ABSTRACT
Pseudomonas aeruginosa plays a major role in many chronic infections. Its ability to readily form biofilms contributes to its success as an opportunistic pathogen and its resistance/tolerance to antimicrobial/antibiotic therapy. A low-molecular-weight alginate oligomer (OligoG CF-5/20) derived from marine algae has previously been shown to impair motility in P. aeruginosa biofilms and disrupt pseudomonal biofilm assembly. As these bacterial phenotypes are regulated by quorum sensing (QS), we hypothesized that OligoG CF-5/20 may induce alterations in QS signaling in P. aeruginosa. QS regulation was studied by using Chromobacterium violaceum CV026 biosensor assays that showed a significant reduction in acyl homoserine lactone (AHL) production following OligoG CF-5/20 treatment (≥2%; P < 0.05). This effect was confirmed by liquid chromatography-mass spectrometry analysis of C4-AHL and 3-oxo-C12-AHL production (≥2%; P < 0.05). Moreover, quantitative PCR showed that reduced expression of both the las and rhl systems was induced following 24 h of treatment with OligoG CF-5/20 (≥0.2%; P < 0.05). Circular dichroism spectroscopy indicated that these alterations were not due to steric interaction between the AHL and OligoG CF-5/20. Confocal laser scanning microscopy (CLSM) and COMSTAT image analysis demonstrated that OligoG CF-5/20-treated biofilms had a dose-dependent decrease in biomass that was associated with inhibition of extracellular DNA synthesis (≥0.5%; P < 0.05). These changes correlated with alterations in the extracellular production of the pseudomonal virulence factors pyocyanin, rhamnolipids, elastase, and total protease (P < 0.05). The ability of OligoG CF-5/20 to modify QS signaling in P. aeruginosa PAO1 may influence critical downstream functions such as virulence factor production and biofilm formation.
KEYWORDS: OligoG CF-5/20, Pseudomonas aeruginosa, alginate, biofilms, homoserine lactone, natural antimicrobial products, quorum-sensing inhibitor
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic and nosocomial human pathogen that can cause extensive tissue damage through the production of virulence factors and toxins, e.g., pyocyanin and proteases (1). The dynamic genome of P. aeruginosa is highly adaptable, enabling it to adjust to a wide range of environmental conditions (2, 3). This versatility allows it to colonize diverse physiological niches, including the respiratory tract, genitourinary tract, and wounds.
The sodium alginate oligomer OligoG CF-5/20, produced from the brown seaweed Laminaria hyperborea, has been shown to potentiate (enhance) antimicrobial efficacy, perturb multidrug-resistant (MDR) bacteria (4–7), and inhibit biofilm formation in a broad range of organisms (8). Furthermore, it has also previously been shown to inhibit swarming and “twitching” motility, exhibiting a significant effect on bacterial flagellum- and pilus-mediated chemotaxis (4, 5). Although OligoG CF-5/20 is known to have cation-chelating properties, the precise mechanism of action in mediating this diverse range of effects remains unclear.
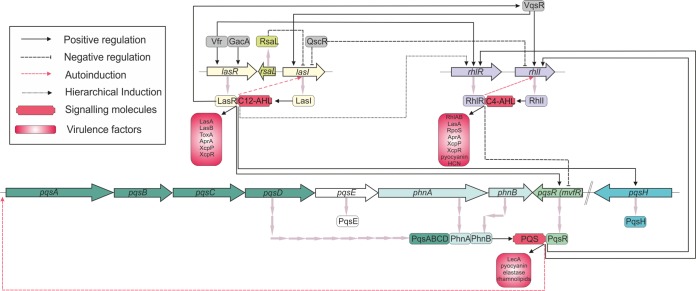
Quorum sensing (QS) is a cell density-dependent system of communication between local populations of bacterial cells, regulating and coordinating their gene expression by using diffusible signaling molecules (9, 10). In Gram-negative pathogens (in particular P. aeruginosa), QS is regulated by acyl homoserine lactones (AHLs) produced by a transcriptional regulator based on the LuxR/LuxI-type QS system that was first characterized in Vibrio fischeri (11). The regulation of QS in Pseudomonas spp. is both subtle and complex (12) (Fig. 1). P. aeruginosa has four QS systems, two AHL-mediated systems, one 2-heptyl-3-hydroxy-4-quinolone-mediated system known as the Pseudomonas quinolone signal (PQS) system, and the more recently identified integrated QS system (IQS) (13). The AHL systems in P. aeruginosa are known as the lasI-lasR and rhlI-rhlR systems, and the transcriptional regulators LasR and RhlR regulate the production of the signaling molecules (autoinducers) N-(3-oxododecanoyl)-l-AHL (3-oxo-C12-AHL) and N-butyryl-l-AHL (C4-AHL), respectively. The QS system in P. aeruginosa is regulated by an interlinked, hierarchical mechanism where lasR–3-oxo-C12-AHL induces the expression of lasI, as well as rhlR-rhlI and the PQS system. Disruption of the IQS signal can effectively paralyze the PQS and rhl QS systems (14). A number of additional regulators of this QS system exist at both the transcriptional and posttranscriptional levels, including the global activator GacA and the regulator Vfr (13). In addition, QS also regulates key cellular processes such as promotion of extracellular DNA (eDNA) release, RNA transcription and translation, cellular division, and amino acid synthesis.
FIG 1.
Schematic diagram of the P. aeruginosa virulence regulatory network showing the three major QS signaling pathways, namely, the AHL Las and Rhl operons and the 2-heptyl-3-hydroxy-4-quinolone PQS operon.
Global gene expression analysis of the QS systems in P. aeruginosa has shown that 6 to 10% of the genome is regulated through the las and rhl systems (12, 15). QS plays a role in swarming motility, biofilm development, and expression of antibiotic efflux pumps (16), as well as virulence factor production. P. aeruginosa QS-activated virulence factors include proteases, e.g., elastase, pyocyanin, lectins, rhamnolipids, and toxins. Such virulence factors can affect biofilm formation and maintenance, as well as swarming motility. Their regulation is complex, with numerous intrinsic and environmental factors involved, such as cell number, composition of the extracellular polymeric substance (EPS), matrix density, and oxygen availability. However, the production of pyocyanin, proteases, and rhamnolipids reflects optimal QS signaling (17). Pyocyanin is a blue secondary metabolite produced by P. aeruginosa evident in the sputum of infected cystic fibrosis (CF) patients (18). As a zwitterion, at a physiological pH, it can readily penetrate biological membranes, inducing host cell necrosis and inflammation both directly (e.g., interleukin-8) and indirectly via cellular damage (19). Importantly, in the context of biofilm persistence in vivo, pyocyanin induces the deposition of eDNA, which is a major component of biofilm EPS, being essential for biofilm formation and stability (20). Production of both pyocyanin and eDNA is mediated by AHL and PQS molecules, as well as by flagella and type IV pili (21, 22).
The regulation of QS in P. aeruginosa is sensitive to, and modulated by, growth and environmental conditions, which impact significantly on the timing of lasI, lasR, rhlI, and rhlR expression (9, 23). The complexity of this QS system in P. aeruginosa is thought to be one of the main factors responsible for its selective adaption and environmental versatility (24). The QS system also affords selective “fitness” advantages in human disease. For example, QS signaling molecules produced by P. aeruginosa are also recognized by Burkholderia cepacia, resulting in synergistic interactions in mixed-species biofilms (25), thereby potentially increasing the virulence of both species in the CF lung. Moreover, the expression of AHL and PQS molecules has been shown to affect the mammalian host-pathogen response (26), with 3-oxo-C12-AHL and PQS having anti-inflammatory and proapoptotic effects on murine fibroblasts and human lung epithelial cells at concentrations of <10 μM (27).
Rhamnolipids are bacterial glycolipid surfactants composed of a rhamnose glycosyl head and a 3-(hydroxyalkanoyloxy)alkanoic acid fatty acid tail. Rhamnolipid expression plays a crucial role in microbial motility, hydrophobic uptake, and biofilm formation on host surfaces. Proteases (including the zinc-dependent metalloproteinase elastase) also play an important role in the pathogenicity of Pseudomonas spp., facilitating invasion and destruction of host tissue (28). Rhamnolipid production is regulated by the P. aeruginosa QS regulator rhlR, while elastase and protease activities are regulated by the rhlI-rhlR system.
QS inhibitors that impede QS pathways in microorganisms are an attractive target for antimicrobial therapy development. We hypothesized that the antibiotic susceptibility, motility, and biofilm assembly modifications induced in P. aeruginosa by OligoG CF-5/20 might relate to alterations in the regulation of lasI-lasR and rhlI-rhlR and studied this in vitro.
RESULTS
OligoG CF-5/20 inhibits P. aeruginosa PAO1 growth and reduces violacein induction and inhibition of the Chromobacterium violaceum biosensor CV026.
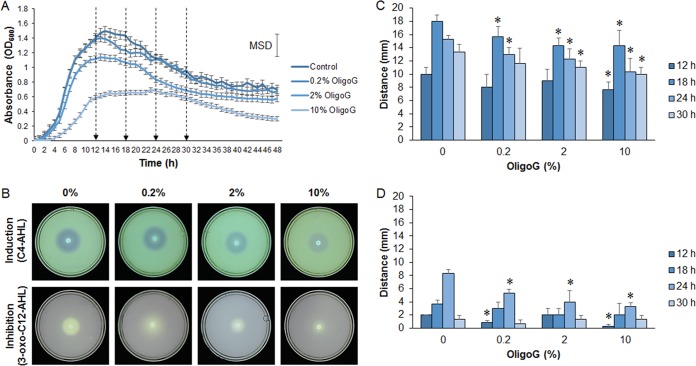
The effect of OligoG CF-5/20 on the growth of P. aeruginosa PAO1 was examined by using growth curves. OligoG CF-5/20 at concentrations of ≥2% was found to significantly reduce the growth of P. aeruginosa PAO1 (minimum significant difference [MSD] = 0.154; P < 0.01; Fig. 2A). These growth curve data were used to determine the time points (12, 18, 24, and 30 h) employed in the subsequent time course study.
FIG 2.
Effect of OligoG CF-5/20 on the growth of P. aeruginosa PAO1 and the production of signaling molecules with the biosensor C. violaceum CV026. (A) Growth curves of P. aeruginosa PAO1 treated with OligoG CF-5/20 showing four specific sampling times (12, 18, 24, and 30 h) for AHL extractions (arrows). (B to D) Well diffusion time course assays detecting AHLs from 24-h (B) or 12-, 18-, 24-, and 30-h (C, D) extracts of P. aeruginosa cultures treated with OligoG CF-5/20 (0.2, 2, or 10%). Induction (zone of coloration) (B, C) and inhibition (zone of clearing) (B, D) of violacein synthesis in C. violaceum CV026 showing changes in C4- and 3-oxo-C12-AHL production following OligoG CF-5/20 treatment. The average from three replicates ± the standard deviation is shown. *, P < 0.05. Differences in culture biomass (at ≥2% OligoG CF-5/20) were corrected according to dry weight.
A time course study was undertaken by using induction or inhibition of violacein in the C. violaceum biosensor strain CV026 as an indicator of QS signaling (C4-AHL and 3-oxo-C12-AHL, respectively) following treatment with OligoG CF-5/20 (Fig. 2B to D). Untreated controls showed distinct differences in P. aeruginosa PAO1 AHL production with time, which were maximal at 18 h for C4-AHL induction and at 24 h for 3-oxo-C12-AHL inhibition. OligoG CF-5/20-treated samples showed a significant reduction in C4-AHL, particularly at 18 and 24 h due to 0.2% OligoG CF-5/20 and at 30 h due to 2% OligoG CF-5/20 (Fig. 2B and C). Measurement of zones of clearing indicated that OligoG CF-5/20 had less of an effect on 3-oxo-C12-AHL inhibition than on C4-AHL induction (zone of coloration). Violacein inhibition was significantly reduced at the 24-h time point by all of the OligoG CF-5/20 concentrations used in comparison to the control level (P < 0.05; Fig. 2B and D).
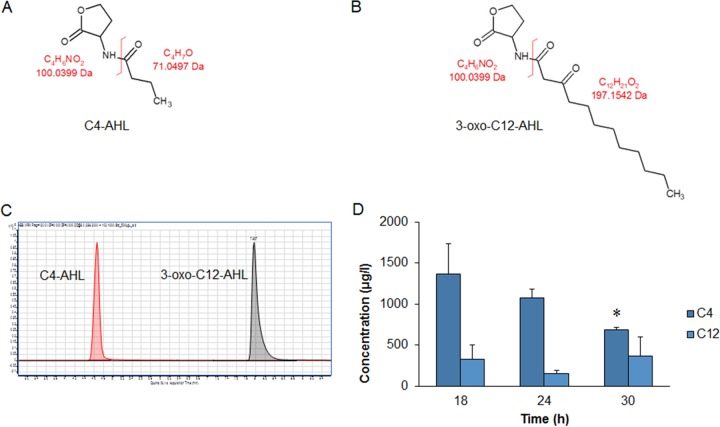
C4-AHL and 3-oxo-C12-AHL can be detected by LC-MS.
For a more accurate determination of AHL concentrations, a preliminary analysis of C4-AHL (Fig. 3A) and 3-oxo-C12-AHL (Fig. 3B) by liquid chromatography (LC)-mass spectrometry (MS) (Fig. 3C) was undertaken from an initial time course monitoring PAO1 growth at 18, 24, and 30 h. LC-MS demonstrated a time-dependent decrease in C4-AHL (Fig. 3D) that was significantly different at 30 h (P < 0.05). Conversely, levels of 3-oxo-C12-AHL were considerably lower (up to 6-fold) and did not demonstrate time-dependent decreases.
FIG 3.
Method development for detection and quantification of AHLs. Panels: A, structure of C4-AHL; B, structure of 3-oxo-C12-AHL; C, C4-AHL and 3-oxo-C12-AHL LC-MS peaks; D, initial time course showing LC-MS quantification of AHL concentrations (μg/liter) from P. aeruginosa PAO1 grown in MH broth at different time points (18, 24, and 30 h). The average from three replicates ± the standard deviation is shown. *, P < 0.05. Differences in culture biomass (at ≥2% OligoG CF-5/20) were corrected according to dry weight.
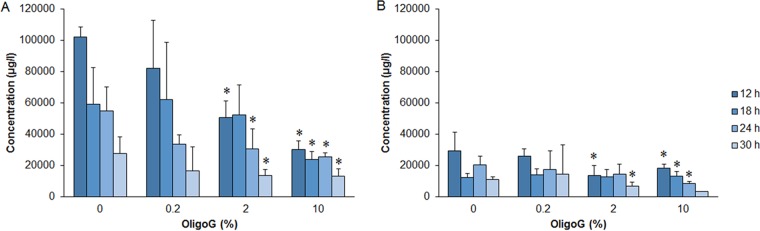
LC-MS shows time-dependent decreases in AHL production following OligoG CF-5/20 treatment.
A subsequent time course of OligoG CF-5/20-treated PAO1 (growth at 12, 18, 24, and 30 h) demonstrated significant reductions in C4-AHL production at all time points at ≥2% OligoG CF-5/20 (Fig. 4A), the exception being 18 h at 2% OligoG CF-5/20, which was not significant. A similar significant reduction was seen for 3-oxo-C12-AHL (Fig. 4B; P < 0.05) in comparison to the untreated control (except for 18 and 24 h at 2% OligoG CF-5/20), although much lower overall levels (up to 29.5 mg/liter) than those obtained with C4-AHL (up to 102.3 mg/liter) were detected (Fig. 4).
FIG 4.
Effect of OligoG CF-5/20 on AHL concentrations (μg/liter) determined by LC-MS at different time points (12, 18, 24, and 30 h) in P. aeruginosa PAO1 grown in MH broth with or without OligoG CF-5/20 (0.2, 2, or 10%). Panels: A, C4-AHL; B, 3-Oxo-C12-AHL. The average from three replicates ± the standard deviation is shown. *, P < 0.05. Differences in culture biomass (at ≥2% OligoG CF-5/20) were corrected according to dry weight.
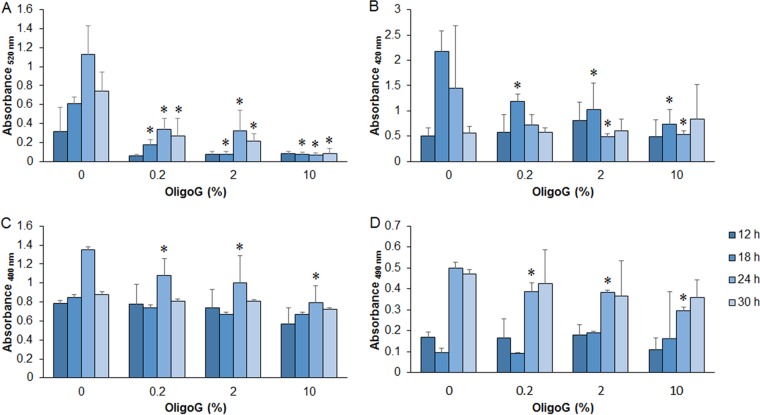
OligoG CF-5/20 reduces extracellular virulence factor production in P. aeruginosa PAO1.
As the biosensor analysis showed that OligoG CF-5/20 affected bacterial signaling, the production of virulence factors, regulated in P. aeruginosa PAO1 by QS, was investigated. OligoG CF-5/20 (≥0.2%) significantly reduced the amount of pyocyanin at all time points of ≥18 h (Fig. 5A; P < 0.05). However, for rhamnolipid production, a significant reduction was observed only at 18 h (at all of the OligoG CF-5/20 concentrations tested) or 24 h (at ≥2% OligoG CF-5/20; P < 0.05), with no significant change seen at either 12 or 30 h (Fig. 5B). In contrast, a significant reduction in total protease (Fig. 5C) and elastase (Fig. 5D) production was seen at ≥0.2% OligoG CF-5/20, and then only at the 24-h time point (P < 0.05).
FIG 5.
Extracellular virulence factor production by P. aeruginosa from 12-, 18-, 24-, and 30-h cell-free culture supernatants treated with OligoG CF-5/20 (0.2, 2, or 10%). Panels: A, pyocyanin; B, rhamnolipids; C, total protease; D, elastase. The average from four replicates ± the standard deviation is shown. *, P < 0.05. Differences in culture biomass (at ≥2% OligoG CF-5/20) were corrected according to dry weight.
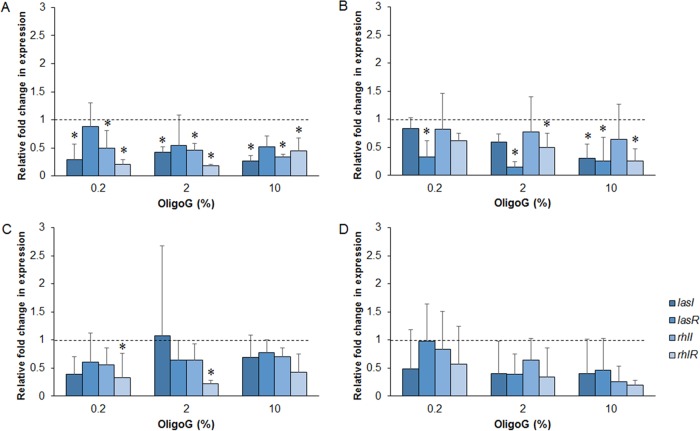
OligoG CF-5/20 reduces the expression of QS genes.
Phenotypic studies were confirmed by genotypic analysis by quantitative PCR (qPCR). Temporal expression of QS genes following OligoG CF-5/20 treatment was observed (Fig. 6). Significant reductions in the expression of lasI, rhlI, and rhlR at 12 h (Fig. 6A); lasI, lasR, and rhlR at 18 h (Fig. 6B); and rhlR at 24 h (Fig. 6B and C, respectively; P < 0.05) were evident, which were significant for lasI, rhlI, and rhlR at 12 h and lasR at 18 h at all three concentrations of OligoG CF-5/20 tested. No significant effect of OligoG CF-5/20 on AHL expression was detected by qPCR at the 30-h time point (Fig. 6D).
FIG 6.
Relative fold change in gene expression compared to untreated control by qPCR of the lasI-lasR and rhlI-rhlR genes from 12-, 18-, 24-, and 30-h cultures of P. aeruginosa PAO1 treated with OligoG CF-5/20 (0.2, 2, or 10%). Panels: A, 12 h; B, 18 h; C, 24 h; D, 30 h. The average from three replicates ± the standard deviation is shown. *, P < 0.05. Differences in culture biomass (at ≥2% OligoG CF-5/20) were corrected according to dry weight.
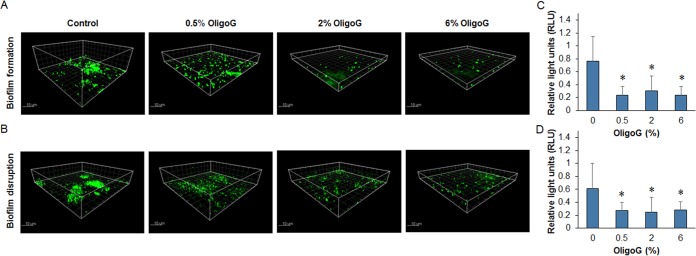
CLSM shows that OligoG CF-5/20 reduces eDNA production and behaves similarly to QS inhibitors against biofilms of P. aeruginosa PAO1.
Confocal laser scanning microscopy (CLSM) imaging of TOTO-1 nucleic acid-stained 24-h biofilms demonstrated that OligoG CF-5/20 (≥0.5%) induced a significant decrease in eDNA production after treatment (P > 0.05) (Fig. 7). This was evident in biofilms grown in the presence of OligoG CF-5/20 (biofilm formation studies) and for 24-h biofilms subsequently treated with OligoG CF-5/20 for a further 24 h (biofilm disruption studies). Although fluorescence analysis of CLSM imaging did not appear to show a dose-dependent decrease in eDNA production (Fig. 7), a dose-dependent decrease was evident at ≥2% OligoG CF-5/20 in a direct analysis of treated biofilm samples (Fig. 8).
FIG 7.
CLSM of P. aeruginosa PAO1 biofilms treated with OligoG CF-5/20 (0.5, 2, or 10%) and stained with nucleic acid-specific TOTO-1 (green). (A) Biofilm formation assay. Biofilms were grown for 24 h in the presence of OligoG CF-5/20. (B) Biofilm disruption assay. (C, D) Twenty-four-hour established biofilms subsequently treated for 24 h with OligoG CF-5/20 are shown with corresponding fluorescence intensities. The average from three replicates ± the standard deviation is shown. *, P < 0.05.
FIG 8.
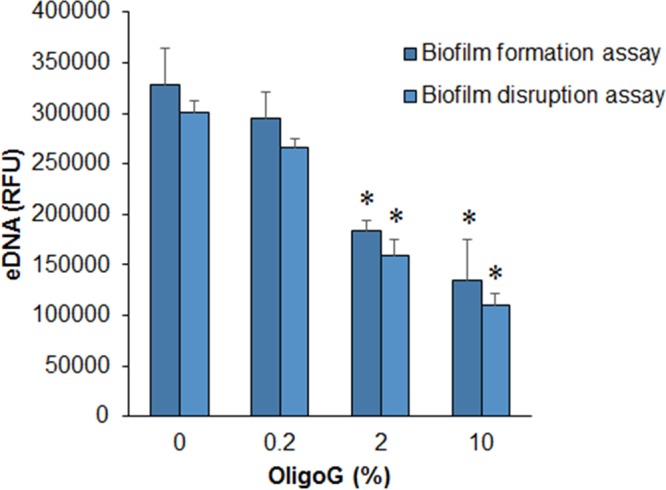
Determination of eDNA concentration. The effect of OligoG CF-5/20 (0.2, 2, or 10%) on the relative eDNA concentration in P. aeruginosa biofilms was measured. In a biofilm formation assay, biofilms were grown for 24 h in the presence of OligoG CF-5/20. In a biofilm disruption assay, 24-h established biofilms were subsequently treated for 24 h with OligoG CF-5/20. The average from three replicates ± the standard deviation is shown. *, P < 0.05. Differences in culture biomass (at ≥2% OligoG CF-5/20) were corrected according to dry weight. RFU, relative fluorescence units.
The structural alterations induced in biofilms by OligoG CF-5/20 were compared to the effects of the QS inhibitors 2(5H)-furanone and N-decanoyl cyclopentylamide (C10-CPA) (29, 30) by using LIVE/DEAD staining (Fig. S1A) showing that the effects of OligoG CF-5/20 resembled the inhibition induced by the other AHL-dependent QS inhibitors tested (see Fig. S1B and C in the supplemental material).
CD showed that OligoG CF-5/20 does not interact directly with AHLs.
Circular dichroism (CD) spectroscopy rapidly determines protein and polypeptide secondary structure and has previously been shown to have excellent comparability with 1H nuclear magnetic resonance (NMR) spectroscopy in determining alginate M/G residue ratios (31). CD was used here to confirm that the effects of OligoG CF-5/20 were not due to simple physical interaction with the AHL molecules. The CD signal of OligoG CF-5/20 titrated with C4-AHL or 3-oxo-C12-AHL showed no substantial change (Fig. S2). The minima of the spectra around 210 nm, revealing the orientation of the alginate carboxy groups and thus directly indicative of the conformation of OligoG CF-5/20 (32), appeared to be unaffected by either of the two AHLs. The ellipticities recorded at 208 nm (after addition of AHLs at their maximum concentrations over ∼1 h) suggested that kinetic effects were not responsible for the absence of a signal (insets in Fig. S2).
DISCUSSION
This study confirms that OligoG CF-5/20 affects global regulatory QS signaling in P. aeruginosa PAO1, as was hypothesized following the original observations on bacterial motility (4, 7). The biosensor strain C. violaceum CV026 demonstrated that OligoG CF-5/20 reduced C4-AHL and 3-oxo-C12-AHL production in P. aeruginosa PAO1 (as shown by QS induction and inhibition, respectively) in a time- and dose-dependent manner. This was further confirmed by LC-MS and qPCR and the fact that OligoG CF-5/20 also had a significant effect on the production of other virulence factors such as pyocyanin, rhamnolipid, total protease, and elastase. The dose-dependent nature of the observed inhibition suggested that OligoG CF-5/20 does not simply act as an AHL receptor antagonist by binding to the receptor, thereby effectively “blocking” all AHL binding. Furthermore, the CD analysis excluded the possibility that the observed alterations in QS signaling molecules and virulence factor expression were the result of simple steric interactions between the oligosaccharide and the AHL signaling molecules in the biofilm system.
The LC-MS data demonstrated the complex, time-dependent nature of virulence factor production by P. aeruginosa with optimal (maximum) production of both AHLs (C4-AHL and 3-oxo-C12-AHL) at 12 h (equivalent to late exponential/early stationary growth phase). These findings are in keeping with previous studies that showed that, while AHL production peaks during exponential growth, C4-AHL levels decrease as stationary phase is attained (12). The finding here, that 3-oxo-C12-AHL levels remained relatively constant if the medium was sufficiently buffered to avoid alkali-mediated lactonolysis, is consistent with that of Yates et al. (33).
Las and Rhl are regulated by the LuxR family of transcriptional regulators (LasR and RhlR), making their expression extremely sensitive to environmental conditions, e.g., hypoxia, pH, and hydrodynamic shear (which are important in biofilm infections). AHL production has been shown to vary significantly under different environmental growth conditions, especially under nutrient limitation, with higher AHL expression observed in minimal or diluted medium than in nutrient medium (9). In addition, both las and rhl were expressed earlier in nutrient-limited medium (in early to mid-log phase) than in nutrient medium (in early stationary phase). Interestingly, these phenomena were unrelated to cell density, which is usually considered a prerequisite for QS expression. Comparing 46 different experimental conditions, Duan and Surette (9) showed that the individual dominance of the las-and-rhl system reflected environmental conditions. LasR mutants are commonly found in both clinical and environmental isolates, indicating autonomous regulation of these integrated systems (34, 35). Transcription of Las and Rhl may also occur independently, permitting further “fine-tuning” of each system in vivo. This may, in part, explain the independent (and distinct) responses to OligoG CF-5/20 treatment observed in C4-AHL and 3-oxo-C12-AHL production in the time course experiments.
Swarming is a complex form of motility and is consequently influenced by a large number of different genes. Rhamnolipids are known to modulate the intricate swarming motility patterns of P. aeruginosa (36). Therefore, it was perhaps unsurprising that, as OligoG CF-5/20 was previously shown to affect the swarming motility of Proteus and P. aeruginosa (references 4 and 7, respectively), rhamnolipid production should also be affected by OligoG CF-5/20. Importantly, QS regulation of rhamnolipids and swarming motility contribute to P. aeruginosa biofilm dispersal and therefore help to explain the dramatic effect of OligoG CF-5/20 on both biofilm formation and disruption of established biofilms previously described (4, 7). In support of this notion, a range of mini-Tn5 insertion, “swarming-negative” P. aeruginosa mutants exhibited impaired biofilm formation (37), confirming the link between the two phenotypes. The finding here, of more significant inhibition of pyocyanin and rhamnolipid production by OligoG CF-5/20 (compared to the effects on elastase and total protease production) may relate to differential expression of the different QS pathways. The three best-characterized QS signaling systems in P. aeruginosa are believed to be sequentially activated in “nutrient-rich” media, with LasR sitting at the top of the temporal cascade and AHLs (las and rhl) being released in the early exponential phase and PQS being released in the late exponential phase of growth (38).
Las and Rhl control both biofilm formation and expression of virulence factors in P. aeruginosa (11). The LasR–3-oxo-C12-AHL complex activates the transcription of target genes, including those encoding virulence factors such as elastase, proteases, and exotoxin. In contrast, RhlR–C4-AHL activates target genes, including those encoding elastase, proteases, pyocyanin, and siderophores (39). There appears to be a considerable overlap of the virulence factors these regulons control (9). The finding that the inhibition of pyocyanin/rhamnolipid production was more evident throughout the time course of the experiment than that of protease and elastase may be a reflection of OligoG CF-5/20 differentially affecting the Rhl QS system to a greater extent than the Las system.
The intrinsically high levels of antimicrobial resistance typically seen in P. aeruginosa are due to its low permeability and multidrug efflux systems, four of which contribute significantly to innate antibiotic resistance. Khan et al. (4) demonstrated that OligoG CF-5/20 increased the potentiation of antibiotics against MDR bacteria (up to 128-fold). The authors established that this did not relate simply to permeabilization of the pseudomonal lipopolysaccharide cell wall or targeting of the multidrug efflux pump MexAB-OrpM, suggesting that the QS inhibition observed here with OligoG CF-5/20 involves a mechanism other than inhibition of AHL efflux pumps in P. aeruginosa. Instead, the OligoG CF-5/20-induced reduction in AHL production more likely reflects an effect further “upstream,” e.g., on bacterial two-component system (TCS) signal transduction pathways (40) by which means bacteria are able to detect and produce a response to environmental changes.
TCSs are composed of an inner membrane-bound “sensor,” generally a histidine kinase (which detects environmental stimuli), and a response regulator (which modulates the response). There are many TCSs in P. aeruginosa, and these are recognized to play a role in the regulation of bacterial virulence, biofilm formation, and antibiotic susceptibility, factors known to be influenced by OligoG CF-5/20, although the precise links between TCSs and QS are still poorly understood (41). At least three TCSs (BfiSR, MifR, and BfmSR) are thought to be involved in the activation of biofilm formation (42). The recently published BfmS-BfmR-RhlR TCS has been shown to be key to regulation of the rhl QS pathway in P. aeruginosa (43) modulating the expression of biofilm formation and virulence. Interestingly, deletion of the sensor gene bfmS was shown to cause inhibition of the rhl QS system, with bfmR playing a central role in biofilm maturation. In addition, it has also recently been suggested that BfmRS is involved the development of virulence during bacterial adaptation to the CF lung (43, 44). Interestingly, AlgR (another key P. aeruginosa transcriptional response regulator) also appears to play an essential role in bacterial virulence and motility (45).
The chemical composition of the EPS represents a formidable “barrier” to diffusion and contributes to resistance to antibiotic and antimicrobial therapy (46). The physical disruption of the biofilm structure and alterations in eDNA distribution within the pseudomonal biofilms (induced by OligoG CF-5/20) were perhaps unsurprising, as QS and pyocyanin have an important regulatory role in eDNA synthesis. Pyocyanin induces eDNA release, with biofilms formed by QS mutants known to possess less eDNA than wild-type biofilms and to be more susceptible to chemical disruption (21, 47). Our results further confirm these findings, where OligoG CF-5/20 treatment of P. aeruginosa PAO1 resulted in significant decreases in pyocyanin and eDNA production.
Virulence-targeted antibacterials, which effectively “disarm” pathogenic bacteria, have received considerable attention (48), although many have proved to be short-lived because of issues with toxicity and the acquisition of bacterial resistance. Resistance to furanone in P. aeruginosa can be selected for in vitro, as well as being found in clinical isolates (49). In contrast to many of the previously described therapeutic modalities, OligoG CF-5/20, which targets bacterial virulence as a QS antagonist, shows considerable promise. Phase I and IIa human studies failed to demonstrate toxicity. Moreover, extended in vitro serial passage in the presence of OligoG CF-5/20 has failed to demonstrate the acquisition of bacterial resistance (4).
As QS inhibitors target specific pathogenicity traits such as virulence determinants, there has been considerable interest in their use as novel anti-infective therapies (50) both by screening for novel compounds (51) and by targeted synthesis of new ligands (52). Similar to OligoG CF-5/20, the QS inhibitors furanone and C10-CPA have previously been shown to impede AHL-mediated QS in P. aeruginosa, leading to an altered biofilm architecture and reduced virulence factor production, as well as enhanced bacterial detachment and antibiotic susceptibility (references 53 and 30, respectively). Predictably, the QS inhibition effects seen with OligoG CF-5/20 appear to more closely resemble those of the C10-CPA tested here, interfering as it does with both the las and rhl QS systems, unlike furanone, which was predominantly found to perturb the las system. The dose-response effects and effects on bacterial growth observed in this study suggest that although OligoG CF-5/20 does not act as a true QS inhibitor, it does act as a QS antagonist, affecting signaling pathways in P. aeruginosa, with expression of Las and Rhl QS pathways altered in a dose-dependent manner following OligoG CF-/20 treatment. This also proposes a mechanistic rationale for the previously described antibiofilm properties of this novel antimicrobial agent that is currently in human clinical trials.
MATERIALS AND METHODS
Alginate oligosaccharides.
The low-molecular-weight alginate oligosaccharide used in the study, OligoG CF-5/20 (number average molecular weight [Mn] of 2,800), was prepared, purified, and characterized as previously described (4).
Growth curves.
Overnight cultures of P. aeruginosa PAO1 grown in tryptone soya broth (TSB; Oxoid) at 37°C at 120 rpm were diluted 1:100 in Mueller-Hinton (MH) broth with or without OligoG CF-5/20 (0.2, 2, or 10%). The growth of P. aeruginosa PAO1 aerobically at 37°C was monitored for 48 h. The optical density at 600 nm (OD600) was measured every hour in a FLUOstar Optima plate reader (BMG LABTECH). One-way analysis of variance (ANOVA) and the Tukey-Kramer posttest were used, and the MSD was generated.
C. violaceum CV026 biosensor strain.
C. violaceum CV026 is unable to produce the purple pigment violacein without an external source of AHLs; therefore, violacein production is induced by AHLs with a carbon chain length of C4 to C8, whereas inhibition of violacein production can also occur when using AHLs with a greater carbon chain length (C10 to C14). Both P. aeruginosa AHLs can therefore be detected by using the C. violaceum CV026 strain: C4-AHL by induction of violacein production and 3-oxo-C12-AHL by inhibition of violacein production.
AHL extraction.
Cell-free supernatants were collected, and equivalent volumes of ethyl acetate (acidified by supplementation with 0.5% formic acid) were added. Following mixing for 30 s, the phases were allowed to separate and the top layer was collected; this was repeated three times. The combined ethyl acetate fractions were evaporated, and the precipitate was resuspended in 1 ml of distilled H2O (54). Samples were used immediately or freeze-dried and stored at −20°C until required.
Screening of AHL extracts by using CV026 induction and inhibition assays.
AHL extracts were tested by using the C. violaceum CV026 biosensor strain in a well diffusion assay (55). C. violaceum CV026 was grown for 16 h at 30°C in LB supplemented with kanamycin (50 μg/ml). This overnight culture was incorporated into LB agar plates (1.2%) by dilution (1:100). In addition, induction plates also contained kanamycin (50 μg/ml) and inhibition plates contained both kanamycin and C10 AHL (50 nM) (17248; Sigma-Aldrich, Pool, United Kingdom). A well (6 mm diameter) was made in the center of each solidified agar plate. Test AHL extracts (or controls) were then added to the well (with adjustment with distilled H2O according to the dry weight of the PAO1 culture used). The plates were then incubated at 30°C for 48 h. Distances of violacein induction or inhibition, as determined by the extent of purple coloration or the zone of clearing, respectively, were then measured in millimeters.
Cell-free culture supernatant.
Cultures of P. aeruginosa PAO1 were grown for 12, 18, 24, and 30 h and prepared as described previously for growth curve determination. MH broth was selected, as nutrient-limited medium has been shown to enhance AHL production (9). Cells were harvested (3,900 × g, 20 min, 4°C) and washed three times with ice-cold 0.9% NaCl and dried at 80°C for 24 h. In each case, differences in culture biomass (at OligoG CF-5/20 concentrations of >2%) from cell-free culture supernatants used for the screening of AHLs and the extraction of all of the virulence factors were corrected by normalization according to dry weight.
Quantitation of extracellular virulence factors.
Pyocyanin was extracted from the culture supernatant (700 μl) with chloroform at a ratio of 3:2 and re-extracted with 150 μl of 0.2 M HCl, and the absorbance was read at 520 nm (17). Rhamnolipids were extracted from culture supernatant with ethyl acetate at a 1:1 ratio, vortexed for 15 s, and centrifuged (10,000 × g, 4°C, 5 min). The upper layer was removed, and ethyl acetate extraction was repeated three times for each sample. The combined upper layer was left to evaporate overnight, and then 900 μl of orcinol reagent (0.19% orcinol in 53% H2SO4) was added to the precipitate, which was incubated at 80°C for 30 min before the absorbance at 420 nm was read (51). Protease activity was determined with a 2% azocasein solution prepared in 50 mM phosphate-buffered saline (PBS), pH 7. The substrate and culture supernatant were incubated at 37°C at a 1:1 ratio for 1 h in a reaction mixture volume of 400 μl. The reaction was stopped by the addition of 500 μl of 10% trichloroacetic acid, and the reaction mixture was centrifuged at 8,000 × g for 5 min to remove residual azocasein. The absorbance at 400 nm was read (17). Elastase extraction employed 200 μl of elastin Congo red solution (5 mg/ml in 0.1 M Tris-HCl [pH 8] and 1 mM CaCl2), which was incubated with 600 μl of cell-free culture supernatant at 37°C for 3 h at 200 rpm. The mixture was then centrifuged at 3,000 × g for 10 min, and the absorbance at 490 nm was read (17).
High-performance LC-QQQ-MS.
AHLs were extracted as described above and freeze-dried until required. Freeze-dried samples were reconstituted in 200 μl of acetonitrile (ACN) with 0.1% acetic acid and the internal standard umbelliferone at 7.2 ng/ml. Samples were vortexed and centrifuged (100 × g, 4°C, 10 min) (16), and supernatants were filtered through a 0.4-μm syringe filter (Phenomenex, United Kingdom); this was performed twice to increase metabolite extraction. Samples were kept on ice throughout the extraction procedure prior to being subjected to LC–triple-quadrupole MS (LC-QQQ-MS). Samples (5 μl) were loaded onto a C18 XDB Eclipse (1.8 μm, 4.6 by 50 mm) reverse-phase column (Agilent Technologies, Palo Alto, CA). Samples were quantified with a 1200 series high-performance liquid chromatograph (Agilent Technologies, USA) coupled to a 6410B enhanced-sensitivity QQQ mass spectrometer (Agilent Technologies, USA). For detection in the positive-ion mode, mobile phase A was 5 mM ammonium acetate in water modified with 0.1% acetic acid and B was ACN containing 0.1% acetic acid. The column was equilibrated in 2% B before increasing in a linear fashion to 100% over 6 min, with 100% B being maintained for a further 2 min before column re-equilibration. The column temperature was maintained at 35°C for the duration with a flow rate of 0.3 ml/min. Source parameters were as follows: temperature, 350°C; gas flow, 10 liters/min; nebulizer pressure, 35 lb/in2; capillary voltage, 4 kV. Data were analyzed with Agilent MassHunter QQQ Quantitative Analysis software (version B.07.00). Peak areas were normalized to the internal standard umbelliferone, and concentrations were calculated by using standard concentration curves offset against blank values (the average peak areas for the blanks).
RNA extraction for RT-qPCR.
RNA was extracted from 24-h cultures of P. aeruginosa PAO1 grown at 37°C in MH broth with or without OligoG CF-5/20 (0.2, 2, or 10%). Cultures were harvested (2,000 × g, 10 min), resuspended, adjusted to 1.0 × 108 CFU/ml in PBS, centrifuged (12,000 × g, 2 min), resuspended in 0.5 ml of RNAlater, and stored at −20°C until required. Cells were pelleted (12,000 × g, 2 min) and resuspended with lysis buffer (RLT buffer; QIAgen, Crawley, United Kingdom) containing 1% (vol/vol) β-mercaptoethanol. Cell debris was pelleted via centrifugation (12,000 × g, 2 min), the resulting supernatants were moved to fresh tubes, and phenol-chloroform-isoamyl alcohol (25:24:1) was used to acquire total nucleic acid. Total RNA was recovered after DNase I treatment with the RNeasy minikit (Qiagen) in accordance with the manufacturer's instructions. Gel electrophoresis was used to check the purity and integrity of the total RNA, the RNA concentration was measured spectrophotometrically, and an additional purity check was performed with an absorbance ratio of 260/280 nm (NanoVue; GE Healthcare, Little Chalfont, United Kingdom) and standardized to 300 ng/ml. Reverse transcription reaction mixtures for cDNA synthesis included 300 ng of a total RNA template and 1 μl of random primer at 50 μg/ml, and molecular grade water was added to give a final reaction mixture volume of 10 μl. Real-time qPCR (RT-qPCR) was performed in triplicate with the NanoScript2 RT kit (Primer Design, Southampton, United Kingdom) and a final annealing step of 5 min at 65°C, after which point the samples were cooled on ice. Annealed samples were then added to the extension mixture; 4 μl of 4× nanoScript2 buffer, 1 μl of a deoxynucleoside triphosphate mixture (10 mM), 1.5 μl of NanoScript2 enzyme (Primer Design, Southampton, United Kingdom), and 2.5 μl of molecular grade water in a final volume of 20 μl were incubated at 25°C for 5 min and then at 42°C for 20 min.
RT-qPCR for analysis of gene expression.
RT-qPCR for analysis of the expression of QS genes was carried out with the primers presented in Table 1. Primer specificity was tested on genomic DNA. RT-qPCR was performed in triplicate with three replicate samples and an ABI 7000 instrument (Life Technologies, United Kingdom). Each reaction mixture contained 2 μl of cDNA, 12.5 μl of (2×) SYBR green PCR master mix (PrecisionPLUS Master Mix; Primer Design, Southampton, United Kingdom), and each primer at 10 mM and was made up to 25 μl with highly purified water (Qiagen). The thermal cycler profile was initial denaturation at 95°C for 2 min and 40 cycles of denaturation at 95°C for 15 s, primer annealing at 58°C for 15 s, and extension at 72°C for 30 s. A final extension at 72°C for 2 min was performed, followed by cooling at 4°C. A dissociation step at 60°C was used to generate a melting curve for verification of the amplified product. After RT-qPCR, the threshold was adjusted in accordance with the amplification curves of all of the genes evaluated. Comparison of groups was based on the cycle number at which both the target and the average of endogenous control genes (rpsL and proD) attained threshold cycle (CT) fluorescence. Analysis of relative gene expression was achieved by the ΔΔCT method (56).
TABLE 1.
Genes and primers used for qPCR in this study
| Gene | Primer sequence (5′–3′) |
Forward primer |
Reverse primer |
Product size (bp) | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Forward | Reverse | BPa | GCb | TMc | BP | GC | TM | |||
| lasI | TGTTCAAGGAGCGCAAAGG | ATGGCGAAACGGCTGAGTT | 19 | 52.6 | 62.4 | 19 | 52.6 | 63 | 244 | 58 |
| lasR | AGCGACCTTGGATTCTCGAAG | CGAAGAACTCGTGCTGCTTTC | 21 | 52.4 | 63 | 21 | 52.4 | 62.5 | 226 | 58 |
| rhlI | TGCTCTCTGAATCGCTGGAA | GTTTGCGGATGGTCGAACTG | 20 | 50 | 59.1 | 20 | 55 | 59.83 | 154 | 58 |
| rhlR | TTGCTGAGCGTGCTTTCC | AGGATGATGGCGATTTCCC | 19 | 52.6 | 62.6 | 19 | 52.6 | 62.1 | 228 | 58 |
| rpsLd | CCTCGTACATCGGTGGTGAAG | CCCTGCTTACGGTCTTTGACAC | 21 | 57.1 | 62.8 | 22 | 54.5 | 63.1 | 148 | 59 |
| proDd | GGGCGAAGAAGGAAATGGTC | CAGGTGGCGTAGGTAGAGAA | 20 | 55 | 63.1 | 20 | 55 | 58 | 178 | 60 |
BP, primer length (base pairs).
GC, G-C content of primer.
TM, melting temperature of primer.
Reference/endogenous control gene.
eDNA determination of P. aeruginosa PAO1 biofilms treated with OligoG CF-5/20 with a nucleic acid-specific, cell-impermeant, fluorescent TOTO-1 stain.
The effect of OligoG CF-5/20 on the formation of 24-h P. aeruginosa PAO1 biofilms was tested. For this, adjusted P. aeruginosa PAO1 cultures (107 CFU/ml) were diluted 1:10 in MH broth with or without OligoG CF-5/20 (0.5, 2, or 6%, wt/vol) and then incubated in Whatman 96-well glass-bottom plates at 37°C for 24 h with gentle agitation prior to staining. The effect of OligoG CF-5/20 on established 24-h biofilms was also tested to look at its effect on biofilm disruption. For this, biofilms were grown without OligoG CF-5/20 treatment with adjusted P. aeruginosa PAO1 cultures (107 CFU/ml) diluted 1:10 in MH broth. After 24 h of incubation, half of the supernatant was removed and replaced with fresh MH broth with or without OligoG CF-5/20 (0.5, 2, or 6%, wt/vol) and the samples were incubated for a further 24 h before staining. After OligoG CF-5/20 treatment, the supernatant was removed and biofilms were stained with TOTO-1 (Thermo Fisher) for 25 min. Biofilm samples were imaged with a Leica TCS SP5 confocal system with a 63× lens.
For fluorescence determination of eDNA, biofilms were homogenized by vigorous pipetting and the resulting supernatant was filtered (0.2 μm). Culture purity was confirmed by plating a loopful of supernatant on nonselective blood agar. Supernatants were stained with TOTO-1 at room temperature for 35 min, and fluorescence was measured at excitation and emission wavelengths of ∼514 and 533 nm on a FLUOstar Optima plate reader (BMG LABTECH) (47).
Synthesis of C10-CPA.
Decanoyl chloride (1 mol equivalent, 0.544 ml, 0.500 g, 2.6 mmol) was added dropwise to a stirring solution of cyclopentylamine (2 mol equivalent, 0.513 ml, 0.443 g, 5.2 mmol) in anhydrous dichloromethane (5 ml) under a nitrogen atmosphere. The reaction mixture was stirred for 6 h, and then the solvent was evaporated under reduced pressure. The residue was redissolved in 20 ml of diethyl ether and washed with a solution of water, 5% NaHCO3, 0.2 M HCl, and saturated NaCl. The organic layer was dried over MgSO4 and concentrated to furnish the N-cyclopentyldecanamide as a white solid that was confirmed by 1H NMR, 13C NMR, and electrospray ionization MS (30).
CLSM imaging of P. aeruginosa biofilms treated with QS inhibitors.
Overnight cultures of P. aeruginosa PAO1 grown in TSB were adjusted to 107 CFU/ml, and the adjusted culture was diluted 1:10 in MH broth in glass-bottom 96-well plates. Biofilms of P. aeruginosa PAO1 were grown for 24 h while being treated (with gentle rocking) with the known AHL QS inhibitor 2(5H)-furanone (283754; Sigma-Aldrich, Pool, United Kingdom) at 1.25 or 2.5 μg · ml−1 (14.9 or 29.7 μM, respectively) (29, 30) or C10-CPA at 100 or 250 μM (30). Untreated and OligoG CF-5/20-treated biofilms were used as controls. Planktonic cells/supernatant were removed before the biofilms were stained with the 6% LIVE/DEAD BacLight bacterial viability kit (Invitrogen, Paisley, United Kingdom) in PBS, incubated in the dark (10 min), and imaged with a Leica TCS SP5 confocal system with a ×63 lens.
CD spectroscopy.
To evaluate whether AHLs influence the conformation of OligoG CF-5/20, CD spectra were recorded on an Aviv 215 instrument (Aviv Biomedical Inc., Lakewood, NJ) from 260 to 200 nm with a 1-nm bandwidth and a 0.5-cm quartz cell at 37°C. OligoG CF-5/20 was dissolved in 100 mM NaCl–10 mM Tris-HCl (pH 7.5) at a concentration of 0.5 mg/ml, and either C4-AHL or 3-oxo-C12-AHL (09014 and 09945; Sigma-Aldrich, Pool, United Kingdom) was added stepwise from a 1-mg/ml stock solution. Buffer baselines and the intrinsic AHL spectra were subtracted, and spectra were corrected for dilution. Data are presented as mean residue weight ellipticities [ϴ]MRW assuming a relative molecular mass of 194 g/mol for the OligoG CF-5/20 monosaccharides.
Statistical analysis.
Microsoft Excel was used to perform statistical analysis, including one-way ANOVA with the Tukey-Kramer posttest, and the MSD was calculated by the Tukey-Kramer method (57). P < 0.05 was considered significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Paul Williams (University of Nottingham) for C. violaceum strain CV026, Shree Patel for technical support for the CLSM, and Deborah Salmon for the LC-MS investigation.
This study was supported by funding from the Research Council of Norway (228542/O30) and AlgiPharma AS, Sandvika, Norway. P.D.R. is an employee of AlgiPharma AS; D.W.T. and K.E.H. have previously had direct research funding from AlgiPharma AS. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02318-17.
REFERENCES
- 1.Rada B, Leto TL. 2013. Pyocyanin effects on respiratory epithelium: relevance in Pseudomonas aeruginosa airway infections. Trends Microbiol 21:73–81. doi: 10.1016/j.tim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW. 2011. Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35:652–680. doi: 10.1111/j.1574-6976.2011.00269.x. [DOI] [PubMed] [Google Scholar]
- 3.Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, Olavarietta R, Doud M, Smith RS, Montgomery P, White JR, Godfrey PA, Kodira C, Birren B, Galagan JE, Lory S. 2008. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A 105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan S, Tondervik A, Sletta H, Klinkenberg G, Emanuel C, Onsøyen E, Myrvold R, Howe RA, Walsh TR, Hill KE, Thomas DW. 2012. Overcoming drug resistance with alginate oligosaccharides able to potentiate the action of selected antibiotics. Antimicrob Agents Chemother 56:5134–5141. doi: 10.1128/AAC.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell LC, Sowedan A, Khan S, Wright CJ, Hawkins K, Onsøyen E, Myrvold R, Hill KE, Thomas DW. 2013. The effect of alginate oligosaccharides on the mechanical properties of Gram-negative biofilms. Biofouling 29:413–421. doi: 10.1080/08927014.2013.777954. [DOI] [PubMed] [Google Scholar]
- 6.Roberts JL, Khan S, Emanuel C, Powell LC, Pritchard MF, Onsøyen E, Myrvold R, Thomas DW, Hill KE. 2013. An in vitro study of alginate oligomer therapies on oral biofilms. J Dent 41:892–899. doi: 10.1016/j.jdent.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 7.Powell LC, Pritchard MF, Emanuel C, Onsøyen E, Rye PD, Wright CJ, Hill KE, Thomas DW. 2014. A nanoscale characterization of the interaction of a novel alginate oligomer with the cell surface and motility of Pseudomonas aeruginosa. Am J Respir Cell Mol Biol 50:483–492. doi: 10.1165/rcmb.2013-0287OC. [DOI] [PubMed] [Google Scholar]
- 8.Tøndervik A, Sletta H, Klinkenberg G, Emanuel C, Powell LC, Pritchard MF, Khan S, Craine KM, Onsøyen E, Rye PD, Wright C, Thomas DW, Hill KE. 2014. Alginate oligosaccharides inhibit fungal cell growth and potentiate the activity of antifungals against Candida and Aspergillus spp. PLoS One 9:e112518. doi: 10.1371/journal.pone.0112518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan K, Surette MG. 2007. Environmental regulation of Pseudomonas aeruginosa PAO1 Las and Rhl quorum-sensing systems. J Bacteriol 189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuqua WC, Winans SC. 1994. A LuxR-LuxI type regulatory system activates Agrobacterium Ti plasmid conjugal transfer in the presence of a plant tumor metabolite. J Bacteriol 176:2796–2806. doi: 10.1128/jb.176.10.2796-2806.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet 43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Zhang L. 2015. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang L-H. 2013. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol 9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 15.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams P, Camara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol 12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 17.Sarabhai S, Sharma P, Capalash N. 2013. Ellagic acid derivatives from Terminalia chebula Retz. downregulate the expression of quorum sensing genes to attenuate Pseudomonas aeruginosa PAO1 virulence. PLoS One 8:e53441. doi: 10.1371/journal.pone.0053441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caldwell CC, Chen Y, Goetzmann HS, Hao Y, Borchers MT, Hassett DJ, Young LR, Mavrodi D, Thomashow L, Lau GW. 2009. Pseudomonas aeruginosa exotoxin pyocyanin causes cystic fibrosis airway pathogenesis. Am J Pathol 175:2473–2488. doi: 10.2353/ajpath.2009.090166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau GW, Ran H, Kong F, Hassett DJ, Mavrodi D. 2004. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun 72:4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand J, Klausen M, Givskov M, Whitchurch C, Engel J, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol 10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 21.Allesen-Holm M, Barken KB, Yang L, Klausen M, Webb JS, Kjelleberg S, Molin S, Givskov M, Tolker-Nielsen T. 2006. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol 59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Gao Q, Chen W, Qin H, Hengzhuang W, Chen Y, Yang L, Zhang G. 2013. Regulation of pqs quorum sensing via catabolite repression control in Pseudomonas aeruginosa. Microbiology 159:1931–1936. doi: 10.1099/mic.0.066266-0. [DOI] [PubMed] [Google Scholar]
- 23.Dekimpe V, Deziel E. 2009. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 155:712–723. doi: 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- 24.Juhas M, Eberl L, Tummler B. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ Microbiol 7:459–471. doi: 10.1111/j.1462-2920.2005.00769.x. [DOI] [PubMed] [Google Scholar]
- 25.Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Høiby N, Givskov M, Molin S, Eberl L. 2001. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- 26.Cooley M, Chhabra SR, Williams P. 2008. N-Acylhomoserine lactone-mediated quorum sensing: a twist in the tail and a blow for host immunity. Chem Biol 15:1141–1147. doi: 10.1016/j.chembiol.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Jahoor A, Patel R, Bryan A, Do C, Krier J, Watters C, Wahli W, Li G, Williams SC, Rumbaugh KP. 2008. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol 190:4408–4415. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stehling EG, daSilveira WD, Leite DS. 2008. Study of biological characteristics of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis and from patients with extra-pulmonary infections. Braz J Infect Dis 12:86–88. doi: 10.1590/S1413-86702008000100018. [DOI] [PubMed] [Google Scholar]
- 29.Ponnusamy K, Paul D, Kim YS, Kweon JH. 2010. 2(5H)-Furanone: a prospective strategy for biofouling-control in membrane biofilm bacteria by quorum sensing inhibition. Braz J Microbiol 41:227–234. doi: 10.1590/S1517-83822010000100032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. 2007. Inhibition of quorum sensing in Pseudomonas aeruginosa by N-acyl cyclopentylamides. Appl Environ Microbiol 73:3183–3188. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donati I, Gamini A, Skjak-Braek G, Vetere A, Campa C, Coslovi A, Paoletti S. 2003. Determination of the diadic composition of alginate by means of circular dichroism: a fast and accurate improved method. Carbohydr Res 338:1139–1142. doi: 10.1016/S0008-6215(03)00094-6. [DOI] [PubMed] [Google Scholar]
- 32.Morris ER, Rees DA, Sanderson GR, Thom D. 1975. Conformation and circular dichroism of uronic acid residues in glycosides and polysaccharides. J Chem Soc Perkin Trans 2 13:1418–1425. doi: 10.1039/p29750001418. [DOI] [Google Scholar]
- 33.Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Camara M, Smith H, Williams P. 2002. N-Acylhomoserine lactones undergo lactonolysis in a pH-, temperature-, and acyl chain length-dependent manner during growth of Yersinia pseudotuberculosis and Pseudomonas aeruginosa. Infect Immun 70:5635–5646. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Soyza A, Hall AJ, Mahenthiralingam E, Drevinek P, Kaca W, Drulis-Kawa Z, Stoitsova SR, Toth V, Coenye T, Zlosnik JEA, Burns JL, Sá-Correia I, De Vos D, Pirnay J-P, Kidd TJ, Reid D, Manos J, Klockgether J, Wiehlmann L, Tummler B, McClean S, Winstanley C. 2013. Developing an international Pseudomonas aeruginosa reference panel. Microbiologyopen 2:1010–1023. doi: 10.1002/mbo3.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SL, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caiazza NC, Shanks RM, O'Toole GA. 2005. Rhamnolipids modulate swarming motility patterns of Pseudomonas aeruginosa. J Bacteriol 187:7351–7361. doi: 10.1128/JB.187.21.7351-7361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overhage J, Lewenza S, Marr AK, Hancock RE. 2007. Identification of genes involved in swarming motility using a Pseudomonas aeruginosa PAO1 mini-Tn5-lux mutant library. J Bacteriol 189:2164–2169. doi: 10.1128/JB.01623-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lépine F, Déziel E, Milot S, Rahme LG. 2003. A stable isotope dilution assay for the quantification of the Pseudomonas quinolone signal in Pseudomonas aeruginosa cultures. Biochim Biophys Acta 1622:36–41. doi: 10.1016/S0304-4165(03)00103-X. [DOI] [PubMed] [Google Scholar]
- 39.Schuster M, Greenberg EP. 2006. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol 296:73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 40.Worthington RJ, Blackledge MS, Melander C. 2013. Small-molecule inhibition of bacterial two-component systems to combat antibiotic resistance and virulence. Future Med Chem 5:1265–1284. doi: 10.4155/fmc.13.58. [DOI] [PubMed] [Google Scholar]
- 41.Beier D, Gross R. 2006. Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9:143–152. doi: 10.1016/j.mib.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 42.Balasubramanian D, Schneper L, Kumari H, Mathee K. 2013. A dynamic and intricate regulatory network determines Pseudomonas aeruginosa virulence. Nucleic Acids Res 41:1–20. doi: 10.1093/nar/gks1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Q, Wang Y, Chen F, Xia Y, Lou J, Zhang X, Yang N, Sun X, Zhang Q, Zhuo C, Huang X, Deng X, Yang C-G, Ye Y, Zhao J, Wu M, Lan L. 2014. A novel signal transduction pathway that modulates rhl quorum sensing and bacterial virulence in Pseudomonas aeruginosa. PLoS Pathog 10:e1004340. doi: 10.1371/journal.ppat.1004340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Son MS, Matthews WJ Jr, Kang Y, Nguyen DT, Hoang TT. 2007. In vivo evidence of Pseudomonas aeruginosa nutrient acquisition and pathogenesis in the lungs of cystic fibrosis patients. Infect Immun 75:5313–5324. doi: 10.1128/IAI.01807-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okkotsu Y, Tieku P, Fitzsimmons LF, Churchill ME, Schurr MJ. 2013. Pseudomonas aeruginosa AlgR phosphorylation modulates rhamnolipid production and motility. J Bacteriol 195:5499–5515. doi: 10.1128/JB.00726-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Q, Ma LZ. 2013. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci 14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Das T, Manefield M. 2012. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS One 7:e46718. doi: 10.1371/journal.pone.0046718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruer S, Pinotsis N, Steadman D, Waksman G, Remaut H. 2015. Virulence-targeted antibacterials: concept, promise, and susceptibility to resistance mechanisms. Chem Biol Drug Des 86:379–399. doi: 10.1111/cbdd.12517. [DOI] [PubMed] [Google Scholar]
- 49.Maeda T, Garcia-Contreras R, Pu M, Sheng L, Garcia LR, Tomas M, Wood TK. 2012. Quorum quenching quandary: resistance to antivirulence compounds. ISME J 6:493–501. doi: 10.1038/ismej.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janssens JC, De Keersmaecker SC, De Vos DE, Vanderleyden J. 2008. Small molecules for interference with cell-cell-communication systems in Gram-negative bacteria. Curr Med Chem 15:2144–2156. doi: 10.2174/092986708785747580. [DOI] [PubMed] [Google Scholar]
- 51.Welsh MA, Eibergen NR, Moore JD, Blackwell HE. 2015. Small molecule disruption of quorum sensing cross-regulation in Pseudomonas aeruginosa causes major and unexpected alterations to virulence phenotypes. J Am Chem Soc 137:1510–1519. doi: 10.1021/ja5110798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore JD, Rossi FM, Welsh MA, Nyffeler KE, Blackwell HE. 2015. A comparative analysis of synthetic quorum sensing modulators in Pseudomonas aeruginosa: new insights into mechanism, active efflux susceptibility, phenotypic response, and next-generation ligand design. J Am Chem Soc 137:14626–14639. doi: 10.1021/jacs.5b06728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Mølin S, Eberl L, Steinberg P, Kjelleberg S, Høiby N, Givskov M. 2003. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smyth TJP, Perfumo A, Marchant R, Banat IM. 2010. Isolation and analysis of low-molecular-weight microbial glycolipids, p 3705–3723. In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany. [Google Scholar]
- 55.Ravn L, Christensen AB, Molin S, Givskov M, Gram L. 2001. Methods for detecting acylated homoserine lactones produced by Gram negative bacteria and their application in studies of AHL-production kinetics. J Microbiol Methods 44:239–251. doi: 10.1016/S0167-7012(01)00217-2. [DOI] [PubMed] [Google Scholar]
- 56.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 57.Fry JC. 1997. One-way analysis of variance, p 3–39. In Fry JC. (ed), Biological data analysis: a practical approach. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 58.Wang EW, Jung JY, Pashia ME, Nason R, Scholnick S, Chole RA. 2005. Otopathogenic Pseudomonas aeruginosa strains as competent biofilm formers. Arch Otolaryngol Head Neck Surg 131:983–989. doi: 10.1001/archotol.131.11.983. [DOI] [PubMed] [Google Scholar]
- 59.Wang L, Zhang C, Gong F, Li H, Xie X, Xia C, Chen J, Song Y, Shen A, Song J. 2011. Influence of Pseudomonas aeruginosa pvdQ gene on altering antibiotic susceptibility under swarming conditions. Curr Microbiol 63:377–386. doi: 10.1007/s00284-011-9979-0. [DOI] [PubMed] [Google Scholar]
- 60.Savli H, Karadenizli A, Kolayli F, Gundes S, Ozbek U, Vahaboglu H. 2003. Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J Med Microbiol 52:403–408. doi: 10.1099/jmm.0.05132-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.