ABSTRACT
Next-generation sequencing of 6 mcr-1-harboring Escherichia coli and Klebsiella pneumoniae isolates collected from a tertiary care hospital in China revealed significant sequence variations in the regions flanking the mcr-1 gene. While sequence variations significantly affected the expression and promoter activity of mcr-1, the mcr-1 gene expression levels did not correlate with the in vitro colistin resistance levels, which warrants further in-depth investigations.
KEYWORDS: colistin, genomics, plasmids, promoter, gene expression
TEXT
Following the initial report (1), the plasmid-mediated colistin resistance gene mcr-1 has been found in over 35 countries on 5 continents, among various bacterial species (2, 3). Meanwhile, mcr-1 has been identified in plasmids from different incompatibility groups, including IncX4, IncI2, IncHI1, IncHI2, IncFI, IncFII, IncP, IncI2-FIB, IncX1-X2, and IncX3-X4 groups (4). Comparative sequence analysis showed that mcr-1 is harbored on an ∼2,600-bp mcr-1-pap2 cassette in different plasmids and the immediate downstream sequences of mcr-1 are commonly conserved, whereas the upstream mcr-1 sequences are more diverse, even among plasmids from the same incompatibility group (4, 5). Previous studies on mcr-1 plasmids focused largely on plasmid sequence analysis, but the effects of sequence variations on mcr-1 gene expression and susceptibility were largely unexplored. Here we conducted a comprehensive molecular characterization of mcr-1-harboring Enterobacteriaceae isolates collected from a tertiary care hospital in southern China in 2015 to early 2016, using next-generation sequencing, gene expression, and promoter activity analyses.
A total of 905 nonduplicated clinical isolates (569 Escherichia coli isolates, 278 Klebsiella pneumoniae isolates, 36 Enterobacter cloacae isolates, and 22 Citrobacter freundii isolates) collected from a tertiary hospital in Guangdong Province in China between 2015 and March 2016 were randomly selected for molecular screening for mcr-1, using the primers described elsewhere (1). Among the 905 clinical isolates, 1 K. pneumoniae isolate (GZ49271) and 5 E. coli isolates (GZ49260, GZ49263, GZ49266, GZ49269, and GZ49273) were found to harbor mcr-1. The 6 isolates were isolated from different types of clinical specimens (urine, blood, and cervical swab specimens) in 2015 (n = 4) or 2016 (n = 2) (see Table S1 in the supplemental material). The overall prevalence of mcr-1 among E. coli isolates was 0.88% (5/569 isolates) and that among K. pneumoniae isolates was 0.36% (1/278 isolates). All 6 patients had a history of previous hospitalization but without exposure to colistin, and none had a history of recent overseas travel. All 6 isolates were resistant to colistin but susceptible to imipenem and meropenem, with various profiles of resistance to other antibiotics (Table S1). Among them, GZ49260, GZ49266, and GZ49269 were resistant to multiple antimicrobials, including ceftriaxone and aztreonam.
The mcr-1 genes from all isolates except GZ49263 were successfully transferred to recipient E. coli J53AZ-R strains by conjugation. Electroporation of plasmids extracted from GZ49263 into E. coli DH5α was also unsuccessful; therefore, the subsequent complete plasmid sequence characterization focused on the other 5 isolates. The genomic DNA from the 6 clinical isolates and 5 mcr-1 harboring plasmids from the E. coli J53AZ-R transconjugants were sequenced using next-generation sequencing (Illumina NextSeq 500), as described previously (6). In silico multilocus sequence typing (MLST) analysis (7) showed that the five E. coli strains belonged to five different sequence types (STs), i.e., ST744, ST7542, ST156, ST4204, and ST1196, while K. pneumoniae GZ49271 was an ST976 strain (Table S1). In silico mining of acquired resistance genes (8) identified the presence of mcr-1 in all 6 isolates and several other antimicrobial resistance genes in GZ49260, GZ49263, GZ49266, GZ49269, and GZ49271 (Table S1). Notably, GZ49260, GZ49266, and GZ49269 carried the extended-spectrum β-lactamase gene blaCTX-M and, among them, GZ49269 harbored two different blaCTX-M variants (blaCTX-M-14 and blaCTX-M-55). In contrast, mcr-1 was the only resistance gene found in E. coli GZ49273.
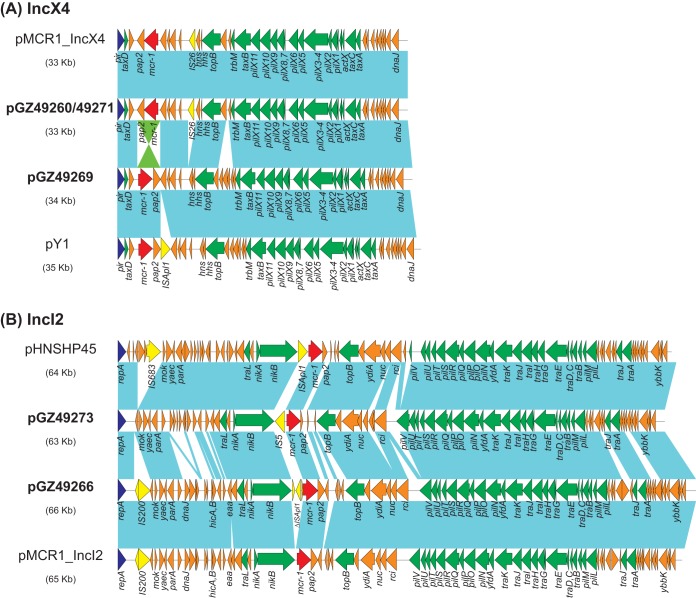
Sequence analysis showed that the five plasmids belonged to two plasmid replicon types, namely, IncX4 (pGZ49260, pGZ49269, and pGZ49271) and IncI2 (pGZ49273 and pGZ49266) types. Two IncX4 plasmids, from E. coli GZ49260 and K. pneumoniae GZ49271 (pGZ49260 and pGZ49271, respectively), were highly similar (5 single-nucleotide variations), with the same size of 33,309 bp and GC contents of 42%, suggesting horizontal transfer of the same mcr-1 plasmids into two different species. BLAST-n results showed that they had high levels of sequence homology (>99.9% identity, with 100% query coverage) to a number of completely sequenced mcr-1-harboring plasmids, including pCSZ4 (GenBank accession no. KX711706) (5) and pMCR1_IncX4 (GenBank accession no. KU761327) (9) from China. In contrast, the mcr-1 plasmid from E. coli GZ49269 (pGZ49269) was 33,858 bp in length, with a GC content of 42%, showing the best match (>99.9% identity, with 100% query coverage) to plasmid pPY1 (GenBank accession no. KX711708), which was isolated from an E. coli strain cultured from a pork specimen in China (5). Of note, the mcr-1-pap2 cassette in pGZ49269 had an inverted orientation, in comparison to pGZ49260 and pGZ49271, which are similar to plasmid pY1 except that pY1 carries an ISApl1 insertion downstream of the mcr-1-pap2 cassette (Fig. 1A).
FIG 1.
Plasmid structures. (A) Comparison of IncX4 plasmids pMCR1_IncX4 (GenBank accession no. KU761326), pGZ49260 (GenBank accession no. MG210937), pGZ49269 (GenBank accession no. MG210939), and pY1 (GenBank accession no. KX711708). (B) Comparison of IncI2 plasmids pHNSHP45 (GenBank accession no. KP347127), pMCR1_IncI2 (GenBank accession no. KU761327), pGZ49273 (GenBank accession no. MG210940), and pGZ49266 (GenBank accession no. MG210938). Colored arrows indicate open reading frames, with dark blue, yellow, green, red, and orange arrows representing replication genes, mobile elements, plasmid transfer genes, the mcr-1 gene, and plasmid backbone genes, respectively. Blue shading denotes regions of shared homology among different plasmids, while green shading indicates inverted shared homology.
The two IncI2 plasmids, pGZ49273 and pGZ49266, were 66,183 and 62,701 bp in length, respectively, with the same GC content of 43%. The overall backbones of the two plasmids were similar to those of several other mcr-1 IncI2 plasmids, including the prototype pHNSHP45 (GenBank accession no. KP347127) (1) and pMCR1_IncI2 (GenBank accession no. KU761326) (9) (Fig. 1B). In comparison to pHNSHP45 and pMCR1_IncI2, the main variable regions in pGZ49273 and pGZ49266 were the mcr-1-pap2 cassette upstream sequences. An ISApl1 element was inserted upstream of mcr-1 in pHNSHP45; in contrast, ISApl1 was absent in the corresponding region in pMCR1_IncI2. Interestingly, novel variants of insertion sequences were identified upstream of mcr-1 in pGZ49273 and pGZ49266. An IS5 element was inserted 62 bp upstream of mcr-1 in pGZ49273 and was located between the −10 and −35 promoter sequences, as reported previously (10). Interestingly, three truncated ISApl1 elements (446, 443, and 234 bp) were found to be integrated 60 bp upstream of mcr-1 in pGZ49266, disrupting the −35 promoter region. It is not clear how the mosaic structures of the three truncated ISApl1 elements were generated, and we speculate that they could be the result of multiple recombination events.
A 4,737-bp mcr-1-harboring element in GZ49263 was resolved using the combination of de novo assembly and regular PCR with Sanger sequencing. The mcr-1 in GZ49263 is harbored by a transposon-like element, with mcr-1 being flanked by two directly repeated ISApl1 elements. This region is nearly identical (1 SNP difference) to that of the corresponding region (nucleotides 21,539 to 26,275) in plasmid pSCC4 (GenBank accession no. CP021078), which was isolated from a Citrobacter braakii strain from a chicken in China.
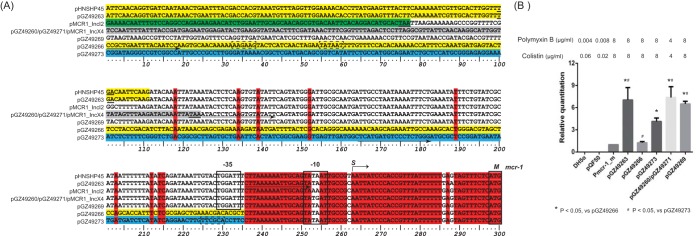
Sequence comparisons of the 5′ untranslated region of the mcr-1 gene among all six genomes identified a 62-bp conserved sequence, encompassing the previously reported −10 promoter (Fig. 2A). However, the −35 promoter sequence regions were variable. pGZ49263, pGZ49260, pGZ49271, and pGZ49269 carried −35 and −10 promoter sequences similar to those of the mcr-1 prototype plasmid pHNSHP45. In contrast, the original −35 promoters in pGZ49266 and pGZ49273 were disrupted due to insertion of a partial ISApl1 element and IS5. BPROM promoter predication software (Softberry, Inc.) identified two putative promoter regions in pGZ49266 and pGZ49273. In pGZ49273, a putative −35 promoter sequence (TTCGCA) was found 67 bp upstream of the start codon, located at the 3′ inverted reverse repeat of IS5 (Fig. 2A). In pGZ49266, a new set of −35 (AAGAAG) and −10 (TATAAT) sequences were identified 237 bp upstream of the start codon of mcr-1, located within the partial ISApl1 sequence (Fig. 2A).
FIG 2.
(A) Sequence analyses of the mcr-1 promoter region. Red shading denotes identical residues, yellow shading shows ISApl1 or partial ISApl1 sequences, and blue shading indicates IS5 sequences. The putative −35 and −10 promoter sequences in pGZ49266 and pGZ49273 are shown in dashed boxes. The cloning primer sequences of Pmcr-1_m, pGZ49260/pGZ49271, pGZ49266, and pGZ49273 are shown as underlining arrows. The primer sequences of pGZ49263 and pGZ49269 are located upstream of this region. S, transcription start site; M, methionine and translation initiation site. (B). Levels of mcr-1 gene expression in E. coli DH5α strains with different plasmid promoter sequence constructs (pQF50). The MIC values for colistin and polymyxin B are shown above the graph.
We cloned each promoter sequence and the full-length mcr-1 gene into a pQF50 expression plasmid in an E. coli DH5α strain and evaluated the mcr-1 expression using real-time reverse transcription (RT)-PCR (Table S2). The 62-bp conserved sequence and the full-length mcr-1 were cloned from pGZ49263 (Pmcr-1_m), and the strain was used as the baseline control strain for quantitative RT-PCR (qRT-PCR) (the expression level was taken as 1) (Fig. 2B). The results showed that the E. coli DH5α constructs with the mcr-1 promoter regions from pGZ49263, pGZ49260, pGZ49271, and pGZ49269 had the highest mcr-1 expression levels (P < 0.05, compared with pGZ49266 and pGZ49273 promoter clones) (Fig. 2B), consistent with the promoter sequence analysis, in which the four strains harbored promoter regions similar to that of the prototype plasmid pHNSHP45. In contrast, the pGZ49273 promoter clone had lower mcr-1 expression, while the pGZ49266 clone had the lowest mcr-1 expression levels, suggesting that the promoter sequence changes associated with partial ISApl1 rearrangement and IS5 insertion, respectively, had significant effects on mcr-1 expression. Furthermore, we evaluated the promoter activity using a promoterless lacZ reporter system, which demonstrated results similar to those of the mcr-1 qRT-PCR analysis, in that pGZ49273 and pGZ49266 promoter clones had significantly lower β-galactosidase activities than did the other four plasmid promoters (pGZ49263, pGZ49260/pGZ49271, and pGZ49269) (data not shown). Interestingly, despite the differences in mcr-1 expression and promoter activity described above, the MIC values of pQF50 E. coli DH5α constructs with different promoter sequences, including the baseline control clone, were similar, with values of 4 to 8 μg/ml for colistin and polymyxin (Fig. 2B).
In this study, we demonstrated that mcr-1-harboring plasmids possessed significant sequence variation, especially in the regions flanking the mcr-1 gene. The sequence variation could significantly affect the promoter activity and mcr-1 expression, but the mcr-1 gene expression did not correlate with in vitro colistin resistance levels. MCR-1 acts as a phosphoethanolamine transferase and modifies the chemical structure of the lipid A moiety on bacterial lipopolysaccharide (LPS) through the addition of phosphoethanolamine, which reduces the negative charge and the binding affinity for colistin and ultimately produces colistin resistance. One explanation for the lack of correlation of mcr-1 expression levels and MICs could be that colistin is sensitive to structure modifications of lipid A and a minimal phosphoethanolamine modification (as a result of low-level mcr-1 expression) may be sufficient to increase colistin MIC levels. Further studies, such as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) analysis of these clones with different promoter sequences, are needed to resolve the structural changes caused by phosphoethanolamine additions and to identify the correlations with MICs.
Accession number(s).
The complete nucleotide plasmid sequences were deposited in GenBank under accession numbers MG210937 to MG210940. The raw whole-genome sequencing data from this study were submitted to NCBI under BioProject record number PRJNA354234.
Supplementary Material
ACKNOWLEDGMENTS
The study was supported in part by the National Natural Science Foundation of China (grants 81572058, 81672081, and 81772249), the Guangdong Natural Science Foundation (grant 2014A030313143), and the State Education Ministry Scientific Research Foundation for the Returned Overseas Chinese Scholars. This work was in part supported by grants from the National Institute of Allergy and Infectious Diseases (grant R01AI090155 to B.N.K. and grant R21AI117338 to L.C.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00018-18.
REFERENCES
- 1.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz S, Johnson AP. 2016. Transferable resistance to colistin: a new but old threat. J Antimicrob Chemother 71:2066–2070. doi: 10.1093/jac/dkw274. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Li Z, Lin J, Wang X, Deng X, Feng Y. 2016. Complex dissemination of the diversified mcr-1-harbouring plasmids in Escherichia coli of different sequence types. Oncotarget 7:82112–82122. doi: 10.18632/oncotarget.12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun J, Fang LX, Wu Z, Deng H, Yang RS, Li XP, Li SM, Liao XP, Feng Y, Liu YH. 2017. Genetic analysis of the IncX4 plasmids: implications for a unique pattern in the mcr-1 acquisition. Sci Rep 7:424. doi: 10.1038/s41598-017-00095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eilertson B, Chen L, Chavda KD, Kreiswirth BN. 2017. Genomic characterization of two KPC-producing Klebsiella isolates collected in 1997 in New York City. Antimicrob Agents Chemother 61:e02458-16. doi: 10.1128/AAC.02458-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li A, Yang Y, Miao M, Chavda KD, Mediavilla JR, Xie X, Feng P, Tang YW, Kreiswirth BN, Chen L, Du H. 2016. Complete sequences of mcr-1-harboring plasmids from extended-spectrum-β-lactamase- and carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 60:4351–4354. doi: 10.1128/AAC.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poirel L, Kieffer N, Brink A, Coetze J, Jayol A, Nordmann P. 2016. Genetic features of MCR-1-producing colistin-resistant Escherichia coli isolates in South Africa. Antimicrob Agents Chemother 60:4394–4397. doi: 10.1128/AAC.00444-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.