ABSTRACT
Vancomycin taper regimens are commonly used for the treatment of recurrent Clostridium difficile infections. One rationale for tapering and pulsing of the dose at the end of therapy is to reduce the selective pressure of vancomycin on the indigenous intestinal microbiota. Here, we used a mouse model to test the hypothesis that the indigenous microbiota that provide colonization resistance against C. difficile and vancomycin-resistant enterococci (VRE) is repopulated during tapering courses of vancomycin. Mice were treated orally with vancomycin daily for 10 days, vancomycin in a tapering dose for 42 days, fidaxomicin for 10 days, or saline. To assess colonization resistance, subsets of mice were challenged with 104 CFU of C. difficile or VRE at multiple time points during and after completion of treatment. The impact of the treatments on the microbiome was measured by cultures, real-time PCR for selected anaerobic bacteria, and deep sequencing. Vancomycin taper-treated mice developed alterations of the microbiota and disruption of colonization resistance that was persistent 18 days after treatment. In contrast, mice treated with a 10-day course of vancomycin exhibited recovery of the microbiota and of colonization resistance by 15 days after treatment, and fidaxomicin-treated mice maintained intact colonization resistance. These findings demonstrate that alteration of the indigenous microbiota responsible for colonization resistance to C. difficile and VRE persist during and after completion of tapering courses of vancomycin.
KEYWORDS: colonization resistance, oral vancomycin
INTRODUCTION
Although effective treatments are available for Clostridium difficile infection (CDI), as many as 20 to 30% of patients who respond to initial treatment with vancomycin or metronidazole develop recurrent CDI, usually within 1 to 4 weeks of completing treatment (1). The risk of recurrent CDI is even higher in patients who have already had one or more recurrences (1). Several mechanisms may contribute to recurrences of CDI. First, the absence of an adequate immune response to C. difficile toxin is associated with recurrence (2). Second, persistent detection of C. difficile in stool at the end of treatment was associated with increased frequency of relapse of CDI (3). Finally, persistent disruption of the indigenous colonic microbiota after completion of CDI treatment predisposes to recurrence (4–10). Based on these mechanisms, strategies that provide passive immunity to C. difficile toxin or that preserve or restore the microbiota have been developed and have been effective in reducing recurrences (11–13).
Although new approaches for management of recurrent CDI are promising, oral vancomycin administered as a tapered and/or pulsed regimen remains the most common treatment strategy for second or greater recurrences of CDI (14–16). These regimens typically include a 10- to 14-day course of oral vancomycin at a dose of 125 mg four times per day, followed by a tapering dose over 2 weeks, followed by “pulsed” dosing with 125 mg once every 2 or 3 days for 2 to 8 weeks (14–16). Standard vancomycin regimens achieve persistent high concentrations in stool (1,000 to 2,000 μg/g stool) and cause marked disruption of the intestinal microbiota (4–9, 17). It has been proposed that tapered and pulsed regimens might reduce the risk for recurrence because the prolonged course provides more time for clearance of C. difficile and the pulsed dosing results in lower vancomycin concentrations and antibiotic-free periods that may allow restoration of the indigenous microbiota (3). Given that vancomycin has relatively limited activity against many Gram-negative anaerobes (e.g., Bacteroides fragilis group [MIC90 of 128 μg/ml] and Prevotella spp. [MIC90 of 256 μg/ml]) (18), it is plausible that lower concentrations and antibiotic-free periods might allow recovery during tapered and pulsed regimens. However, it is not known whether the administration of vancomycin in a tapered and pulsed regimen does allow recovery of the microbiota. In addition, no randomized trials have been reported that compared recurrence in CDI patients receiving vancomycin taper and pulse regimens versus standard vancomycin regimens. In a retrospective analysis of patients participating in the placebo arm of a randomized trial of Saccharomyces boulardii for prevention of CDI recurrence, tapered and pulsed vancomycin regimens had significantly fewer recurrences than one of the three standard vancomycin regimens (1 g/day vancomycin) (3). However, in a recent propensity score-matched analysis, vancomycin taper regimens did not provide benefit over vancomycin regimens without taper in preventing additional CDI recurrence in patients with a first or second recurrence of CDI (19).
Here, we used a mouse model to test the hypothesis that the intestinal microbiota that provide colonization resistance to C. difficile recover when vancomycin is administered as a tapered and pulsed regimen. For comparison, we evaluated the impact of treatment with a 10-day course of vancomycin or fidaxomicin. Fidaxomicin was chosen as a comparator because it is a treatment option for patients with recurrent CDI that has been shown to cause less adverse impact on the indigenous intestinal microbiota than standard regimens of vancomycin (5, 20).
RESULTS
Concentrations of vancomycin in stool.
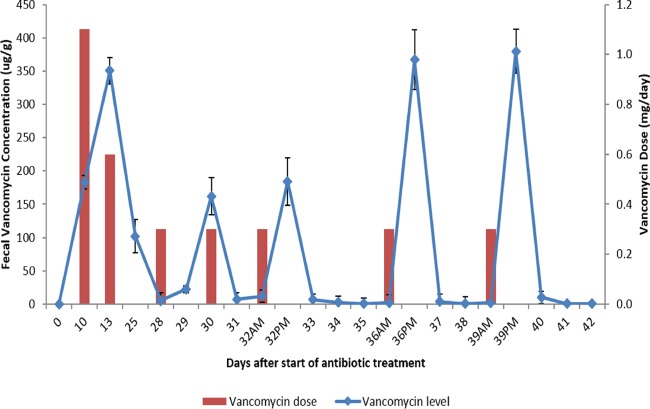
Figure 1 shows concentrations of vancomycin in stool of mice treated with a 10-day course of vancomycin, followed by the tapered and pulsed vancomycin regimen. The mean concentrations of vancomycin on days 10 and 13 of treatment were 183 and 350 μg/g of stool, respectively. During the pulse phase of the vancomycin taper, vancomycin was present at 161 to 380 μg/g of stool 24 h after each dose, but was undetectable at 2 to 3 days after dosing.
FIG 1.
Concentrations of vancomycin in stool of mice treated with a 10-day course of vancomycin, followed by the tapered and pulsed vancomycin regimen. Mean concentrations of vancomycin (n = 5 mice) are shown at each time point. The bars show the vancomycin dose in mg per day. Error bars show the standard errors.
Colonization resistance to C. difficile and VRE.
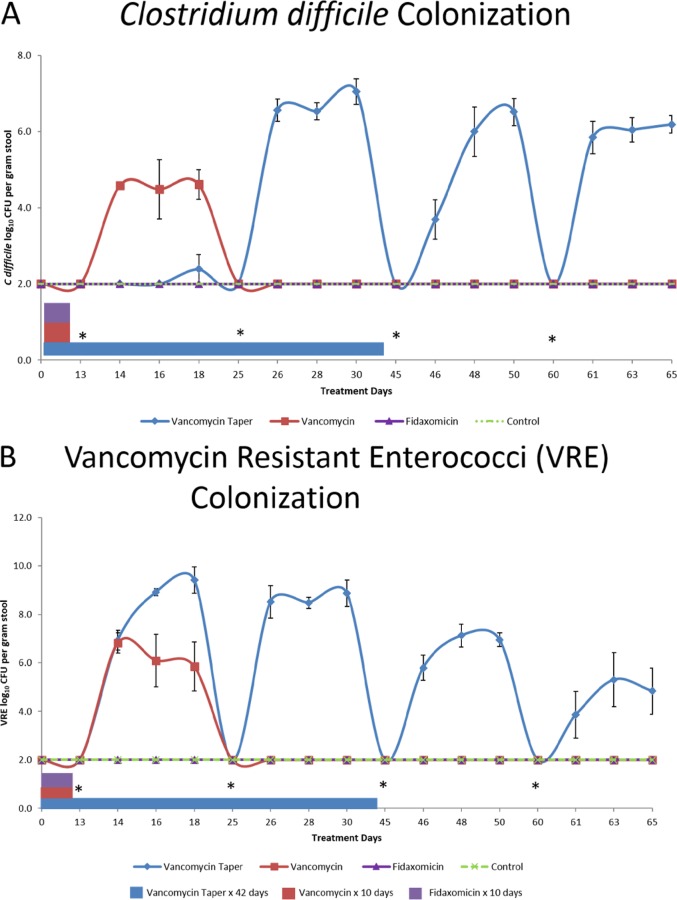
Figure 2 shows the effects of antibiotic treatment on colonization resistance to C. difficile (Fig. 2A) and VRE (Fig. 2B). Saline controls did not develop a greater than 1 log increase in concentration of either pathogen with any challenge. The fidaxomicin-treated mice maintained intact colonization resistance to both pathogens. Mice receiving the 10-day course of vancomycin developed loss of colonization resistance to VRE and C. difficile, at day 13 (3 days after the last vancomycin dose), but colonization resistance was intact on days 25, 45, and 60. The vancomycin taper and pulse-treated mice developed loss of colonization resistance to VRE on days 13, 25, 45, and 60 and to C. difficile on days 25, 45, and 60.
FIG 2.
Effect of antibiotic treatment on in vivo colonization resistance to Clostridium difficile (A) and vancomycin-resistant enterococci (VRE) (B). Mice received oral antibiotic treatment or sterile water (control) for the durations shown, and subgroups (five per group) were challenged by gastric gavage with 10,000 CFU of C. difficile spores or VRE on days 13, 25, 45, and 60 after the initial antibiotic dose. Treatment days indicate days after the start of antibiotic treatment. Error bars show standard error. *, inoculation of C. difficile spores or VRE.
Effect of the antibiotics on indigenous facultative Gram-negative bacilli by culture.
In addition to increasing susceptibility to colonization by exogenous microorganisms, antibiotic-induced alteration of colonization resistance results in overgrowth of indigenous components of the intestinal microbiota that are resistant to the administered antibiotics (21). Because vancomycin and fidaxomicin do not have significant activity against indigenous facultative Gram-negative bacilli, we measured concentrations of this component of the microbiota as a secondary measure of whether colonization resistance was intact. Figure 3 shows the effect of antibiotic treatment on the concentrations of facultative Gram-negative bacilli by culture. In comparison to saline controls, fidaxomicin did not promote overgrowth of Gram-negative bacilli (P ≥ 0.88), whereas oral vancomycin for 10 days promoted significant overgrowth on days 5, 10, and 13 (P < 0.01), but not on days 18, 25, 45, and 60 (P ≥ 0.08). In comparison to controls, mice treated with the vancomycin taper and pulse developed significant overgrowth of Gram-negative bacilli that persisted through day 60 (18 days after the last dose of vancomycin) (P < 0.01). All Gram-negative bacilli recovered from the highest plated dilution were lactose-fermenting organisms and all 10 isolates subjected to identification were Escherichia coli.
FIG 3.
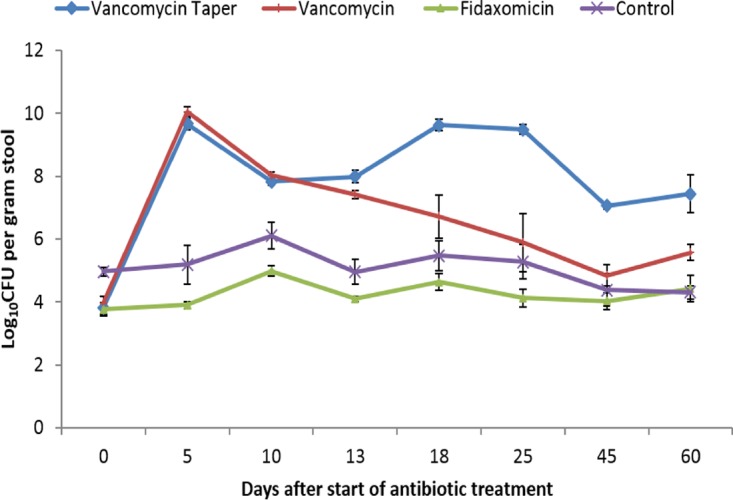
Effect of antibiotic treatment on the concentrations of facultative Gram-negative bacilli in stool by culture. Mice received oral vancomycin for 10 days, vancomycin taper and pulsed regimen for 42 days, fidaxomicin for 10 days, or sterile water (control). Results for five mice from each treatment group are shown at each time point. Error bars show the standard errors. *, P < 0.05.
Effect of antibiotic treatment on the indigenous microbiota by real-time PCR and deep-sequencing analysis.
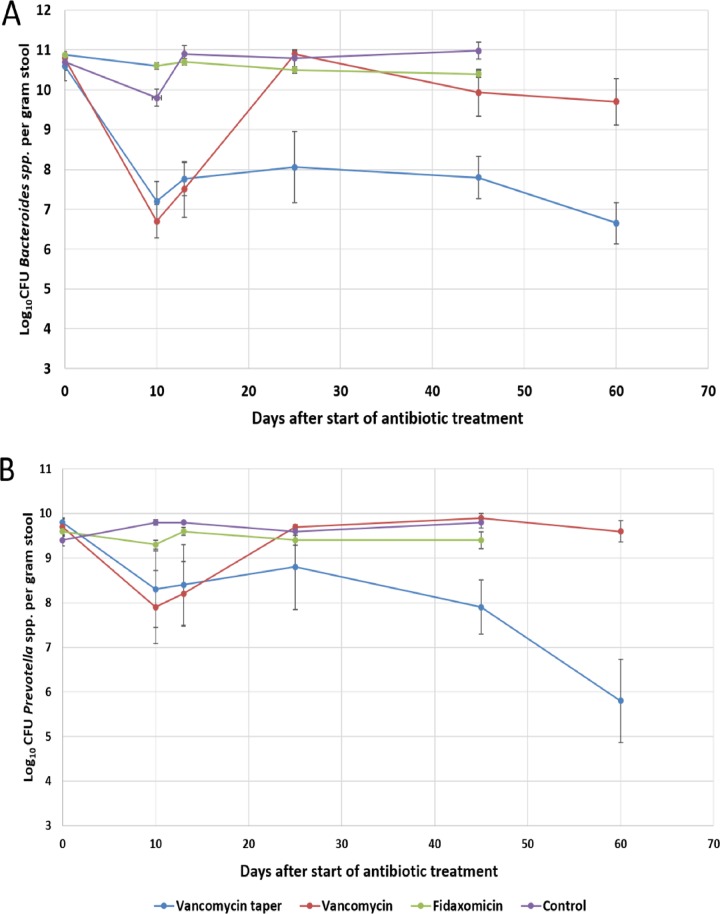
In comparison to saline controls, vancomycin treatment for 10 days significantly reduced the concentrations of Bacteroides spp. and Prevotella spp. detected by real-time PCR on days 10 and 13 (P < 0.01), but not on day 25 (P ≥ 0.8) (Fig. 4); the vancomycin taper and pulse regimen resulted in significant reductions in each of the anaerobes that persisted through day 60 (P < 0.01). Fidaxomicin did not cause significant reductions in either of the anaerobic species tested by PCR (P > 0.05).
FIG 4.
Effect of antibiotic treatment on the concentrations of Bacteroides spp. (A) and Prevotella spp. (B) in the stool specimens of mice as determined by real-time PCR. Results for five mice from each treatment group are shown at each time point. Error bars represent the standard errors. *, P < 0.05.
Figure 5 shows the relative proportions of the different taxa in the treatment groups before, during, and after treatment based on deep-sequencing analysis. We chose to present the analysis at phylum level due to inability to obtain sufficient resolution at lower taxonomic levels. Prior to treatment (day 0), each of the groups had similar distributions of the bacterial groups with >50% Bacteroidetes, 27 to 34% Firmicutes, and <10% Proteobacteria. For the vancomycin group, the proportion of Proteobacteria increased on day 10, while Bacteroidetes and Firmicutes were suppressed; by day 25, the relative proportions of the bacterial groups returned to pretreatment levels. For the vancomycin taper group, the proportion of Proteobacteria increased on day 10, while Bacteroidetes and Firmicutes were suppressed. However, in contrast to the vancomycin group, Bacteroidetes remained suppressed in the vancomycin taper group through day 45, and Proteobacteria and Firmicutes were suppressed through day 25. Fidaxomicin treatment did not cause significant alteration of the microbiota.
FIG 5.
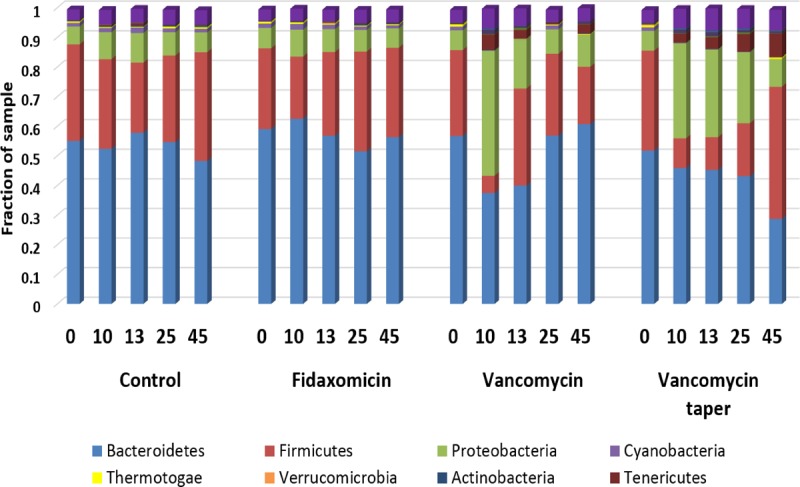
Comparison of stool microbiota of mice (n = 3 per group at each time point) by 16S deep-sequencing analysis before, during, and after treatment with oral vancomycin for 10 days, vancomycin taper and pulsed regimen for 42 days, fidaxomicin for 10 days, or sterile water (control). The relative abundances of the major bacterial phyla are shown at each time point.
Using permutation-based multivariate analysis of variance of the Bray-Curtis dissimilarity matrix to test effects of treatment and time from baseline, significant differences across treatment groups were detected (P = 0.01). There was not a significant effect of days from baseline or an interaction between days and treatment. The Shannon diversity index of each sample was calculated at the phylum level. Although variability in diversity increased over time, there were no differences in mean diversity indices for time or treatment.
DISCUSSION
Several studies have demonstrated that oral vancomycin treatment causes profound disruption of the intestinal microbiota of humans and mice that may persist for days to weeks after discontinuation of treatment (4–9). This disruption of the microbiota results in loss of colonization resistance to C. difficile and other pathogens, including VRE and multidrug-resistant Gram-negative bacilli (7–10). Our findings expand upon these previous studies by demonstrating that disruption of the intestinal microbiota that provide colonization resistance to C. difficile and VRE does not recover when a 10-day course of oral vancomycin is followed by a tapered and pulsed regimen. Moreover, alteration of the microbiota and of colonization resistance persisted for at least 18 days after completion of the taper. In contrast, colonization resistance recovered by 15 days after completion of a 10-day course of oral vancomycin and was not affected by fidaxomicin.
Our results have several important implications for the management of recurrent CDI. First, patients receiving vancomycin tapers may be at high risk for recurrence if they are exposed to spores after the taper or if small numbers of spores persist in intestinal tract during the taper. Administering a relatively long taper could potentially be beneficial to provide sufficient time to eliminate residual spores from the intestinal tract. However, Sirbu et al. (15) reported similar cure rates in patients receiving vancomycin taper regimens of <10 and >10 weeks in total duration. Efforts to minimize the risk for reexposure to spores after the taper may also be beneficial (e.g., environmental disinfection and patient hand hygiene and bathing). Second, because fidaxomicin did not alter colonization resistance, further studies are needed to determine the efficacy of this agent in patients with second or greater recurrences of CDI. In a small case series, administration of fidaxomicin as a chaser or taper was effective in treating patients with multiple recurrences of CDI (21). In addition, an extended and pulsed regimen of fidaxomicin was more effective than a standard vancomycin course in achieving sustained clinical cure in CDI patients (22). Finally, in current vancomycin taper and pulse regimens, vancomycin is administered at a dose of 125 mg which resulted in transient high levels of vancomycin above the MIC90 of many Gram-negative anaerobes in stool of mice during the pulse phase of treatment (18). It is possible that lower doses of vancomycin might be sufficient to maintain suppression of C. difficile while allowing some components of the indigenous anaerobic microbiota to recover.
Our study has some limitations. First, findings in healthy mice may differ from findings in humans given differences in diet, microbiota, and intestinal transit time. However, previous studies have demonstrated similar results for mice and patients treated with standard doses of vancomycin (4–10). In addition, tapering vancomycin regimens also resulted in a prolonged deleterious effect on the indigenous microbiota in an in vitro human gut model (23). Second, it is possible that patients may vary in the ability of their microbiota to recover during a vancomycin taper. Third, we only studied one strain each of C. difficile and VRE. We have previously shown that other VRE and C. difficile strains give similar results in our mouse model (6, 8, 9; unpublished data). Fourth, we studied a 6-week taper and pulsed regimen and cannot exclude the possibility that longer tapers might allow more recovery of the microbiota. Fifth, the dose of fidaxomicin was lower than the vancomycin dose and resulted in lower peak concentrations in stool. However, we have previously demonstrated that a higher fidaxomicin dose (75 mg/kg) also does not alter colonization resistance in mice (9). Sixth, we measured vancomycin concentrations in stool specimens and cannot exclude the possibility that levels differ substantially in the intestinal tract. Finally, factors independent of the recovery of the microbiota may impact the likelihood that tapered and pulsed vancomycin regimens may result in the clinical cure of CDI.
In summary, we found that a vancomycin tapered and pulsed regimen caused persistent disruption of the intestinal microbiota that provide colonization resistance to C. difficile and VRE in mice. Future studies are needed to examine the impact of vancomycin tapered and pulsed regimens in patients. In addition, there is a need to identify treatment strategies that allow the indigenous microbiota of patients with multiple recurrences to recover during therapy.
MATERIALS AND METHODS
Pathogens studied.
E. faecium C68 is a previously described VanB-type clinical VRE isolate (8, 9). For C68, the MICs of vancomycin and fidaxomicin are 0.5 and 1 μg/ml, respectively. VA17 is an epidemic North American pulsed-field gel electrophoresis type 1 (NAP1) C. difficile strain. For VA17, the MICs of vancomycin and fidaxomicin are 0.25 and 0.125 μg/ml, respectively. C. difficile spores were prepared as previously described (24).
Bioassay for antibiotic concentrations.
The concentration of vancomycin in stool was determined by an agar diffusion assay with Clostridium perfringens as the indicator strain (6).
Quantification of stool pathogens.
VRE and C. difficile concentrations in stool specimens were quantified as described elsewhere (6, 8, 9). Stool specimens were emulsified in 5-fold (wt/vol) prereduced phosphate-buffered saline. To quantify VRE and C. difficile, serially diluted aliquots were inoculated onto Enterococcosel agar (Becton Dickinson, Cockeysville, MD) containing vancomycin 20 μg/ml and prereduced cycloserine-cefoxitin-brucella agar containing 0.1% taurocholic acid and 5 mg/ml lysozyme (C. difficile brucella agar), respectively. The number of CFU of each pathogen per gram of sample was calculated.
Antibiotic dose and duration.
The doses of the antibiotics were fidaxomicin (30 mg/kg/day or 0.9 mg/day) for 10 days, vancomycin (37.5 mg/kg/day or 1.125 mg/day) for 10 days, and vancomycin administered as a taper and pulsed regimen for 42 days (37.5 mg/kg/day for 10 days, 18.75 mg/kg/day for 7 days, 9.38 mg/kg/day for 7 days, 9.38 mg/kg/day every other day for three doses, and 9.38 mg/kg/day every 3 days for three doses). The dose of vancomycin was based on dose-finding experiments that demonstrated that maximal fecal peak levels were 1,370 μg/g of stool at 4 to 8 h postdosing, providing levels within the range measured in stool of humans receiving oral vancomycin (i.e., 500 to 2,000 μg/g of stool) (8). The dose of fidaxomicin was 80% of the dose of vancomycin because the human dose of fidaxomicin is 80% of the daily dose of vancomycin (i.e., 400 mg versus 500 mg per day). Previous dose-finding experiments demonstrated that this fidaxomicin dose resulted in a measured maximal peak fecal concentration of 920 μg/g of stool of fidaxomicin+OP-1118 (9).
Effect of antibiotic treatment on colonization resistance to C. difficile and VRE.
The Animal Care Committee of the Cleveland Veterans Affairs Medical Center approved the experimental protocol. Female CF-1 outbred white mice (100 total, including 25 in each antibiotic group and 25 controls) weighing ∼30 g (Harlan Sprague-Dawley, Indianapolis, IN) were housed in individual cages. The antibiotics were administered once daily on Bacon Yummie food treats (Bio-Serve, Prospect, CT); 10 μl of solution containing the antibiotic dose was pipetted onto the food treat and allowed to air dry prior to placement in the cage bottom for consumption. Control mice received food treats inoculated with 10 μl of sterile water for 10 days. For all experiments, cages were changed daily during and after treatment to reduce reexposure to contaminated bedding and cage material.
To assess the effects of antibiotic exposure on colonization resistance, subgroups of mice in each treatment group were challenged at baseline and during or after the treatment courses with 104 CFU of VA 17 C. difficile spores (n = 5 mice at each time point) or VRE C68 (n = 5 mice at each time point) in 0.2 ml of sterile water by oral gavage. The subgroups of mice were challenged on days 13 (3 days after completion of the 10-day course of fidaxomicin or vancomycin), 25 (15 days after completion of the 10-day treatment course and 1 day after a 9.38-mg dose in the vancomycin taper group), 45 (35 days after the 10-day course and 3 days after completion of the vancomycin taper), and 60 (18 days after completion of the vancomycin taper) after the first antibiotic dose. Concentrations of C. difficile or VRE were measured in stool specimens collected before and on days 1, 3, and 5 after challenge with the pathogens. Loss of colonization resistance due to antibiotic treatment was defined as an increase in stool concentration of the pathogens of greater than 1 log10 CFU in comparison to saline controls.
Effect of the antibiotics on the intestinal microbiota.
Stool samples collected at baseline and on days 13, 25, 45, and 60 after the first antibiotic dose were used to determine the effect of the antibiotics on the microbiota. Quantitative cultures for facultative and aerobic Gram-negative bacilli were performed for five mice per group at each time point by plating serially diluted specimens onto MacConkey agar (Difco Laboratories, Detroit, MI). Organisms recovered on MacConkey agar were characterized as lactose-fermenting or non-lactose-fermenting Gram-negative bacilli, and for a subset of 10 plates a colony growing at the highest dilution was subjected to identification using standard methods.
Real-time PCR was used to monitor the effect of antibiotic treatment on the concentration of Bacteroides spp. and Prevotella spp. for five mice per group at each time point as previously described (8). Purified template DNA from Bacteroides fragilis (American Type Culture Collection [ATCC] 25825) and Prevotella oris (ATCC 33573) was used for melting-curve analysis and to generate standard curves for each primer set using 10-fold serial dilutions of DNA ranging from 10 to 10−6 ng. The PCR results were used to calculate CFU per gram of stool as described by Louie et al. (5).
Microbiome analysis was completed for three mice per group at each time point by deep-sequencing bacterial 16S rRNA gene amplicons (25). Briefly, DNA was extracted from ∼200 mg of feces using the QIAamp DNA stool minikit (Qiagen, Frederick, MD) according to the manufacturer's instructions with lysis conditions optimized to increase the ratio of nonhuman to human DNA. The V3-V4 region of the bacterial 16S rRNA gene, defined as the most promising bacterial primer pair for deep-sequencing-based diversity (25), was PCR amplified, and single amplicons of ∼460 bp were visualized by electrophoresis in 1% agarose gels and cleaned up using AMPure XP beads (Agencourt AMPure XP). Dual indices (barcodes) and Illumina sequencing adapters were added to the amplicons using the Nextera XT Index kit (Illumina, Inc., San Diego, CA), followed by DNA purification (Agencourt AMPure XP). Individual barcoded DNA samples were then quantified (Qubit 2.0; Thermo Fisher Scientific), normalized, and pooled. Multiplexed libraries, including 5% PhiX as an internal control, were diluted to 20 pM and denatured with NaOH prior to sequencing on the MiSeq system (Illumina) using the MiSeq reagent Kit v3 600 cycle (2 × 300 bp; Illumina). All 66 samples were multiplexed into a single Miseq sequencing run, generating over 52.9 million quality reads, distributed homogenously among all samples (mean, 441,790 reads; interquartile range, 369,237 to 520,492 reads).
Indexed reads were demultiplexed to generate sample-specific fastq files, which were mapped and aligned against a set of 16S rRNA reference sequences, allowing taxonomic classification based on an Illumina-curated version of the Greengenes database (http://greengenes.lbl.gov/) using the 16S Metagenomics app v.1.0.1 (Illumina, Inc.). The algorithm is a high-performance implementation of the Ribosomal Database Project as previously described (26). The total number of reads that passed quality filtering were clustered into operational taxonomic units (OTU) using a 97% similarity threshold. OTU with a number of sequences lower than 0.005% of the total number of sequences generated were discarded, and those remaining were summarized to get taxonomies.
Statistical analysis.
One-way analysis of variance was performed to compare concentrations of organisms among treatment groups. P values were adjusted for multiple comparisons using the Scheffe correction. Relative abundance by phylum was calculated for each mouse sample. Abundance data were further aggregated across mice by treatment and day, and bar plot representations were generated to show the top eight microbial groups at the phylum level for each treatment-day group. Analyses were performed with R 3.4.3 (27) using the vegan package v2.4.
ACKNOWLEDGMENTS
This study was supported by the Department of Veterans Affairs; by a grant from Merck and Co., Inc., Kenilworth, NJ; and by the CWRU/UH Center for AIDS Research (P30 AI036219).
The authors declare that no competing interests exist.
REFERENCES
- 1.Johnson J. 2009. Recurrent Clostridium difficile infection: a review of risk factors, treatments, and outcomes. J Infect 58:403–410. doi: 10.1016/j.jinf.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 2.Kyne L, Warny M, Qamar A, Kelly CR. 2000. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 3.McFarland LV, Elmer GW, Surawicz CM. 2002. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am J Gastroenterol 97:1769–1775. doi: 10.1111/j.1572-0241.2002.05839.x. [DOI] [PubMed] [Google Scholar]
- 4.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 5.Louie TJ, Cannon K, Byrne B, Emery J, Ward L, Eyben M, Krulicki W. 2012. Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis 55(Suppl 2):S132–S142. doi: 10.1093/cid/cis338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abujamel T, Cadnum JL, Jury LA, Sunkesula VC, Kundrapu S, Jump RL, Stintzi AC, Donskey CJ. 2013. Defining the vulnerable period for reestablishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS One 8:e76269. doi: 10.1371/journal.pone.0076269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis BB, Buffie CG, Carter RA, Leiner I, Toussaint NC, Miller LC, Gobourne A, Ling L, Pamer EG. 2015. Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J Infect Dis 212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande A, Hurless K, Cadnum JL, Chesnel L, Gao L, Chan L, Kundrapu S, Polinkovsky A, Donskey CJ. 2016. Effect of surotomycin, a novel cyclic lipopeptide antibiotic, on intestinal colonization with vancomycin-resistant enterococci and Klebsiella pneumoniae in mice. Antimicrob Agents Chemother 60:3333–3339. doi: 10.1128/AAC.02904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshpande A, Hurless K, Cadnum JL, Chesnel L, Gao L, Chan L, Kundrapu S, Polinkovsky A, Donskey CJ. 2016. Effect of fidaxomicin versus vancomycin on susceptibility to intestinal colonization with vancomycin-resistant enterococci and Klebsiella pneumoniae in mice. Antimicrob Agents Chemother 60:3988–3993. doi: 10.1128/AAC.02590-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson S, Homann SR, Bettin KM, Quick JN, Clabots CR, Peterson LR, Gerding DN. 1992. Treatment of asymptomatic Clostridium difficile carriers (fecal excretors) with vancomycin or metronidazole. A randomized, placebo-controlled trial. Ann Intern Med 117:297–302. [DOI] [PubMed] [Google Scholar]
- 11.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 12.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, Speelman P, Dijkgraaf MG, Keller JJ. 2013. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 13.Wilcox MH, Gerding DN, Poxton IR, Kelly C, Nathan R, Birch T, Cornely OA, Rahav G, Bouza E, Lee C, Jenkin G, Jensen W, Kim YS, Yoshida J, Gabryelski L, Pedley A, Eves K, Tipping R, Guris D, Kartsonis N, Dorr MB; MODIFY I, Investigators MODIFY II. 2017. Bezlotoxumab for prevention of recurrent Clostridium difficile infection. N Engl J Med 376:305–317. doi: 10.1056/NEJMoa1602615. [DOI] [PubMed] [Google Scholar]
- 14.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH; Society for Healthcare Epidemiology of America; Infectious Diseases Society of America. 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 15.Sirbu BD, Soriano MM, Manzo C, Lum J, Gerding DN, Johnson S. 2017. Vancomycin taper and pulsed regimen with careful follow up for patients with recurrent Clostridium difficile infection. Clin Infect Dis 65:1396–1399. doi: 10.1093/cid/cix529. [DOI] [PubMed] [Google Scholar]
- 16.Hota SS, Sales V, Tomlinson G, Salpeter MJ, McGeer A, Coburn B, Guttman DS, Low DE, Poutanen SM. 2017. Oral vancomycin followed by fecal transplantation versus tapering oral vancomycin treatment for recurrent Clostridium difficile infection: an open-label, randomized controlled trial. Clin Infect Dis 64:265–271. doi: 10.1093/cid/ciw731. [DOI] [PubMed] [Google Scholar]
- 17.Edlund C, Barkholt L, Olsson-Liljequist B, Nord CE. 1997. Effect of vancomycin on intestinal flora of patients who previously received antimicrobial therapy. Clin Infect Dis 25:729–732. doi: 10.1086/513755. [DOI] [PubMed] [Google Scholar]
- 18.Citron DM, Tyrrell KL, Vreni Merriam C, Goldstein EJC. 2012. In vitro activities of CB-183,315, vancomycin, and metronidazole against 556 strains of Clostridium difficile, 445 other intestinal anaerobes, and 56 Enterobacteriaceae species. Antimicrob Agents Chemother 56:1613–1615. doi: 10.1128/AAC.05655-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soriano MM, Danziger LH, Gerding DN, Johnson S. 2014. Novel fidaxomicin treatment regimens for patients with multiple Clostridium difficile infection recurrences that are refractory to standard therapies. Open Forum Infect Dis 1:ofu069. doi: 10.1093/ofid/ofu069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donskey CJ. 2004. The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis 39:219–226. doi: 10.1086/422002. [DOI] [PubMed] [Google Scholar]
- 21.Cornely OA, Vehreschild MJ, Adomakoh N, Georgopoli A, Karas A, Kazeem G, Guery B. 2017. Subgroup analyses of the EXTEND Study: a randomised, controlled, open-label phase III/IV study comparing the efficacy of extended-pulsed fidaxomicin with standard vancomycin therapy for sustained clinical cure of Clostridium difficile infection in an older population. ID Week, abstr 1873. [Google Scholar]
- 22.Crowther GS, Chilton CH, Longshaw C, Todhunter SL, Ewin D, Vernon J, Karas A, Wilcox MH. 2016. Efficacy of vancomycin extended-dosing regimens for treatment of simulated Clostridium difficile infection within an in vitro human gut model. J Antimicrob Chemother 71:986–991. doi: 10.1093/jac/dkv453. [DOI] [PubMed] [Google Scholar]
- 23.Nerandzic MM, Donskey CJ. 2009. Effective and reduced-cost modified selective medium for isolation of Clostridium difficile. J Clin Microbiol 47:397–400. doi: 10.1128/JCM.01591-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Illumina. 2013. 16S metagenomic sequencing library preparation. Illumina, San Diego, CA: https://www.illumina.com/content/dam/illumina-support/documents/documentation/chemistry_documentation/16s/16s-metagenomic-library-prep-guide-15044223-b.pdf. [Google Scholar]
- 25.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glöckner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zapala MA, Schork NJ. 2006. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci U S A 103:19430–19435. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jari O, Guillaume B, Roeland K, Pierre L, Peter R, O'Hara R, Gavin L, Peter S, Henry H, Helene W. 2013. Vegan: community ecology R package, version 2.0-10 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]