ABSTRACT
Aspergillus section Terrei is a species complex currently comprised of 14 cryptic species whose prevalence in clinical samples as well as antifungal susceptibility are poorly known. The aims of this study were to investigate A. Terrei clinical isolates at the species level and to perform antifungal susceptibility analyses by reference and commercial methods. Eighty-two clinical A. Terrei isolates were collected from 8 French university hospitals. Molecular identification was performed by sequencing parts of beta-tubulin and calmodulin genes. MICs or minimum effective concentrations (MECs) were determined for 8 antifungal drugs using both EUCAST broth microdilution (BMD) methods and concentration gradient strips (CGS). Among the 79 A. Terrei isolates, A. terreus stricto sensu (n = 61), A. citrinoterreus (n = 13), A. hortai (n = 3), and A. alabamensis (n = 2) were identified. All strains had MICs of ≥1 mg/liter for amphotericin B, except for two isolates (both A. hortai) that had MICs of 0.25 mg/liter. Four A. terreus isolates were resistant to at least one azole drug, including one with pan-azole resistance, yet no mutation in the CYP51A gene was found. All strains had low MECs for the three echinocandins. The essential agreements (EAs) between BMD and CGS were >90%, except for those of amphotericin B (79.7%) and itraconazole (73.4%). Isolates belonging to the A. section Terrei identified in clinical samples show wider species diversity beyond the known A. terreus sensu stricto. Azole resistance inside the section Terrei is uncommon and is not related to CYP51A mutations here. Finally, CGS is an interesting alternative for routine antifungal susceptibility testing.
KEYWORDS: Aspergillus section Terrei, Aspergillus terreus, molecular identification, antifungal susceptibility testing, Etest, EUCAST
INTRODUCTION
Invasive aspergillosis (IA) is becoming a serious threat and a leading cause of morbidity and mortality in immunocompromised patients. A. fumigatus is the most common species found in clinical samples of patients suffering from IA; however, other Aspergillus spp. can also be the cause of IA (1, 2). Among the five most common is A. terreus, which is difficult to manage and is often associated with increased mortality because of its intrinsic resistance to amphotericin B (3, 4).
A. terreus is found worldwide in the environment and belongs to the section Terrei (5). On the whole, 14 cryptic species that are phenotypically indistinguishable belong to the section Terrei (5, 6). However, only a few of them, namely, A. terreus stricto sensu, A. citrinoterreus, A. alabamensis, A. hortai, and A. niveus, have been reported in human diseases (7–10). The prevalence of these cryptic species in clinical samples is poorly known.
In recent years, a striking emergence of azole resistance has been described in Aspergillus species (11, 12). Azole resistance is mainly related to mutations in the CYP51A gene and in its promoter, yet there might be other mechanisms (13, 14). The intrinsic resistance of A. terreus to amphotericin B puts it at very high risk of multidrug resistance. Nevertheless, only a few azole-resistant A. terreus isolates have been reported so far (15).
The broth microdilution method (BMD) based on the EUCAST or CLSI guidelines is currently the reference method for antifungal susceptibility testing (AFST) of Aspergillus species (16, 17). Clinical breakpoints and epidemiological cutoffs (ECOFFs) have been determined by EUCAST and can be used to detect resistance (18). However, BMD is time-consuming and requires expertise. As such, many clinical microbiology laboratories routinely apply alternative methods, such as concentration gradient strips (CGS), to assess the in vitro susceptibility of their isolates. Few studies have assessed the correlation between these two methods in the susceptibility tests of A. terreus, and even when it was examined, it was not done so for all drugs (19–21). Therefore, interpreting MICs obtained by CGS is difficult.
In this study, we investigated 79 morphologically identified A. Terrei French clinical isolates. We aimed to identify them molecularly by sequencing parts of the beta-tubulin and calmodulin genes and to assess their susceptibility to 8 antifungal drugs by CGS and BMD according to the EUCAST guidelines.
RESULTS
Isolates and patients.
Seventy-nine non-temporally related A. Terrei isolates were collected from 50 patients from 8 university hospitals (more than 1,000 beds each, except one with 850 beds), the majority of whom were immunocompromised patients, including hematopoietic stem cell, renal, cardiac, liver, and pulmonary transplant patients. The origin of isolates was mainly respiratory samples (n = 70; 88.6%). Other isolates were taken from deep samples (n = 4; 5.1%), such as cutaneous biopsy specimen, joint fluid, and intravascular thrombus, or from superficial samples (n = 5; 6.3%), such as nail, external ear, and stools. Sex ratio was 1.5 (30 males and 20 females) with a mean age of 54 years. Underlying diseases were mainly bronchopulmonary diseases (n = 22; 44%), like cystic fibrosis (n = 15) and chronic obstructive pulmonary disease (COPD; n = 7), followed by hematological malignancy (n = 7; 14%), solid-organ transplant (n = 4; 8%), solid malignancy (n = 3; 6%), local trauma (n = 2; 4%), and other conditions (n = 6; 12%). Three patients (6%) had no underlying disease, and there were no data for 3 patients (6%). The clinical forms consisted of 5 cases of invasive aspergillosis, 1 case of onychomycosis, and 44 cases of colonization (Table 1).
TABLE 1.
Origin of isolates and patient characteristics according to the cryptic species of section Terrei
| Species | No. of isolates | No. of hospitals involved | Origin of sample [no. (%)] |
No. of patients | Sex ratio (no. male/no. female) |
Median age (yr) | Underlying disease/condition [no. (%)] |
Aspergillus disease (no.) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Respiratory | Deep | Superficial | Bronchopulmonary disease | Hematological malignancy | Solid-organ transplant | Other conditions | Invasive aspergillosis | Onychomycosis | Colonization | ||||||
| A. terreus sensu stricto | 61 | 7/8 | 55 (90.2) | 4 (6.5) | 2 (3.3) | 36 | 19/17 | 54 | 20 (55.6) | 4 (11.1) | 3 (8.3) | 9 (25) | 5 | 0 | 31 |
| A. citrinoterreus | 13 | 5/8 | 11 (84.6) | 0 | 2 (15.4) | 10 | 9/1 | 56 | 1 (10) | 2 (20) | 1 (10) | 6 (60) | 0 | 0 | 10 |
| A. hortai | 3 | 3/8 | 2 (66.7) | 0 | 1 (33.3) | 3 | 1/2 | 54 | 0 | 1 (33.3) | 0 | 2 (66.7) | 0 | 1 | 2 |
| A. alabamensis | 2 | 1/8 | 2 (100) | 0 | 0 | 1 | 1/0 | 23 | 1 (100) | 0 | 0 | 0 | 0 | 0 | 1 |
| All Aspergillus Terrei | 79 | 8 | 70 (88.6) | 4 (5.1) | 5 (6.3) | 50 | 30/20 | 54 | 22 (44) | 7 (14) | 4 (8) | 17 (34) | 5 | 1 | 44 |
Molecular identification.
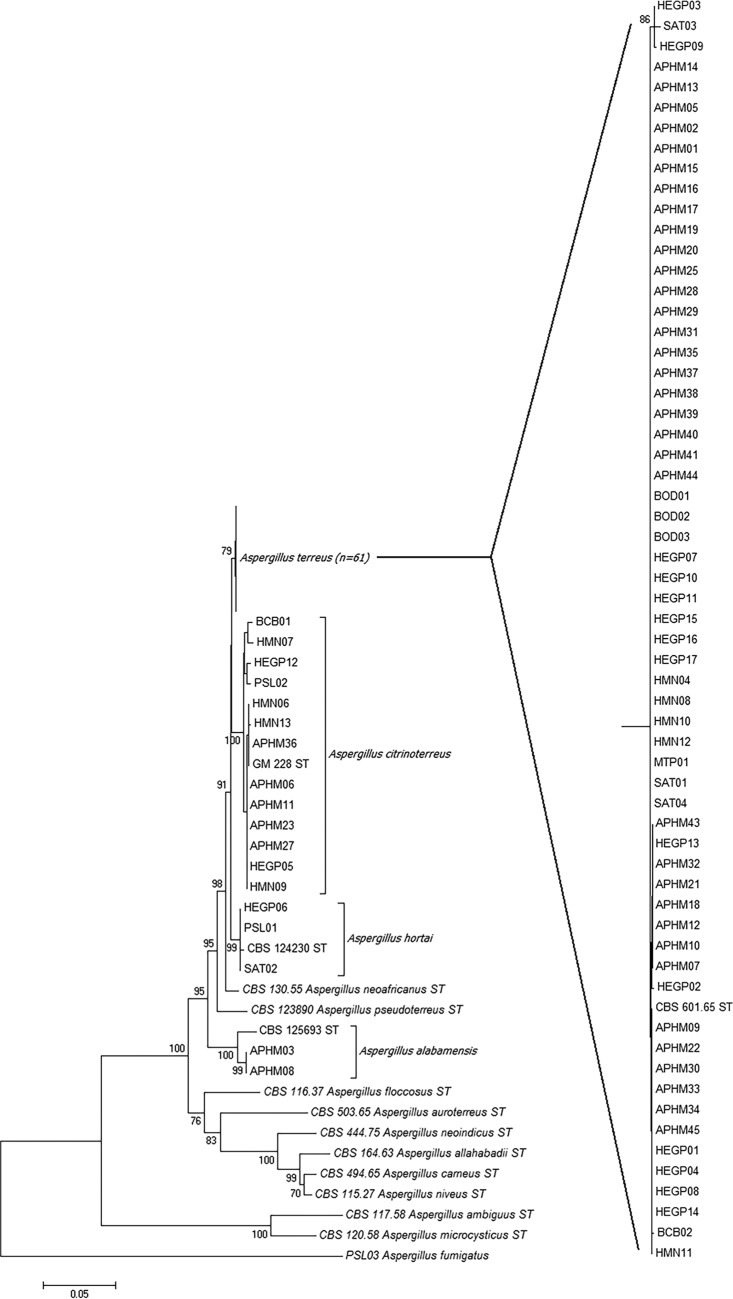
Among the 79 A. Terrei isolates, A. terreus sensu stricto was the most common species (n = 61; 77.2%), found in 7 of the 8 participating centers, and was responsible for the 5 cases of invasive aspergillosis. A. citrinoterreus was the second most frequent species and represented 13 isolates (16.5%). The two other cryptic species were identified as A. hortai (3 isolates, 3.8%) and A. alabamensis (2 isolates, 2.5%) (Fig. 1). Both calmodulin and β-tubulin genes were able to identify the isolates to the species level and gave the same results.
FIG 1.
Neighbor-joining tree obtained from the analysis of combined beta-tubulin and calmodulin data set. Numbers above the nodes represent bootstrapping values generated from 1,000 replicates using a Kimura 2-parameter model. Only values above 70% are indicated. Branch lengths are proportional to phylogenetic distances.
Antifungal susceptibility testing by EUCAST method.
Geometric mean MIC, MIC50/50% minimum effective concentration (MEC50), MIC90/MEC90, and ranges for the 79 isolates are presented in Table 2.
TABLE 2.
Results of in vitro antifungal susceptibility test for the 82 A. Terrei isolates by EUCAST and CGS methodsa
| Species and drug | EUCAST method |
CGS method |
EA (%) | CA (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Range (mg/liter) | GM (mg/liter) | MIC50/MEC50 (mg/liter) | MIC90/MEC90 (mg/liter) | Range (mg/liter) | GM (mg/liter) | MIC50/MEC50 (mg/liter) | MIC90/MEC90 (mg/liter) | |||
| A. terreus sensu stricto (n = 61) | ||||||||||
| AMB | 1–8 | 2.482 | 2 | 4 | 0.25–4 | 1.023 | 1 | 2 | 82 | 91.8 |
| ITC | 0.016–8 | 0.092 | 0.06 | 0.25 | 0.125–2 | 0.381 | 0.5 | 0.5 | 68.9 | 98.4 |
| VRC | 0.25–8 | 0.664 | 0.5 | 1 | 0.06–16 | 0.363 | 0.5 | 1 | 93.4 | 100 |
| POS | 0.016–0.5 | 0.063 | 0.06 | 0.125 | 0.016–0.5 | 0.13 | 0.125 | 0.25 | 96.7 | 73.8 |
| ISA | 0.25–4 | 0.517 | 0.5 | 1 | 0.06–4 | 0.310 | 0.25 | 0.5 | 100 | 96.7 |
| CAS | 0.03–0.25 | 0.071 | 0.06 | 0.125 | 0.016–0.125 | 0.028 | 0.03 | 0.06 | 96.7 | NA |
| MFG | <0.016 | <0.016 | <0.016 | <0.016 | <0.016–0.03 | <0.016 | <0.016 | 0.016 | 100 | NA |
| AFG | <0.016–0.03 | <0.016 | <0.016 | <0.016 | <0.016 | <0.016 | <0.016 | <0.016 | 100 | NA |
| A. citrinoterreus (n = 13) | ||||||||||
| AMB | 2–8 | 4.219 | 4 | 8 | 0.5–4 | 1.238 | 1 | 2 | 69.2 | 69.2 |
| ITC | 0.016–0.125 | 0.069 | 0.06 | 0.125 | 0.016–0.5 | 0.154 | 0.25 | 0.25 | 100 | 100 |
| VRC | 0.25–1 | 0.404 | 0.5 | 0.5 | 0.06–0.25 | 0.139 | 0.125 | 0.25 | 92.3 | 100 |
| POS | 0.016–0.062 | 0.033 | 0.031 | 0.062 | 0.008–0.125 | 0.058 | 0.06 | 0.125 | 100 | 100 |
| ISA | 0.125–1 | 0.293 | 0.25 | 1 | 0.03–0.25 | 0.131 | 0.125 | 0.25 | 84.6 | 100 |
| CAS | 0.031–0.25 | 0.086 | 0.125 | 0.125 | 0.016–0.25 | 0.068 | 0.06 | 0.25 | 100 | NA |
| MFG | <0.016–0.03 | <0.016 | <0.016 | <0.016 | <0.016–0.03 | <0.016 | <0.016 | 0.016 | 100 | NA |
| AFG | <0.016 | <0.016 | <0.016 | <0.016 | <0.016–0.016 | <0.016 | <0.016 | <0.016 | 100 | NA |
| A. hortai (n = 3) | ||||||||||
| AMB | 0.25–4 | NA | NA | NA | 0.5–2 | NA | NA | NA | 66.7 | 100 |
| ITC | 0.016–0.03 | NA | NA | NA | 0.125–0.5 | NA | NA | NA | 33.3 | 100 |
| VRC | 0.25 | NA | NA | NA | 0.125–0.5 | NA | NA | NA | 100 | 100 |
| POS | 0.016–0.03 | NA | NA | NA | 0.03–0.25 | NA | NA | NA | 66.7 | 66.7 |
| ISA | 0.25 | NA | NA | NA | 0.25 | NA | NA | NA | 100 | 100 |
| CAS | 0.06–0.25 | NA | NA | NA | 0.016–0.125 | NA | NA | NA | 66.7 | NA |
| MFG | <0.016 | NA | NA | NA | <0.016 | NA | NA | NA | 100 | NA |
| AFG | <0.016 | NA | NA | NA | <0.016 | NA | NA | NA | 100 | NA |
| A. alabamensis (n = 2) | ||||||||||
| AMB | >8 | NA | NA | NA | 32 | NA | NA | NA | 100 | 100 |
| ITC | 0.125–0.25 | NA | NA | NA | 0.5 | NA | NA | NA | 100 | 100 |
| VRC | 1 | NA | NA | NA | 0.5–1 | NA | NA | NA | 100 | 100 |
| POS | 0.06–0.125 | NA | NA | NA | 0.125–0.25 | NA | NA | NA | 100 | 50 |
| ISA | 0.25–1 | NA | NA | NA | 0.5 | NA | NA | NA | 100 | 100 |
| CAS | 0.06 | NA | NA | NA | 0.125 | NA | NA | NA | 100 | NA |
| MFG | <0.016 | NA | NA | NA | <0.016 | NA | NA | NA | 100 | NA |
| AFG | <0.016 | NA | NA | NA | <0.016–0.016 | NA | NA | NA | 100 | NA |
| All A. Terrei (n = 79) | ||||||||||
| AMB | 0.25–8 | 2.648 | 2 | 8 | 0.25–32 | 1.161 | 1 | 2 | 79.7 | 88.6 |
| ITC | 0.016–8 | 0.084 | 0.06 | 0.25 | 0.016–2 | 0.322 | 0.5 | 0.5 | 73.4 | 98.7 |
| VRC | 0.25–8 | 0.596 | 0.5 | 1 | 0.06–16 | 0.311 | 0.25 | 1 | 93.7 | 100 |
| POS | 0.016–0.5 | 0.055 | 0.06 | 0.125 | 0.008–0.5 | 0.112 | 0.125 | 0.25 | 96.2 | 77.2 |
| ISA | 0.125–4 | 0.458 | 0.5 | 1 | 0.03–4 | 0.270 | 0.25 | 0.5 | 97.5 | 97.4 |
| CAS | 0.031–0.25 | 0.074 | 0.06 | 0.125 | 0.016–0.25 | 0.034 | 0.03 | 0.125 | 96.2 | NA |
| MFG | <0.016–0.03 | <0.016 | <0.016 | <0.016 | <0.016–0.016 | <0.016 | <0.016 | <0.016 | 100 | NA |
| AFG | <0.016–0.03 | <0.016 | <0.016 | <0.016 | <0.016–0.03 | <0.016 | <0.016 | 0.016 | 100 | NA |
GM, geometric mean; EA, essential agreement; CA, categorical agreement; NA, not available.
Amphotericin B susceptibility testing by the BMD method resulted in MICs above 1 mg/liter for all isolates except two, which had MICs of 0.25 mg/liter. Interestingly, these isolates were both A. hortai; however, the third A. hortai isolate had a MIC of 4 mg/liter. Eleven isolates had a MIC above the ECOFF (MIC of 8 mg/liter by EUCAST), including the two A. alabamensis isolates, which showed the highest MICs for amphotericin B by the CGS method (MIC of >32 mg/liter). The last 9 isolates with MICs above the ECOFF were five A. terreus sensu stricto and four A. citrinoterreus isolates (Table 2).
Concerning antifungal susceptibility testing by the BMD method for azole drugs, no differences were found between the different cryptic species. However, four isolates either had MICs above the ECOFF (MIC of 4 to 8 mg/liter for voriconazole and 2 to 4 mg/liter for isavuconazole) or were resistant (MIC of 8 mg/liter for itraconazole and 0.5 mg/liter for posaconazole) to at least one azole drug, e.g., one isolate showed resistance to a pan-azole. These four isolates were all A. terreus sensu stricto.
As for echinocandin drugs, all isolates had low MECs for the three echinocandins by both BMD and Etest methods (MECs of <0.25 mg/liter for caspofungin and <0.031 mg/liter for micafungin and anidulafungin).
MIC/MEC comparison between EUCAST and CGS methods.
Considering all A. Terrei species, MIC50/MEC50 and MIC90/MEC90, as determined by the two methods, were identical (within ±2 dilutions) for all drugs, except for the MIC50 of itraconazole, which had a higher value with the CGS method (Table 2). The EA values were above 90% for all antifungal drugs, except for those for amphotericin B and itraconazole, which had EA values of 79.7% and 73.4%, respectively. For amphotericin B, the MICs were higher by the BMD method (P < 0.001 by Wilcoxon test), whereas for itraconazole the MICs were significantly higher by the CGS method (P < 0.001 by Wilcoxon test). The CA values were above 90% for itraconazole, voriconazole, and isavuconazole but were only 88.6% for amphotericin B and 77.2% for posaconazole and were not available for echinocandins.
When we separately considered the two cryptic species (A. terreus sensu stricto and A. citrinoterreus) with a sufficient number of isolates for each, the EAs were acceptable (>90%) for all drugs except amphotericin B and itraconazole for A. terreus sensu stricto and amphotericin B and isavuconazole for A. citrinoterreus (Table 2).
Comparison of visual and spectrophotometric readings for EUCAST MIC values.
For azole drugs and amphotericin B, the spectrophotometric reading was made for 69 of the 79 A. Terrei isolates. MIC50 and MIC90, determined visually and by the spectrophotometer, were identical (within ±2 dilutions) for all drugs. Similarly, the EAs were above 90% for all drugs.
CYP51A sequencing.
The CYP51A gene and its promoter were sequenced for the four isolates that were resistant in vitro to at least one azole drug. The same sequencing was made for 11 other susceptible isolates as controls. Of note, all of these isolates were A. terreus sensu stricto. No mutation in the amino acid sequence was detected in the four resistant isolates or in nine of the susceptible isolates. For the remaining two susceptible isolates sequenced for CYP51A and its promoter, we found polymorphism in the CYP51A amino acid sequence; one isolate harbored E313K mutation and the other A249G mutation.
DISCUSSION
In recent years, the development of fungal identification protocols based on molecular methods has increased our knowledge on Aspergillus species epidemiology. Among the main Aspergillus species implicated in human diseases (like A. fumigatus, A. flavus, and A. niger), A. terreus belongs to the section Terrei (5). This section is currently composed of 14 recognized cryptic species (6). To our knowledge, little research work was directed to study the epidemiology inside the section Terrei, and the prevalence of the cryptic species involved in human disease is poorly known (7, 8, 22). However, it may be useful to identify Aspergillus at the species level in clinical samples to improve patient management or to deepen knowledge of the local epidemiology. Several of these cryptic species can be the main species isolated in clinical samples of patients with IA or exhibit resistance to antifungal drugs. For example, within the section Fumigati, A. lentulus has been shown to be intrinsically resistant to voriconazole (23). Moreover, within the section Nigri, A. niger sensu stricto alone stands for 7 to 58% of clinical isolates, whereas A. tubingensis and A. awamori account for 17.8 to 76.2% and 16.7 to 55.6%, respectively (24–27).
In the present study, we investigated the molecular identification and in vitro antifungal susceptibility of 79 clinical isolates identified initially as A. terreus. The results show that A. terreus sensu stricto is the most common species, and that almost 25% of the isolates represent the three other cryptic species: A. citrinoterreus, A. hortai, and A. alabamensis. Moreover, the newly described (6) A. citrinoterreus is the second most common species (16.5%). These four cryptic species have already been involved in human invasive aspergillosis (6, 8, 9), but interestingly, in our study the five cases of invasive aspergillosis were caused only by A. terreus sensu stricto. The remaining isolates were mainly responsible for colonization in patients suffering from chronic bronchopulmonary diseases (cystic fibrosis and COPD).
Our A. Terrei isolates come from two main distinct French geographical areas: Paris (north of France) and Marseille (south of France). No difference was found in the species distribution between these two areas, although the incidence might differ between the cities, as was already reported (28).
A. terreus is known to be intrinsically resistant to amphotericin B (29). However, our results indicate that this pattern of susceptibility is dependent on the cryptic species inside the Terrei section. For instance, A. hortai seems to have the lowest MICs, and two of its isolates had MICs of <1 mg/liter. This is consistent with previous observations where 6 to 38% of the A. Terrei isolates had MICs of <1 mg/liter (7, 29, 30). However, in the Risslegger et al. study, the low-MIC isolates all were A. terreus sensu stricto (7). In contrast, A. alabamensis had the highest MIC and corresponds to the only two isolates with MICs of >32 mg/liter by the CGS method. Such observations need to be confirmed by using a larger number of isolates for each species.
Azole resistance in Aspergillus species is an emerging issue in recent years (11, 12). This phenomenon is well studied for A. fumigatus and is mainly related to mutations in the CYP51A gene and its promoter (14, 31, 32). A unique case of pan-azole-resistant A. terreus with a mutated CYP51A has already been reported (15). Among our 79 A. Terrei isolates, four were resistant to at least one azole drug, including one with pan-azole resistance. Unfortunately, we did not find mutation in the CYP51A gene or its promoter in these four isolates. We think that other mechanisms, already described in other Aspergillus species, like increased drug efflux or overexpression of the target, stand behind the azole resistance of these isolates (13, 31). Interestingly, we detected polymorphism in the CYP51A amino acid sequence in two of our susceptible control isolates; one harbored an E313K mutation and the other an A249G mutation without association with higher MICs for azole drugs.
ECOFFs and clinical breakpoints used to categorize isolates as wild type/non-wild type or susceptible/resistant are only defined for the reference BMD method (18). Given that BMD requires experience and is not practicable for all laboratories, the CGS method often replaces it. Several studies have already assessed the correlation between CLSI and CGS methods for A. terreus, but this has never been the case between EUCAST and CGS methods. Therefore, in the present study we compared, for the first time, EUCAST and CGS methods for AFST of our 79 A. Terrei isolates. The EAs were acceptable (i.e., ≥90%) for all antifungal drugs except itraconazole and amphotericin B, which had EAs of 73.4% and 79.7%, respectively. The same finding on amphotericin B was already described between CLSI and CGS in previous studies, with EAs ranging from 16% to 75% (19, 33). However, a good agreement for this drug was also shown with EAs of 100% (34, 35). Regarding azole drugs, previous comparison studies between CLSI and CGS methods showed various results ranging from EAs of 100% for voriconazole (19), itraconazole (35), posaconazole (34), and isavuconazole (36) to less acceptable EAs for itraconazole (EA of 88%) (33) and posaconazole (EA of 64%) (19). Consequently, our results show that the CGS method could be a good alternative to BMD, allowing the use of EUCAST ECOFF and breakpoints for all drugs but itraconazole and amphotericin B. For the latter, low MICs obtained by CGS should be interpreted with caution in these amphotericin B intrinsically resistant species.
EUCAST guidelines recommend reading MICs of azole and amphotericin B in filamentous fungi visually (17). However, this method is subject to variations and errors, as it is operator dependent. It has been shown previously that spectrophotometric reading for AFST of Aspergillus spp. was reliable (37, 38). More recently, authors have assessed the use of this spectrophotometric reading in AFST of A. fumigatus using a 490-nm wavelength (39). They found good agreement between this spectrophotometric and visual reading, so they proposed additional investigations on other Aspergillus species, hence their suggestion to consider the use of a spectrometer in practice. Our results showed the same good agreement for our A. Terrei isolates, with EAs above 90% for all tested drugs, adding more evidence to the use of spectrophotometric readings in AFST of Aspergillus species.
In conclusion, A. terreus isolates found in clinical samples show several species besides A. terreus sensu stricto. The antifungal susceptibility testing demonstrated a low rate of azole resistance inside the section Terrei and was not related to CYP51A mutations. Amphotericin B MICs showed significant differences between the isolates, which could be due to species identification. Finally, CGS could be a good alternative to EUCAST BMD for AFST of A. Terrei isolates.
MATERIALS AND METHODS
Isolates and patients.
Phenotypically identified A. Terrei clinical isolates were collected retrospectively over a 13-year period (2003 to 2015) from 8 French university hospitals, of which five are in the Parisian area and 3 in the south of France (Marseille, Montpellier, and Bordeaux). For each isolate, age, sex, underlying disease of the patient, site of isolation, and clinical form of aspergillosis were registered. The study was approved by the local Ethical Committee, and the database was declared to the Commission Nationale de l'Informatique et des Libertés (CNIL) (no. 1699340).
Molecular identification.
Molecular identification within the Terrei section was performed by sequencing part of the calmodulin and β-tubulin genes, as described in previous studies (40). Briefly, complete genomic DNA was extracted from a mature subculture on Sabouraud-chloramphenicol-gentamicin agar using a QIAamp DNA blood minikit (Qiagen Sciences Ing., Courtaboeuf, France) after a step of beading in a MagNA Lyser instrument (Roche Diagnostics, Meylan, France). Primers used for the amplification were 5′-TGGTGCCGCTTTCTGGTA-3′ and 5′-AAGTTGTCGGGACGGAATAG-3′ for β-tubulin and 5′-GTAGCGCAGCGGCCAGTCCGAGTACAAGGARGCCTTC-3′ and 5′-CAGGGCGCAGCGATGACCCGATRGAGGTCATRACGTGG-3′ for calmodulin. Obtained DNA sequences were analyzed using Seqscape v2.5 (Applied Biosystems) and were compared with A. Terrei GenBank and MycoBank database sequences. Phylogenetic analyses were also performed for each gene individually and with the concatenated sequences of the two genes using MEGA 6.0 software. Neighbor-joining trees were built using the Kimura two-parameter model with 1,000 bootstraps replications and included sequences of the type strain of each cryptic species inside the Terrei section.
Antifungal susceptibility testing.
Antifungal susceptibility testing of each isolate was performed for 8 drugs: itraconazole (Sigma-Aldrich, Saint-Quentin Fallavier, France), voriconazole (Sigma-Aldrich), posaconazole (MSD, Kenilworth, NJ), isavuconazole (Basilea Pharmaceutica International Ltd., Basel, Switzerland), amphotericin B (Sigma-Aldrich), caspofungin (Sigma-Aldrich), anidulafungin (Pfizer New York, NY), and micafungin (Astellas Pharma Inc., Tokyo, Japan). The BMD method was used.
BMD was performed according to the EUCAST guidelines (17). Final drug concentration ranges were 0.016 to 8 mg/liter for each drug. Quality control strains C. parapsilosis ATCC 22019 and C. krusei ATCC 6258 were included, but MICs were read after 24 h of incubation. For azoles and amphotericin B, MICs were also read by a spectrophotometer at 550 nm, using a 90% growth inhibition endpoint, and compared with the drug-free control well.
For the agar diffusion method, we used MIC test strips (Liofilchem, Roseto degli Abruzzi, Italy) for isavuconazole and Etest (bioMérieux, Marcy l'Etoile, France) for the other seven drugs. A spore suspension adjusted to a 0.5 McFarland standard in sterile water was prepared and inoculated on RPMI medium plates supplemented with 2% glucose. MICs and MECs were determined after incubation at 37°C for 48 h. Values obtained with the CGS method were adjusted to the next higher concentration matching the 2-fold dilution scheme of the BMD method.
Essential agreement (EA) between BMD and CGS methods, and between the visual and spectrophotometric readings for the EUCAST method, was considered to have been achieved when the MIC/MEC values were within ±2 dilutions. EA values above 90% were considered acceptable.
Isolates were categorized as wild type/non-wild type or as susceptible/resistant by comparing MIC values with ECOFFs and clinical breakpoints (18).
CYP51A sequencing.
For azole-resistant and control isolates, the whole CYP51A gene and its promoter were amplified using 4 couples of in-house primers (Table 3). The 4 obtained partial sequences were analyzed and assembled using SeqScape. The generated sequence was then translated and aligned to those of gene ATEG05917 from the reference strain NIH2624 and set for amino acid sequence analysis.
TABLE 3.
Primers used in this study to sequence gene CYP51A and its promoter
| Fragment | Sequence |
|---|---|
| 1 | |
| Forward | 5′-GTAGCGCAGCGGCCAGTGGTGGGAGAACTTTCGTTCTA-3′ |
| Reverse | 5′-CAGGGCGCAGCGATGACTGTACACCTCTTCTGCGTTGAC-3′ |
| 2 | |
| Forward | 5′-GTAGCGCAGCGGCCAGTTCATCCTCAACGGCAAACTC-3′ |
| Reverse | 5′-CAGGGCGCAGCGATGACAGATCATGTCGGAGTCGGAG-3′ |
| 3 | |
| Forward | 5′-GTAGCGCAGCGGCCAGTTCTACATGGACATCATCCGC-3′ |
| Reverse | 5′-CAGGGCGCAGCGATGACCATCATCATCAGGTTCCGCT-3′ |
| 4 | |
| Forward | 5′-GTAGCGCAGCGGCCAGTTTCCTAACGCTAGTCGCTGG-3′ |
| Reverse | 5′-CAGGGCGCAGCGATGACGCCGTTCTTACGCCTTGTT-3′ |
Accession number(s).
The obtained sequences were submitted to GenBank under accession numbers MH006731 to MH006809 and MH006810 to MH006888.
ACKNOWLEDGMENTS
We gratefully acknowledge Suhad Assad for her critical linguistic review and Chloé Guillot for her technical assistance.
This research received no specific grant from any funding agency in the public, commercial, or nonprofit sectors. The data were obtained as part of routine work at the University Hospital of Créteil, Créteil, France.
During the past 5 years, E.D. has received research grants from MSD and Gilead; travel grants from Gilead, MSD, Pfizer, and Astellas; and speaker's fees from Gilead, MSD, and Astellas. F.B. received grants from Astellas and MSD and payment for lectures from Merck. A.F. has received payment for lectures from Merck and travel grants from Astellas, Gilead, Merck, and Pfizer. A.F. has also been a consultant for Pfizer. E.D. has received grants from Gilead, Ferrer, and Bio-Rad and payment for lectures from Gilead, MSD, and Schering. I.A. has received travel grants from Gilead, MSD, Pfizer, and Astellas and speaker's fees from MSD. S.I., A.C.N., S.R., J.M.C., C.H., C.B., S.H., N.B., and R.P. have no conflicts of interest to declare.
REFERENCES
- 1.Kontoyiannis DP, Marr KA, Park BJ, Alexander BD, Anaissie EJ, Walsh TJ, Ito J, Andes DR, Baddley JW, Brown JM, Brumble LM, Freifeld AG, Hadley S, Herwaldt LA, Kauffman CA, Knapp K, Lyon GM, Morrison VA, Papanicolaou G, Patterson TF, Perl TM, Schuster MG, Walker R, Wannemuehler KA, Wingard JR, Chiller TM, Pappas PG. 2010. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001-2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis 50:1091–1100. doi: 10.1086/651263. [DOI] [PubMed] [Google Scholar]
- 2.Steinbach WJ, Marr KA, Anaissie EJ, Azie N, Quan SP, Meier-Kriesche HU, Apewokin S, Horn DL. 2012. Clinical epidemiology of 960 patients with invasive aspergillosis from the PATH Alliance registry. J Infect 65:453–464. doi: 10.1016/j.jinf.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Hachem R, Gomes MZ, El Helou G, El Zakhem A, Kassis C, Ramos E, Jiang Y, Chaftari AM, Raad II. 2014. Invasive aspergillosis caused by Aspergillus terreus: an emerging opportunistic infection with poor outcome independent of azole therapy. J Antimicrob Chemother 69:3148–3155. doi: 10.1093/jac/dku241. [DOI] [PubMed] [Google Scholar]
- 4.Baddley JW, Pappas PG, Smith AC, Moser SA. 2003. Epidemiology of Aspergillus terreus at a university hospital. J Clin Microbiol 41:5525–5529. doi: 10.1128/JCM.41.12.5525-5529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samson RA, Peterson SW, Frisvad JC, Varga J. 2011. New species in Aspergillus section Terrei. Stud Mycol 69:39–55. doi: 10.3114/sim.2011.69.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guinea J, Sandoval-Denis M, Escribano P, Pelaez T, Guarro J, Bouza E. 2015. Aspergillus citrinoterreus, a new species of section Terrei isolated from samples of patients with nonhematological predisposing conditions. J Clin Microbiol 53:611–617. doi: 10.1128/JCM.03088-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Risslegger B, Zoran T, Lackner M, Aigner M, Sanchez-Reus F, Rezusta A, Chowdhary A, Taj-Aldeen SJ, Arendrup MC, Oliveri S, Kontoyiannis DP, Alastruey-Izquierdo A, Lagrou K, Lo Cascio G, Meis JF, Buzina W, Farina C, Drogari-Apiranthitou M, Grancini A, Tortorano AM, Willinger B, Hamprecht A, Johnson E, Klingspor L, Arsic-Arsenijevic V, Cornely OA, Meletiadis J, Prammer W, Tullio V, Vehreschild JJ, Trovato L, Lewis RE, Segal E, Rath PM, Hamal P, Rodriguez-Iglesias M, Roilides E, Arikan-Akdagli S, Chakrabarti A, Colombo AL, Fernandez MS, Martin-Gomez MT, Badali H, Petrikkos G, Klimko N, Heimann SM, Houbraken J, Uzun O, Edlinger M, Fuente S, Lass-Flörl C. 2017. A prospective international Aspergillus terreus survey: an EFISG, ISHAM and ECMM joint study. Clin Microbiol Infect 23:776. doi: 10.1016/j.cmi.2017.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Kathuria S, Sharma C, Singh PK, Agarwal P, Agarwal K, Hagen F, Meis JF, Chowdhary A. 2015. Molecular epidemiology and in-vitro antifungal susceptibility of Aspergillus terreus species complex isolates in Delhi, India: evidence of genetic diversity by amplified fragment length polymorphism and microsatellite typing. PLoS One 10:e0118997. doi: 10.1371/journal.pone.0118997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balajee SA, Baddley JW, Peterson SW, Nickle D, Varga J, Boey A, Lass-Florl C, Frisvad JC, Samson RA, ISHAM Working Group on A. terreus. 2009. Aspergillus alabamensis, a new clinically relevant species in the section Terrei. Eukaryot Cell 8:713–722. doi: 10.1128/EC.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auberger J, Lass-Florl C, Clausen J, Bellmann R, Buzina W, Gastl G, Nachbaur D. 2008. First case of breakthrough pulmonary Aspergillus niveus infection in a patient after allogeneic hematopoietic stem cell transplantation. Diagn Microbiol Infect Dis 62:336–339. doi: 10.1016/j.diagmicrobio.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 11.van der Linden JW, Arendrup MC, Warris A, Lagrou K, Pelloux H, Hauser PM, Chryssanthou E, Mellado E, Kidd SE, Tortorano AM, Dannaoui E, Gaustad P, Baddley JW, Uekotter A, Lass-Florl C, Klimko N, Moore CB, Denning DW, Pasqualotto AC, Kibbler C, Arikan-Akdagli S, Andes D, Meletiadis J, Naumiuk L, Nucci M, Melchers WJ, Verweij PE. 2015. Prospective multicenter international surveillance of azole resistance in Aspergillus fumigatus. Emerg Infect Dis 21:1041–1044. doi: 10.3201/eid2106.140717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verweij PE, Chowdhary A, Melchers WJ, Meis JF. 2016. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis 62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Camps SM, Dutilh BE, Arendrup MC, Rijs AJ, Snelders E, Huynen MA, Verweij PE, Melchers WJ. 2012. Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 7:e50034. doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowdhary A, Sharma C, Hagen F, Meis JF. 2014. Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms. Future Microbiol 9:697–711. doi: 10.2217/fmb.14.27. [DOI] [PubMed] [Google Scholar]
- 15.Arendrup MC, Jensen RH, Grif K, Skov M, Pressler T, Johansen HK, Lass-Florl C. 2012. In vivo emergence of Aspergillus terreus with reduced azole susceptibility and a Cyp51a M217I alteration. J Infect Dis 206:981–985. doi: 10.1093/infdis/jis442. [DOI] [PubMed] [Google Scholar]
- 16.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed, CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 17.Arendrup MC, Guinea J, Cuenca-Estrella M, Meletiadis J, Mouton JW, Lagrou K, Howard SJ, Subcommittee on Antifungal Susceptibility Testing (AFST) of the ESCMID European Committee for Antimicrobial Susceptibility Testing (EUCAST). 2017. Method for the determination of broth dilution minimum inhibitory concentrations of antifungal agents for conidia forming moulds (version 9.3.1). http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Files/EUCAST_E_Def_9_3_1_Mould_testing__definitive.pdf. [Google Scholar]
- 18.European Committee on Antimicrobial Susceptibility Testing. 2017. Antifungal agents breakpoints for interpretation of MICs (version 8.1). http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/AFST/Clinical_breakpoints/Antifungal_breakpoints_v_8.1_March_2017.pdf.
- 19.Lamoth F, Alexander BD. 2015. Comparing Etest and broth microdilution for antifungal susceptibility testing of the most-relevant pathogenic molds. J Clin Microbiol 53:3176–3181. doi: 10.1128/JCM.00925-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heo MS, Shin JH, Choi MJ, Park YJ, Lee HS, Koo SH, Lee WG, Kim SH, Shin MG, Suh SP, Ryang DW. 2015. Molecular identification and amphotericin B susceptibility testing of clinical isolates of Aspergillus from 11 hospitals in Korea. Ann Lab Med 35:602–610. doi: 10.3343/alm.2015.35.6.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martos AI, Romero A, Gonzalez MT, Gonzalez A, Serrano C, Castro C, Peman J, Canton E, Martin-Mazuelos E. 2010. Evaluation of the Etest method for susceptibility testing of Aspergillus spp. and Fusarium spp. to three echinocandins. Med Mycol 48:858–861. doi: 10.3109/13693781003586943. [DOI] [PubMed] [Google Scholar]
- 22.Escribano P, Pelaez T, Recio S, Bouza E, Guinea J. 2012. Characterization of clinical strains of Aspergillus terreus complex: molecular identification and antifungal susceptibility to azoles and amphotericin B. Clin Microbiol Infect 18:E24–E26. doi: 10.1111/j.1469-0691.2011.03714.x. [DOI] [PubMed] [Google Scholar]
- 23.Balajee SA, Gribskov JL, Hanley E, Nickle D, Marr KA. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot Cell 4:625–632. doi: 10.1128/EC.4.3.625-632.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iatta R, Nuccio F, Immediato D, Mosca A, De Carlo C, Miragliotta G, Parisi A, Crescenzo G, Otranto D, Cafarchia C. 2016. Species distribution and in vitro azole susceptibility of Aspergillus section Nigri isolates from clinical and environmental settings. J Clin Microbiol 54:2365–2372. doi: 10.1128/JCM.01075-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alcazar-Fuoli L, Mellado E, Alastruey-Izquierdo A, Cuenca-Estrella M, Rodriguez-Tudela JL. 2009. Species identification and antifungal susceptibility patterns of species belonging to Aspergillus section Nigri. Antimicrob Agents Chemother 53:4514–4517. doi: 10.1128/AAC.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badali H, Fakhim H, Zarei F, Nabili M, Vaezi A, Poorzad N, Dolatabadi S, Mirhendi H. 2016. In vitro activities of five antifungal drugs against opportunistic agents of Aspergillus Nigri complex. Mycopathologia 181:235–240. doi: 10.1007/s11046-015-9968-0. [DOI] [PubMed] [Google Scholar]
- 27.Howard SJ, Harrison E, Bowyer P, Varga J, Denning DW. 2011. Cryptic species and azole resistance in the Aspergillus niger complex. Antimicrob Agents Chemother 55:4802–4809. doi: 10.1128/AAC.00304-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lass-Florl C, Grif K, Kontoyiannis DP. 2007. Molecular typing of Aspergillus terreus isolates collected in Houston, Texas, and Innsbruck, Austria: evidence of great genetic diversity. J Clin Microbiol 45:2686–2690. doi: 10.1128/JCM.00917-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lass-Florl C, Alastruey-Izquierdo A, Cuenca-Estrella M, Perkhofer S, Rodriguez-Tudela JL. 2009. In vitro activities of various antifungal drugs against Aspergillus terreus: global assessment using the methodology of the European committee on antimicrobial susceptibility testing. Antimicrob Agents Chemother 53:794–795. doi: 10.1128/AAC.00335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Won EJ, Choi MJ, Shin JH, Park YJ, Byun SA, Jung JS, Kim SH, Shin MG, Suh SP. 2017. Diversity of clinical isolates of Aspergillus terreus in antifungal susceptibilities, genotypes and virulence in Galleria mellonella model: comparison between respiratory and ear isolates. PLoS One 12:e0186086. doi: 10.1371/journal.pone.0186086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hagiwara D, Watanabe A, Kamei K, Goldman GH. 2016. Epidemiological and genomic landscape of azole resistance mechanisms in Aspergillus fungi. Front Microbiol 7:1382. doi: 10.3389/fmicb.2016.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellado E, Garcia-Effron G, Alcazar-Fuoli L, Melchers WJ, Verweij PE, Cuenca-Estrella M, Rodriguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. doi: 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espinel-Ingroff A. 2001. Comparison of the E-test with the NCCLS M38-P method for antifungal susceptibility testing of common and emerging pathogenic filamentous fungi. J Clin Microbiol 39:1360–1367. doi: 10.1128/JCM.39.4.1360-1367.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Espinel-Ingroff A. 2006. Comparison of three commercial assays and a modified disk diffusion assay with two broth microdilution reference assays for testing zygomycetes, Aspergillus spp., Candida spp., and Cryptococcus neoformans with posaconazole and amphotericin B. J Clin Microbiol 44:3616–3622. doi: 10.1128/JCM.01187-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szekely A, Johnson EM, Warnock DW. 1999. Comparison of E-test and broth microdilution methods for antifungal drug susceptibility testing of molds. J Clin Microbiol 37:1480–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guinea J, Pelaez T, Recio S, Torres-Narbona M, Bouza E. 2008. In vitro antifungal activities of isavuconazole (BAL4815), voriconazole, and fluconazole against 1,007 isolates of zygomycete, Candida, Aspergillus, Fusarium, and Scedosporium species. Antimicrob Agents Chemother 52:1396–1400. doi: 10.1128/AAC.01512-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dannaoui E, Persat F, Monier MF, Borel E, Piens MA, Picot S. 1999. Use of spectrophotometric reading for in vitro antifungal susceptibility testing of Aspergillus spp. Can J Microbiol 45:871–874. [PubMed] [Google Scholar]
- 38.Dannaoui E, Lortholary O, Dromer F. 2004. In vitro evaluation of double and triple combinations of antifungal drugs against Aspergillus fumigatus and Aspergillus terreus. Antimicrob Agents Chemother 48:970–978. doi: 10.1128/AAC.48.3.970-978.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meletiadis J, Leth Mortensen K, Verweij PE, Mouton JW, Arendrup MC. 2017. Spectrophotometric reading of EUCAST antifungal susceptibility testing of Aspergillus fumigatus. Clin Microbiol Infect 23:98–103. doi: 10.1016/j.cmi.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsube S, Szigeti G, Yaguchi T, Frisvad JC. 2014. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]