ABSTRACT
In a carbapenem-resistant Escherichia coli clinical isolate of sequence type 167, two copies of blaNDM-5 were found on a 144,225-bp IncF self-transmissible plasmid of the F36:A4:B− type. Both blaNDM-5 genes were located in 11,065-bp regions flanked by two copies of IS26. The two regions were identical in sequence but were present at different locations on the plasmid, suggesting a duplication of the same region. This study highlights the complex genetic contexts of blaNDM-5.
KEYWORDS: carbapenem resistance, plasmids, NDM, Escherichia coli, carbapenemase, carbapenems
TEXT
New Delhi metallo-β-lactamase (NDM) is a type of carbapenem-hydrolyzing enzyme (carbapenemase) with the ability to hydrolyze all β-lactams except monobactams (1), and it represents a serious challenge for treatment of bacterial infections, infection control, and public health. To date, there are 21 variants of NDM, including NDM-5, one of the most common variants encountered in the Enterobacteriaceae (2–5). The NDM-5-encoding gene, blaNDM-5, usually exists in a single copy on plasmids. However, we have found the peculiar presence of two copies of blaNDM-5 on a single plasmid within an Escherichia coli clinical isolate, which is reported here.
E. coli strain SCEC020007 was recovered from the urine of a female outpatient with a urinary tract infection in October 2016 in China. The strain was resistant to amikacin (MIC, >512 μg/ml), ceftazidime (>512 μg/ml), ceftazidime-avibactam (>512/4 μg/ml), ciprofloxacin (256 μg/ml), imipenem (64 μg/ml), meropenem (256 μg/ml), piperacillin-tazobactam (>512/4 μg/ml), and trimethoprim-sulfamethoxazole (128/2,432 μg/ml) and intermediate to aztreonam (8 μg/ml) but was susceptible to colistin (2 μg/ml) and tigecycline (0.25 μg/ml), as determined using the broth dilution method of the Clinical and Laboratory Standards Institute (6). As there are no breakpoints of colistin and tigecycline from CLSI, those defined by EUCAST (http://www.eucast.org/) were applied.
A draft genome sequence of the strain was generated on the Illumina HiSeq X10 platform, which generated 5,557,833 clean reads and 1.67 Gb of clean bases. A total of 113 contigs (102 >1,000 bp; N50, 126,680 bp) with a 50.76% GC content were de novo assembled using SPAdes (7). Strain SCEC020007 belonged to phylogenetic group A, determined using PCR as described previously (8), and sequence type 167 (ST167) was determined by using the genomic sequence to query the E. coli multilocus sequence typing database (http://enterobase.warwick.ac.uk/species/index/ecoli). Antimicrobial resistance genes were identified from genome sequences using the ABRicate program (https://github.com/tseemann/abricate) to query the ResFinder database (http://genomicepidemiology.org/). Strain SCEC020007 had 9 antimicrobial resistance genes mediating resistance to aminoglycosides (aadA2, aadA5, and rmtB), β-lactams (blaNDM-5 and blaTEM-1), tetracycline [tet(A)], sulfonamides (sul1), and trimethoprim (dfrA12 and dfrA17). Plasmid replicon types within strain SCEC020007 were determined using the PlasmidFinder tool (http://genomicepidemiology.org/). Surprisingly, strain SCEC020007 had an IncFIA, an IncFII, and an IncB/O/K/Z replicon, but no IncX3, which is the common replicon type of plasmids associated with blaNDM-5.
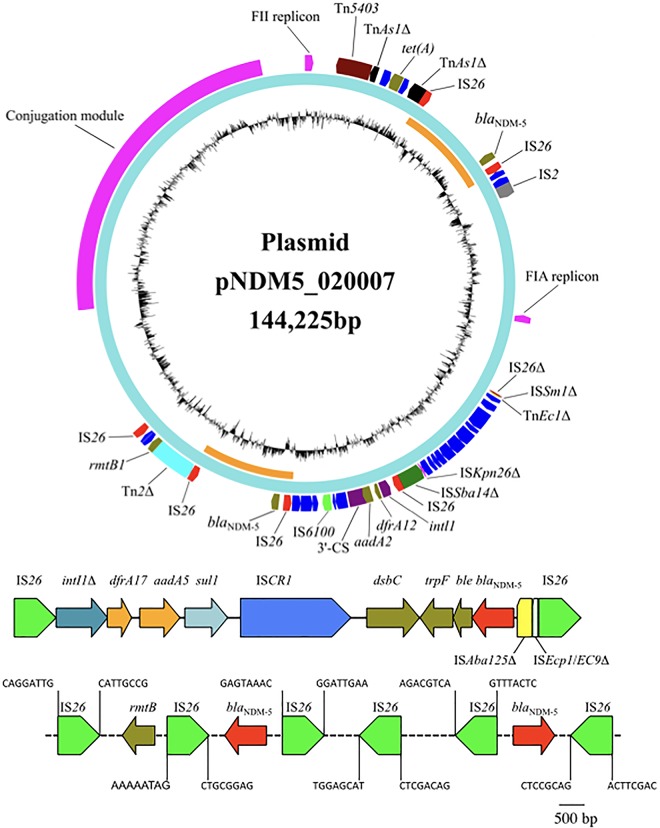
To untangle the genetic context of blaNDM-5, strain SCEC020007 was subjected to sequencing using the long-read MinION Sequencer (Nanopore, Oxford, UK). MinION sequencing generated 66,354 reads and 524,460,566 nucleotides (mean size, 7,905 bp; N50, 12,429 bp; mean read quality, 9). A de novo hybrid assembly of both short Illumina reads and long MinION reads was constructed using Unicycler v0.4.3 (9) in conservative mode for increased accuracy. The complete circular contigs generated were then corrected using Plion v1.22 (10) with Illumina reads for several rounds until no change was detected. The hybrid assembly of Illumina and MinION reads revealed that strain SCEC020007 had a 4.8-Mb circular chromosome, a 144,225-bp plasmid (designated pNDM5_020007) containing IncFIA and FII replicons, and an 84,952-bp plasmid with an IncB/O/K/Z replicon (designated pBOKZ_020007). Surprisingly, there were two copies of blaNDM-5 in strain SCEC020007, both of which were present on pNDM5_020007. Both blaNDM-5 genes were located in 11,065-bp regions flanked by two copies of IS26. The two regions were identical in sequence but were present at different locations on pNDM5_020007 (Fig. 1), suggesting that the 11,065-bp region is duplicated. The presence of the two blaNDM-5 genes and their locations on pNDM5_020007 were confirmed by PCR. The 11,065-bp region contained a complex class 1 integron with a dfrA17-aadA5 cassette array and ISCR1 (insertion sequence common region 1), which is truncated by IS26 at its 5′ conserved segment; a 69-bp remnant of cutA1 (encoding an ion-tolerant protein); dsbC (encoding an oxidoreductase); trpF (encoding a phosphoribosyl anthranilate isomerase); ble (mediating bleomycin resistance); blaNDM-5, a truncated ISAba125; and a truncated ISEcp1/ISEc9 element (Fig. 1). The coexistence of two blaNDM-5 genes has not been reported before, but the coexistence of two blaNDM-1 genes has been described previously (11, 12). Two tandem copies of blaNDM-1 genes have been found in the chromosomes of an ST167 E. coli in China (11) and a Pseudomonas aeruginosa strain in Serbia (12). In both cases, the tandem copies of blaNDM-1 are associated with ISCR1 but not with IS26. It is known that ISCR1 uses the rolling circle mechanism for transposition and may generate tandem duplication of its mobilized sequence via homologous recombination (13). However, the duplication of the 11,065-bp region carrying blaNDM-5 on pNDM5_020007 is not tandem, suggesting that the duplication might not result from the action of ISCR1, but could be mediated by IS26. Two copies of IS26 could form a composite transposon able to mobilize the intervening genetic components. However, no 8-bp direct target repeats, which are characteristics of IS26 transposition, were present flanking any of the IS26-bracketed regions (Fig. 1), suggesting that homologous recombination had occurred. The exact mechanism for the duplication of such a large region warrants further studies.
FIG 1.
pNDM5_020007 and the genetic context of blaNDM-5. The two 11,065-bp blaNDM-5-containing regions bracketed by IS26 are indicated by orange circles in the map of pNDM5_020007 and are shown in detail in the middle. Δ represents truncated genes or mobile genetic elements. The 8-bp target sequences flanking IS26 are shown at bottom.
Assembly based on Illumina reads alone generated only a single contig containing blaNDM-5 and was unable to reveal that there were actually two identical copies of the same contig. This imposes difficulties for completing the blaNDM-5-carrying plasmid sequence by conventional methods, including by PCR and Sanger sequencing to close gaps between contigs. In contrast, MinION sequencing was able to resolve the copy numbers of genes and contigs and their exact position on the plasmid relative to each other.
Plasmid multilocus sequence typing (pMLST) was performed using the pMLST tool (https://cge.cbs.dtu.dk/services/pMLST/). pNDM5_020007 belongs to the F36:A4:B− type. pNDM5_020007 has the closest similarity (97% coverage and 99% identity) to a 149.5-kb unnamed plasmid (GenBank accession no. CP023871) from E. coli strain FDAARGOS_434, which was recovered from a human rectal swab in British Colombia, Canada, in 2014. This unnamed plasmid also carries blaNDM-5 (a single copy) and belongs to the F36:A4:B− type. Backbones of pNDM5_020007 and the unnamed plasmid of strain FDAARGOS_434 are almost identical, suggesting that they might have originated from a common plasmid. Conjugation experiments were carried out in broth and on filters, with the azide-resistant E. coli strain J53 as the recipient. pNDM5_020007 could be transferred by conjugation, suggesting that it is self-transmissible. MICs of imipenem and carbapenem against the E. coli J53 transconjugant carrying pNDM5_020007 were 8 and 16 μg/ml, respectively, which were identical to those against the E. coli J53 transconjugant carrying an IncX3 plasmid with a single copy of blaNDM-5 from a local strain (14). Although the two plasmids belonged to different replicon types, it appears that the two copies of blaNDM-5 did not alter the activity of carbapenems compared with that of a plasmid with a single copy.
In conclusion, we identified the presence of two blaNDM-5 genes on an F36:A4:B− self-transmissible plasmid. The coexistence of two blaNDM-5 genes was due to the duplication of an IS26-bracketed region containing ISCR1.
Accession number(s).
The complete sequences of pBOKZ_020007 and pNDM5_020007 and the chromosomal sequence of strain SCEC020007 have been deposited in GenBank under the accession no. CP025625, CP025626, and CP025627, respectively.
ACKNOWLEDGMENTS
This work was supported by grants from the National Natural Science Foundation of China (projects 81222025, 81572030, and 81772233) and a joint grant from the National Natural Science Foundation of China (project 81661130159) and the Newton Advanced Fellowship, Royal Society, United Kingdom (project NA150363).
REFERENCES
- 1.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske CG, Irfan S, Krishnan P, Kumar AV, Maharjan S, Mushtaq S, Noorie T, Paterson DL, Pearson A, Perry C, Pike R, Rao B, Ray U, Sarma JB, Sharma M, Sheridan E, Thirunarayan MA, Turton J, Upadhyay S, Warner M, Welfare W, Livermore DM, Woodford N. 2010. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 10:597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hornsey M, Phee L, Wareham DW. 2011. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother 55:5952–5954. doi: 10.1128/AAC.05108-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iovleva A, Doi Y. 2017. Carbapenem-resistant Enterobacteriaceae. Clin Lab Med 37:303–315. doi: 10.1016/j.cll.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215:S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, Li Y, Liao K, Chen S. 2017. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine 19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; twenty-third informational supplement. M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 7.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen P, Yi M, Fu Y, Ruan Z, Du X, Yu Y, Xie X. 2017. Detection of an Escherichia coli sequence type 167 strain with two tandem copies of blaNDM-1 in the chromosome. J Clin Microbiol 55:199–205. doi: 10.1128/JCM.01581-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jovcic B, Lepsanovic Z, Begovic J, Rakonjac B, Perovanovic J, Topisirovic L, Kojic M. 2013. The clinical isolate Pseudomonas aeruginosa MMA83 carries two copies of the blaNDM-1 gene in a novel genetic context. Antimicrob Agents Chemother 57:3405–3407. doi: 10.1128/AAC.02312-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toleman MA, Bennett PM, Walsh TR. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol Mol Biol Rev 70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Feng Y, Zhang X, McNally A, Zong Z. 2017. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 61:e01757-17. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]