ABSTRACT
Clostridium difficile infection (CDI), a common cause of hospital-acquired infections, typically occurs after disruption of the normal gut microbiome by broad-spectrum antibiotics. Fidaxomicin is a narrow-spectrum antibiotic that demonstrates a reduced impact on the normal gut microbiota and is approved for the treatment of CDI. To further explore the benefits of this property, we used a murine model to examine the effects of fidaxomicin versus vancomycin on gut microbiota and susceptibility to C. difficile colonization while tracking microbiota recovery over time. Mice were exposed to fidaxomicin or vancomycin by oral gavage for 3 days and subsequently challenged with C. difficile spores at predetermined time points up to 21 days postexposure to antibiotics. Fecal samples were subsequently collected for analysis. Twenty-four hours postchallenge, mice were euthanized and the colon contents harvested. The microbiota was characterized using 16S rRNA gene sequencing. All fidaxomicin-exposed mice (except for one at day 8) were resistant to C. difficile colonization. However, 9 of 15 vancomycin-exposed mice were susceptible to C. difficile colonization until day 12. All vancomycin-exposed mice recovered colonization resistance by day 16. Bacterial diversity was similar prior to antibiotic exposure in both arms and decreased substantially after exposure. A shift in taxonomic structure and composition occurred after both exposures; however, the shift was greater in vancomycin-exposed than in fidaxomicin-exposed mice. In summary, compared with vancomycin, fidaxomicin exposure had less impact on microbiota composition, promoted faster microbial recovery, and had less impact on the loss of C. difficile colonization resistance.
KEYWORDS: Clostridium difficile, diarrhea, fidaxomicin, recurrences, vancomycin
INTRODUCTION
Clostridium difficile infection (CDI) remains a leading cause of hospital-acquired diarrhea (1, 2) and is one of the most common causes of hospital-acquired infections (3). The incidence of CDI has increased over the last decade, more than doubling in the United States (1, 4).
The highest incidence of CDI occurs after the normal gut microbiome has been disrupted by broad-spectrum antibiotic treatment for other infections (5). Meta-analyses have identified clindamycin, cephalosporin, and fluoroquinolone antibiotic use to be the greatest risk factor of CDI (6–8). A lack of bacterial community richness and diversity after antibiotic treatment allows colonization and overgrowth of pathogenic species, such as C. difficile (9).
After CDI has resolved, posttreatment recurrences can occur, and the rate of recurrence after standard antibiotic treatment is rising (10–13). CDI recurs in 20 to 25% of patients after the first episode (14, 15).
Following an initial recurrence, ∼38 to 65% of patients experience further recurrence episodes (16, 17). The primary cause of recurrence is linked to nonoptimal recovery of the protective healthy microbiota (18, 19). In mice, exposure to antibiotics changes the gut microbiome, with a reduction in the predominant bacterial phyla, including Bacteroidetes and Firmicutes, and an increase in Proteobacteria, which make up only 2% of the healthy gut microbiome (20). Similarly, analysis of fecal microbiota composition from patients with CDI shows decreased microbially diverse populations in those with recurrent CDI versus in those presenting with an initial incident of recurrence (21). Thus, the recovery of a healthy and diverse microbiome after antibiotic treatment may reduce CDI recurrence (19).
For many years, two drugs were used primarily for CDI treatment, metronidazole and vancomycin (22). Metronidazole is a nitroimidazole antibiotic that inhibits nucleic acid synthesis by disrupting the DNA of microbial cells (23). Metronidazole is highly active against Gram-negative anaerobic bacteria, such as Bacteroides fragilis, and Gram-positive anaerobic bacteria, including C. difficile. It is considered appropriate for use in mild-to-moderate first-time cases of CDI (22, 24). Vancomycin is a large hydrophilic antibiotic used to treat Gram-positive bacteria; it is used in both mild-to-moderate and more severe cases of CDI (22). While these antibiotics suppress the growth and proliferation of C. difficile, they also disrupt the resident bacterial population in the colon that provides a barrier to CDI (25).
Fidaxomicin is a narrow-spectrum first-in-class macrolide antibacterial drug indicated for use in adults (≥18 years of age) for the treatment of C. difficile-associated diarrhea (CDAD). Fidaxomicin has been shown to have little disruptive effect on the major Gram-negative and Gram-positive species of patients with CDI, including B. fragilis, clostridial clusters XIVa and IV, and Bifidobacterium spp., meaning that commensal gut bacteria are preserved in vivo upon treatment (26, 27).
To further explore the preservation of commensal bacteria by fidaxomicin and the associated lack of disruption of natural resistance against CDI, we utilized a murine model. The aims of this study were 3-fold: to evaluate the effect of a 3-day treatment with either vancomycin or fidaxomicin on the normal murine gut microbiota, to determine whether susceptibility to C. difficile colonization increased posttreatment, and to assess the recovery of the microbiome over time postexposure initiation.
RESULTS
Microbiological outcomes.
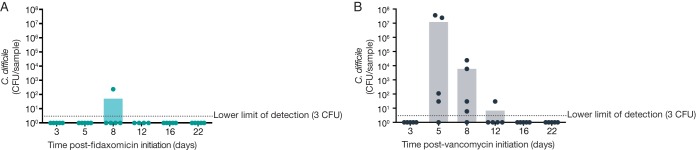
In general, a reduced loss of microbiota-mediated C. difficile colonization resistance was observed from the fidaxomicin-exposed mice compared with vancomycin-exposed mice (Fig. 1). On day 3, all fidaxomicin- and vancomycin-exposed mice had C. difficile CFU numbers below the lower limit of quantification, likely due to insufficient time to grow or to inhibitory levels of antibiotic remaining in the gastrointestinal tract.
FIG 1.
Susceptibility to C. difficile colonization following antibiotic regimens. (A) Response to C. difficile challenge in fidaxomicin-exposed mice. (B) Response to C. difficile challenge in vancomycin-exposed mice. Dots represent data from individual mice; bars represent mean values.
On study days 5, 8, 12, 16, and 22, all fidaxomicin-exposed mice, except for one on day 8, were still resistant to C. difficile colonization. Vancomycin-exposed mice lost resistance to C. difficile colonization between days 5 and 12; 4 out of 5 mice were susceptible to C. difficile challenge on days 5 and 8, and one out of five mice was still susceptible to challenge by day 12. All mice in the vancomycin group had recovered colonization resistance by day 16 (2 weeks postcessation of antibiotics).
Impact on intestinal microbiota and diversity.
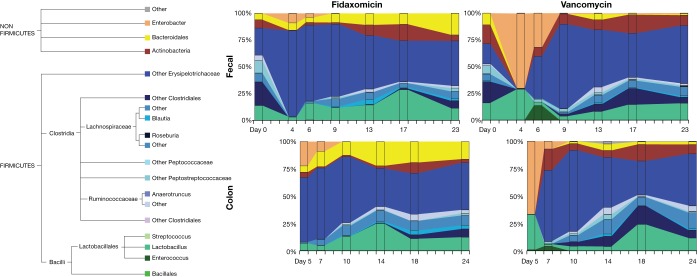
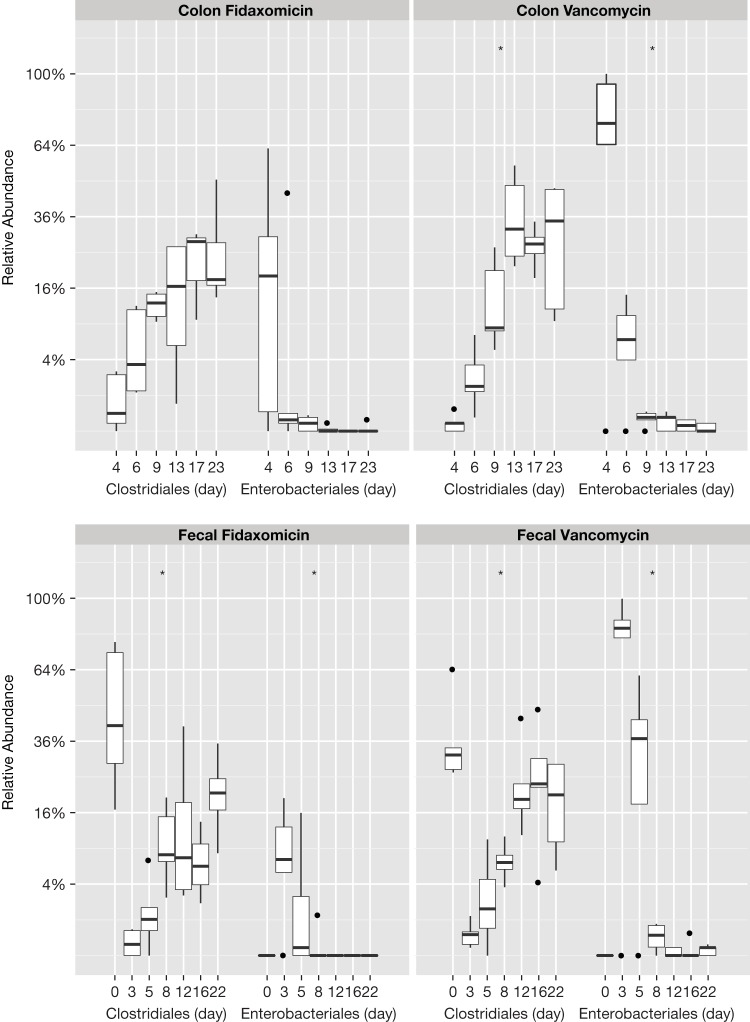
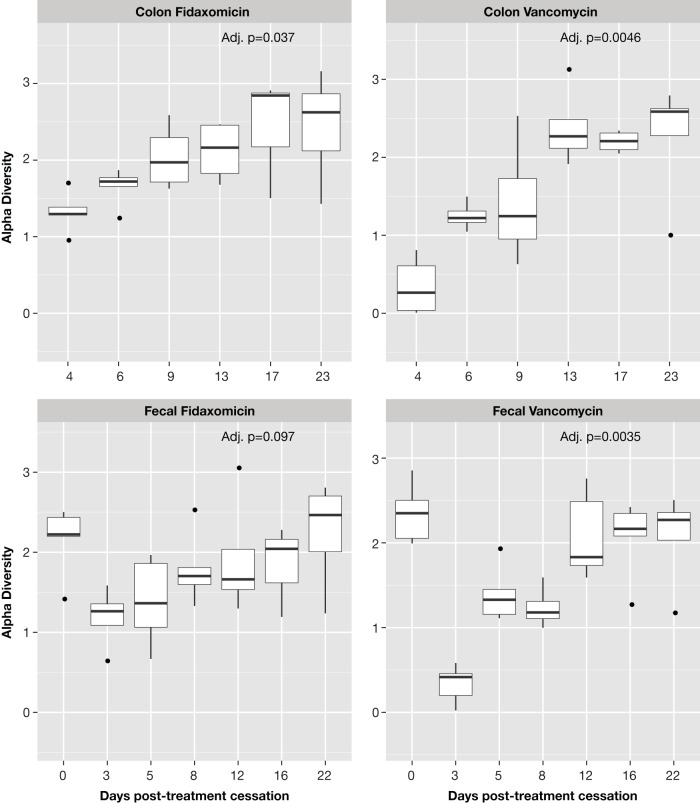
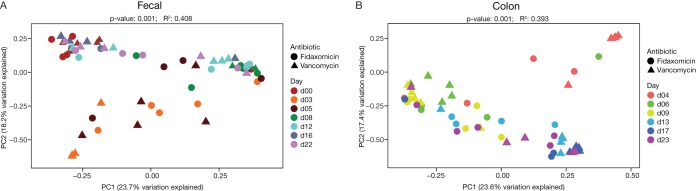
Fecal bacterial diversity was similar before antibiotic exposure in both arms but decreased substantially after exposure (Fig. 2; see also Table S1 in the supplemental material). A shift in taxonomic structure and composition occurred after exposure to both antibiotics (Fig. 3). Fecal samples from mice exposed to either of the antibiotics showed an increase in Enterobacteriales and a decrease in Clostridiales immediately after antibiotic cessation. This shift was greater in vancomycin-exposed than in fidaxomicin-exposed mice. Immediately after antibiotic cessation, fecal (assessed on day 3) and colonic (assessed on day 4) samples showed that operational taxonomic units (OTUs) for Clostridiales in both exposure arms had increased. Enterobacteriales started to decrease after the cessation of fidaxomicin exposure; however, the trend toward recovery was delayed in vancomycin-exposed mice. Exposure to either antibiotic resulted in a reduction in the richness and evenness of the bacterial community, as evidenced by the Shannon diversity index scores calculated from fecal and colonic samples. This difference was less pronounced in mice exposed to fidaxomicin than in mice exposed to vancomycin (Fig. 4). During the recovery of the microbiota following the administration of either antibiotic, there was a lack of expansion of Enterococcus OTUs and a rapid expansion of various other orders of bacteria. Principal coordinates were generated to explain maximum aggregate variance of samples in each exposure arm. The coordinates indicated that the early vancomycin samples clustered separately from the majority of other samples (Fig. 5). To evaluate the similarity in composition between samples, Jaccard distances were generated, and statistically significant differences in the structure of the fecal (P < 0.01, F-statistic = 3.41) and colon (P < 0.01, F-statistic = 2.95) microbiomes were observed after fidaxomicin and vancomycin treatments.
FIG 2.
Fecal and colon microbiota composition following antibiotic regimens. Taxonomy is described to the most detailed level possible.
FIG 3.
Tukey box-and-whisker plot showing fecal and colon taxonomic abundance following antibiotic regimens. Boxes represent Q1/Q3 limits; whiskers are ±1.5 standard deviation (SD). Dots represent data points outside the ±1.5 standard deviation (SD), and the horizontal lines are the statistical medians.
FIG 4.
Tukey box-and-whisker plot showing biodiversity of fecal and colon microbiota following antibiotic regimens (Shannon diversity index scores). Shannon diversity indices of fecal and colon bacterial communities collected at successive time points following antibiotic regimen, calculated from 16S rRNA gene sequence analyses. Boxes represent Q1/Q3 limits; whiskers are ±1.5 standard deviation (SD). Dots represent data points outside the ±1.5 SD, and the horizontal lines are the statistical medians. Adj. p, adjusted P value.
FIG 5.
Principal-coordinate analysis plots of Jaccard distances of fecal (A) and colon (B) microbiota generated from 16S rRNA gene sequence analysis following antibiotic regimens. Axis labels indicate the percentage of variance explained by the respective principal coordinate axis. PC1, principal coordinate 1; PC2, principal coordinate 2.
DISCUSSION
In this study, the influence of fidaxomicin or vancomycin on susceptibility to C. difficile gut colonization and microbially diverse populations was evaluated in a murine model. All mice but one exposed to fidaxomicin remained resistant to C. difficile colonization postexposure to antibiotics, which correlated with reduced disruption and rapid recovery of the intestinal microbiota. Exposure to either fidaxomicin or vancomycin resulted in a loss of microbially diverse populations and a shift in the structure and composition of the microbiota.
The magnitude and specificity of the effect on microbiota for each treatment differed; for example, in mice exposed to vancomycin, a larger percentage of the microbiota present after exposure in both colon and fecal samples was of the enterococcal clade than in the gut microbiota of animals exposed to fidaxomicin. This observation is in agreement with previous fidaxomicin animal studies in which exposure to fidaxomicin led to minimal microbiota dysbiosis and did not promote colonization by vancomycin-resistant enterococci and Klebsiella pneumoniae, whereas exposure to vancomycin did promote colonization by these bacteria (20, 28).
The lesser impact of fidaxomicin on the microbiota correlates with prior observations in patients with CDI (26). The link between the reduced susceptibility to C. difficile challenge and exposure to fidaxomicin versus vancomycin supports the observations in clinical trials where fidaxomicin has demonstrated decreased CDI recurrence rates compared with vancomycin (19, 26, 27).
With regard to the clinical setting, the preservation of species able to restrict C. difficile overgrowth upon exposure to fidaxomicin may allow for a faster recovery of the microbiota in patients with CDI, returning the patient to a state supportive of lowering recurrence after successful treatment. A narrow-spectrum antibiotic preserving or supporting recovery of a microbiome that will suppress the growth of colonizing species is important when considering the recent emergence of vancomycin-resistant enterococci, which can cause further problems when treating patients with CDI with vancomycin (20, 28, 29).
A potential limitation of the study was that the concentration of fidaxomicin was not measured following exposure. The lack of impact on colonization resistance in mice, particularly at the earliest time points investigated, could be explained by antibiotics persisting at inhibitory levels in the gut postexposure. Vancomycin can persist in patients with CDI for several days at therapeutic concentrations after the cessation of treatment (30). Similarly, fidaxomicin is known to bind to C. difficile spores in vitro (31), although this mechanism is unlikely to be applicable to our model, as there were no spores prior to C. difficile challenge. Alternatively, residual fidaxomicin in the colon may have a continuing effect on the gut microbiota. An additional limitation of the study was the relatively short treatment duration of fidaxomicin and vancomycin assessed, compared with clinical treatment norms.
While both fidaxomicin and vancomycin alter gut microbiota composition and structure, the gut microbiota in mice exposed to fidaxomicin recovered more quickly than in those exposed to vancomycin. Furthermore, fidaxomicin had a lower impact on the composition and structure of the gut microbiota. Mice exposed to fidaxomicin proved to maintain colonization resistance to subsequent spore challenges, whereas mice exposed to vancomycin and then subsequently challenged with C. difficile spores remained susceptible to colonization until day 10.
MATERIALS AND METHODS
Mouse husbandry.
All experiments were performed with wild-type female C57BL/6 mice, age 6 to 8 weeks (Jackson Laboratories, ME, USA). The mice were housed in the specific pathogen-free facility at Memorial Sloan Kettering's Animal Resource Center, fed irradiated feed, and provided with acidified water. Cages were changed at least once a week. The experiments were performed in compliance with Memorial Sloan Kettering's institutional guidelines and were approved by its Institutional Animal Care and Use Committee.
Antibiotic administration and study design.
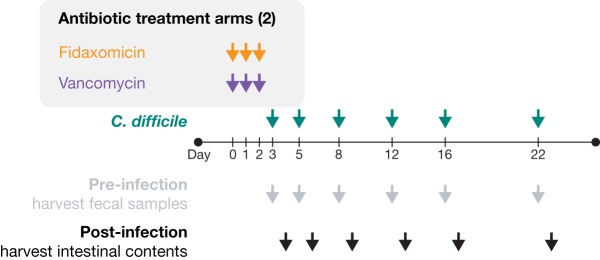
A total of 30 mice per experimental arm were exposed to 1.125 mg/day vancomycin (37.5 mg/kg in water) or 0.9 mg/day fidaxomicin (30 mg/kg in Labrasol) by oral gavage (200 μl on days 0, 1, and 2) (Fig. 6). The antibiotics were given at biologically relevant doses, as previously described (20). Each antibiotic arm consisted of 5 independently housed colonies of 6 mice during antibiotic exposure. Following the course of antibiotic exposure, a single mouse from each colony was removed, housed separately, and challenged with 1 × 103 C. difficile VPI 10463 (American Type Culture Collection 43255) spores in phosphate-buffered saline by oral gavage on days 3, 5, 8, 12, 16, and 22, as previously described (32). Fecal content, collected from the cage floor, was analyzed on the same day of C. difficile challenge. Colonic samples were recovered from sacrificed mice 1 day after challenge on days 4, 6, 9, 13, 17, and 23. Samples were immediately flash-frozen and DNA extracted as previously described (33). The samples (approximately 100 mg) were suspended in 500 μl of extraction buffer (200 mmol/liter Tris [pH 8.0], 200 mmol/liter sodium chloride, 20 mol/liter EDTA), and 200 μl of 20% sodium dodecyl sulfate, 500 μl of phenol-chloroform-isoamyl alcohol (24:24:1), and with 500 μl of 0.1-mm-diameter zirconia/silica beads (BioSpec Products, OK, USA). Bacterial cells were lysed by bead beating (BioSpec Products) for 2 min, and DNA was isolated with 2 rounds of phenol-chloroform-isoamyl alcohol extraction. Postextraction, DNA was precipitated in ethanol and resuspended in 200 μl of Tris-EDTA buffer with 100-μg/ml RNase. The DNA sample was then purified with QIAamp Mini spin columns (Qiagen, MD, USA).
FIG 6.
Study design. Mice were exposed to fidaxomicin (30 mg/kg) or vancomycin (37.5 mg/kg) by oral gavage once daily for 3 consecutive days (days 0, 1, and 2); on days 3, 5, 8, 12, 16, and 22 (post-antibiotic exposure initiation), and 1 mouse was removed from each colony, housed separately, and challenged with 1 × 103 C. difficile spores of strain VPI 10463 by oral gavage. Twenty-four hours later, the separated mice were sacrificed, and the colon contents were collected.
Sample analysis.
The susceptibility of mice to C. difficile colonization was determined by selective culture of fecal and colonic contents. Colonization resistance was defined as the C. difficile CFU count being under the lower limit of detection (<3 CFU). Fecal and colon sample 16S rRNA genes were analyzed via Illumina MiSeq (CA, USA) to determine intestinal microbiota composition. The broad-range bacterial 16S primers 517F (5′-GCCAGCAGCCGCGCTAA-3′) and 798R (5′-AGGGTATCTAATCCT-3′) were used at 0.2-mmol/liter concentrations with the DyNAmo SYBR green quantitative PCR kit (Finnzymes, Thermo Fisher Scientific, MA, USA) quantifying amplifications for library normalization. Sample amplification was compared with standard curves to quantify the 16S rRNA gene copy number. The cycling program was as follows: 95°C for 15 min, followed by 40 cycles of 94°C for 15 s, 52°C for 30 s, and 72°C for 30 s, and finished with 95°C for 15 min, 60°C for 1 min, and 95°C for 15 min (33). The 16S rRNA gene V4-V5 sequences were processed and clustered into OTUs with >97% sequence homology. Amplicons of the V4-V5 region of the 16S rRNA gene were amplified and sequenced with the Illumina MiSeq platform, as previously described (32).
Sequence reads were merged using a minimum overlap of 50 bp and filter of 0.5% expected error. The merged reads were stripped from primer sequences and then oriented identically. The resulting data set was analyzed using the UPARSE pipeline (34). Reads were dereplicated, sorted, and clustered at 97% sequence identity. Singletons were removed from centroid consideration. Centroids or OTUs were aligned to the SILVA version 123 (35) database using USEARCH (36) to determine taxonomies. OTU-based bacterial richness and evenness were estimated by calculating the Shannon diversity index. Jaccard distances were calculated, and principal coordinates for each exposure arm were generated from 16S rRNA gene V4-V5 sequences.
Data and statistical analysis.
The OTU table was used for downstream analyses using the Agile Toolkit for Incisive Microbial Analyses (ATIMA) visualization toolkit, which was developed at the Alkek Center for Metagenomics and Microbiome Research. ATIMA is a standalone tool for analyzing and visualizing microbiome data sets built in R (37) combining publicly available packages (i.e., APE and VEGAN) (38) and purpose-written code to import sample data and identify trends in taxa abundance, alpha diversity, and beta diversity with sample metadata. The data set was rarefied to 5,012 reads per sample, and the significance of categorical variables was determined using the nonparametric Kruskal-Wallis test (39) for comparing 3 or more categories. Correlation between 2 continuous variables was determined with R's base “lm” function for linear regression models, where P values indicate the probability that the slope of the regression line is zero. Principal-coordinate analysis (PCoA) plots were constructed with Jaccard distances and employed the Monte Carlo permutation test (40) to estimate P values. All P values were adjusted for multiple comparisons with the Benjamini-Hochberg false-discovery rate (FDR) algorithm (41).
Supplementary Material
ACKNOWLEDGMENTS
We thank E. G. Pamer of Memorial Sloan Kettering Cancer Center, NY, USA, and C. G. Buffie of Weill-Cornell Medical College, NY, USA, for their technical contributions. Medical writing assistance was provided by Corey Monteith of Complete Medical Communications, San Francisco, CA, USA.
The medical writing assistance and overall funding for this research was provided by Merck & Co., Inc., Kenilworth, NJ, USA.
N.J.A., J.L.C., M.C.W., and J.F.P. own shares in Diversigen, Inc., TX, USA. L.C. is an employee of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., and may own stock and/or stock options.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02112-17.
REFERENCES
- 1.Reveles KR, Lee GC, Boyd NK, Frei CR. 2014. The rise in Clostridium difficile infection incidence among hospitalized adults in the United States: 2001–2010. Am J Infect Control 42:1028–1032. doi: 10.1016/j.ajic.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Lessa FC, Gould CV, McDonald LC. 2012. Current status of Clostridium difficile infection epidemiology. Clin Infect Dis 55(Suppl 2):S65–S70. doi: 10.1093/cid/cis319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khanna S, Pardi DS. 2012. Clostridium difficile infection: new insights into management. Mayo Clin Proc 87:1106–1117. doi: 10.1016/j.mayocp.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanna S, Pardi DS. 2016. Clinical implications of antibiotic impact on gastrointestinal microbiota and Clostridium difficile infection. Expert Rev Gastroenterol Hepatol 10:1–8. doi: 10.1586/17474124.2016.1120157. [DOI] [PubMed] [Google Scholar]
- 5.Ofosu A. 2016. Clostridium difficile infection: a review of current and emerging therapies. Ann Gastroenterol 29:147–154. doi: 10.20524/aog.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slimings C, Riley TV. 2014. Antibiotics and hospital-acquired Clostridium difficile infection: update of systematic review and meta-analysis. J Antimicrob Chemother 69:881–891. doi: 10.1093/jac/dkt477. [DOI] [PubMed] [Google Scholar]
- 7.Brown KA, Khanafer N, Daneman N, Fisman DN. 2013. Meta-analysis of antibiotics and the risk of community-associated Clostridium difficile infection. Antimicrob Agents Chemother 57:2326–2332. doi: 10.1128/AAC.02176-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deshpande A, Pasupuleti V, Thota P, Pant C, Rolston DD, Sferra TJ, Hernandez AV, Donskey CJ. 2013. Community-associated Clostridium difficile infection and antibiotics: a meta-analysis. J Antimicrob Chemother 68:1951–1961. doi: 10.1093/jac/dkt129. [DOI] [PubMed] [Google Scholar]
- 9.Ley RE, Peterson DA, Gordon JI. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Maroo S, Lamont JT. 2006. Recurrent Clostridium difficile. Gastroenterology 130:1311–1316. doi: 10.1053/j.gastro.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 11.Pépin J, Saheb N, Coulombe MA, Alary ME, Corriveau MP, Authier S, Leblanc M, Rivard G, Bettez M, Primeau V, Nguyen M, Jacob CE, Lanthier L. 2005. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis 41:1254–1260. doi: 10.1086/496986. [DOI] [PubMed] [Google Scholar]
- 12.Ma GK, Brensinger CM, Wu Q, Lewis JD. 2017. Increasing incidence of multiply recurrent Clostridium difficile infection in the United States: a cohort study. Ann Intern Med 167:152–158. doi: 10.7326/M16-2733. [DOI] [PubMed] [Google Scholar]
- 13.Saini SD, Waljee AK. 2017. Fool me thrice: the evolving story of multiply recurrent Clostridium difficile infection. Ann Intern Med 167:202–203. doi: 10.7326/M17-1565. [DOI] [PubMed] [Google Scholar]
- 14.Kelly CP, LaMont JT. 2008. Clostridium difficile–more difficult than ever. N Engl J Med 359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 15.Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, Harmsen WS, Zinsmeister AR. 2012. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol 107:89–95. doi: 10.1038/ajg.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McFarland LV, Elmer GW, Surawicz CM. 2002. Breaking the cycle: treatment strategies for 163 cases of recurrent Clostridium difficile disease. Am J Gastroenterol 97:1769–1775. doi: 10.1111/j.1572-0241.2002.05839.x. [DOI] [PubMed] [Google Scholar]
- 17.Sheitoyan-Pesant C, Abou Chakra CN, Pepin J, Marcil-Héguy A, Nault V, Valiquette L. 2016. Clinical and healthcare burden of multiple recurrences of Clostridium difficile infection. Clin Infect Dis 62:574–580. doi: 10.1093/cid/civ958. [DOI] [PubMed] [Google Scholar]
- 18.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 19.Louie TJ, Cannon K, Byrne B, Emery J, Ward L, Eyben M, Krulicki W. 2012. Fidaxomicin preserves the intestinal microbiome during and after treatment of Clostridium difficile infection (CDI) and reduces both toxin reexpression and recurrence of CDI. Clin Infect Dis 55(Suppl 2):S132–S142. doi: 10.1093/cid/cis338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deshpande A, Hurless K, Cadnum JL, Chesnel L, Gao L, Chan L, Kundrapu S, Polinkovsky A, Donskey CJ. 2016. Effect of fidaxomicin versus vancomycin on susceptibility to intestinal colonization with vancomycin-resistant enterococci and Klebsiella pneumoniae in mice. Antimicrob Agents Chemother 60:3988–3993. doi: 10.1128/AAC.02590-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM, Young VB. 2008. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 22.Leffler DA, Lamont JT. 2015. Clostridium difficile infection. N Engl J Med 373:287–288. doi: 10.1056/NEJMc1506004. [DOI] [PubMed] [Google Scholar]
- 23.Neu HC, Gootz TD. 1996. Antimicrobial Chemotherapy. In Baron S. (ed), Medical microbiology, 4th ed University of Texas Medical Branch at Galveston, Galveston, TX. [PubMed] [Google Scholar]
- 24.Brook I, Wexler HM, Goldstein EJ. 2013. Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin Microbiol Rev 26:526–546. doi: 10.1128/CMR.00086-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullane KM, Gorbach S. 2011. Fidaxomicin: first-in-class macrocyclic antibiotic. Expert Rev Anti Infect Ther 9:767–777. doi: 10.1586/eri.11.53. [DOI] [PubMed] [Google Scholar]
- 26.Tannock GW, Munro K, Taylor C, Lawley B, Young W, Byrne B, Emery J, Louie T. 2010. A new macrocyclic antibiotic, fidaxomicin (OPT-80), causes less alteration to the bowel microbiota of Clostridium difficile-infected patients than does vancomycin. Microbiology 156:3354–3359. doi: 10.1099/mic.0.042010-0. [DOI] [PubMed] [Google Scholar]
- 27.Louie TJ, Miller MA, Mullane KM, Weiss K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK, PT-80-003 Clinical Study Group. 2011. Fidaxomicin versus vancomycin for Clostridium difficile infection. N Engl J Med 364:422–431. doi: 10.1056/NEJMoa0910812. [DOI] [PubMed] [Google Scholar]
- 28.Nerandzic MM, Mullane K, Miller MA, Babakhani F, Donskey CJ. 2012. Reduced acquisition and overgrowth of vancomycin-resistant enterococci and Candida species in patients treated with fidaxomicin versus vancomycin for Clostridium difficile infection. Clin Infect Dis 55(Suppl 2):S121–S126. doi: 10.1093/cid/cis440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donskey CJ, Chowdhry TK, Hecker MT, Hoyen CK, Hanrahan JA, Hujer AM, Hutton-Thomas RA, Whalen CC, Bonomo RA, Rice LB. 2000. Effect of antibiotic therapy on the density of vancomycin-resistant enterococci in the stool of colonized patients. N Engl J Med 343:1925–1932. doi: 10.1056/NEJM200012283432604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abujamel T, Cadnum JL, Jury LA, Sunkesula VC, Kundrapu S, Jump RL, Stintzi AC, Donskey CJ. 2013. Defining the vulnerable period for re-establishment of Clostridium difficile colonization after treatment of C. difficile infection with oral vancomycin or metronidazole. PLoS One 8:e76269. doi: 10.1371/journal.pone.0076269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chilton CH, Crowther GS, Ashwin H, Longshaw CM, Wilcox MH. 2016. Association of fidaxomicin with C. difficile spores: effects of persistence on subsequent spore recovery, outgrowth and toxin production. PLoS One 11:e0161200. doi: 10.1371/journal.pone.0161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis BB, Buffie CG, Carter RA, Leiner I, Toussaint NC, Miller LC, Gobourne A, Ling L, Pamer EG. 2015. Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J Infect Dis 212:1656–1665. doi: 10.1093/infdis/jiv256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 35.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 37.Core Team R 2014. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 38.Paradis E, Claude J, Strimmer K. 2004. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 39.Kruskal WH, Wallis WA. 1952. Use of ranks in one-criterion variance analysis. J Am Stat Assoc 47:583–621. doi: 10.1080/01621459.1952.10483441. [DOI] [Google Scholar]
- 40.Dwass M. 1957. Modified randomization tests for nonparametric hypotheses. Ann Math Stat 28:181–187. doi: 10.1214/aoms/1177707045. [DOI] [Google Scholar]
- 41.Benjamini YHY. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.