ABSTRACT
The molecular mechanisms of tolerance and persistence associated with several compounds in Acinetobacter baumannii clinical isolates are unknown. Using transcriptomic and phenotypic studies, we found a link between mechanisms of bacterial tolerance to chlorhexidine and the development of persistence in the presence of imipenem in an A. baumannii strain belonging to clinical clone ST-2 (OXA-24 β-lactamase and AbkAB toxin-antitoxin [TA] system carried in a plasmid). Interestingly, the strain A. baumannii ATCC 17978 (AbkAB TA system from plasmid) showed persistence in the presence of imipenem and chlorhexidine.
KEYWORDS: tolerance, persistence, chlorhexidine, imipenem, Acinetobacter, toxin-antitoxin
TEXT
The importance of preventing the development of tolerance and/or persistence has recently been highlighted as a new strategy for delaying the emergence of resistance (1–4). In this context, it is essential to distinguish between bacterial resistance, tolerance, and persistence (5). Resistance refers to the ability of bacterial populations to grow at the same rate in the presence of antibiotic-induced or environmental stress. Tolerance is the ability of a bacterial population to grow slowly in response to stress. Finally, persistence is the latent state of a bacterial subpopulation, which is activated under certain conditions (5).
Several bacterial tolerance mechanisms develop during stress and antibiotic exposure (6). These mechanisms include (p)ppGpp signaling accumulation, reactive oxygen species (ROS) and SOS responses, bacterial communication (quorum sensing), efflux pumps, and energy metabolism (6).
Carbapenem-resistant Acinetobacter baumannii (CRAb) is currently a major source of nosocomial infections and is considered a highly successful human pathogen (7). Among the different mechanisms associated with carbapenem resistance in A. baumannii, the production of acquired carbapenem-hydrolyzing class D β-lactamases (CHDLs) and class B metallo-β-lactamases (MBLs) has been widely studied (8). On the other hand, the main mechanisms of development of persister cells in the presence of antibiotics (such as imipenem [IMP]) involve toxin-antitoxin (TA) modules (6, 9).
Studies about molecular mechanisms of tolerance and persistence from A. baumannii strains in response to several compounds are scarce. In this study, we used transcriptomic and phenotypic assays to analyze the tolerance and persistence mechanisms of A. baumannii isolates in response to chlorhexidine and imipenem (resistance and susceptibility to carbapenems).
In a previous work of the REIPI-GEIH Ab-2010 project (10), we worked with the A. baumannii clinical strains Ab-2_clon_2010 (belonging to clone ST-2) and Ab-2_clon_2010-CHLX, which showed the absence of an increase of MICs to antibiotics after exposure to subinhibitory concentrations of chlorhexidine digluconate (CHLX) (0.25× MIC) during 4 weeks (see Table S1 in the supplemental material). The genome of this Ab-2_clon_2010 strain, together with 17 other clinical strains from this ST-2 clone, were sequenced by Lopez et al. (11) in the Umbrella GenBank BioProject number PRJNA422585. All strains from this ST-2 clone belonged to the REIPI-GEIH Ab-2010 project and had a plasmid with the blaOXA24/40 β-lactamase gene (conferring resistance to carbapenems), as well as the abkAB genes from a toxin-antitoxin system (12). RNA assays by transcriptomics had a number of reads assigned to the different genes and were analyzed using the EdgeR and DESeq2 packages and reverse transcription PCR (RT-PCR) techniques using UPLs Probe (see Table S3 in the supplemental material; Roche, Germany) of both clinical isolates (DNase-treated RNA of Ab-2_clon_2010 and Ab-2_clon_2010-CHLX) (GenBank BioProject number PRJNA433173 and GEO series number GSE110207), the results of which are shown in Table S2 and Fig. S1 and S2 in the supplemental material.
The results showed the activation of tolerance molecular mechanisms (known as “tolerome”) in response to chlorhexidine in strain Ab-2_clon_2010-CHLX (Table 1). In relation with the tolerome, in the strain Ab-2_clon_2010-CHLX, we observed overexpression (1.5- to 6-fold change [FC]) of genes encoding the AdeABC, arsenite, and AceI chlorhexidine efflux pumps (10, 13–16). Some of these additional protective mechanisms, such as the production of efflux pumps, may also reduce the effective concentration of the antibiotic, which increases the MIC and results in a mixed phenotype of resistance and tolerance (5). We also observed an increase in the expression of genes involved in tetracycline and aminoglycoside resistance (FC, 3.4 to 6). The genes with the highest level of overexpression in this study were those carried by the AbATCC329 plasmid (PMMCU3p), such as OXA24/40 β-lactamase, DNA replication protein, and OriV (FC, 5.2 to 12) (12) (Table 1). Interestingly, the gene expression FCs of abkA (antitoxin gene) and abkB (toxin gene) from this plasmid were 0. 63 and 1.25, respectively. In addition, we observed the overexpression of genes associated with molecular mechanisms of bacterial tolerance (FC, 3.5 to 10), namely, the CsuA/BABCDE operon (17, 18), the CydAB operon (cytochrome d ubiquinol oxidase complex) (19, 20, 21), the taurine operon complex (taurine metabolism/electron carrier activity) (22, 23), and finally, regulatory genes involved in the quorum-sensing (QS) system, i.e., abaR and abaI (Table 1) (22–25).
TABLE 1.
Mechanisms of bacterial tolerance to chlorhexidine in strain Ab-2_clon_2010-CHLX, revealed by transcriptomic studiesa
| GenBankb protein accession no. | Gene expression fold change determined by: |
Functional description | Defense mechanism (reference no.) | Tolerome type (reference no.) | |
|---|---|---|---|---|---|
| DESeq2 | EdgeR | ||||
| ODA53993.1 | 6.933753475 | 6.982635042 | AdeA protein | AdeABC system (RND-type) (10) | Transporter/efflux pump (5) |
| ODA53994.1 | 6.149907892 | 6.175526694 | AdeB protein | ||
| ODA53995.1 | 4.257153566 | 4.270842036 | AdeC protein | ||
| ODA55718.1 | 6.119321647 | 6.133494454 | Tetracycline resistance protein | MFS system | |
| ODA54617.1 | 5.377292457 | 7.227172031 | Arsenite efflux pump | ACR3 system (13) | |
| ODA56577.1 | 3.498098186 | 3.528728206 | Aminoglycoside phosphotransferase | APT family | |
| ODA54814.1 | 3.605781331 | 3.649808635 | Chlorexidine efflux pump | AceI system (16) | |
| ODA56167.1 | 5.265550668 | 7.054151044 | MFS transporter | MFS system | |
| ODA53764.1 | 12.16763575 | 14.92175121 | OXA 24/40 β-lactamase | AbATCC329p/pMMCU3 | Plasmid (5) |
| ODA53763.1 | 8.975633873 | 11.30715263 | DNA replication protein A | ||
| ODA53762.1 | 5.273985066 | 5.329333593 | RepB family plasmid replication initiator | ||
| ODA54084.1 | 3.511019975 | 3.547062538 | CsuA protein | CsuABCDE (17, 18) | Biofilm (14) |
| ODA54083.1 | 3.199749378 | 3.259685195 | CsuB protein | ||
| ODA54082.1 | 2.575094974 | 2.584527435 | CsuC protein | ||
| ODA54081.1 | 2.810613341 | 2.819199271 | CsuD protein | ||
| ODA54080.1 | 2.782552791 | 2.791313686 | CsuE protein | ||
| ODA53940.1 | 2.037734523 | 2.053934504 | Cytochrome b | Cytochrome operon (19–21) | Stress oxidative (ROS) (21) |
| ODA57053.1 | 2.173049691 | 2.184371809 | Cytochrome bd biosynthesis protein | ||
| ODA56663.1 | 2.405101873 | 2.428897655 | Sodium/proline symporter | ||
| ODA56171.1 | 10.75708444 | 13.32903693 | Cytochrome bd biosynthesis protein | ||
| ODA56172.1 | 10.35093438 | 12.86380541 | Cytochrome d ubiquinol oxidase subunit | ||
| ODA54604.1 | 10.07652823 | 12.56398102 | Taurine ABC transporter substrate-binding | Taurine transporter (22, 23) | Electron transport |
| ODA54605.1 | 9.758316312 | 12.21616998 | Taurine transporter-binding subunit (TauB) | ||
| ODA54606.1 | 8.966908008 | 11.30350134 | Taurine ABC transporter permease (TauC) | ||
| ODA54607.1 | 10.85324686 | 13.44475271 | Taurine dioxygenase (TauD) | ||
| ODA55153.1 | −6.486154998 | −6.530626555 | Hypothetical protein | Replication | ppGpp network (28)c |
| ODA54592.1 | 0.932475218 | 0.929842277 | DNA polymerase I | ||
| ODA54625.1 | 0.931688577 | 0.929428817 | DNA polymerase III subunit alpha | ||
| ODA54730.1 | −1.77207536 | −1.816317901 | Response regulator | ||
| ODA55878.1 | 0.500078184 | 0.506140675 | 50S ribosomal protein L17 | ||
| ODA55763.1 | 0.438241011 | 0.436148678 | RNA polymerase subunit omega | ||
| ODA55654.1 | 0.582523178 | 0.580921263 | 50S ribosomal protein L7/L12 | ||
| ODA55933.1 | −0.523115918 | −0.531876133 | ATP synthase subunit beta | ATP metabolism | Energy production (31, 32)c |
| ODA55935.1 | −0.570647356 | −0.579254835 | ATP synthase subunit alpha | ||
| ODA54585.1 | 0.422390483 | 0.418615357 | Transcription termination factor rho | ||
The relative expression (expressed as fold change [FC]) of abaI (3.05) and abaR (2.88) genes, determined by RT-PCR, indicated activation of the quorum-sensing system.
Sanger sequencing of these genes from the Ab-2_clon_2010-CHLX strain, as well as of the regulatory genes adeR and adeS, did not show mutations with respect to the sequence of strain Ab-2_clon_2010.
Genes that belonged to the ppGpp network and energy production categories showed downregulation (FC, <1).
We used the Kyoto Encyclopedia of Genes and Genomes (KEGG) tool to analyze those genes that were downregulated (FC, ≤0.5-fold) in Ab-2_clon_2010-CHLX. We studied two metabolic pathways. The first one was the ppGpp network (KEGG accession numbers ec00230 2.7.7.6 and ec00230 2.7.7.7) involving RNA polymerases, DNA polymerases, and finally, 50S ribosomal protein. The ppGpp network is mediated by a variety of RelA/SpoT homologue (RSH) proteins with a nucleotidyl transferase domain, with some displaying only synthetic or hydrolytic activities, and others displaying both (Rel) (26, 27). Accumulation of (p)ppGpp affects resource-consuming cell processes, such as replication, transcription, and translation. Furthermore, (p)ppGpp is thought to bind RNA polymerase and alter the transcriptional profile, decreasing the synthesis of translational machinery (such as rRNA and tRNA) and increasing transcription of the biosynthetic gene (28). Additionally, initiation of new rounds of replication is inhibited, and the cell cycle arrests until nutrient conditions improve (29). Translational GTPases involved in protein biosynthesis are also affected by ppGpp, with initiation factor 2 (IF2) being the main target (30). Although these proteins are scarcely known in A. baumannii, in this study we describe RelA-SpoT-homologous (RSH) proteins associated with these functions that show repression in Ab-2_clon_2010-CHLX isolate. The second metabolic pathway studied was that of oxidative phosphorylation (KEGG accession no. ec00190 3.6.3.14, ATP phosphohydrolases), in which the alpha/beta ATP synthase subunit and transcription termination factor rho were downregulated. Finally, energy production by ATP metabolism has been associated with the development of tolerant cells in Escherichia coli (31). Moreover, Wang et al., described how genes mapped in this pathway have an important role in the survival of clinical strains of Staphylococcus aureus (32).
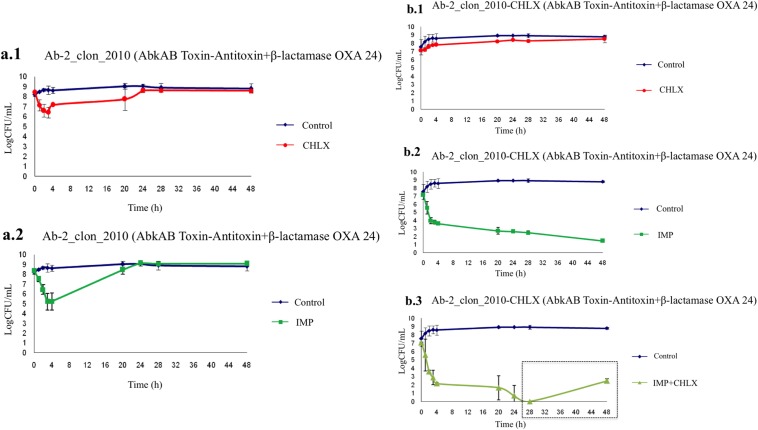
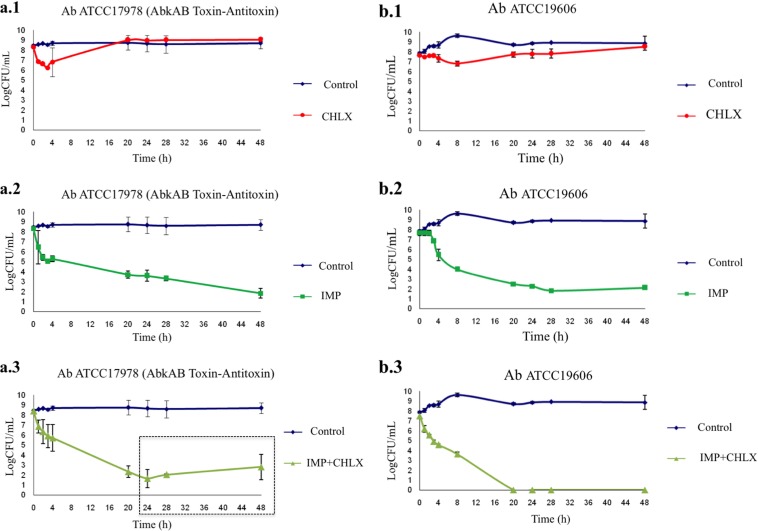
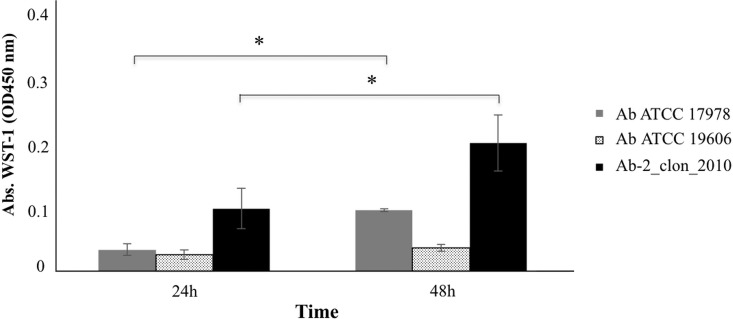
The time-kill curves for strains Ab-2_clon_2010 and Ab-2_clon_2010-CHLX were performed following the indications of Hofsteenge and colleagues (33) in low-nutrient Luria-Bertani broth (LN-LB; 2 g/liter tryptone, 1 g/liter yeast extract, and 5 g/liter NaCl) (13, 16). The cultures were incubated for 4 h to ensure logarithmic growth, and CHLX (0.25× MIC) and IMP (10× MIC) were then added alone or in combination to the cultures. We observed a lower growth rate of the Ab-2_clon_2010 strain in the presence of CHLX than in its absence (Fig. 1), as well as faster growth rate in the presence of IMP. Interestingly, the time-kill curves for isolate Ab-2_clon_2010-CHLX showed a massive killing in the presence of IMP (Fig. 1). The results of RT-PCR analysis confirmed a lower expression of OXA24/40 β-lactamase and abkA antitoxin genes (FC, 0.06 and 0.04, respectively) in Ab-2_clon_2010-CHLX, as well as overexpression of the abkB toxin gene (FC, 2.77) relative to that in Ab-2_clon_2010 (known as the persistome) (34–36). This toxin protein belongs to the AbkAB toxin-antitoxin module in the AbATCC329p/pMMCU3 plasmid (12). Mosqueda et al. located the AbkB/AbkA TA system (the so-called SplTA) in the most prevalent plasmids (GenBank KJ534568 and KJ534569) found in clinical isolates of A. baumannii (12, 37). Finally, we observed regrowth of persister cells in the Ab-2_clon_2010-CHLX isolate grown in the presence of IMP+CHLX for 28 h (Fig. 1). Moreover, we used two A. baumannii ATCC isolates as controls (both susceptible to carbapenems) whose complete genomes have been sequenced, A. baumannii strain ATCC 17978 (which harbors the AbkAB toxin-antitoxin system encoded by plasmid pAB2, GenBank number CP000523.1) and A. baumannii strain ATCC 19606 (which does not have this AbkAB toxin-antitoxin system). In Fig. 2, we observed that in the A. baumannii strain ATCC 17978, there was a reactivation of growth in the presence of IMP+CHLX for 28 h, in contrast to the lack of growth of the A. baumannii ATCC 19606 under the same conditions. These results of regrowth in the A. baumannii strain ATCC 17978 and in Ab-2_clon_2010-CHLX with IMP (10× MIC) and CHLX (0.25× MIC) at 48 h were confirmed by enzymatic analysis using the cell proliferation reagent WST-1 protocol (Roche, Germany) and calculating the serial dilutions of each culture (CFU/ml; Fig. 3).
FIG 1.
Time-kill curves in the presence of biocides (CHLX) and antibiotics (IMP) in Ab-2_clon_2010 (carbapenem-resistant) and Ab-2_clon_2010-CHLX isolates. Box in panel b.3, regrowth is due to putative reactivation of persister cells.
FIG 2.
Time-kill curves in the presence of antibiotics (IMP) and biocides (CHLX) in susceptible A. baumannii ATCC strains. (a) A. baumannii strain ATCC 17978, which harbors the plasmid with the AbKA/AbkB toxin-antitoxin system (positive control); (b) A. baumannii ATCC 19606 strain without this AbKA/AbkB toxin-antitoxin system (negative control). Box in panel a.3, regrowth is due to putative reactivation of persister cells.
FIG 3.
Enzymatic activity by colorimetric assay (WST-1-based) of the isolates A. baumannii ATCC 17978, A. baumannii ATCC 19606, and A. baumannii Ab-2_clon_2010-CHLX in the presence of IMP and CHLX. The x axis represents absorbance (optical density at 450 nm [OD450]), and the y axis represents time (h). *, P < 0.05 (Student's t test).
In conclusion, this is the first study describing the important link between mechanisms of bacterial tolerance and persistence under chlorhexidine and imipenem pressure in a clinical isolate of A. baumannii ST-2 harboring the blaOXA 24/40 β-lactamase gene and abKA/abkB genes (toxin-antitoxin system) in a plasmid. The study of these mechanisms (bacterial tolerance and persistence) is key to the development of new anti-infective treatments which will allow for the eradication of multidrug resistant pathogens.
Accession number(s).
The whole-genome sequence (WGS) studies of GEIH-2010 isolate Ab-2_clon_2010 comprise part of the II Spanish Multicenter Study. GEIH-REIPI A. baumannii 2000 to 2010 project (umbrella GenBank BioProject number PRJNA422585), as well as the transcriptomic results shown in GenBank BioProject number PRJNA433173 (GEO series number GSE110207). The WGSs of the A. baumannii strain ATCC 17978 complete genome and A. baumannii strain ATCC 19606 complete genome are deposited under GenBank accession numbers CP018664.1 and GG704575.1, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by grants PI13/02390 and PI16/01163, awarded to M.T., and by grant PI11-02046, awarded to F.F.-C., within the State Plan for R+D+I 2013–2016 (National Plan for Scientific Research, Technological Development and Innovation 2008–2011) and cofinanced by the ISCIII-Deputy General Directorate of Evaluation and Promotion of Research—European Regional Development Fund “A Way of Making Europe” and the Instituto de Salud Carlos III FEDER, Spanish Network for the Research in Infectious Diseases (REIPI, grants RD12/0015/0010 and RD16/0016/0001), as well as by the Study Group on Mechanisms of Action and Resistance to Antimicrobials, GEMARA (SEIMC). M.T. was financially supported by the Miguel Servet Research Programme (SERGAS and ISCIII). L.F.-G. was financially supported by a predoctoral fellowship from the Xunta de Galicia (GAIN, Axencia de Innovación).
We declare that we have no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00250-18.
REFERENCES
- 1.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 2.Cohen NR, Lobritz MA, Collins JJ. 2013. Microbial persistence and the road to drug resistance. Cell Host Microbe 13:632–642. doi: 10.1016/j.chom.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol 5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 4.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131. [DOI] [PubMed] [Google Scholar]
- 5.Brauner A, Fridman O, Gefen O, Balaban NQ. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 6.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 7.Vila J, Pachon J. 2011. Acinetobacter baumannii resistant to everything: what should we do? Clin Microbiol Infect 17:955–956. doi: 10.1111/j.1469-0691.2011.03566.x. [DOI] [PubMed] [Google Scholar]
- 8.Gupta V. 2008. Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin Invest Drugs 17:131–143. doi: 10.1517/13543784.17.2.131. [DOI] [PubMed] [Google Scholar]
- 9.Wood TK, Knabel SJ, Kwan BW. 2013. Bacterial persister cell formation and dormancy. Appl Environ Microbiol 79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernández-Cuenca F, Tomás M, Caballero-Moyano FJ, Bou G, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual Á; Spanish Group of Nosocomial Infections (GEIH) from the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC) and the Spanish Network for Research in Infectious Diseases (REIPI); Spanish Group of Nosocomial Infections GEIH from the Spanish Society of Clinical Microbiology and Infectious Diseases SEIMC and the Spanish Network for Research in Infectious Diseases REIPI . 2015. Reduced susceptibility to biocides in Acinetobacter baumannii: association with resistance to antimicrobials, epidemiological behaviour, biological cost and effect on the expression of genes encoding porins and efflux pumps. J Antimicrob Chemother 70:3222–3229. doi: 10.1093/jac/dkv262. [DOI] [PubMed] [Google Scholar]
- 11.López M, Mayer C, Fernández-García L, Blasco L, Muras A, Ruiz FM, Bou G, Otero A, Tomás M; GEIH-GEMARA (SEIMC). 2017. Quorum sensing network in clinical strains of A. baumannii: AidA is a new quorum quenching enzyme. PLoS One 12:e0174454. doi: 10.1371/journal.pone.0174454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mosqueda N, Gato E, Roca I, López M, de Alegría CR, Fernández Cuenca F, Martínez-Martínez L, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Vila J, Bou G, Tomás M; GEIH-GEMARA (SEIMC) and REIPI. 2014. Characterization of plasmids carrying the blaOXA-24/40 carbapenemase gene and the genes encoding the AbkA/AbkB proteins of a toxin/antitoxin system. J Antimicrob Chemother 69:2629–2633. doi: 10.1093/jac/dku179. [DOI] [PubMed] [Google Scholar]
- 13.Rumbo C, Gato E, López M, Ruiz de Alegría C, Fernández-Cuenca F, Martínez-Martínez L, Vila J, Pachón J, Cisneros JM, Rodríguez-Baño J, Pascual A, Bou G, Tomás M; Spanish Group of Nosocomial Infections and Mechanisms of Action and Resistance to Antimicrobials (GEIH-GEMARA); Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC); Spanish Network for Research in Infectious Diseases (REIPI) . 2013. Contribution of efflux pumps, porins, and β-lactamases to multidrug resistance in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 57:5247–5257. doi: 10.1128/AAC.00730-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Acker H, Van Dijck P, Coenye T. 2014. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol 22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Langevin AM, Dunlop MJ. 2017. Stress introduction rate alters the benefit of AcrAB-TolC efflux pumps. J Bacteriol 200:e00525-. doi: 10.1128/JB.00525-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hassan KA, Liu Q, Henderson PJ, Paulsen IT. 2015. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 6:e01982-. doi: 10.1128/mBio.01982-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomaras AP, Dorsey CW, Edelmann RE, Actis LA. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 18.Houari A, Di Martino P. 2007. Effect of chlorhexidine and benzalkonium chloride on bacterial biofilm formation. Lett Appl Microbiol 45:652–656. doi: 10.1111/j.1472-765X.2007.02249.x. [DOI] [PubMed] [Google Scholar]
- 19.Braoudaki M, Hilton AC. 2004. Adaptive resistance to biocides in Salmonella enterica and Escherichia coli O157 and cross-resistance to antimicrobial agents. J Clin Microbiol 42:73–78. doi: 10.1128/JCM.42.1.73-78.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turner AK, Barber LZ, Muhammad S, Jones MA, Lovell MA, Hulme S, Barrow PA. 2003. Contribution of proton-translocating proteins to the virulence of Salmonella enterica serovars Typhimurium, Gallinarum, and Dublin in chickens and mice. Infect Immun 71:3392–3401. doi: 10.1128/IAI.71.6.3392-3401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant SS, Hung DT. 2013. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 4:273–283. doi: 10.4161/viru.23987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tara Rema PM, Lawrence JR, Vidovic S, Leppard GG, Reid M, Korber DR. 2015. Proteomic analyses of chlorhexidine tolerance mechanisms in Delftia acidovorans biofilms. mSphere 1:e00017-. doi: 10.1128/mSphere.00017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Seeve C, Pierson LS, Pierson EA. 2013. Transcriptome profiling reveals links between ParS/ParR, MexEF-OprN, and quorum sensing in the regulation of adaptation and virulence in Pseudomonas aeruginosa. BMC Genomics 14:618. doi: 10.1186/1471-2164-14-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clemmer KM, Bonomo RA, Rather PN. 2011. Genetic analysis of surface motility in Acinetobacter baumannii. Microbiology 157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luo LM, Wu LJ, Xiao YL, Zhao D, Chen ZX, Kang M, Zhang Q, Xie Y. 2015. Enhancing pili assembly and biofilm formation in Acinetobacter baumannii ATCC19606 using non-native acyl-homoserine lactones. BMC Microbiol 15:62. doi: 10.1186/s12866-015-0397-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avarbock A, Avarbock D, Teh JS, Buckstein M, Wang ZM, Rubin H. 2005. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44:9913–9923. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 27.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cashel M. 1969. The control of ribonucleic acid synthesis in Escherichia coli. IV. Relevance of unusual phosphorylated compounds from amino acid-starved stringent strains. J Biol Chem 244:3133–3141. [PubMed] [Google Scholar]
- 29.Chatterji D, Fujita N, Ishihama A. 1998. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells 3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 30.Chatterji D, Ojha AK. 2001. Revisiting the stringent response, ppGpp and starvation signaling. Curr Opin Microbiol 4:160–165. doi: 10.1016/S1369-5274(00)00182-X. [DOI] [PubMed] [Google Scholar]
- 31.Keren I, Shah D, Spoering A, Kaldalu N, Lewis K. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J Bacteriol 186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W, Chen J, Chen G, Du X, Cui P, Wu J, Zhao J, Wu N, Zhang W, Li M, Zhang Y. 2015. Transposon mutagenesis identifies novel genes associated with Staphylococcus aureus persister formation. Front Microbiol 6:1437. doi: 10.3389/fmicb.2015.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofsteenge N, Van Nimwegen E, Silander OK. 2013. Quantitative analysis of persister fractions suggests different mechanisms of formation among environmental isolates of E. coli. BMC Microbiol 13:25. doi: 10.1186/1471-2180-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rotem E, Loinger A, Ronin I, Levin-Reisman I, Gabay C, Shoresh N, Biham O, Balaban NQ. 2010. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci U S A 107:12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butt A, Higman VA, Williams C, Crump MP, Hemsley CM, Harmer N, Titball RW. 2014. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochem J 459:333–344. doi: 10.1042/BJ20140073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dörr T, Vulić M, Lewis K. 2010. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol 8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, Tomas M. 2016. Toxin-antitoxin systems in clinical pathogens. Toxins (Basel) 8:E227. doi: 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.