ABSTRACT
Rapid diagnostic tests (RDTs) have revolutionized the management of Gram-negative bacteremia by allowing antimicrobial stewardship teams the ability to escalate therapy and improve patient outcomes through timely organism identification and detection of certain resistance determinants. However, given the complex nature of Gram-negative resistance, stewardship teams are left without clear direction for how to respond when resistance determinants are absent, as the safety of de-escalation in this setting is unknown. The primary purpose of this analysis was to determine the negative predictive values (NPVs) of resistance marker absence for predicting susceptibility in target bug-drug scenarios at two geographically distinct institutions. A total of 1,046 Gram-negative bloodstream isolates that were analyzed with the Verigene BC-GN platform were assessed. Except for Pseudomonas aeruginosa, the absence of resistance determinants as reported by the RDT largely predicted susceptibility to target antibiotics at both institutions. NPVs for ceftriaxone susceptibility in Escherichia coli and Klebsiella pneumoniae in the absence of either CTX-M or a carbapenemase gene were 98% and 93 to 94%, respectively. Similar results were seen with other target bug-drug scenarios, with NPVs of 94 to 100% demonstrated at both institutions, with the exception of P. aeruginosa, for which NPVs were poor, likely due to the more complex nature of resistance in this pathogen. The results of this study show that clinicians at both institutions should have confidence in de-escalation in the absence of resistance determinant detection by Verigene BC-GN testing, and the methodology described within this article can serve as a blueprint for other stewardship programs to employ at their institutions to optimize management of Gram-negative bacteremia.
KEYWORDS: Gram negative, Verigene, bacteremia, film array, rapid diagnostic, stewardship
INTRODUCTION
Molecular rapid diagnostic tests (RDTs) have shown the ability to decrease time to appropriate therapy for patients with bloodstream infections (BSIs), leading to decreases in both mortality and length of stay of survivors (1). This decrease in time to appropriate therapy is driven by earlier organism and key resistance determinant identification coupled with initiation of therapy in patients who are not on antimicrobials or escalation of therapy in those who are deemed to be on inadequate regimens. This improvement can be most marked for Gram-negative (GN) BSIs, for which, due to complexities in and prevalence of resistance determinants, common empirical therapies (such as ceftriaxone or an antipseudomonal beta-lactam) are less likely to have in vitro activity against the identified organism. In contrast, for Gram-positive BSIs, empirical therapy is often vancomycin, which is likely to at least provide in vitro activity against most organisms, although it may not be the ideal agent for definitive use.
The Verigene BC-GN platform is a multiplex microarray platform that can detect organisms to the genus level for four genera (Acinetobacter spp., Citrobacter spp., Proteus spp., and Enterobacter spp.), as well as four organisms to the species level (Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Pseudomonas aeruginosa), within 2.5 h from the time the blood culture bottle is flagged positive and Gram staining is performed (2). Additionally, this platform can identify six beta-lactamase genes (the extended-spectrum beta-lactamase [ESBL] CTX-M as well as the carbapenemases KPC, VIM, IMP, NDM, and OXA) (2). When Verigene BC-GN testing identifies a Gram-negative organism and/or beta-lactamase gene, stewardship personnel can escalate therapy, decrease the time to appropriate therapy, and ultimately improve outcomes. A recent quasi-experimental study by Rivard and colleagues demonstrated that the coupling of this technology with active stewardship intervention was able to decrease the time to effective therapy by 15.7 h (8.8 versus 24.5 h; P = 0.03) for patients on inappropriate therapy at the time of Gram stain identification (3). In the entire postimplementation cohort, patients in the intervention group had decreased lengths of stay, with a median of a 2-day decrease (7 versus 9 days; P = 0.001) (3).
Given the complexity of Gram-negative resistance and the incomplete information provided through current RDTs, stewardship teams are left without clear direction for intervention when an organism is identified in the absence of resistance determinants. For example, if an institution has a high rate of ESBL-producing organisms and the Verigene BC-GN result shows a K. pneumoniae isolate that is CTX-M negative, stewardship personnel might be hesitant to de-escalate therapy to a third-generation cephalosporin (or to continue ceftriaxone and not escalate to a carbapenem) given that resistance could still exist due to either non-CTX-M-type ESBLs, plasmid-mediated AmpC beta-lactamases, or other mechanisms of resistance not detectable by RDTs. This often leads to a hesitance to de-escalate or modify therapy in these settings, and patients commonly remain on or are placed on therapy in accordance with their local antibiogram data to ensure appropriate coverage of the identified organism, limiting the clinical impact of RDTs. In fact, in the aforementioned analysis by Rivard and colleagues, the median time to de-escalation from the Verigene BC-GN result was a modest 4-h reduction over the time for the control arm, for which no RDT was available (49.9 versus 54.4 h; P = 0.01) (3). This analysis shows the need for enhanced strategies to inform de-escalation in these settings.
To date, one analysis, by Rödel and colleagues, attempted to provide treatment recommendations that combined Verigene test results with final susceptibility information to give preferred therapeutic options based on presence/absence of resistance determinants (4). This analysis suggested high negative predictive values (NPVs) for prediction of resistance to target antimicrobials if resistance genes were absent (i.e., if resistance genes were not present, susceptibility to the target agent was seen). However, extremely small numbers, particularly for GN organisms, limited the study. In the analysis, detection of the CTX-M marker gene identified 10 of the 13 ceftriaxone-resistant E. coli/K. pneumoniae isolates, resulting in an NPV of 93% when this marker was absent (i.e., susceptibility to ceftriaxone was 93% when the CTX-M result was negative). This finding is somewhat limited by the fact that only 55 E. coli or K. pneumoniae isolates were included in the analysis, and no more than 6 of any other target Gram-negative organisms were included in the study. Furthermore, given that mechanisms of resistance to target Gram-negative antimicrobials can vary from region to region, the external generalizability of these data remains unclear.
Given the importance of limiting unnecessary exposure to broad-spectrum antimicrobial therapy, strategies are needed to determine if de-escalation (or a lack of escalation) can safely be performed if the results from the Verigene BC-GN platform show an absence of resistance determinants. Based on the current absence of evidence and strategies to aid clinicians' decision-making in these scenarios, the primary aim of the present analysis was to determine the ability of Verigene BC-GN organism identification and resistance determinant presence/absence to predict antimicrobial susceptibility among target Gram-negative organisms in order to better direct antimicrobial stewardship programs. Furthermore, given known variations in local epidemiology throughout various regions of the country, the potential external validity of this process was analyzed by comparison of two institutions in different geographic areas.
RESULTS
Microbiology results.
Over the study period, 1,046 positive Gram-negative blood cultures were analyzed, including 765 at the Detroit Medical Center (DMC) and 281 at the University of Maryland Medical Center (UMMC). The organism breakdown differed by institution (Table 1); although E. coli (n = 489) and K. pneumoniae (n = 197) were the two most prevalent organisms at both institutions, E. coli was significantly more common at the DMC (50% versus 37%; P < 0.001), whereas P. aeruginosa was more common at UMMC (15% versus 7%; P < 0.001). At the DMC, 103 (13%) isolates were positive for a resistance determinant, with 89 CTX-M, 6 KPC, and 8 OXA enzymes identified. At UMMC, resistance determinants were seen in 34 (12%) isolates and consisted of 24 CTX-M, 5 KPC, and 5 OXA enzymes. There were no significant differences in the presence of any resistance determinant between practice sites.
TABLE 1.
Identification of Gram-negative organisms at each institution by Verigene BC-GN testinga
| Organism | No. (%) of isolates |
|
|---|---|---|
| DMC (n = 765) | UMMC (n = 281) | |
| E. coli | 384 (50) | 105 (37) |
| K. pneumoniae | 140 (18) | 57 (20) |
| K. oxytoca | 23 (3) | 11 (4) |
| Proteus spp. | 57 (8) | 11 (4) |
| Enterobacter spp. | 61 (8) | 34 (12) |
| Citrobacter spp. | 10 (1) | 6 (2) |
| Pseudomonas spp. | 51 (7) | 43 (15) |
| Acinetobacter spp. | 39 (5) | 14 (5) |
DMC, Detroit Medical Center; UMMC, University of Maryland Medical Center.
Performance of Verigene BC-GN in predicting susceptibility in target bug-drug scenarios.
Table 2 describes the rate of resistance to the target antimicrobial agent for the identified GN organism, frequency at which resistance to the target agent was identified by the resistance determinants identified by Verigene BC-GN, sensitivity, specificity, positive predictive value (PPV), and NPV for each target bug-drug combination. In general, the identification of resistance determinants by Verigene BC-GN performed very well at detecting the presence or absence of resistance to key antimicrobial agents. These results were consistent across both institutions. The one notable exception was P. aeruginosa, for which Verigene BC-GN testing failed to detect any resistant isolates.
TABLE 2.
Verigene BC-GN analytic results by bug-drug combinationa
| Institution and target organism | n | Target drug | Resistance marker | No. (%) of resistant isolates | No. of isolates identified by Verigene | SN (%) | SP (%) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|---|
| Detroit Medical Center | |||||||||
| E. coli | 384 | Ceftriaxone | Any (CTX-M)b | 63 (16) | 56 | 89 | 99 | 97 | 98 |
| K. pneumoniae | 140 | Ceftriaxone | Any | 39 (28) | 33 | 85 | 99 | 97 | 94 |
| 140 | Ertapenem | KPC | 7 (5) | 6 | 86 | 100 | 100 | 99 | |
| K. oxytoca | 23 | Ceftriaxone | Any (CTX-M)b | 2 (9) | 1 | 50 | 100 | 100 | 95 |
| Proteus spp. | 57 | Ceftriaxone | Any (CTX-M)b | 7 (12) | 4 | 57 | 100 | 100 | 94 |
| Enterobacter spp. | 61 | Cefepime | Any (CTX-M)b | 3 (5) | 1 | 33 | 100 | 100 | 97 |
| Citrobacter spp. | 10 | Cefepime | Any | 0 | 0 | 100 | |||
| Acinetobacter spp. | 39 | Meropenem | OXA | 10 (26) | 8 | 80 | 93 | 80 | 93 |
| Pseudomonas spp. | 51 | Cefepime | Any | 6 (12) | 0 | 88 | |||
| University of Maryland Medical Center | |||||||||
| E. coli | 105 | Ceftriaxone | Any (CTX-M)b | 15 (14) | 14 | 93 | 100 | 100 | 99 |
| K. pneumoniae | 57 | Ceftriaxone | Any | 15 (26) | 12 | 80 | 100 | 100 | 93 |
| 57 | Ertapenem | KPC | 5 (9) | 5 | 100 | 100 | 100 | 100 | |
| K. oxytoca | 9 | Ceftriaxone | Any | 0 | 0 | 100 | |||
| Proteus spp. | 11 | Ceftriaxone | Any | 0 | 0 | 100 | |||
| Enterobacter spp. | 34 | Cefepime | Any (CTX-M)b | 3 (9) | 3 | 100 | 100 | 100 | 100 |
| Citrobacter spp. | 6 | Cefepime | Any | 0 | 0 | 100 | |||
| Acinetobacter spp. | 14 | Meropenem | OXA | 5 (36) | 5 | 100 | 100 | 100 | 100 |
| Pseudomonas spp. | 43 | Piperacillin-tazobactam | Any | 15 (35) | 0 | 65 |
NPV, negative predictive value; PPV, positive predictive value; SN, sensitivity; SP, specificity. The cells for SN, SP, and PPV were left blank if either no resistance to the target drug was seen in study isolates or Verigene did not identify any resistance for the target drug.
While the presence of any resistance marker was tested as a marker of ceftriaxone resistance in E. coli, K.oxytoca, and Proteus spp. or cefepime resistance in Enterobacter spp., only CTX-M was identified in this study for each of these species.
At the DMC, ceftriaxone resistance was present in 16% of E. coli isolates, and 56/63 (89%) of these were predicted by the presence of the CTX-M resistance determinant. Therefore, the NPV of the absence of this determinant was 98%, meaning that 98% of E. coli isolates were susceptible to ceftriaxone if CTX-M was not detected. Similarly, at UMMC, ceftriaxone resistance was seen in 14% of E. coli isolates, 14/15 (93%) of which were predicted by Verigene BC-GN detection of CTX-M, giving an NPV of 99%. At the DMC, ceftriaxone resistance was seen in 28% of K. pneumoniae isolates, with 33/39 (85%) of these detected by Verigene BC-GN testing (positive for either CTX-M or KPC), for an NPV of 94%. At the DMC, ertapenem resistance was seen in 5% of K. pneumoniae isolates, and 6/7 (86%) of these were detected by Verigene BC-GN, for an NPV of 99% in the absence of the KPC marker. Similar results for ceftriaxone and ertapenem resistance in K. pneumoniae were seen at UMMC. The rate of ceftriaxone resistance was 26%, with 12/15 (80%) resistant isolates detected by Verigene BC-GN, for an NPV of 93%. Ertapenem resistance was seen in 9% of K. pneumoniae isolates, all of which (5/5 isolates) were detected by Verigene BC-GN, for an NPV of 100%.
Similar findings were demonstrated at both institutions for Proteus spp., K. oxytoca, Citrobacter spp., and Enterobacter spp., with NPVs ranging from 94 to 100% at the DMC and an NPV of 100% for each of these organisms at UMMC for assessing resistance to target agents in the absence of key resistance determinants (Table 2). For Acinetobacter baumannii, the presence/absence of detection of the OXA genes by Verigene BC-GN was predictive of meropenem/imipenem susceptibility at each site. At the DMC, of the 26% carbapenem-resistant A. baumannii (CRAB) isolates, 8/10 (80%) were detected by Verigene BC-GN, while at UMMC all of the 36% CRAB isolates were detected, leading to NPVs of 93% and 100%, respectively.
Antibiograms as a function of organism identified and presence or absence of resistance determinants.
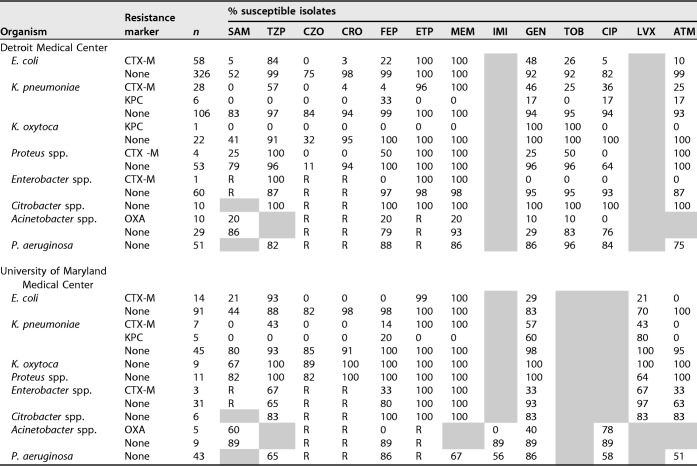
Table 3 describes susceptibilities to common antimicrobials for each organism as a function of the presence or absence of resistance determinants for the DMC and UMMC. As demonstrated by the NPVs described above, high degrees of ceftriaxone susceptibility were seen in target organisms at both institutions in the absence of resistance determinants. In the absence of resistance markers, susceptibilities of E. coli, K. pneumoniae, K. oxytoca, and Proteus mirabilis to ceftriaxone were high at both institutions, ranging from 94 to 98% at the DMC and from 91 to 100% at UMMC. Importantly, cefazolin susceptibility was significantly lower for both E. coli (75 to 82%) and K. pneumoniae (84 to 85%) isolates, arguing against reliance on cefazolin prior to the return of susceptibility information. Interestingly, there were large discordances between the two institutions with regard to cefazolin susceptibility in K. oxytoca (89% at UMMC versus 32% at the DMC) and P. mirabilis (82% at UMMC versus 11% at the DMC) in the absence of a resistance determinant. These significant discordances are likely due to Vitek limitations at UMMC, where the low end of the MIC range for testing cefazolin is ≤4 mg/liter, which is higher than the updated CLSI susceptibility breakpoint employed at the DMC.
TABLE 3.
Antibiograms as a function of organism and resistance determinantsa
SAM, ampicillin-sulbactam; TZP, piperacillin-tazobactam; CZO, cefazolin; CRO, ceftriaxone; FEP, cefepime; ETP, ertapenem; MEM, meropenem; IMI, imipenem; GEN, gentamicin; TOB, tobramycin; CIP, ciprofloxacin; LVX, levofloxacin; ATM, aztreonam; R, intrinsic organism resistance. Shaded boxes indicate that the antibiotic was not tested.
For Enterobacter and Citrobacter spp., susceptibilities to cefepime in the absence of resistance determinants were high, at 97 and 100% at the DMC and 80 and 100% at UMMC. The difference in Enterobacter susceptibilities between the two sites is related to differing breakpoints utilized at the institutions (i.e., the DMC uses the CLSI breakpoint of 8 mg/liter, whereas UMMC uses the EUCAST breakpoint of 1 mg/liter). Ertapenem susceptibilities were similarly high in these organisms. For A. baumannii, the presence or absence of OXA reflected susceptibility to both meropenem and ampicillin-sulbactam at each site. In the absence of OXA, susceptibilities to these two agents were 93 and 86% at the DMC and 89% (for each) at UMMC. For P. aeruginosa, no resistance determinants were identified, and as is common for this organism, susceptibilities differed between sites.
The antibiograms in Table 3 display important susceptibility differences in non-beta-lactam antimicrobials and highlight interhospital differences as a function of the presence/absence of beta-lactamase genes. For example, in E. coli, the presence of CTX-M was associated with decreases in susceptibility to gentamicin and ciprofloxacin of 44 and 77%, respectively, at the DMC and with decreases in susceptibility to gentamicin and levofloxacin of 55% and 42%, respectively, at UMMC. The likely explanation for this difference is the known coexistence of resistance determinants to other agents on plasmids encoding CTX-M.
DISCUSSION
Rapid diagnostic tests for organism and resistance determinant identification in GN BSIs have revolutionized the management of patients with these infections and clearly demonstrated the opportunity for antimicrobial stewardship programs to escalate therapy, improve the time to appropriate therapy, and ultimately decrease both length of hospital stay and mortality (1, 5). However, as evidenced by a recent publication by Rivard and colleagues (3), knowing how to manage patients in cases where organism identification is known but resistance determinant results are negative is much more difficult. Given the known morbidity and mortality of these infections, the rising rates of antimicrobial resistance, and the inability of RDTs to detect all mechanisms of resistance, clinicians are often hesitant to de-escalate therapy based on these incomplete data, and in some instances will escalate therapy to cover the more resistant forms of infection. This is certainly the case at both institutions in this study, where, for example, ESBL rates of 15 to 28% for BSIs with E. coli and K. pneumoniae made withholding carbapenem therapy difficult with unstable patients, even in the absence of the CTX-M gene.
It was in the setting of this conundrum that the current analysis was developed and performed. Encouragingly, for all organisms except P. aeruginosa at both institutions, susceptibility was largely predicted by the presence/absence of the resistance determinants. Even in scenarios where the pretest probability of resistance was high (e.g., ceftriaxone with K. pneumoniae or meropenem with A. baumannii), the absence of the resistance determinant largely removed the doubt surrounding the presence of resistance, with NPVs of approximately 95 to 100%, meaning that only 0 to 5% of isolates will be resistant if the test is negative. To put this into perspective, current guidelines for hospital-acquired/ventilator-associated pneumonia recommend addition of dual Gram-negative empirical therapy only when resistance rates are >10% for the agent being considered for monotherapy (6), and therefore clinicians should feel comfortable de-escalating (or not escalating) therapy based on these results, even for critically ill patients. Importantly, however, these data should not be taken in a vacuum, and coverage for resistant organisms might still be warranted in certain scenarios, such as in patients with a history of resistance or those who are not responding to an antimicrobial that would be predicted to be active in vitro against the organism based on the results from the Verigene BC-GN platform.
To further optimize the management of these patients, antibiograms similar to those displayed in Table 3 can be provided to clinicians to increase awareness of optimal antimicrobial options in each organism-resistance determinant scenario. It is important that antibiograms in some of the organism-resistance determinant scenarios should be interpreted with caution given the relatively small numbers of samples analyzed, which are well below the ≥30 isolates recommend by the CLSI (7). However, given the amount of time it would take to obtain 30 BSI isolates at an institution for each individual organism-resistance determinant scenario, it is not realistic to wait for identification of such a large number of patients, and we still recommend that such findings be shared with clinicians to better drive care, but with a word of caution about the small numbers of samples.
As mentioned above, it is important that Verigene BC-GN testing was unable to predict resistance to both first-line agents (cefepime and piperacillin-tazobactam) in P. aeruginosa. This is not surprising, as resistance to these two agents in Pseudomonas aeruginosa is most commonly driven by chromosomally encoded AmpC beta-lactamases and/or a variety of efflux pumps, none of which are detected by Verigene BC-GN, and thus local antibiogram data still need to drive clinical decisions for this organism (8).
One interesting finding was that despite the completion of this analysis in two geographically distinct regions of the United States, the results were strikingly consistent. Despite this finding, we caution against extrapolation of these results to other institutions. Local epidemiology, particularly with regard to ESBL genes/ceftriaxone resistance, can vary greatly across regions and is unknown for most institutions. Therefore, although it is encouraging that the results in these two institutions were similar, it would be inappropriate to apply these findings or develop an algorithm elsewhere based on the results presented here without validation based on local institutional data. However, the process described here can easily be performed within one's institution.
There are important limitations to this analysis that warrant comment. First, despite the large sample sizes at both institutions, the results of this analysis represent only one time frame. As local, regional, and national mechanisms of resistance can change over time, it is important to continually perform such analyses to ensure continued applicability of the findings and to modify treatment algorithms accordingly. Additionally, as this was a retrospective analysis, no interventions were made based on the findings in this study. Therefore, the impact of this strategy, while theoretically beneficial in regard to antimicrobial usage, resistance, and adverse events, is still unclear, and future analyses should address outcomes, including antimicrobial usage outcomes, based on these interventions.
Additionally, it is important that this analysis was performed only on isolates for which the organism identification results from the Verigene BC-GN platform matched those of traditional microbiological methods. There are known limitations of Verigene BC-GN. While its sensitivity and specificity are both high (≥95%), the system is not absolute, and both false-negative results and organism misidentification can occur (2, 9). Furthermore, failure to identify all organisms in a polymicrobial BSI is a significant shortcoming of the system (2). Recent data from our group demonstrated that the Verigene BC-GN platform missed one or more pathogen in 36/56 (64%) polymicrobial GN BSIs (10). Importantly, however, in that analysis it was determined that the failure to identify the second organism would potentially lead to inappropriate de-escalation in only <2% of overall patients with GN BSIs. Regardless, these limitations should be considered in interpreting these results (10).
Finally, the resistance marker-antimicrobial agent pairs chosen for analysis were those that were deemed to be relevant based on local susceptibility and epidemiology data at the two practice sites. Therefore, our findings and interpretations might not be the most important questions at other sites. However, the methodology described herein can be modified to meet the needs of a given institution. For example, if cefazolin and ceftriaxone susceptibilities are similar among E. coli and K. pneumoniae isolates within an institution, the CTX-M analyses can be modified for their ability to predict cefazolin susceptibility. Additionally, in a region where OXA-48 or NDM is endemic, the analyses presented here can be modified to assess those resistance markers where we assessed KPC. Finally, if institutions are more comfortable with use of third-generation cephalosporins for treatment of potential chromosomal ampC-carrying pathogens, ceftriaxone can be substituted where we analyzed cefepime.
In conclusion, this analysis demonstrates that at both the DMC and UMMC, treatment decisions (both escalation and de-escalation) can be made based on the results of Verigene BC-GN testing for all organisms detected, with the exception of P. aeruginosa. This has important stewardship implications with regard to limiting unnecessary broad-spectrum antibiotic usage and potential collateral damage associated with it. While it must be stressed that the findings of this analysis should not be applied to other institutions given differences in local epidemiology, this report provides a blueprint that stewardship programs can utilize at their institutions to assess how Verigene BC-GN performs in these scenarios, to develop treatment algorithms based on the results, and to provide antibiogram data to clinicians as a function of presence/absence of resistance determinants to assist in optimal antimicrobial selection. This type of analysis will allow stewards to optimize the management of GN BSI and, ultimately, patient outcomes at their institution.
MATERIALS AND METHODS
Study design and setting.
This was a multicenter retrospective microbiological review of all blood cultures positive for at least one Gram-negative organism that underwent testing with the Verigene BC-GN platform as part of the routine clinical testing by the microbiology lab at either the DMC or UMMC from June 2015 to July 2016. In order to be included in the analysis, the same organism had to be identified by both Verigene BC-GN and traditional microbiology methods. Additionally, susceptibility testing had to have been performed for the target GN organism. Isolates were excluded from analysis if an off-panel organism (e.g., Serratia spp.) was identified. Duplicate isolates (or multiple isolates from the same patient) were eligible for this analysis if they met institution rules for reexamination by Verigene BC-GN. At the DMC, this consisted of GN isolates identified >7 days after the index organism, and at UMMC this consisted of GN isolates that were morphologically different from the index organism during the same hospital admission. The institutional review board (IRB) at the University of Maryland approved this study, and the IRB at the Detroit Medical Center and Wayne State University determined that it was exempt from review.
Microbiological analysis.
At the DMC, blood cultures were collected using Bactec Lytic/10 Anaerobic and/or Bactec Plus Aerobic/F culture vials and incubated on a Bactec FX (Becton, Dickinson and Co., Sparks, MD) system. At UMMC, blood cultures were collected using BacTAlert FA and/or FN bottles and incubated on a BacTAlert 3D (bioMérieux, Durham, NC) system. If organisms were detected by these systems, Gram staining was performed, and if GN rods were identified, Verigene BC-GN testing was performed. Traditional clinical microbiology techniques consisted of matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) analysis (Bruker, Billerica, MA) for identification and BD Phoenix testing (Becton, Dickinson and Co., Sparks, MD) for susceptibility testing at the DMC and Vitek 2 testing (bioMérieux, Durham, NC) for organism identification and susceptibility testing for Enterobacteriaceae spp. and Kirby-Bauer disc diffusion testing for susceptibility testing of non-lactose-fermenting GN organisms at UMMC. Both the DMC and UMMC utilized CLSI breakpoints for the interpretation of susceptibilities for all causative organisms (11), with the exception of UMMC utilizing EUCAST breakpoints for cefepime and aztreonam for Enterobacteriaceae (12). Furthermore, due to panel limitations with the Vitek 2 system, isolates at UMMC were considered susceptible to cefazolin at MICs of ≤4 mg/liter, whereas at the DMC the CLSI breakpoint (2 mg/liter) was utilized (11).
Data analysis.
The primary objective of this analysis was to compare the results of Verigene BC-GN testing with those of conventional antimicrobial susceptibility testing at each site. Specifically, the aim was to determine the sensitivity, specificity, PPV, and NPV for organism identification and the presence/absence of beta-lactamase genes detected by Verigene BC-GN for antimicrobial susceptibility testing to target antimicrobials at each institution. The ultimate goal of this assessment was to be able to give a degree of confidence of antimicrobial susceptibility in the absence of target resistance genes in order to assess the feasibility of de-escalation (or lack of escalation) in cases where resistance determinants are not detected.
For the primary objective, clinically relevant resistance marker-antimicrobial agent pairs were analyzed for the different GN organisms. For the analyses of enteric Enterobacteriaceae (E. coli, K. pneumoniae, and Proteus spp.), the presence/absence of any resistance gene (for either CTX-M or a carbapenemase) was assessed for the ability to predict ceftriaxone susceptibility. KPC was the only carbapenemase identified in Enterobacteriaceae in both institutions, and therefore the presence/absence of KPC was assessed for the ability to predict ertapenem susceptibility. Given that both institutions recommend avoidance of third-generation cephalosporins for BSIs caused by Enterobacter or Citrobacter spp. due to the potential presence of and selection for chromosomally encoded AmpC beta-lactamases, the presence/absence of any resistance gene (for either CTX-M or a carbapenemase) was utilized to assess the predictive ability for susceptibility of these organisms to cefepime.
As OXA was the only carbapenemase present in A. baumannii at both institutions, the presence/absence of OXA was assessed for the ability to predict meropenem/imipenem susceptibility in Acinetobacter spp. For P. aeruginosa, the presence/absence of any resistance gene was assessed for the ability to predict susceptibility to either cefepime or piperacillin-tazobactam. As the two institutions utilize different “workhorse” antipseudomonal agents, the DMC assessed predictive ability for cefepime susceptibility, whereas UMMC assessed predictive ability for piperacillin-tazobactam susceptibility.
Statistical analyses included comparisons between institutions with regard to causative organism and frequencies of resistance determinants. Dichotomous variables were compared using the Pearson chi-square test or the Fisher exact test, as appropriate. A P value of ≤0.05 was considered statistically significant. Study data were collected and managed using Microsoft Excel. Statistical analysis was completed using SPSS, version 23.0 (IBM Corp., Armonk, NY).
Antibiogram construction.
To further direct care, antibiograms were developed in order to display percentages of antimicrobial susceptibility of target organisms to key antimicrobial agents as a function of resistance gene presence/absence (Table 3). This was first accomplished by determining the percentage of isolates of each pathogen susceptible to each antimicrobial agent when no resistance determinant was identified and displaying susceptibility rates in this setting. This process was then repeated for each organism-resistance determinant pair that was identified in the analysis. Although standard antibiogram rules recommend against reporting antibiogram data when <30 isolates exist for a given scenario (7), this rule was ignored for this analysis in order to give clinicians as much information as possible.
Antimicrobial agents included in the analyses were those deemed important to the individual sites, based on treatment preferences, formulary decisions, and testing methodologies. Shaded cells in the antibiograms represent either scenarios where automated susceptibility methodologies block results for organism-drug combinations (e.g., piperacillin-tazobactam and A. baumannii) or those where a study institution (UMMC) utilizes different testing methodologies and agent selection for target bug-drug scenarios (i.e., ciprofloxacin for nonfermenters and levofloxacin for fermenters). Furthermore, “R” is used in situations where intrinsic resistance mechanisms make therapy questionable regardless of the susceptibility pattern displayed (e.g., first- to third-generation cephalosporins and potential chromosomal ampC-carrying organisms).
REFERENCES
- 1.Timbrook TT, Morton JB, McConeghy KW, Caffrey AR, Mylonakis E, LaPlante KL. 2017. The effect of molecular rapid diagnostic testing on clinical outcomes in bloodstream infections: a systematic review and meta-analysis. Clin Infect Dis 64:15–23. doi: 10.1093/cid/ciw649. [DOI] [PubMed] [Google Scholar]
- 2.Nanosphere, Inc. 2014. Verigene Gram-negative blood culture nucleic acid test (BC-GN) package insert 027-00039-01, rev B. Nanosphere Inc, Northbrook, IL. [Google Scholar]
- 3.Rivard KR, Athans V, Lam SW, Gordon SM, Procop GW, Richter SS, Neuner E. 2017. Impact of antimicrobial stewardship and rapid microarray testing on patients with Gram-negative bacteremia. Eur J Clin Microbiol Infect Dis 36:1879–1887. doi: 10.1007/s10096-017-3008-6. [DOI] [PubMed] [Google Scholar]
- 4.Rödel J, Karrasch M, Edel B, Stoll S, Bohnert J, Löffler B, Saupe A, Pfister W. 2016. Antibiotic treatment algorithm development based on a microarray nucleic acid assay for rapid bacterial identification and resistance determination from positive blood cultures. Diagn Microbiol Infect Dis 84:252–257. doi: 10.1016/j.diagmicrobio.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 5.Bookstaver PB, Nimmich EB, Smith TJ III, Justo JA, Kohn J, Hammer KL, Troficanto C, Albrecht HA, Al-Hasan MN. 2017. Cumulative effect of an antimicrobial stewardship and rapid diagnostic testing bundle on early streamlining of antimicrobial therapy in Gram-negative bloodstream infections. Antimicrob Agents Chemother 61:e00189-17. doi: 10.1128/AAC.00189-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalil AC, Metersky ML, Klompas M, Muscedere J, Sweeney DA, Palmer LB, Napolitano LM, O'Grady NP, Bartlett JG, Carratalà J, El Solh AA, Ewig S, Fey PD, File TM Jr, Restrepo MI, Roberts JA, Waterer GW, Cruse P, Knight SL, Brozek JL. 2016. Management of adults with hospital-acquired and ventilator-associated pneumonia: 2016 clinical practice guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin Infect Dis 63:e61–e111. doi: 10.1093/cid/ciw353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CLSI. 2014. Analysis and presentation of cumulative antimicrobial susceptibility test data; approved guideline, 4th ed CLSI document M39-A4. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Livermore DM. 2002. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin Infect Dis 34:634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- 9.Bork JT, Leekha S, Heil EL, Zhao L, Badamas R, Johnson JK. 2015. Rapid testing using the Verigene Gram-negative blood culture nucleic acid test in combination with antimicrobial stewardship intervention against Gram-negative bacteremia. Antimicrob Agents Chemother 59:1588–1595. doi: 10.1128/AAC.04259-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claeys KC, Heil EL, Pogue JM, Lephart PL, Johnson JK. 2018. The Verigene dilemma: gram-negative polymicrobial bloodstream infections and clinical decision making. Diagn Microbiol Infect Dis 2018:S0732-8893(18)30017-8. doi: 10.1016/j.diagmicrobio.2018.01.012. [DOI] [PubMed] [Google Scholar]
- 11.CLSI. 2018. Performance standards for antimicrobial susceptibility testing, 28th ed CLSI supplement M100. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 12.European Committee on Antimicrobial Susceptibility Testing. 2018. Breakpoint tables for interpretation of MICs and zone diameters, version 8.0. http://www.eucast.org/clinical_breakpoints/.