in the SB258585 and SB399885 curves obtained at 20 μM and 100 μM was not statistically significant.
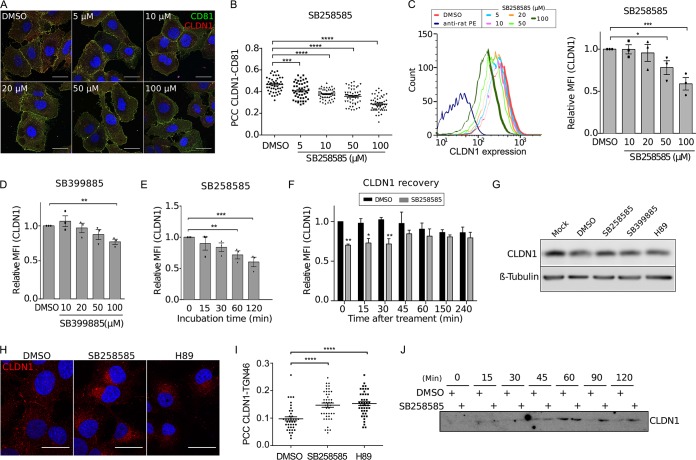
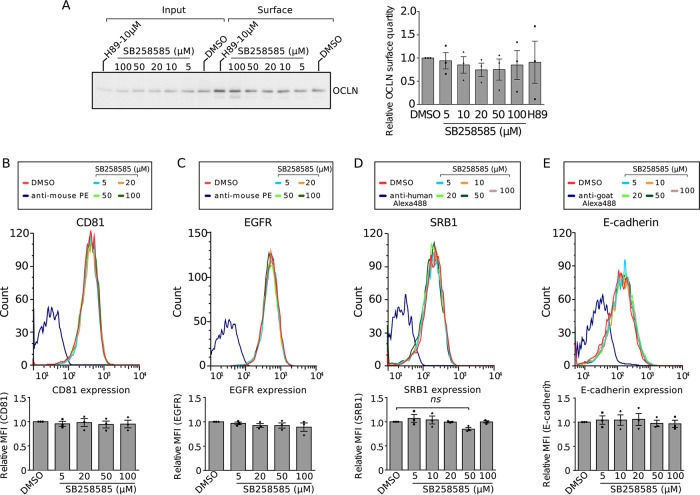
Knowing that CD81-CLDN1 interaction is necessary for clathrin-dependent endocytosis of HCV particles (8), we analyzed the effect of these piperazinylbenzenesulfonamides on CD81-CLDN1 colocalization. A dose-dependent decrease of CD81-CLDN1 colocalization was observed after treatment with SB258585, as confirmed by determining the Pearson correlation coefficient (Fig. 4A and B). Furthermore, we observed a decrease of cell surface CLDN1 expression by flow cytometry analyses of noninfected (Fig. 4C) and infected (data not shown) cells treated with SB258585. This could explain the alteration in CD81-CLDN1 colocalization. The same phenotype was also observed in cells treated with SB399885 (Fig. 4D). Furthermore, kinetic analyses revealed that the decrease of CLDN1 cell surface expression after SB258585 treatment was quite rapid and reversible (Fig. 4E and F). It is worth noting that this compound had no effect on the distribution of other major HCV entry coreceptors, including CD81 (Fig. 5).
FIG 4.
SB258585 alters CLDN1 recycling, causing its intracellular accumulation. (A) Huh-7 cells were treated for 2 h with DMSO or increasing concentrations of SB258585. Cell surface expression of CD81 and CLDN1 was analyzed by immunofluorescence assay. Images were taken using a Zeiss LSM-880 microscope and a 63× objective. (B) Pearson correlation coefficients (PCCs) were calculated for cell surface ROIs for at least 40 different cells for each condition. (C) Huh-7 cells were treated for 2 h with DMSO or increasing concentrations of SB258585, and CLDN1 expression was analyzed by flow cytometry. Curves from a representative experiment are shown. Mean fluorescence intensities (MFI) relative to that for the DMSO-treated condition are also presented. (D) Huh-7 cells were treated for 2 h with DMSO or increasing concentrations of SB399885, and CLDN1 expression was analyzed by flow cytometry. (E) Huh-7 cells were incubated with SB258585 (100 μM) for the indicated periods. CLDN1 present at the cell surface was quantified by flow cytometry. (F) Huh-7 cells were treated for 2 h with SB258585 (100 μM). The drug was then removed and replaced by DMEM for the indicated times. Cytometry analyses were performed to quantify CLDN1 at the cell surface. For panels D to F, mean fluorescence intensities relative to those for the DMSO-treated condition are shown. (G) Huh-7 cells were treated for 2 h with DMSO, SB258585 (100 μM), SB399885 (100 μM), or H89 (10 μM). The total quantity of CLDN1 was assessed by Western blotting. β-Tubulin was used as a loading control. (H) Huh-7 cells were treated for 2 h with DMSO, SB258585 (100 μM), or H89 (10 μM). CLDN1 subcellular localization was determined by immunofluorescence assay after membrane permeabilization. Images were taken with a 63× objective. (I) TGN46 was stained concomitantly with CLDN1, and PCCs were calculated for intracellular CLDN1-TGN46 colocalization for >35 cells for each condition. (J) After surface biotinylation, Huh-7 cells were incubated at 37°C with DMSO or SB258585 (100 μM) for the indicated times. Biotin remaining at the cell surface was cleaved by use of glutathione. The amount of internalized CLDN1 was determined by Western blotting after pulldown of biotin-labeled proteins with streptavidin-agarose beads. A representative Western blot (n = 3) is presented. All results are presented as means ± SEM (n = 3). One-way ANOVA (B to E and I) or two-way ANOVA (F) followed by the Dunnett or Bonferroni posttest was performed for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001. Bars = 30 μm.
FIG 5.
SB258585 does not alter surface localization of the other main HCV entry factors. (A) Huh-7 cells were treated for 2 h with DMSO or increasing concentrations of SB258585. Surface biotinylation followed by biotin immunoprecipitation was performed, and a representative Western blot for OCLN is shown. The quantity of biotinylated OCLN was determined from Western blots by use of Fiji (n = 3). (B to E) Huh-7 cells were treated for 2 h with DMSO or increasing concentrations of SB258585. CD81 (B), EGFR (C), SRB1 (D), and E-cadherin (E) expression levels were analyzed by flow cytometry. Curves from a representative experiment are shown. Mean fluorescence intensities relative to those for the DMSO-treated condition are presented. All results are presented as means ± SEM (n = 3). One-way ANOVA followed by the Dunnett posttest was performed for statistical analysis. ns, nonsignificant.
Alteration of cell surface expression of CLDN1 mediated by SB258585 is due to intracellular accumulation of CLDN1.
Decreased CLDN1 cell surface expression may be due to either CLDN1 degradation or intracellular accumulation. To determine which was the case in our study, we first analyzed the total level of CLDN1 by Western blotting. Treatment of Huh-7 cells with SB258585 and SB399885 did not affect the amount of CLDN1 (Fig. 4G), indicating that these compounds do not induce CLDN1 degradation. Instead, immunofluorescence analyses (IFA) showed an intracellular accumulation of CLDN1 in the presence of SB258585 (Fig. 4H), which was confirmed by an increased colocalization with the intracellular markers TGN46 (Fig. 4I), ERGIC53, and GM130 (data not shown). This intracellular accumulation may have been due to either an increase in CLDN1 endocytosis or an alteration in CLDN1 export. To elucidate which was true, we used an internalization assay based on cell surface biotinylation to determine the kinetics of CLDN1 endocytosis in the presence or absence of the drug. As shown in Fig. 4J, CLDN1 endocytosis was maximal after 1 h. The protein amounts were comparable for DMSO- and SB258585-treated cells, suggesting no alteration of CLDN1 endocytosis. Conversely, at later time points, biotinylated CLDN1 disappeared from DMSO-treated cells, suggesting that it had either been degraded or been recycled to the cell surface, while it remained stored intracellularly in the presence of the compound. This is in agreement with the accumulation of CLDN1 detected by immunofluorescence assay (Fig. 4H). Since no change in total CLDN1 level was observed between DMSO and SB258585 treatments (Fig. 4G), it is unlikely that endocytosed CLDN1 is degraded. Rather, CLDN1 must be recycled to the plasma membrane. The accumulation of intracellular CLDN1 observed after SB258585 treatment (Fig. 4H to J) suggests a defect of CLDN1 recycling and explains the decrease of CLDN1 at the cell surface.
PKA inhibition affects a late HCV entry step and regulates CLDN1 trafficking.
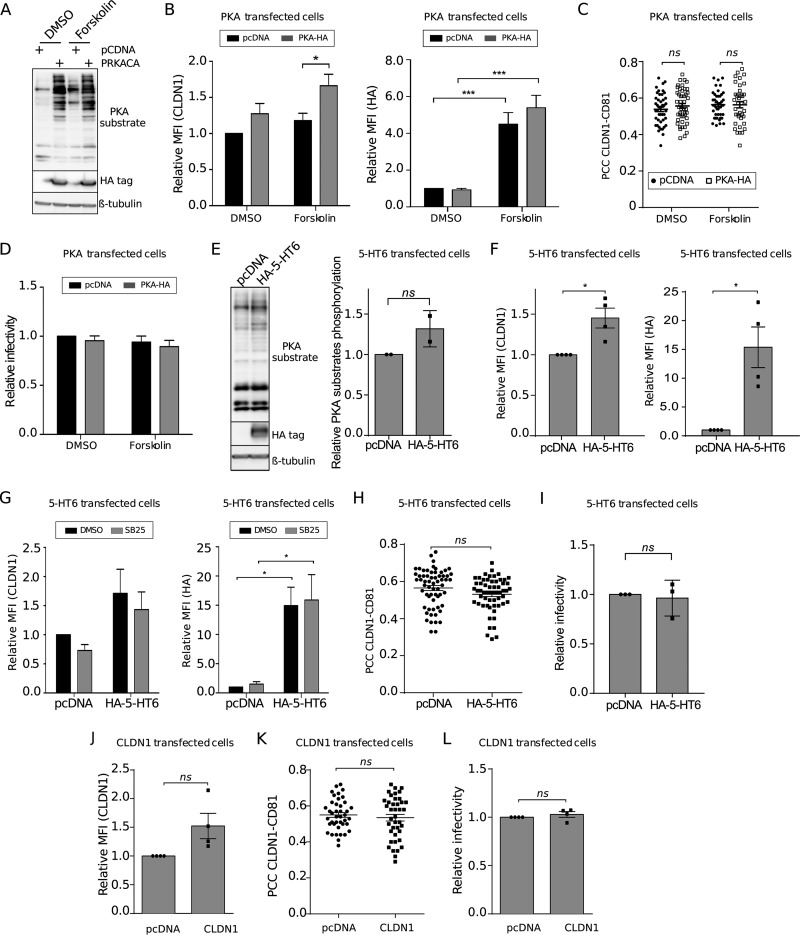
To further confirm whether SB258585's inhibitory effect on HCV could be mediated by the PKA downstream pathway, we tested the effect of the PKA inhibitor H89 on HCV infection. As reported by Farquhar and coworkers (29), we confirmed a dose-dependent inhibitory effect of this compound on both HCVpp and HCVcc of strain JFH1 (genotype 2a) at nontoxic concentrations (Fig. 6A to C). We then used a time-of-addition assay with increasing concentrations of H89, and our data indicated an inhibitory effect at a late step of HCV entry (Fig. 6D). Furthermore, as observed for the two piperazinylbenzenesulfonamides, H89 treatment also induced a decrease of cell surface expression of CLDN1 (Fig. 6E), without affecting its total level of expression (Fig. 4G) but rather resulting in its intracellular accumulation (Fig. 4H to I). In addition, transfection of an exogenous catalytic subunit of PKA (PRKACA) followed by stimulation of PKA with the activator of adenylyl cyclase forskolin also increased the phosphorylation levels of PKA substrates (Fig. 7A). Interestingly, PKA stimulation caused an increase in cell surface expression of CLDN1 but no increase in CLDN1-CD81 colocalization (Fig. 7B and C). In agreement with these data, no increase in HCV infection was observed after activation of PKA (Fig. 7D).
FIG 6.
Inhibition of the PKA signaling pathway downregulates HCV entry and CLDN1 cell surface localization. (A) Huh-7 cells were treated for 2 h with H89 at different concentrations. An MTS assay was performed at 30 h posttreatment in order to evaluate the cell toxicity of the compound. (B) Huh-7 cells were treated for 3 h with H89 at different concentrations, along with infection by HCVpp JFH1 and RD114pp. At 48 h postinfection, cells were lysed and luciferase activity measured in order to quantify the infection. (C) Huh-7 cells were treated with H89 at different concentrations, in accordance with the schematized kinetics, along with HCVcc JFH1 infection. Infection was quantified at 30 h postinfection by immunofluorescence assay. (D) After 1 h of viral attachment at 4°C, Huh-7 cells were shifted to 37°C and treated with H89 (10 μM), following 15-min kinetics for 2 h. Proteinase K (50 μg/ml) and bafilomycin A (25 nM) were used as controls for early and late entry steps, respectively. Infection was quantified at 30 h postinfection by immunofluorescence assay, and the value for each time point was normalized to that for the corresponding DMSO condition. (E) Huh-7 cells were treated for 2 h with DMSO or H89 (10 μM). Cell surface CLDN1 was analyzed by flow cytometry. Curves from a representative experiment and mean fluorescence intensities relative to that for the DMSO-treated condition are shown. Results are presented as means ± SEM (n = 3 [A to C and E] and n = 2 [D]). Two-tailed Student's t test (E) or one-way (A) or two-way (B and C) ANOVA followed by the Dunnett or Bonferroni posttest was performed for statistical analysis. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001.
FIG 7.
PKA activation increases CLDN1 localization at the cell surface, without increasing HCV entry or CLDN1-CD81 colocalization. (A to D) Huh-7 cells were transfected with pcDNA or pcDNA-PRKACA for 48 h. Cells were treated for 2 h with DMSO or forskolin (20 μM), as indicated. (A) A representative Western blot (n = 3) is presented in order to show the phosphorylation of PKA substrates. β-Tubulin was used as a loading control, and PKA transfection was checked by HA immunoblotting. (B) Cytometry analysis was performed to quantify CLDN1 cell surface localization and PKA-HA transfection. Mean fluorescence intensities relative to that for the DMSO-treated condition are shown (n = 3). (C) CLDN1 and CD81 were analyzed by immunofluorescence assay, and PCCs were calculated after confocal image acquisition for cell surface ROIs for at least 40 different cells for each condition. (D) Transfected cells were treated for 2 h with DMSO or forskolin (20 μM), together with HCVcc infection. Infection was quantified at 30 h postinfection by immunofluorescence assay. (E to I) Huh-7 cells were transfected with pcDNA or pcDNA-5-HT6 for 48 h. (E) A representative Western blot (n = 2) and relative quantification of the total phosphorylation of PKA substrates normalized to the loading control (β-tubulin) are presented. HA–5-HT6 transfection was checked by HA immunoblotting. (F and G) Cell surface CLDN1 and HA–5-HT6 were immunolabeled and quantified by flow cytometry. Mean fluorescence intensities relative to that for the DMSO-treated condition are shown. For panel G, transfected Huh-7 cells were treated for 2 h with SB258585 (100 μM) before labeling for flow cytometry analyses. (H) CLDN1 and CD81 immunostaining followed by confocal microscopy allowed for the calculation of PCCs for cell surface ROIs for at least 40 different cells for each condition. (I) Transfected Huh-7 cells were infected with HCVcc for 2 h. Infection was quantified at 30 h postinfection by immunofluorescence assay. (J to L) Huh-7 cells were transfected with pcDNA or pcDNA-CLDN1 for 48 h. Cell surface CLDN1 was immunolabeled and quantified by flow cytometry. (J) Mean fluorescence intensities relative to that for the DMSO-treated condition are shown. (K) CLDN1 and CD81 immunostaining followed by confocal microscopy allowed for the calculation of PCCs for cell surface ROIs for at least 40 different cells for each condition. (L) Transfected cells were infected with HCVcc at 48 h posttransfection. Infection was quantified at 30 h postinfection by immunofluorescence assay. Results are presented as means ± SEM (n = 3 [C, D, H, and I] or n = 4 [B, F, G, and J to L] independent experiments). Two-tailed Student's t test (E, F, and H to L) or two-way ANOVA (B, D, and G) followed by the Bonferroni posttest was performed for statistical analysis. *, P < 0.05; ***, P < 0.001; ns, nonsignificant.
Similar experiments were performed to activate the PKA pathway by overexpression of a GPCR through exogenous transfection of a plasmid encoding 5-HT6 in Huh-7 cells. The increased phosphorylation levels of PKA substrates (Fig. 7E) suggested that the exogenous receptor is functional and able to activate the PKA downstream pathway. As observed after PKA transfection, overexpression of 5-HT6 led to an increase in CLDN1 cell surface expression (Fig. 7F), as also observed upon SB258585 treatment (Fig. 7G). However, this increase in CLDN1 cell surface localization did not correlate with an increase in CLDN1-CD81 colocalization (Fig. 7H) or HCV infectivity (Fig. 7I).
Together these data strongly suggest that PKA regulates CLDN1 localization at the cell surface. However, its activation does not seem to be sufficient to mediate CLDN1 interaction with CD81, which is necessary for HCV entry. Similarly, an increase of CLDN1 cell surface expression without modification of CLDN1-CD81 interaction and HCV infection was observed after overexpression of CLDN1 (Fig. 7J to L). The failure to enhance HCV infection upon CLDN1 overexpression is in accordance with previous data (5). Together these results suggest that decreasing CLDN1 at the plasma membrane has a strong impact on HCV entry, while increasing CLDN1 alone has no effect, probably due to a lack of recruitment to its main partner, CD81.
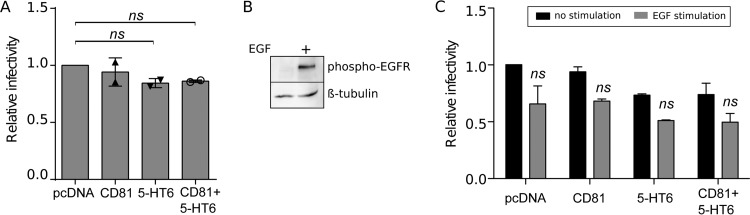
One potential explanation for the failure to increase CLDN1-CD81 colocalization is the possibility of a limited availability of endogenous CD81. To address this hypothesis, we overexpressed this tetraspanin concomitantly with the GPCR 5-HT6. However, no enhancement of HCV infection was observed (Fig. 8A), suggesting that increasing the available quantities of both coreceptors was not sufficient to enhance their interaction and, consequently, viral entry.
FIG 8.
An increase of CLDN1 localization at the cell surface is not sufficient to increase HCV entry upon either CD81 overexpression or EGF stimulation. (A) Huh-7 cells were transfected with pcDNA, HA–5-HT6, or CD81-YFP alone or in combination, as indicated. At 48 h posttransfection, cells were infected with HCVcc JFH1. At 30 h postinfection, cells were fixed and the infection rate was determined by IFA. (B) After 2 h of starvation, Huh-7 cells were kept nonstimulated or stimulated for 1 h with EGF (1 μg/ml). Western blotting was performed to verify the activation of EGFR through phosphorylation. β-Tubulin was used as a loading control. (C) Huh-7 cells transfected for 48 h with pcDNA, HA–5-HT6, or CD81-YFP, alone or in combination, after 2 h of starvation were treated for 1 h with EGF (1 μg/ml) and infected for 2 h with HCVcc JFH1 concomitantly with EGF (1 μg/ml). At 30 h postinfection, cells were fixed and the infection rate was determined by IFA. Results are presented as means ± SEM (n = 2 [A and C]). One-way (A) or two-way (C) ANOVA followed by the Dunnett or Bonferroni posttest was performed for statistical analysis. ns, nonsignificant.
Epidermal growth factor receptor (EGFR) signaling has been described as important for CLDN1-CD81 interaction (30, 31). To verify if our phenotype was due to an insufficient activation of this receptor and its downstream pathway, we treated Huh-7 cells with EGF. Although the stimulation induced the phosphorylation of EGFR and thus its activation (Fig. 8B), no enhancement of infectivity, but rather a slight decrease, was observed in our hands after the increase of cell surface CLDN1 induced by transfection of the 5-HT6 GPCR (Fig. 8C). Therefore, an additional signal upstream of EGFR is likely necessary to relocalize CLDN1 to the site of interaction with CD81.
The C-terminal region of CLDN1 is not involved in PKA-mediated inhibition.
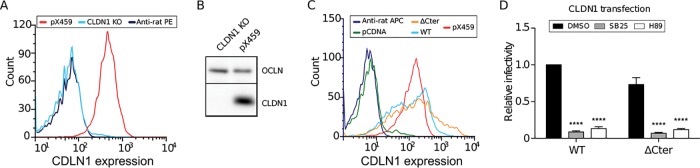
Predicted phosphorylation sites in the cytosolic domains of CLDN1 are present only in the C-terminal region of CLDN1 (32) and may be responsible for its localization. A mutant in which the cytosolic C terminus of CLDN1 was deleted was previously described to remain functional for HCV entry (5). We therefore used this mutant to determine if the effect of 5-HT6 activation antagonists is maintained if the CLDN1 C-terminal region is deleted. For this purpose, we generated a clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 CLDN1 knockout (KO) cell line (Fig. 9A and B), and we reexpressed wild-type or C-terminally deleted CLDN1 (ΔCter) in this cell line (Fig. 9C and D). After verification that the transfection levels and surface localization of the two constructs were comparable (Fig. 9C), we infected the cells with HCV in the presence or absence of SB258585. As shown in Fig. 9D, the absence of the C terminus of CLDN1 did not alter the sensitivity of HCV infection to SB258585 or H89, suggesting that the CLDN1 C-terminal cytosolic tail is not a direct target for PKA activity and is not required for its recycling.
FIG 9.
Deletion of the CLDN1 C-terminal region does not alter HCV sensitivity to SB258585 and H89. Flow cytometry (A) and Western blotting (B) were performed on CLDN1 CRISPR/Cas9 and control pX459 cells in order to evaluate CLDN1 silencing. OCLN was used as a loading control for the Western blot. (C and D) CLDN1 CRISPR/Cas9 cells were transfected with pcDNA 3.1, wild-type CLDN1 (CLDN1 WT), or CLDN1 ΔCter. At 48 h posttransfection, cells were labeled for CLDN1 at the cell surface and analyzed by flow cytometry (C) or infected with HCVcc JFH1 (D). At 30 h postinfection, cells were fixed and the infection rate was determined by IFA. Results are presented as means ± SEM (n = 3). Two-way ANOVA (D) followed by the Bonferroni posttest was performed for statistical analysis. ****, P < 0.001.
DISCUSSION
HCV entry is a highly complex multistep process involving a series of cellular proteins. Trafficking of these entry factors provides an additional level of complexity in the regulation of HCV entry. In the present study, by screening a chemical compound library, we identified an antagonist of 5-HT6, SB258585, as a novel inhibitor of the HCV entry process. This drug and another 5-HT6 antagonist, SB399885, with which it shares similarities in structure, inhibit a postbinding step of HCV entry in a 5-HT6-independent manner. These two drugs induced a decrease in recycling of the HCV entry coreceptor CLDN1 and its colocalization with CD81. The resulting reduction of CLDN1-CD81 interaction, known to be critical for HCV internalization, likely explains the drop of HCV infectivity. Treatment with these drugs showed a reduction in PKA activity, and the involvement of this kinase-related pathway was confirmed to regulate CLDN1 localization and a late HCV entry step.
The attempt to detect 5-HT6 in liver biopsy specimens and hepatocytes was unsuccessful. This failure to detect 5-HT6 mRNA is supported by other reports (33). In addition, SB258585 and SB399885 are supposed to inhibit 5-HT6 in a highly specific way at nanomolar concentrations. Therefore, the fact that in our hands the inhibition is observed only at micromolar concentrations supports the idea of an effect on another target, for which the drug shows less affinity. However, although the major target of these drugs in our model is unlikely to be 5-HT6, these chemical compounds inhibit PKA activity, suggesting an inhibitory effect on one or several other GPCRs, likely associated with a Gα stimulatory protein. According to Stitch 5.0 prediction (17), SB258585 shows an affinity for other 5-HT receptors, including the GPCR coupled to the Gαs 5-HT7, recently shown to play a role in liver fibrosis (33). Unfortunately, no data are available concerning SB399885, although the common structure may suggest similar low-affinity targets.
PKA likely controls the export of CLDN1. Indeed, a downregulation of PKA activity reduces the cell surface expression of CLDN1, whereas its activation shows a direct increase of this tight junction protein at the cell surface, confirming a direct correlation between the PKA pathway and CLDN1 subcellular localization. The 5-HT6 receptor is a GPCR associated with a Gα stimulatory protein and is known to activate the PKA pathway (34). Therefore, it is not surprising that its overexpression stimulates PKA signaling and the export of the tight junction protein CLDN1, similar to what happens upon direct PKA transfection or activation. Therefore, this experiment reinforces the hypothesis of a role for GPCRs in regulating PKA signaling in hepatocytes. This is in line with a previous report showing that PKA inhibitors affect HCV entry (29).
Activation of the PKA pathway by either forskolin treatment, overexpression of the exogenous GPCR 5-HT6, or overexpression of the PKA catalytic subunit itself increased CLDN1 export at the surface or rescued its surface localization in cells treated with SB258585. However, this was not sufficient to increase the basal level or to rescue HCV infection. Similarly, overexpression of CLDN1 led to an increase in CLDN1 expression at the cell surface without increasing HCV entry. In our case, in addition to analyzing CLDN1 expression at the cell surface and HCV infection, we also monitored CD81-CLDN1 colocalization, which was not rescued despite reexport of CLDN1 at the cell surface. Thus, there seems to be a disconnect between the level of CLDN1 expressed at the cell surface and CD81-CLDN1 interaction. These observations are in accordance with Farquhar's work (29), in which treatment with forskolin did not enhance HCVcc entry. Since CLDN1 interaction with the tetraspanin CD81 is necessary to mediate HCV endocytosis (6, 7), this may explain the lack of rescue of HCV infection. Why CLDN1-CD81 colocalization is not restored upon PKA activation remains unclear. One possibility is that in rescue experiments, despite the restoration of cell surface expression of CLDN1, CD81 is no longer available to interact with CLDN1. The interaction between these two proteins is described to be regulated by the EGF receptor through HRas signaling (30, 31). However, neither overexpression of CD81 concomitantly with the GPCR 5-HT6 nor EGF stimulation restored the phenotype. This suggests that an additional signal upstream of EGFR is needed to target CLDN1 to the site of interaction with CD81. Further investigations are therefore required to better characterize the nature and dynamics of the interaction between CLDN1 and CD81. However, we cannot exclude the possibility that this defect in a late HCV entry step is not due only to the decreased interaction between CLDN1 and CD81. In addition to its involvement during exocytosis, there is in fact evidence of a role for the cAMP/PKA pathway during endocytosis (reviewed in reference 35). Therefore, the defect in HCV entry may be associated not only with the CLDN1 localization change but also with a general perturbation of membrane trafficking directly affecting HCV endocytosis. Further investigations are needed in order to elucidate the mechanism connecting PKA pathway activation and endocytosis to better clarify its additional involvement in HCV entry.
Regulation of CLDN1 trafficking in hepatocytes remains poorly characterized. However, the CLDN1 C-terminal cytosolic tail presents several potential sites of posttranslational modification, including phosphorylation and ubiquitination, which may play a role in its trafficking (36). Indeed, other reports indicate a role for the phosphorylation of the CLDN1 cytosolic tail in the regulation of its subcellular localization in nonhepatic cells (37–39) or through a competition between phosphorylation and ubiquitination (40), as described for other members of the CLDN family (41, 42). However, in our experimental work, the absence of the C terminus of CLDN1 did not alter the sensitivity of HCV infection to 5-HT6 and PKA antagonists, excluding a role for direct phosphorylation on CLDN1. Therefore, an additional, as-yet-unidentified intermediate protein, likely phosphorylated by PKA, is probably involved in regulation of CLDN1 localization in the hepatocyte. However, cAMP/PKA pathway activation has been shown to stimulate exocytosis of several proteins toward both apical and basolateral surfaces of hepatocytes (reviewed in reference 35), suggesting a conserved mechanism of export.
Several compounds targeting other GPCRs were identified as HCV inhibitors in our screen. Previous studies described antihistamines, neuroleptic drugs, and other ligands of GPCRs as inhibitors of HCV entry (16, 43, 44). In the future, it would be interesting to test if the viral resistance mutations against any of these drugs eventually confer cross-resistance to piperazinylbenzenesulfonamides. Moreover, the identification of compounds targeting GPCRs in HCV screens may be in accordance with data from a recent chemical compound screen performed on Ebola and Marburg viruses that identified the involvement of several antagonists of GPCRs, including 5-HT receptors, as inhibitors of a postbinding step of viral entry (45). In addition, connections between 5-HT receptors and viral infections have already been documented for several viruses, such as reovirus, JC polyomavirus, chikungunya virus, and mouse hepatitis virus, especially for postbinding steps of viral entry (46–49). Several chemical compounds targeting 5-HT receptors were identified as HCV inhibitors in other HCS screens (50, 51). This suggests that a possible common mechanism, potentially affected by drugs targeting 5-HT receptors, might be used by different viruses. In this context, CLDN1 might not be the only protein whose trafficking is altered by modulation of a 5-HT pathway. Interestingly, in immune cells treated with an inhibitor of 5-HT uptake, the HIV entry receptor and coreceptor were also observed to be downregulated at the cell surface (52); however, the mechanism was not investigated. Further studies are necessary in order to better clarify if there actually is a common pathway influencing the entry of all these viruses that may represent a potential drug target for wide-range antiviral development.
In summary, we identified SB258585 and SB399885, antagonists of the 5-HT6 receptor, as inhibitors of a postbinding step of HCV entry. Although we confirmed the absence of 5-HT6 in liver and hepatic cells, we proved an inhibition of PKA activity as a consequence of the treatment, suggesting an off-target effect of these drugs on another GPCR. We thus confirmed an involvement of PKA in a postbinding step of HCV entry, showing that modulation of PKA activity regulates CLDN1 localization through regulation of CLDN1 recycling toward the cell surface. We thus suggest a role for PKA in the liver, as a regulator of CLDN1 export, which affects the HCV entry process and influences viruses of the six major genotypes. Finally, our work reinforces the hypothesis of a common pathway, shared by several enveloped viruses, involving PKA-dependent signaling in late steps of viral entry.
MATERIALS AND METHODS
Cell culture.
Huh-7 and Huh-7.5 cells have been described previously (53, 54) and were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Experiments were performed in DMEM supplemented with 5% FBS. Huh-7 and Huh-7.5 cells were authenticated by the company Multiplexion, which performed a multiplex human cell line authentication (MCA) test.
For experiments using forskolin and EGF, cells were preincubated for 2 h in serum-free medium before incubation with the compound. DMEM, Opti-MEM, phosphate-buffered saline (PBS), goat serum, and FBS were purchased from Life Technologies. PHHs were purchased from Biopredic and cultured at 37°C in supplemented hepatocyte basal medium (Clonetics HCM Bullet kit; Lonza).
Viruses.
Cell culture-derived HCV (HCVcc) was produced by electroporation of Huh-7 cells with in vitro-transcribed RNA of a modified JFH1 plasmid kindly provided by T. Wakita (National Institute of Infectious Diseases, Tokyo, Japan) (11). This modified JFH1 plasmid bears some titer-enhancing mutations and reconstitution of the A4 epitope in E1 (10). Sequence-confirmed virus stocks of JFH1-based core-NS2 HCV recombinants were generated as described previously (21–27).
Antibodies.
Anti-HCV E1 mouse monoclonal antibody (MAb) A4 was produced in vitro by use of a MiniPerm apparatus (Heraeus) following the manufacturer's protocol. Anti-CD81 MAb 5A6 was kindly provided by S. Levy (Stanford University). Anti-SRB1 antibody C1671 was kindly given by A. Nicosia (Okairos, Italy). The anti-CLDN1 (OM 8A9-A3) MAb was described previously (55). Anti-phospho-Ser/Thr PKA substrate (9621), anti-hemagglutinin (anti-HA) (C29F4), and anti-phospho-EGFR (Tyr1068) (1H12; 2236) mouse MAbs were supplied by Cell Signaling Technology. Anti-EGFR (cocktail R19/48), anti-OCLN (OC-3F10), and anti-CLDN1 (51-9000) were purchased from Life Technologies. Anti-β-tubulin (T5201), anti-HA-tag (16B12), anti-E-cadherin (AF648), anti-TGN46 (AHP500G), anti-GM130 (610822), and anti-ERGIC53 (ALX-804-602) antibodies were obtained from Sigma, Covance, R&D Systems, AbD Serotec, BD Biosciences, and Enzo Life Sciences, respectively. Phycoerythrin (PE)-coupled anti-rat and anti-mouse antibodies were supplied by BD Biosciences. Cy3-conjugated anti-mouse, Alexa 488-conjugated anti-mouse, Alexa 647-conjugated anti-rabbit, horseradish peroxidase (HRP)-conjugated anti-mouse, and HRP-conjugated anti-rabbit were purchased from Jackson ImmunoResearch. Allophycocyanin (APC)-coupled goat anti-rat and anti-mouse antibodies as well as Alexa 488-conjugated anti-goat, Alexa 555-conjugated anti-rat, Alexa 488-conjugated anti-sheep, and Alexa 488-conjugated anti-human antibodies were supplied by Life Technologies.
Chemicals.
SB258585 hydrochloride (sc-361339), SB399885 hydrochloride (sc-204264), H89 dihydrochloride (sc-3537), and forskolin (sc-3562) were purchased from Santa Cruz Biotechnology. Bafilomycin A, proteinase K, DAPI (4′,6-diamidino-2-phenylindole), and puromycin dihydrochloride from Streptomyces alboniger were obtained from Sigma-Aldrich. Mowiol was purchased from Calbiochem. Dimethyl sulfoxide (DMSO), streptavidin-agarose beads from Streptomyces avidinii, and bafilomycin A1 were obtained from Sigma-Aldrich. EGF, EZ-link sulfo-NHS-SS-biotin, Dynabeads sheep anti-rat IgG, and Hoechst 33342 were purchased from Life Technologies.
Primers.
The following primers were used for this study: CLDN1-crispR-F, 5′-CACCGCTTCATTCTCGCCTTCC-3′; CLDN1-crispR-R, 5′-AAACGGAAGGCGAGAATGAAGC-3′; CLDN1-R, 5′-GTGGATCCTCACACGTAGTCTTTCCCGC-3′; CLDN1-F, 5′-TTAAGCTTGCCACCATGGCCAACGCGGGGCTGC-3′; and CLDN1-ΔCter-R, 5′-GTGGATCCTCAACAGGAACAGCAAAGTAGGG-3′.
Plasmids.
pX459-CLDN1 was generated by insertion of the described RNA guides into pSpCas9(BB)-2A-Puro (PX459) (Addgene). pcDNA3.1-CD81-YFP was generated by insertion of the yellow fluorescent protein (YFP) gene sequence into the pcDNA3.1-CD81 vector. pCDNA3.1-CLDN1 and pCDNA 3.1-CLDN1 ΔCter were generated by amplification of the CLDN1 human cDNA contained in the pCMV6-Ac vector (OriGene) by using the CLDN1-F primer and the CLDN1-R and CLDN1-ΔCter-R primers, respectively, and inserted into pCDNA3.1(+) (Invitrogen) by digestion with HindIII and BamH1. The HTR6 N-terminally 3×HA-tagged plasmid was purchased from the UMR cDNA Resource Center, University of Missouri-Rolla (HTR060TN00), and a PRKACA open reading frame (ORF)-expressing mammalian expression plasmid with a C-HA tag was purchased from Sino Biological Inc. (HG11544-CY).
Human biopsy specimens.
For mRNA expression analysis of 17 normal tissue samples (Ambion), total RNA (500 ng) was reverse transcribed using SuperScript II reverse transcriptase (Invitrogen) and random hexamer primers. Using a TaqMan gene expression assay (Life Technologies) for the 5HT-6 gene (Hs00168381_m1) and the RPLP0 gene (Hs99999902_m1) as a control, real-time PCR amplifications were run in duplicate, using LightCycler 480 probe master mix (Roche) and were performed on a LightCycler 480 instrument II (Roche) according to the MIQE guidelines (56). Samples that lacked either a template or reverse transcriptase were used as controls. The relative expression of each gene was calculated according to the 2−ΔCT quantification method, where the ΔCT value was determined by subtracting the average threshold cycle (CT) value for the target gene from the average CT value for the control gene.
HCS analysis.
A total of 1,120 biologically active compounds from the Tocriscreen Total library, at 10 mM in pure DMSO, were dispensed into 384-well μClear black plates (Greiner Bio-One) by use of an Echo 550 nanoliter acoustic liquid handler (LabCyte). Sixteen wells containing pure DMSO were used as controls. Eight hundred Huh-7 cells were seeded in 30 μl in 384-well plates and incubated with each compound at a final concentration of 1, 10, or 20 μM at 37°C and 5% CO2. Sixteen hours later, cells were infected with 30 μl of HCVcc at a multiplicity of infection (MOI) of 0.4 and incubated at 37°C and 5% CO2. After 72 h, 40 μl of the supernatant was harvested in a second set of 384-well μClear black plates containing 1,200 Huh-7 cells by use of a Bravo automated liquid handler (Agilent) and incubated at 37°C and 5% CO2 for 48 h. The infected cells in the two sets of 384-well plates were fixed for 30 min by use of a neutral buffered 10% formalin solution (Sigma-Aldrich) at room temperature. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 3 min at room temperature, and HCV-infected cells were revealed by immunofluorescence assay with anti-E1 MAb A4 (1:1,000 dilution) for 30 min at room temperature. Alexa 488-conjugated goat anti-mouse was used as a secondary antibody (1:500 dilution), and nuclei were labeled with DAPI (1:500 dilution) for 30 min at room temperature. Two fields per well were acquired by using a 20× objective (numerical aperture [NA] = 0.45) as well as an excitation laser (Ex) at 405 nm and an emission filter (Em) at 450 nm for DAPI detection and Ex at 488 nm and Em at 540 nm for virus detection on an InCell 6000 Analyzer automated confocal microscope (GE Healthcare). Images were analyzed using a dedicated image analysis method performed with Columbus software, allowing automated quantification of the number of cells and the percentage of infected cells per well. Compounds reducing the cell number by more than 35% compared to the average for DMSO control wells were considered toxic and were discarded. Compounds decreasing the percentage of infection by at least 45% and whose effect was confirmed upon reinfection were identified as positive hits. The image analysis parameters of Columbus software are available upon request.
JFH1 infection assay.
HCVcc JFH1 infection was quantified as previously described (10).
Core-NS2 HCV recombinant infection assay.
Huh7.5 cells were plated at 7,000 cells/well in poly-d-lysine-coated 96-well plates. The following day, four replicates of the described core-NS2 HCV recombinants were mixed with different concentrations of SB258585 in DMEM supplemented with 10% FBS, along with 8 replicates of virus only (without SB258585), and added to Huh7.5 cells for 3 h. Following this incubation, the cells were washed and full medium was added. After a total of 48 h post-virus inoculation, the numbers of focus-forming units were visualized by HCV-specific immunostaining using NS5A antibody 9E10 (24). Counting was automated using an ImmunoSpot series 5 UV analyzer as described previously (57), and the counts were normalized to the mean count for virus only.
PHH infection.
PHHs (Biopredic) were inoculated for 3 h with a cell culture-adapted JFH1 strain in the presence of increasing concentrations of SB258585, and infection was measured by quantification of intracellular HCV RNA at 24 h postinoculation, as previously described (20).
HCVpp and infection assays.
HCVpp bearing the E1E2 glycoproteins of strain JFH1 (genotype 2a) were produced as previously described (58). RD114pp were produced by use of a plasmid encoding the feline endogenous virus RD114 glycoprotein (59). Huh-7 cells were incubated with pseudotyped particles for 3 h at 37°C concomitant with increasing concentrations of drugs. At 48 h postinfection, firefly luciferase (FLuc) assays were performed following the manufacturer's protocol (Promega).
HCVcc entry assay.
After 2 h of JFH1 HCVcc attachment at 4°C, Huh-7 cells were washed and shifted to 37°C to induce viral internalization. Each drug was added every 15 min during a 2-h kinetics course. Proteinase K (50 μg/ml) and bafilomycin A (25 nM) were included as controls for an early and a late step of viral entry, respectively. Cells treated with proteinase K were washed in cold PBS, incubated with proteinase K for 1 h at 4°C under gentle agitation, and then washed again, and fresh medium was added at 37°C. The infection rate was calculated by immunofluorescence assay (IFA) 30 h later, and the value for each time point was normalized to that for the corresponding DMSO condition.
AdV infection.
A recombinant adenovirus (AdV) expressing green fluorescent protein (GFP) was generated as previously described (10). Huh-7 cells were inoculated with the virus for 2 h at 37°C concomitant with increasing concentrations of SB258585 and cultured for 24 h at 37°C. AdV infections were scored using an Axioplan2 epifluorescence microscope (Zeiss) after fixation and nuclear staining with Hoechst dye.
Viability assay.
Huh-7 cells were incubated for 2 h with increasing concentrations of the indicated compounds. An MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay was performed at 28 h posttreatment, according to the manufacturer's protocol (Promega), in order to evaluate cell viability.
Immunofluorescence assay and quantification of CLDN1 colocalization with markers of subcellular compartments.
Huh-7 cells previously seeded on coverslips were treated with the indicated compounds. After treatment, cells were fixed for 30 min with 3% paraformaldehyde. Cells were permeabilized for 5 min with 0.1% Triton X-100 and saturated for 30 min with PBS containing 10% goat serum. Primary antibodies were incubated for 30 min in PBS-goat serum. After three washes, secondary antibodies and DAPI were added for 30 min. After three more washes, coverslips were mounted on glass slides by use of Mowiol. Images were taken using a Zeiss LSM880 or LSM780 confocal microscope with a 63× oil-immersion objective. Pearson correlation coefficients were calculated using the Fiji coloc2 plugin on regions of interest (ROIs) from single cells, designed on the basis of cell compartment labeling. Each experiment was repeated at least three times, and at least 35 cells for each condition were quantified.
Quantification of CD81-CLDN1 colocalization.
Huh-7 cells were seeded on coverslips and treated with the indicated inhibitor. After treatment, cells were fixed for 30 min with 3% paraformaldehyde. Cells were incubated for 30 min with PBS containing 10% goat serum. Primary antibodies diluted in PBS-goat serum were added for 30 min. After three washes, secondary antibodies and Hoechst dye were added for another 30 min. After three more washes, coverslips were mounted on glass slides by use of Mowiol. Images were taken using a Zeiss LSM880 confocal microscope with a 63× oil-immersion objective. Pearson correlation coefficients were calculated using the Fiji coloc2 plugin on ROIs from single cells, designed on the basis of CD81 surface labeling. Each experiment was repeated at least three times, and a total of at least 35 cells for each condition were quantified.
Flow cytometry.
Huh-7 cells treated with the indicated compound for the indicated period were incubated for 30 min at 4°C with the indicated primary antibody diluted in cold PBS supplemented with 2% bovine serum albumin (BSA). After three washes with cold PBS-BSA, cells were incubated for 30 min at 4°C with secondary antibodies. After three more washes, cells were incubated for another 30 min with cold PBS supplemented with 2 mM EDTA and then resuspended and fixed with a formalin solution at a final concentration of 1.5% formalin. For PKA-HA labeling, cells were permeabilized for 30 min with 5% saponin, and each following step was performed in the presence of 5% saponin. Cells were analyzed using a BD FACSCanto II flow cytometer. Analyses were performed using Weasel and FlowJo software.
Surface biotinylation and endocytosis.
Biotinylation assays were performed as previously described (60). Western blotting was performed in order to detect CLDN1. Proteins were resolved by SDS-PAGE and transferred onto a nitrocellulose membrane. The membrane was immunoblotted with the indicated primary antibody followed by peroxidase-coupled secondary antibodies. Proteins were detected using a LAS3000 machine (Fuji).
cDNA transfection.
Huh-7 cells were seeded in 24-well plates and transfected with 250 ng of DNA by use of Trans-IT LT1 transfection reagent according to the manufacturer's protocol. Either treatment followed by fluorescence-activated cell sorter (FACS) analysis or infection experiments were performed at 48 h posttransfection.
RNA extraction and qRT-PCR.
RNA was extracted from Huh-7 cells by use of a NucleoSpin RNA extraction kit (Macherey-Nagel), and 2 μg was used for cDNA retrotranscription by use of a high-capacity cDNA reverse transcription kit (Life Technologies). A TaqMan predesigned gene expression assay approach (Applied Biosystems) using a TaqMan 6-carboxyfluorescein (FAM)–MGB probe (Hs00168381_m1) was used to perform qPCR in order to quantify HTR6 (50 cycles). The RPLP0 gene (Hs00420895_gH) was used as a housekeeping gene. ΔCT values were calculated as follows: ΔCT = CT(HTR6) − CT(RPLP0).
CLDN1-KO cell line.
Huh-7 cells were seeded in 6-well plates and transfected with 1 μg of pX459-CLDN1 plasmid by use of Trans-IT LT1 reagent (Mirus) according to the manufacturer's protocol. Three days after transfection, cells were selected by use of DMEM supplemented with 2 μg/ml of puromycin. The selection medium was removed after 4 days, and cells were cultured in complete DMEM. The knockout was checked by both Western blotting and flow cytometry analysis. In order to remove the residual population of cells still expressing CLDN1, selection was performed using Dynabeads sheep anti-rat IgG after labeling of the cells with anti-CLDN1 OM 8A9-A3 antibody.
Quantification and statistical analysis.
Data are presented as means ± standard errors of the means (SEM) except where specified differently. Statistical parameters and analyses are reported in the figure legends, with n representing the number of independent experiments. Statistical analyses were performed using GraphPad Prism 7.
Supplementary Material
ACKNOWLEDGMENTS
The Copenhagen Hepatitis C Program (CO-HEP) is a joint venture of the Department of Infectious Diseases and Clinical Research Centre, Hvidovre Hospital, and the Department of Immunology and Microbiology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
We thank F. L. Cosset, C. Rice, and T. Wakita for providing essential reagents. We also thank S. Ung for his help in preparing the figures and L. Linna for manuscript proofreading. We are very grateful to A. Danneels for her precious help during the revisions. Immunofluorescence and flow cytometry analyses were performed with the help of the imaging core facility of the BioImaging Center Lille Nord-de-France.
L.R., J.D., Y.R., L.C., O.S., L.L., C.-H.G., T.A., P.B., J.P., and J.B. designed the experiments. L.R., O.S., A.V., F.H., J.P., C.-H.G., L.F., and S.B. performed the experiments. L.R., O.S., L.L., F.H., J.P., and C.-H.G. analyzed the results. T.F.B. assisted in study design and contributed reagents. L.R. and J.D. wrote the manuscript, and all the authors commented on it.
This work was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) and the ANR through the ERA-NET Infect-ERA program (grant ANR-13-IFEC-0002-01). In addition, this study was supported by research grants from the Danish Council for Independent Research-Medical Sciences and the Novo Nordisk Foundation. The BioImaging Center Lille Nord-de-France is supported by the Equipex IMAGINEX program.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01982-17.
REFERENCES
- 1.Miao Z, Xie Z, Miao J, Ran J, Feng Y, Xia X. 2017. Regulated entry of hepatitis C virus into hepatocytes. Viruses 9:100. doi: 10.3390/v9050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Douam F, Lavillette D, Cosset F-L. 2015. The mechanism of HCV entry into host cells. Prog Mol Biol Transl Sci 129:63–107. doi: 10.1016/bs.pmbts.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Brazzoli M, Bianchi A, Filippini S, Weiner A, Zhu Q, Pizza M, Crotta S. 2008. CD81 is a central regulator of cellular events required for hepatitis C virus infection of human hepatocytes. J Virol 82:8316–8329. doi: 10.1128/JVI.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potel J, Rassam P, Montpellier C, Kaestner L, Werkmeister E, Tews BA, Couturier C, Popescu C-I, Baumert TF, Rubinstein E, Dubuisson J, Milhiet P-E, Cocquerel L. 2013. EWI-2wint promotes CD81 clustering that abrogates hepatitis C virus entry: dynamics and partitioning of CD81. Cell Microbiol 15:1234–1252. doi: 10.1111/cmi.12112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 6.Harris HJ, Farquhar MJ, Mee CJ, Davis C, Reynolds GM, Jennings A, Hu K, Yuan F, Deng H, Hubscher SG, Han JH, Balfe P, McKeating JA. 2008. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J Virol 82:5007–5020. doi: 10.1128/JVI.02286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris HJ, Davis C, Mullins JGL, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, Balfe P, McKeating JA. 2010. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 285:21092–21102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farquhar MJ, Hu K, Harris HJ, Davis C, Brimacombe CL, Fletcher SJ, Baumert TF, Rappoport JZ, Balfe P, McKeating JA. 2012. Hepatitis C virus induces CD81 and claudin-1 endocytosis. J Virol 86:4305–4316. doi: 10.1128/JVI.06996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. 2010. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol 84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich H-G, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesland M, Mingorance L, Chung J, Chisari FV, Gastaminza P. 2013. Sigma-1 receptor regulates early steps of viral RNA replication at the onset of hepatitis C virus infection. J Virol 87:6377–6390. doi: 10.1128/JVI.03557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashida K, Shoji I, Deng L, Jiang D-P, Ide Y-H, Hotta H. 2010. 17Beta-estradiol inhibits the production of infectious particles of hepatitis C virus. Microbiol Immunol 54:684–690. doi: 10.1111/j.1348-0421.2010.00268.x. [DOI] [PubMed] [Google Scholar]
- 14.Ishida H, Ohkawa K, Hosui A, Hiramatsu N, Kanto T, Ueda K, Takehara T, Hayashi N. 2004. Involvement of p38 signaling pathway in interferon-alpha-mediated antiviral activity toward hepatitis C virus. Biochem Biophys Res Commun 321:722–727. doi: 10.1016/j.bbrc.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 15.Mannova P, Beretta L. 2005. Activation of the N-Ras-PI3K-Akt-mTOR pathway by hepatitis C virus: control of cell survival and viral replication. J Virol 79:8742–8749. doi: 10.1128/JVI.79.14.8742-8749.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perin PM, Haid S, Brown RJP, Doerrbecker J, Schulze K, Zeilinger C, von Schaewen M, Heller B, Vercauteren K, Luxenburger E, Baktash YM, Vondran FWR, Speerstra S, Awadh A, Mukhtarov F, Schang LM, Kirschning A, Muller R, Guzman CA, Kaderali L, Randall G, Meuleman P, Ploss A, Pietschmann T. 2016. Flunarizine prevents hepatitis C virus membrane fusion in a genotype-dependent manner by targeting the potential fusion peptide within E1. Hepatology 63:49–62. doi: 10.1002/hep.28111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. 2016. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44:D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marazziti D, Baroni S, Catena Dell'Osso M, Bordi F, Borsini F. 2011. Serotonin receptors of type 6 (5-HT6): what can we expect from them? Curr Med Chem 18:2783–2790. doi: 10.2174/092986711796011283. [DOI] [PubMed] [Google Scholar]
- 19.Murray AJ. 2008. Pharmacological PKA inhibition: all may not be what it seems. Sci Signal 1:re4. doi: 10.1126/scisignal.122re4. [DOI] [PubMed] [Google Scholar]
- 20.Helle F, Brochot E, Fournier C, Descamps V, Izquierdo L, Hoffmann TW, Morel V, Herpe Y-E, Bengrine A, Belouzard S, Wychowski C, Dubuisson J, Francois C, Regimbeau J-M, Castelain S, Duverlie G. 2013. Permissivity of primary human hepatocytes and different hepatoma cell lines to cell culture adapted hepatitis C virus. PLoS One 8:e70809. doi: 10.1371/journal.pone.0070809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlsen THR, Pedersen J, Prentoe JC, Giang E, Keck Z-Y, Mikkelsen LS, Law M, Foung SKH, Bukh J. 2014. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology 60:1551–1562. doi: 10.1002/hep.27298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottwein JM, Scheel TKH, Jensen TB, Lademann JB, Prentoe JC, Knudsen ML, Hoegh AM, Bukh J. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 23.Jensen TB, Gottwein JM, Scheel TKH, Hoegh AM, Eugen-Olsen J, Bukh J. 2008. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J Infect Dis 198:1756–1765. doi: 10.1086/593021. [DOI] [PubMed] [Google Scholar]
- 24.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 25.Pedersen J, Carlsen THR, Prentoe J, Ramirez S, Jensen TB, Forns X, Alter H, Foung SKH, Law M, Gottwein J, Weis N, Bukh J. 2013. Neutralization resistance of hepatitis C virus can be overcome by recombinant human monoclonal antibodies. Hepatology 58:1587–1597. doi: 10.1002/hep.26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheel TKH, Gottwein JM, Jensen TB, Prentoe JC, Hoegh AM, Alter HJ, Eugen-Olsen J, Bukh J. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc Natl Acad Sci U S A 105:997–1002. doi: 10.1073/pnas.0711044105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheel TKH, Gottwein JM, Carlsen THR, Li Y-P, Jensen TB, Spengler U, Weis N, Bukh J. 2011. Efficient culture adaptation of hepatitis C virus recombinants with genotype-specific core-NS2 by using previously identified mutations. J Virol 85:2891–2906. doi: 10.1128/JVI.01605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meertens L, Bertaux C, Dragic T. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol 80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farquhar MJ, Harris HJ, Diskar M, Jones S, Mee CJ, Nielsen SU, Brimacombe CL, Molina S, Toms GL, Maurel P, Howl J, Herberg FW, van Ijzendoorn SCD, Balfe P, McKeating JA. 2008. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J Virol 82:8797–8811. doi: 10.1128/JVI.00592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset F-L, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoël M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M, Durand SC, Duong FHT, Heim MH, Cosset F-L, Hirsch I, Samuel D, Brino L, Zeisel MB, Le Naour F, McKeating JA, Baumert TF. 2013. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 13:302–313. doi: 10.1016/j.chom.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 32.Ahmad W, Shabbiri K, Ijaz B, Asad S, Sarwar MT, Gull S, Kausar H, Fouzia K, Shahid I, Hassan S. 2011. Claudin-1 required for HCV virus entry has high potential for phosphorylation and O-glycosylation. Virol J 8:229. doi: 10.1186/1743-422X-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polat B, Halici Z, Cadirci E, Karakus E, Bayir Y, Albayrak A, Unal D. 2017. Liver 5-HT7 receptors: a novel regulator target of fibrosis and inflammation-induced chronic liver injury in vivo and in vitro. Int Immunopharmacol 43:227–235. doi: 10.1016/j.intimp.2016.12.023. [DOI] [PubMed] [Google Scholar]
- 34.Kohen R, Metcalf MA, Khan N, Druck T, Huebner K, Lachowicz JE, Meltzer HY, Sibley DR, Roth BL, Hamblin MW. 1996. Cloning, characterization, and chromosomal localization of a human 5-HT6 serotonin receptor. J Neurochem 66:47–56. doi: 10.1046/j.1471-4159.1996.66010047.x. [DOI] [PubMed] [Google Scholar]
- 35.Wojtal KA, Hoekstra D, van Ijzendoorn SCD. 2008. cAMP-dependent protein kinase A and the dynamics of epithelial cell surface domains: moving membranes to keep in shape. Bioessays 30:146–155. doi: 10.1002/bies.20705. [DOI] [PubMed] [Google Scholar]
- 36.Offringa R, Huang F. 2013. Phosphorylation-dependent trafficking of plasma membrane proteins in animal and plant cells. J Integr Plant Biol 55:789–808. doi: 10.1111/jipb.12096. [DOI] [PubMed] [Google Scholar]
- 37.French AD, Fiori JL, Camilli TC, Leotlela PD, O'Connell MP, Frank BP, Subaran S, Indig FE, Taub DD, Weeraratna AT. 2009. PKC and PKA phosphorylation affect the subcellular localization of claudin-1 in melanoma cells. Int J Med Sci 6:93–101. doi: 10.7150/ijms.6.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujibe M, Chiba H, Kojima T, Soma T, Wada T, Yamashita T, Sawada N. 2004. Thr203 of claudin-1, a putative phosphorylation site for MAP kinase, is required to promote the barrier function of tight junctions. Exp Cell Res 295:36–47. doi: 10.1016/j.yexcr.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 39.Jian Y, Chen C, Li B, Tian X. 2015. Delocalized Claudin-1 promotes metastasis of human osteosarcoma cells. Biochem Biophys Res Commun 466:356–361. doi: 10.1016/j.bbrc.2015.09.028. [DOI] [PubMed] [Google Scholar]
- 40.Shiomi R, Shigetomi K, Inai T, Sakai M, Ikenouchi J. 2015. CaMKII regulates the strength of the epithelial barrier. Sci Rep 5:13262. doi: 10.1038/srep13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikari A. 2006. Phosphorylation of paracellin-1 at Ser217 by protein kinase A is essential for localization in tight junctions. J Cell Sci 119:1781–1789. doi: 10.1242/jcs.02901. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi S, Iwamoto N, Sasaki H, Ohashi M, Oda Y, Tsukita S, Furuse M. 2009. The E3 ubiquitin ligase LNX1p80 promotes the removal of claudins from tight junctions in MDCK cells. J Cell Sci 122:985–994. doi: 10.1242/jcs.040055. [DOI] [PubMed] [Google Scholar]
- 43.He S, Lin B, Chu V, Hu Z, Hu X, Xiao J, Wang AQ, Schweitzer CJ, Li Q, Imamura M, Hiraga N, Southall N, Ferrer M, Zheng W, Chayama K, Marugan JJ, Liang TJ. 2015. Repurposing of the antihistamine chlorcyclizine and related compounds for treatment of hepatitis C virus infection. Sci Transl Med 7:282ra49. doi: 10.1126/scitranslmed.3010286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu Z, Hu X, He S, Yim HJ, Xiao J, Swaroop M, Tanega C, Zhang Y, Yi G, Kao CC, Marugan J, Ferrer M, Zheng W, Southall N, Liang TJ. 2015. Identification of novel anti-hepatitis C virus agents by a quantitative high throughput screen in a cell-based infection assay. Antiviral Res 124:20–29. doi: 10.1016/j.antiviral.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng H, Lear-Rooney CM, Johansen L, Varhegyi E, Chen ZW, Olinger GG, Rong L. 2015. Inhibition of Ebola and Marburg virus entry by G protein-coupled receptor antagonists. J Virol 89:9932–9938. doi: 10.1128/JVI.01337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Assetta B, Maginnis MS, Gracia Ahufinger I, Haley SA, Gee GV, Nelson CDS, O'Hara BA, Allen Ramdial SA, Atwood WJ. 2013. 5-HT2 receptors facilitate JC polyomavirus entry. J Virol 87:13490–13498. doi: 10.1128/JVI.02252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bialowas S, Hagbom M, Nordgren J, Karlsson T, Sharma S, Magnusson K-E, Svensson L. 2016. Rotavirus and serotonin cross-talk in diarrhoea. PLoS One 11:e0159660. doi: 10.1371/journal.pone.0159660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elphick GF, Querbes W, Jordan JA, Gee GV, Eash S, Manley K, Dugan A, Stanifer M, Bhatnagar A, Kroeze WK, Roth BL, Atwood WJ. 2004. The human polyomavirus, JCV, uses serotonin receptors to infect cells. Science 306:1380–1383. doi: 10.1126/science.1103492. [DOI] [PubMed] [Google Scholar]
- 49.Mainou BA, Ashbrook AW, Smith EC, Dorset DC, Denison MR, Dermody TS. 2015. Serotonin receptor agonist 5-nonyloxytryptamine alters the kinetics of reovirus cell entry. J Virol 89:8701–8712. doi: 10.1128/JVI.00739-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu Z, Lan K-H, He S, Swaroop M, Hu X, Southall N, Zheng W, Liang TJ. 2014. Novel cell-based hepatitis C virus infection assay for quantitative high-throughput screening of anti-hepatitis C virus compounds. Antimicrob Agents Chemother 58:995–1004. doi: 10.1128/AAC.02094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chockalingam K, Simeon RL, Rice CM, Chen Z. 2010. A cell protection screen reveals potent inhibitors of multiple stages of the hepatitis C virus life cycle. Proc Natl Acad Sci U S A 107:3764–3769. doi: 10.1073/pnas.0915117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greeson JM, Gettes DR, Spitsin S, Dubé B, Benton TD, Lynch KG, Douglas SD, Evans DL. 2016. The selective serotonin reuptake inhibitor citalopram decreases human immunodeficiency virus receptor and coreceptor expression in immune cells. Biol Psychiatry 80:33–39. doi: 10.1016/j.biopsych.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 54.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. 1982. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res 42:3858–3863. [PubMed] [Google Scholar]
- 55.Fofana I, Krieger SE, Grunert F, Glauben S, Xiao F, Fafi-Kremer S, Soulier E, Royer C, Thumann C, Mee CJ, McKeating JA, Dragic T, Pessaux P, Stoll-Keller F, Schuster C, Thompson J, Baumert TF. 2010. Monoclonal anti-claudin 1 antibodies prevent hepatitis C virus infection of primary human hepatocytes. Gastroenterology 139:953–964, 964.e1–964.e4. doi: 10.1053/j.gastro.2010.05.073. [DOI] [PubMed] [Google Scholar]
- 56.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 57.Gottwein JM, Scheel TKH, Callendret B, Li Y-P, Eccleston HB, Engle RE, Govindarajan S, Satterfield W, Purcell RH, Walker CM, Bukh J. 2010. Novel infectious cDNA clones of hepatitis C virus genotype 3a (strain S52) and 4a (strain ED43): genetic analyses and in vivo pathogenesis studies. J Virol 84:5277–5293. doi: 10.1128/JVI.02667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartosch B, Dubuisson J, Cosset F-L. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J Exp Med 197:633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandrin V, Boson B, Salmon P, Gay W, Negre D, Le Grand R, Trono D, Cosset F-L. 2002. Lentiviral vectors pseudotyped with a modified RD114 envelope glycoprotein show increased stability in sera and augmented transduction of primary lymphocytes and CD34+ cells derived from human and nonhuman primates. Blood 100:823–832. doi: 10.1182/blood-2001-11-0042. [DOI] [PubMed] [Google Scholar]
- 60.Belouzard S, Delcroix D, Rouille Y. 2004. Low levels of expression of leptin receptor at the cell surface result from constitutive endocytosis and intracellular retention in the biosynthetic pathway. J Biol Chem 279:28499–28508. doi: 10.1074/jbc.M400508200. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z, Tian Y, Machida K, Lai MMC, Luo G, Foung SKH, Ou JJ. 2012. Transient activation of the PI3K-AKT pathway by hepatitis C virus to enhance viral entry. J Biol Chem 287:41922–41930. doi: 10.1074/jbc.M112.414789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valero ML, Sabariegos R, Cimas FJ, Perales C, Domingo E, Sánchez-Prieto R, Mas A. 2016. Hepatitis C virus RNA-dependent RNA polymerase interacts with the Akt/PKB kinase and induces its subcellular relocalization. Antimicrob Agents Chemother 60:3540–3550. doi: 10.1128/AAC.03019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakata K, Hara M, Terada T, Watanabe N, Takaya D, Yaguchi S, Matsumoto T, Matsuura T, Shirouzu M, Yokoyama S, Yamaguchi T, Miyazawa K, Aizaki H, Suzuki T, Wakita T, Imoto M, Kojima S. 2013. HCV NS3 protease enhances liver fibrosis via binding to and activating TGF-β type I receptor. Sci Rep 3:3243. doi: 10.1038/srep03243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takamatsu M, Fujita T, Hotta H. 2001. Suppression of serum starvation-induced apoptosis by hepatitis C virus core protein. Kobe J Med Sci 47:97–112. [PubMed] [Google Scholar]
- 65.Yang S-H, Lee CG, Lee CW, Choi E-J, Yoon SK, Ahn KS, Sung YC. 2002. Hepatitis C virus core inhibits the Fas-mediated p38 mitogen activated kinase signaling pathway in hepatocytes. Mol Cells 13:452–462. [PubMed] [Google Scholar]
- 66.Erhardt A, Hassan M, Heintges T, Häussinger D. 2002. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology 292:272–284. doi: 10.1006/viro.2001.1227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.