ABSTRACT
Wastewater is considered a major source of antibiotic-resistant bacteria released into the environment. Here, we characterized carbapenemase-producing Enterobacteriaceae (CPE) in wastewater by whole-genome analysis. Wastewater samples (n = 40) were collected from municipal wastewater treatment plants and hospital wastewater in Japan and Taiwan. Samples were screened for CPE using selective media, and the obtained isolates were sequenced using an Illumina MiSeq. The isolates (n = 45) included the following microorganisms: Klebsiella quasipneumoniae (n = 12), Escherichia coli (n = 10), Enterobacter cloacae complex (n = 10), Klebsiella pneumoniae (n = 8), Klebsiella variicola (n = 2), Raoultella ornithinolytica (n = 1), Citrobacter freundii (n = 1), and Citrobacter amalonaticus (n = 1). Among the 45 isolates, 38 harbored at least one carbapenemase-encoding gene. Of these, the blaGES (blaGES-5, blaGES-6, and blaGES-24) genes were found in 29 isolates. The genes were situated in novel class 1 integrons, but the integron structures were different between the Japanese (In1439 with blaGES-24 and In1440 with blaGES-5) and Taiwanese (In1441 with blaGES-5 and In1442 with blaGES-6) isolates. Other carbapenemase-encoding genes (blaVIM-1, blaNDM-5, blaIMP-8, blaIMP-19, and blaKPC-2) were found in one to three isolates. Notably, class 1 integrons previously reported among clinical isolates obtained in the same regions as the present study, namely, In477 with blaIMP-19 and In73 with blaIMP-8, were found among the Japanese and Taiwanese isolates, respectively. The results indicate that CPE with various carbapenemase-encoding genes in different genetic contexts were present in biologically treated wastewater, highlighting the need to monitor for antibiotic resistance in wastewater.
KEYWORDS: Enterobacteriaceae, carbapenemases, wastewater, whole-genome sequencing
INTRODUCTION
Contamination of environmental waters by antibiotic-resistant bacteria (ARB) is a global health concern, because environmental waters are used for various purposes, including as sources of drinking water and for irrigation. Previous studies have reported fecal carriage of ARB in both community and clinical settings (1–4), indicating that wastewater released from municipal wastewater treatment plants (WWTPs) and hospitals can be a major source of environmental ARB.
Among ARB, carbapenemase-producing Enterobacteriaceae (CPE) are of great concern because these organisms can be resistant to carbapenems, which are often antimicrobial agents of last resort, and to other commonly used drugs (5). A large variety of carbapenemases have been reported, including those belonging to molecular Ambler class A, class B, and class D (6). Class A serine carbapenemases include members of the SME, IMI, and GES enzymes, as well as clinically important KPC enzymes. Class B metallo-beta-lactamases include IMP, VIM, and NDM types of enzymes. Class D carbapenemases are OXA-type β-lactamases and include OXA-48 and OXA-181. Carbapenem resistance can also be conferred by mechanisms such as overexpression of β-lactamases possessing very weak carbapenemase activity in combination with decreased outer membrane permeability (7). However, resistance conferred by carbapenemases is considered to be more important than these mechanisms because carbapenemase-encoding genes are often located on mobile genetic elements and can be transferred to other bacteria (7).
Some previous studies have reported the occurrence of CPE in wastewater, including KPC-2-producing Klebsiella spp. and Escherichia spp. (8), KPC-2-producing Klebsiella pneumoniae (9), and NDM-1-positive Escherichia coli (10). However, compared with clinical isolates, data are limited for CPE in wastewater with respect to detailed genetic characteristics, such as fine-scale within-species phylogeny and virulence gene profiles. Detailed characterization of environmental CPE is important to better understand the molecular epidemiology and reservoirs of these clinically important microbes.
The present study was conducted to characterize CPE isolates in wastewater collected from municipal WWTPs and hospitals in Japan and Taiwan. Japan and Taiwan are not geographically distant, but the prevalences of carbapenemase-encoding genes in clinical Enterobacteriaceae isolates seem to be different in the two locales. Previous studies reported that Japanese CPE usually carry blaIMP genes (11), whereas Taiwanese CPE carry a variety of carbapenemase-encoding genes, including blaKPC, blaNDM, blaIMP, and blaVIM (12). However, the prevalences of carbapenemase-encoding genes in CPE beyond clinical settings are not well understood in the two regions. In the present study, we performed whole-genome sequencing and analysis on CPE in wastewater to provide genetic information about resistance determinants, genetic contexts of carbapenemase-encoding genes, fine-scale phylogenies, and virulence gene profiles. Such information will help clarify the relatedness of environmental and clinical isolates and the mechanisms of spread of carbapenemase-encoding genes.
RESULTS AND DISCUSSION
Detection of CPE in wastewater.
In total, 40 water samples were collected during the study period. CPE isolates were obtained from samples taken from a municipal WWTP in Taiwan, hospital wastewater in Taiwan, and a municipal WWTP in Japan. No CPE isolates were obtained from hospital wastewater samples in Japan. This may be because samples from the Japanese hospital consisted of untreated wastewater taken from sewer pipes and contained fecal material from fewer persons than the samples from the other locations, which consisted of treated wastewater (a mixture of fecal material from the population). We noted that some colonies showing Enterobacteriaceae profiles on chromID CARBA (bioMérieux, Marcy-l’Étoile, France) plates were oxidase positive, and we also detected such oxidase-positive Enterobacteriaceae-like colonies in samples from which we could not isolate CPE strains, including hospital wastewater samples in Japan. A previous study using chromID CARBA SMART for analyzing environmental samples also reported a high frequency of isolation of oxidase-positive microorganisms, such as Pseudomonas spp. (13). We could not determine the number of CPE in each sample because of the presence of these oxidase-positive Enterobacteriaceae-like colonies.
A total of 46 isolates were obtained. As described above, these isolates were obtained from samples taken from a municipal WWTP and hospital wastewater in Taiwan and a municipal WWTP in Japan, all consisting of biologically treated wastewater. One isolate was identified as redundant (i.e., isolated from the same sample, belonging to the same species and the same sequence type [ST], and carrying the same antimicrobial resistance genes). We excluded this redundant isolate, leaving 3 isolates from municipal WWTP samples from Taiwan, 32 isolates from hospital wastewater samples from Taiwan, and 10 isolates from municipal WWTP samples from Japan for further analysis. The species composition of these 45 isolates was as follows: E. coli, n = 10; Klebsiella spp., n = 22; Enterobacter cloacae complex, n = 10; Citrobacter spp., n = 2; and Raoultella ornithinolytica, n = 1 (Table 1). Species identified by the MALDI (matrix-assisted laser desorption ionization) Biotyper Compass 4.1 (Bruker Daltonics GmbH, Bremen, Germany) and by genome sequence-based methods (average nucleotide identity [ANI] and digital DNA-DNA hybridization [DDH] calculation) were congruent in most cases, but the genome sequence-based methods were more accurate in determining the phylogenetic groups of Klebsiella spp. (KpI, KpII-A, KpII-B, and KpIII) and the E. cloacae complex (phylogenetic groups A to R). Data Set S1 in the supplemental material summarizes the genetic characteristics (sequence type, resistance genes, genetic context of carbapenemase-encoding genes, and plasmid replicons associated with carbapenemase-encoding genes), assembly statistics, and antibiotic susceptibility of each isolate. It should be noted that we did not collect isolates randomly from each plate but selected colonies based on their morphologies to represent the genetic diversity of bacterial strains in a sample. Moreover, only a limited number of colonies were picked from each plate. Therefore, the actual species composition and clonal composition of CPE isolates in the wastewater samples may differ from those that we observed in the present study, and the diversity of CPE is likely to be underestimated.
TABLE 1.
Species composition and carbapenemase-encoding genes of Enterobacteriaceae isolates
| Carbapenemase-encoding gene (n) | Sample typea (n) |
||||
|---|---|---|---|---|---|
| E. coli (n = 10) | Klebsiella spp. (n = 22) | E. cloacae complex (n = 10) | Citrobacter spp. (n = 2) | R. ornithinolytica (n = 1) | |
| blaGES-5 (20) | TH (6) | JW (1), TW (1), TH (10) | TH (1) | JW (1) | |
| blaGES-6 (7) | TH (1) | TW (1), TH (4) | TH (1) | ||
| blaGES-24 (1) | JW (1) | ||||
| blaNDM-5 (3) | TH (3) | ||||
| blaIMP-8 (2) | TH (2) | ||||
| blaIMP-19 (1) | JW (1) | ||||
| blaKPC-2 (2) | TH (1) | TH (1) | |||
| blaVIM-1 (1) | TH (1) | ||||
| blaGES-5 + blaIMP-8 (1) | TH (1) | ||||
| NDb (7) | TW (1) | JW (6) | |||
JW, Japanese municipal WWTP; TW, Taiwanese municipal WWTP; TH, Taiwanese hospital wastewater.
ND, no carbapenemase-encoding genes were detected.
Phenotypic and genotypic resistance.
The carbapenemase-encoding genes detected in the present study were as follows: blaGES-5 (n = 21), blaGES-6 (n = 7), blaIMP-8 (n = 3), blaNDM-5 (n = 3), blaKPC-2 (n = 2), blaGES-24 (n = 1), blaIMP-19 (n = 1), and blaVIM-1 (n = 1) (Table 1). One Citrobacter freundii isolate carried both blaGES-5 and blaIMP-8. Seven isolates did not carry any carbapenemase-encoding genes, but they carried class C β-lactamases. AmpC β-lactamase overproduction and decreased outer membrane protein expression combined with an active efflux pump can contribute to carbapenem resistance in Enterobacteriaceae (7, 14), which may explain the carbapenem resistance in these seven isolates. All but one (blaIMP-19-carrying JSWP033) of the CPE isolates from municipal WWTP samples carried blaGES genes. The apparent high prevalence of blaGES genes among our municipal WWTP isolates indicates that blaGES-harboring microbes may be prevalent in the intestinal tracts of the sampled local populations in Japan and Taiwan, because bacterial isolates in wastewater may serve as representatives of the strains present within the human population in a given locale (15, 16). blaGES-5 was the most prevalent carbapenemase-encoding gene among the Taiwanese hospital wastewater isolates, despite the apparent absence of blaGES among patients in Taiwan (12). A previous study similarly reported that blaGES-5 was prevalent among CPE isolated from hospital wastewater despite the absence of the gene among screening and clinical isolates within the hospital (17). The authors concluded that this may have been because unidentified carriers within the hospital were a reservoir for GES-5 or GES-5-positive Enterobacteriaceae were adapted to the environment and consistently present within the wastewater pipework. These possibilities may also apply to our study. In addition, there is potential for underestimation of the occurrence of blaGES among clinical isolates because MICs of carbapenems tended to be lower for the isolates harboring blaGES and blaGES-harboring isolates can be falsely negative in the Carba NP test, as discussed below. There were also CPE isolates from Taiwanese hospital wastewater carrying blaVIM-1, blaNDM-5, blaIMP-8, and blaKPC-2, findings consistent with a variety of carbapenemases recovered from patients in Taiwan (12).
Carbapenemase activity was confirmed in all isolates harboring blaVIM-1, blaNDM-5, blaIMP-8, blaIMP-19, or blaKPC-2 by the Carba NP test, whereas isolates harboring blaGES genes and isolates without carbapenemase-encoding genes were negative (or invalid for one blaGES-5-harboring isolate) by the Carba NP test (see Data Set S1 in the supplemental material). A previous study reported that Enterobacteriaceae harboring GES-5 can be falsely negative in the Carba NP test (18). MICs of imipenem and meropenem tended to be higher for isolates harboring metallo-beta-lactamase-encoding genes (blaVIM-1, blaNDM-5, blaIMP-8, and blaIMP-19) or blaKPC-2 than for those harboring blaGES genes (see Fig. S1 in the supplemental material). In fact, all but one isolate (blaIMP-8-harboring TTHS028) with these metallo-beta-lactamase-encoding genes or blaKPC-2 were nonsusceptible to imipenem or meropenem, while more than half of the blaGES-harboring isolates were susceptible to both agents. These results may reflect the relatively weak carbapenemase activity of GES enzymes (19). Importantly, all isolates carried resistance genes other than carbapenemase-encoding genes (see Data Set S1 in the supplemental material). Although some genes seem to be chromosomally encoded and core to the species/genus (e.g., fosA, oqxAB, and the β-lactamase genes blaSHV, blaOKP, and blaLEN in KpI, KpII, and KpIII [20]), the observed high prevalence of genes conferring resistance to different classes of antibiotics among CPE is of great concern.
Genetic contexts of carbapenemase-encoding genes.
Among the 38 CPE isolates, we could determine the genetic contexts of carbapenemase-encoding genes in 32 isolates (Fig. 1 to 3; see also Data Set S1 in the supplemental material for the genetic context of the carbapenemase-encoding genes in each isolate). For the remaining six isolates, the genetic contexts could not be determined due to the limitations associated with short-read sequencing.
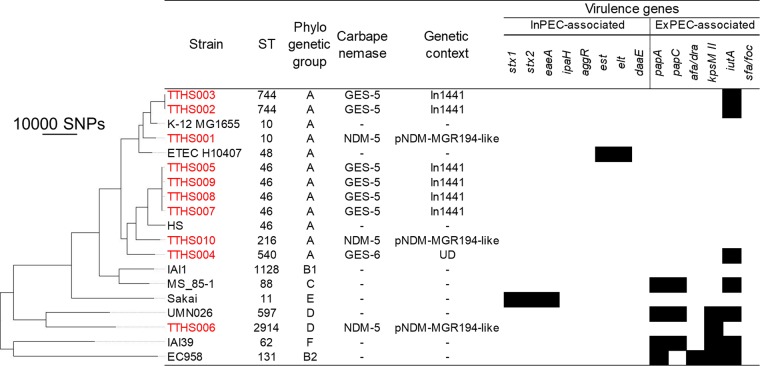
FIG 1.
Phylogenetic tree of E. coli isolates. The tree was visualized using FigTree (http://tree.bio.ed.ac.uk/software/figtree/). In total, 181,286 SNP positions were identified, 107,866 of which occurred in at least 80% of the genomes and were used for the tree construction. UD indicates the genetic context of a carbapenemase-encoding gene was not determined for the isolate. In1441 contained a blaGES-5-blaOXA-17 cassette array. The dashes in the carbapenemase and genetic context columns indicate the absence of carbapenemase-encoding genes. The black blocks represent the presence of a virulence gene. No chromosomal ampC promoter/attenuator mutations that can result in ampC overexpression were found among these E. coli isolates. Strains from Taiwanese hospital wastewater begin with TTHS and are colored red. The other strains are reference strains.
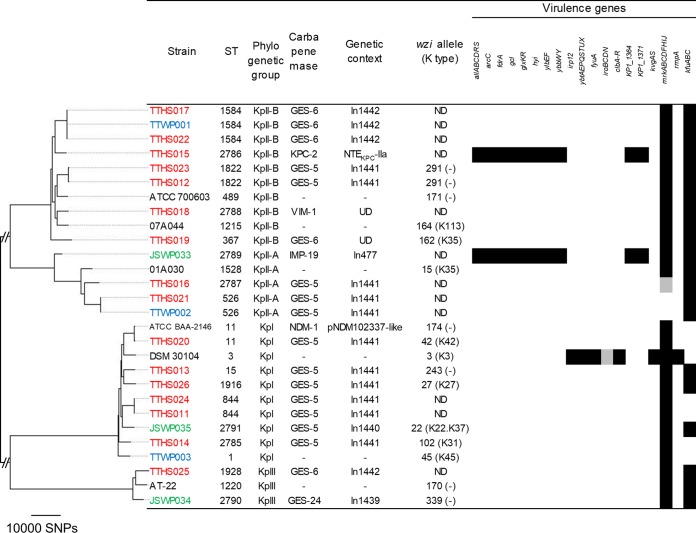
FIG 2.
Phylogenetic tree of Klebsiella isolates. In total, 402,516 SNP positions were identified, 100,591 of which occurred in at least 80% of the genomes and were used for the tree construction. KpI corresponds to K. pneumoniae, KpII-A corresponds to K. quasipneumoniae subsp. quasipneumoniae, KpII-B corresponds to K. quasipneumoniae subsp. similipneumoniae, and KpIII corresponds to K. variicola. ST2785 to ST2791 are novel sequence types found in this study. UD indicates the genetic context of a carbapenemase-encoding gene was not determined for the isolate. In477 and In1439 to In1442 contained the following cassette arrays: blaIMP-19-aacA31-blaOXA-21-aadA1 (In477), blaGES-24-aacA4 (In1439), blaGES-5-aacA31-catB8-aadA5 (In1440), blaGES-5-blaOXA-17 (In1441), and blaGES-6-aacA4-blaOXA-17 (In1442). NTEKPC indicates a blaKPC-bearing non-Tn4401 element. In ATCC BAA-2146, Tn125 was truncated at ISAba125 upstream of blaNDM-1 and at ISCR27. The dashes in the carbapenemase and genetic context columns indicate the absence of carbapenemase-encoding genes. ND indicates the wzi allele was not determined for the isolate. K types were not assigned for some wzi alleles (indicated by the dashes in parentheses), and only the wzi alleles are shown for these isolates. The black blocks represent the presence of a virulence gene/gene cluster. The gray blocks represent the presence of some genes in a gene cluster. Virulence genes of iucABCD, iutA, mceABCDEGHIJ, and rmpA2 were sought but not found. Strains from Taiwanese hospital wastewater begin with TTHS and are colored red. Strains from the Taiwanese municipal WWTP begin with TTWP and are colored blue. Strains from the Japanese municipal WWTP begin with JSWP and are colored green. The other strains are reference strains.
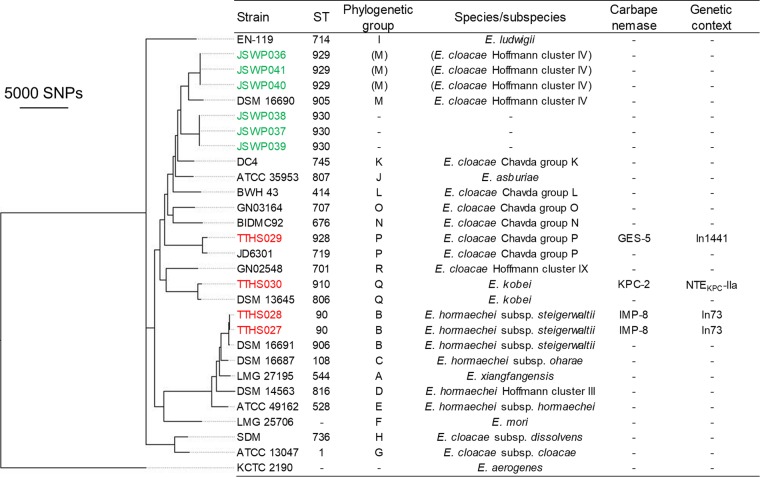
FIG 3.
Phylogenetic tree of E. cloacae complex isolates. In total, 1,321,589 SNP positions were identified, 24,510 of which occurred in at least 80% of the genomes and were used for the tree construction. Enterobacter aerogenes KCTC 2190 was used as an outgroup. Hoffmann clusters are genetic clusters identified by Hoffmann and Roggenkamp based most exhaustively on hsp60 sequences (64). ST928, ST929, and ST930 are novel sequence types identified in the present study. The dashes in the ST, phylogenetic group, and species/subspecies columns indicate that these properties could not be assigned to the corresponding isolates. In73 contained a blaIMP-8-aacA4-catB3 cassette array, and In1441 contained a blaGES-5-blaOXA-17 cassette array. NTEKPC is a blaKPC-bearing non-Tn4401 element. The dashes in the carbapenemase and genetic context columns indicate the absence of carbapenemase-encoding genes. Strains from the Taiwanese hospital wastewater begin with TTHS and are colored red. Strains from the Japanese municipal WWTP begin with JSWP and are colored green. The other strains are reference strains.
blaGES genes were situated within novel class 1 integrons. In1441, with a gene cassette array of blaGES-5-blaOXA-17, was prevalent among E. coli (n = 6), Klebsiella spp. (n = 11), and E. cloacae complex (n = 1) isolates from the Taiwanese municipal WWTP and hospital wastewater. In1442, with a gene cassette array of blaGES-6-aacA4-blaOXA-17, was found in Klebsiella isolates (n = 4) from the Taiwanese municipal WWTP and hospital wastewater. These findings suggest the circulation of the same integron among different species and in both community and clinical settings in Taiwan. In all of the gene cassette arrays mentioned above, the attC site of the blaOXA-17 gene cassette was interrupted by ISPa25, which contains a putative transposase gene (orf4). Interestingly, the nucleotide sequence of this fused gene cassette, containing blaOXA-17 and orf4, shared 100% nucleotide identity with those in previously characterized class 1 integrons of Pseudomonas aeruginosa from Taiwan (21) and K. pneumoniae from Korea (22). On the other hand, different novel integrons without this fused gene cassette were found among blaGES-harboring Japanese isolates, namely, In1439 (cassette array, blaGES-24-aacA4) in one KpIII isolate and In1440 (cassette array, blaGES-5-aacA31-catB8-aadA5) in one KpI isolate. We were able to determine the promoter types and downstream structures in 8 and 12 blaGES-containing integrons, respectively (see also Data Set S1 in the supplemental material). Only strong promoters, namely, PcWTGN-10 (n = 5), PcH2TGN-10 (n = 2), and PcS (n = 1), were detected. Of the 12 integrons for which we could determine downstream structures, all carried 3′-conserved segment (CS) structures immediately downstream of the gene cassettes. Of these, 3′-CS-IS6100 (with or without a 1-bp deletion in IS6100) was the most common (n = 9).
blaIMP genes were situated in previously reported class 1 integrons. Two E. cloacae complex isolates from Taiwanese hospital wastewater carried In73 with a gene cassette array of blaIMP-8-aacA4-catB3. This integron was previously found in a clinical isolate of K. pneumoniae (23) and clinical isolates of the E. cloacae complex (24) obtained in Tainan City, Taiwan. One KpII-A isolate from the Japanese municipal WWTP carried In477 with a gene cassette array of blaIMP-19-aacA31-blaOXA-21-aadA1. This integron was previously found in clinical isolates of Acinetobacter spp. in the Kansai region of Japan (25). These results are interesting because the same integrons were found among clinical isolates obtained in the same regions as the present study. We were able to determine the promoter type and the downstream structure of the blaIMP-19-containing integron of the Japanese isolate (see Data Set S1 in the supplemental material). This integron contained a PcS strong promoter and carried 3′-CS-IS6100 immediately downstream of the gene cassettes.
blaNDM-5 was detected in three E. coli isolates from Taiwanese hospital wastewater. blaNDM-5 was carried by an IS26-like insertion sequence (contig break in the transposase gene)-dsbC-trpF-bleMBL-blaNDM-5-ISAba125 (interrupted by IS5) genetic element. For all three isolates, PlasmidFinder detected an IncX3 replicon in blaNDM-5-containing contigs. Further analysis revealed that the blaNDM-5-containing contigs were highly similar (≥99% sequence identity over ≥90% of the length) to a previously described IncX3 plasmid, pNDM-MGR194 of K. pneumoniae MGR-K194 in India (26). Plasmids that are identical or nearly identical to plasmid pNDM-MGR194 have been reported among E. coli isolates from China, Australia, and Denmark (27).
blaKPC-2 was found in one KpII-B isolate and one E. cloacae complex isolate from Taiwanese hospital wastewater. In both cases, the gene occurred in the context of the blaKPC-bearing non-Tn4401 element IIa (28). An IncP6 replicon was detected in the blaKPC-2-carrying contig of the KpII-B isolate, whereas no plasmid replicons were detected in that of the E. cloacae complex isolate.
Phylogenetic characteristics and virulence potential.
The E. coli isolates were detected only in Taiwanese hospital wastewater and belonged to six different STs (Fig. 1). ST46 (n = 4) was the most prevalent, followed by ST744 (n = 2) (note that these isolates belonging to the same ST were obtained from different samples and not redundant, as described above). Nine isolates belonged to phylogenetic group A, which is usually associated with intestinal pathogenic E. coli (InPEC) or commensals (29). E. coli pathotyping based on the presence of virulence genes indicated that none of our CPE isolates were pathogenic, although three strains carried the extraintestinal pathogenic E. coli (ExPEC)-associated gene iutA and one strain carried another ExPEC-associated gene, kpsM II (carriage of at least two ExPEC-associated genetic markers is needed to assign an isolate to ExPEC).
Isolates classically identified as K. pneumoniae can be divided into KpI (K. pneumoniae), KpII-A (Klebsiella quasipneumoniae subsp. quasipneumoniae), KpII-B (Klebsiella quasipneumoniae subsp. similipneumoniae), and KpIII (Klebsiella variicola) (20, 30). The prevalence of these phylogenetic groups among the Klebsiella isolates in the present study was as follows: KpI, n = 8; KpII-A, n = 4; KpII-B, n = 8; and KpIII, n = 2 (Fig. 2). This prevalence is different from those observed in previous studies analyzing clinical isolates (31, 32), which reported a higher prevalence of KpI than of KpII and KpIII. This may primarily be because KpII and KpIII are associated more frequently with carriage (not being the cause of an infection), whereas KpI is associated with human infection (20, 33). The most prevalent ST was ST1584 (n = 3), followed by ST526 (n = 2), ST844 (n = 2), and ST1822 (n = 2). Clonal overlaps for ST1584 and ST526 were observed between the municipal WWTP and hospital wastewater isolates from Taiwan, suggesting that carbapenemase-producing KpII isolates belonging to these STs may be prevalent in both community and clinical settings in Taiwan. It is worrisome that successful multidrug-resistant clones, namely, ST11 and ST15 (31, 34), were found among CPE isolates in the present study. Among the virulence genes analyzed, the allantoinase gene cluster was found in two of the KpII isolates, kfuABC was found in all of the KpII and KpIII isolates and 38% of the KpI isolates, and mrkABCDFHIJ was found in all the isolates except some of the KpII-A isolates. These results are in agreement with a previous study (20). However, the genes rmpA and rmpA2 and the siderophore clusters, which are significantly associated with invasive human infections among KpI isolates (20), were not detected among the Klebsiella isolates in the present study. The polysaccharide capsule is a key virulence determinant, and the K types K1, K2, K5, K20, K54, and K57 are known to be associated with liver abscesses and other community-acquired invasive infections (35). None of our isolates were identified as these K types by wzi typing.
By employing whole-genome analyses, Chavda et al. determined that E. cloacae complex isolates fall into 18 phylogenetic groups (A to R), each corresponding to a distinct species/subspecies (36). According to the phylogenetic tree in Fig. 3 and ANI and digital DDH analyses, four of our E. cloacae complex isolates could be assigned to 1 of these 18 phylogenetic groups, namely, group B (n = 2), group P (n = 1), or group Q (n = 1). Group B (Enterobacter hormaechei subsp. steigerwaltii) is one of the most prevalent phylogenetic groups among clinical E. cloacae complex isolates (37). Two blaIMP-8-carrying strains (TTHS027 and TTHS028) belonging to this group were assigned to ST90, which is one of the STs found among clinical IMP-producing isolates (38). A fine-scale phylogenetic tree was constructed to gain insights into the phylogeny of the remaining six isolates (see Fig. S2 in the supplemental material). Three isolates (JSWP036, JSWP040, and JSWP041) seem to be closely related to group M (E. cloacae Hoffmann cluster IV), and the ANI values of these three strains and the type strain of group M were 95.8%, just slightly higher than 95%. However, the digital DDH estimate was 64.7%, indicating that the three strains may belong to different species than group M. The other three isolates (JSWP037, JSWP038, and JSWP039) showed ANI values of ≤95% among all of the type or reference strains of 18 phylogenetic groups, indicating that the three strains belong to another, unrecognized group. Further studies are needed to elucidate the phylogeny and determine the species of these six isolates.
Two Citrobacter isolates were obtained in the present study, one belonging to C. freundii and the other belonging to Citrobacter amalonaticus. The C. freundii isolate belonged to ST22. This ST was previously reported among carbapenemase-producing clinical Citrobacter isolates (39). We also detected one R. ornithinolytica isolate. Two recent studies reported carbapenemase-producing Citrobacter spp. and R. ornithinolytica isolates in wastewater and surface water, but the detected isolates carried different types of carbapenemase-encoding genes (blaOXA, blaVIM, and blaNDM) than those in the present study (blaGES and blaIMP) (40, 41). However, it should be noted that these two studies detected carbapenemase-encoding genes by PCR, and blaGES and blaIMP were not investigated. This highlights the strength of the whole-genome-sequencing approach, which enables the detection of all antibiotic resistance genes annotated in the database.
This study has some limitations. The number of isolates characterized (n = 45) is not large, and most isolates (n = 32) were from Taiwanese hospital wastewater (note that E. coli isolates were obtained only from Taiwanese hospital wastewater). Our collection does not represent the prevalence of CPE in wastewater in Japan and Taiwan because our isolates were obtained from a limited number of samples collected over relatively short periods. Moreover, we were not able to determine the genetic surroundings of carbapenemase-encoding genes in some isolates and could not resolve the plasmid structures due to the limitations associated with short reads. Further studies are needed, including a larger number of global isolates obtained from different types of WWTPs and employing long-read sequencing.
In the present study, we characterized, by whole-genome analysis, CPE isolates in wastewater in terms of resistance determinants, genetic contexts of carbapenemase-encoding genes, phylogeny, and virulence potential. The results indicate that CPE isolates with various carbapenemase-encoding genes in different genetic contexts are present in biologically treated wastewater and may disseminate into the environment. This study highlights the need to monitor for antibiotic-resistant bacteria in the environment, not only in clinical settings.
MATERIALS AND METHODS
Sample collection and isolation of CPE.
Ten wastewater samples per location were collected from a municipal WWTP and a hospital in the Kansai region of Japan in October 2015 (42). Samples from the WWTP were collected from effluent from the final settling tanks after biological (activated-sludge) treatment, and samples from the hospital were taken from a sewer system (sewer pipes) and consisted of untreated wastewater. We also collected 10 biologically treated wastewater samples (secondary sedimentation tank effluent) per location from a municipal WWTP and a hospital WWTP in Tainan City, Taiwan, between August and September 2015. All the samples were collected in sterile 50-ml centrifuge tubes or sampling bottles, transported to the laboratory, and processed as soon as possible. A total of 40 water samples were processed by using membrane filtration methods with chromID CARBA agar (bioMérieux, Marcy-l’Étoile, France) for enumeration and isolation of CPE. For each sample, up to four colonies showing E. coli profiles (pink to burgundy) or KESC (Klebsiella-Enterobacter-Serratia-Citrobacter) group profiles (bluish green to bluish gray) on the chromID CARBA plate were isolated, respectively (care was taken to select colonies showing different morphologies, if possible). The isolates were restreaked on fresh chromID CARBA plates and incubated until pure colonies were obtained. Isolates collected in Taiwan were stored in Casitone medium (Eiken, Tokyo, Japan) and transported to the laboratory in Japan. The oxidase test was performed using a cytochrome oxidase test strip (Nissui, Tokyo, Japan) for each isolate, and oxidase-negative isolates were stored at −85°C in 35% glycerol.
Species identification and antibiotic susceptibility testing.
Species identification was performed with a MALDI Biotyper Compass 4.1. Susceptibility to 23 antibiotics was evaluated by microdilution using the dry-plate Eiken assay (Eiken, Tokyo, Japan) according to CLSI guidelines (43). Strains were checked for carbapenemase activity by the Carba NP test (44).
Genome sequencing and assembly.
DNA was extracted from each isolate using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). DNA libraries were prepared using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA) and subsequently sequenced using Illumina MiSeq 300-bp paired-end sequencing technologies to achieve an average depth of coverage of 75. Raw reads generated from each sample were trimmed using ERNE-Filter (45) and assembled using SPAdes v3.10.0 (46). Assemblies were improved using Pilon (47). The assembled contigs were subjected to the analyses described below.
Phylogenetic analysis.
Species identification was also performed for each isolate based on the genomic data. The ANI analysis was performed using JSpeciesWS (48). If an isolate shared an ANI value of >95% with a type strain (or a reference strain if the genome of the type strain was not available) of a certain species belonging to the family Enterobacteriaceae, the isolate was identified as that species (49). Digital DDH values were also calculated using formula 2 on the GGDC website to confirm the results (50).
Isolates identified as E. coli, K. pneumoniae (KpI), K. quasipneumoniae subsp. quasipneumoniae (KpII-A), K. quasipneumoniae subsp. similipneumoniae (KpII-B), K. variicola (KpIII), and E. cloacae complex (phylogenetic groups A to R) were analyzed with kSNP to construct whole-genome single nucleotide polymorphism (SNP)-based within-species/genus phylogenetic trees (51, 52). Selected isolates analyzed in other studies, i.e., isolates analyzed by Kaas et al. and Forde et al. for E. coli (53, 54), isolates analyzed by Brisse et al. and Hudson et al. for Klebsiella spp. (30, 55), and isolates analyzed by Chavda et al. for the E. cloacae complex (36), were also included in the trees to place our isolates in broader phylogenetic contexts. All the trees were parsimony trees and were constructed based on SNP loci occurring in at least 80% of the strains. STs were assigned to isolates using multilocus sequence type (MLST) databases (http://bigsdb.pasteur.fr/klebsiella/; http://pubmlst.org/ecloacae/; http://pubmlst.org/cfreundii/; http://mlst.ucc.ie/mlst/dbs/Ecoli/). E. coli phylogenetic groups were determined as described previously (56).
Identification of antimicrobial resistance determinants and virulence genes.
Antimicrobial resistance genes were detected using the ResFinder antimicrobial resistance gene database (57) (with a threshold of 90% identity and a minimum length of 60%) and NCBI Beta-Lactamase Data Resources. For the E. coli isolates, chromosomal ampC promoter/attenuator mutations were analyzed as described previously (58). Contigs with carbapenemase-encoding genes were manually annotated to determine the genetic contexts of the genes. Integrons were classified according to INTEGRALL (http://integrall.bio.ua.pt/) (59). Pc-P2 promoters in class 1 integrons were analyzed according to a previous study (60). Insertion sequences were identified by using the ISfinder database (61). Plasmid replicons were detected using PlasmidFinder (62) (with a threshold of 80% identity and a minimum length of 60%).
For E. coli isolates, virulence genes were detected as described previously (42), and pathotypes were defined based on the presence of specific virulence genes (63). For Klebsiella isolates, virulence genes were detected using the Institut Pasteur Klebsiella database (http://bigsdb.pasteur.fr/klebsiella), and the K capsular type was determined based on wzi alleles (35). Presence of a virulence gene was arbitrarily defined as >80% sequence identity over >80% of the length of the reference gene.
Accession number(s).
The genome sequence data obtained in the present study have been deposited in the DDBJ Sequence Read Archive database (DDBJ accession number DRA006131). The sequences of novel integrons found in this study are available under the following accession numbers: In1439, LC318533; In1440, LC318534; In1441, LC318535 and LC318536; and In1442, LC318537.
Supplementary Material
ACKNOWLEDGMENTS
We thank the team of curators of the Institut Pasteur MLST and whole-genome MLST databases for curating the data and making them publicly available at http://bigsdb.pasteur.fr. We thank Tohru Miyoshi-Akiyama for curating the Enterobacter cloacae MLST databases. We thank Thomas Jové from INTEGRALL for curating the integrons.
This work was supported by the Kyoto University Foundation, the Kurita Water and Environment Foundation, and the River Fund of the River Foundation, Japan. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02501-17.
REFERENCES
- 1.Ruppe E, Armand-Lefevre L, Estellat C, El-Mniai A, Boussadia Y, Consigny PH, Girard PM, Vittecoq D, Bouchaud O, Pialoux G, Esposito-Farese M, Coignard B, Lucet JC, Andremont A, Matheron S. 2014. Acquisition of carbapenemase-producing Enterobacteriaceae by healthy travellers to India, France, February 2012 to March 2013. Euro Surveill 19:20768. doi: 10.2807/1560-7917.ES2014.19.14.20768. [DOI] [PubMed] [Google Scholar]
- 2.Gijon D, Curiao T, Baquero F, Coque TM, Canton R. 2012. Fecal carriage of carbapenemase-producing Enterobacteriaceae: a hidden reservoir in hospitalized and nonhospitalized patients. J Clin Microbiol 50:1558–1563. doi: 10.1128/JCM.00020-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stromdahl H, Tham J, Melander E, Walder M, Edquist PJ, Odenholt I. 2011. Prevalence of faecal ESBL carriage in the community and in a hospital setting in a county of Southern Sweden. Eur J Clin Microbiol Infect Dis 30:1159–1162. doi: 10.1007/s10096-011-1202-5. [DOI] [PubMed] [Google Scholar]
- 4.Woerther PL, Burdet C, Chachaty E, Andremont A. 2013. Trends in human fecal carriage of extended-spectrum beta-lactamases in the community: toward the globalization of CTX-M. Clin Microbiol Rev 26:744–758. doi: 10.1128/CMR.00023-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi Y, Paterson DL. 2015. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med 36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V, European Network on Carbapenemases. 2012. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect 18:432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang F, Mao D, Zhou H, Luo Y. 2016. Prevalence and fate of carbapenemase genes in a wastewater treatment plant in northern China. PLoS One 11:e0156383. doi: 10.1371/journal.pone.0156383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chagas TP, Seki LM, da Silva DM, Asensi MD. 2011. Occurrence of KPC-2-producing Klebsiella pneumoniae strains in hospital wastewater. J Hosp Infect 77:281. doi: 10.1016/j.jhin.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Mantilla-Calderon D, Jumat MR, Wang T, Ganesan P, Al-Jassim N, Hong PY. 2016. Isolation and characterization of NDM-positive Escherichia coli from municipal wastewater in Jeddah, Saudi Arabia. Antimicrob Agents Chemother 60:5223–5231. doi: 10.1128/AAC.00236-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno Y, Nakamura A, Hashimoto E, Matsutani H, Abe N, Fukuda S, Hisashi K, Komatsu M, Nakamura F. 2017. Molecular epidemiology of carbapenemase-producing Enterobacteriaceae in a primary care hospital in Japan, 2010-2013. J Infect Chemother 23:224–229. doi: 10.1016/j.jiac.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 12.Wang JT, Wu UI, Lauderdale TL, Chen MC, Li SY, Hsu LY, Chang SC. 2015. Carbapenem-nonsusceptible Enterobacteriaceae in Taiwan. PLoS One 10:e0121668. doi: 10.1371/journal.pone.0121668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piedra-Carrasco N, Fabrega A, Calero-Caceres W, Cornejo-Sanchez T, Brown-Jaque M, Mir-Cros A, Muniesa M, Gonzalez-Lopez JJ. 2017. Carbapenemase-producing enterobacteriaceae recovered from a Spanish river ecosystem. PLoS One 12:e0175246. doi: 10.1371/journal.pone.0175246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang FC, Yan JJ, Hung KH, Wu JJ. 2012. Characterization of ertapenem-resistant Enterobacter cloacae in a Taiwanese university hospital. J Clin Microbiol 50:223–226. doi: 10.1128/JCM.01263-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boczek LA, Rice EW, Johnston B, Johnson JR. 2007. Occurrence of antibiotic-resistant uropathogenic Escherichia coli clonal group A in wastewater effluents. Appl Environ Microbiol 73:4180–4184. doi: 10.1128/AEM.02225-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meir-Gruber L, Manor Y, Gefen-Halevi S, Hindiyeh MY, Mileguir F, Azar R, Smollan G, Belausov N, Rahav G, Shamiss A, Mendelson E, Keller N. 2016. Population Screening using sewage reveals pan-resistant bacteria in hospital and community samples. PLoS One 11:e0164873. doi: 10.1371/journal.pone.0164873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White L, Hopkins KL, Meunier D, Perry CL, Pike R, Wilkinson P, Pickup RW, Cheesbrough J, Woodford N. 2016. Carbapenemase-producing Enterobacteriaceae in hospital wastewater: a reservoir that may be unrelated to clinical isolates. J Hosp Infect 93:145–151. doi: 10.1016/j.jhin.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Tijet N, Boyd D, Patel SN, Mulvey MR, Melano RG. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammoudi D, Moubareck CA, Sarkis DK. 2014. How to detect carbapenemase producers?. A literature review of phenotypic and molecular methods J Microbiol Methods 107:106–118. doi: 10.1016/j.mimet.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan JJ, Hsueh PR, Lu JJ, Chang FY, Ko WC, Wu JJ. 2006. Characterization of acquired beta-lactamases and their genetic support in multidrug-resistant Pseudomonas aeruginosa isolates in Taiwan: the prevalence of unusual integrons. J Antimicrob Chemother 58:530–536. doi: 10.1093/jac/dkl266. [DOI] [PubMed] [Google Scholar]
- 22.Bae IK, Lee YN, Jeong SH, Hong SG, Lee JH, Lee SH, Kim HJ, Youn H. 2007. Genetic and biochemical characterization of GES-5, an extended-spectrum class A beta-lactamase from Klebsiella pneumoniae. Diagn Microbiol Infect Dis 58:465–468. doi: 10.1016/j.diagmicrobio.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Yan JJ, Ko WC, Wu JJ. 2001. Identification of a plasmid encoding SHV-12, TEM-1, and a variant of IMP-2 metallo-beta-lactamase, IMP-8, from a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:2368–2371. doi: 10.1128/AAC.45.8.2368-2371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen YT, Liao TL, Liu YM, Lauderdale TL, Yan JJ, Tsai SF. 2009. Mobilization of qnrB2 and ISCR1 in plasmids. Antimicrob Agents Chemother 53:1235–1237. doi: 10.1128/AAC.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto M, Nagao M, Matsumura Y, Matsushima A, Ito Y, Takakura S, Ichiyama S. 2011. Interspecies dissemination of a novel class 1 integron carrying blaIMP-19 among Acinetobacter species in Japan. J Antimicrob Chemother 66:2480–2483. doi: 10.1093/jac/dkr336. [DOI] [PubMed] [Google Scholar]
- 26.Krishnaraju M, Kamatchi C, Jha AK, Devasena N, Vennila R, Sumathi G, Vaidyanathan R. 2015. Complete sequencing of an IncX3 plasmid carrying blaNDM-5 allele reveals an early stage in the dissemination of the blaNDM gene. Indian J Med Microbiol 33:30–38. doi: 10.4103/0255-0857.148373. [DOI] [PubMed] [Google Scholar]
- 27.Zhu YQ, Zhao JY, Xu C, Zhao H, Jia N, Li YN. 2016. Identification of an NDM-5-producing Escherichia coli sequence type 167 in a neonatal patient in China. Sci Rep 6:29934. doi: 10.1038/srep29934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. 2014. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol 22:686–696. doi: 10.1016/j.tim.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohler CD, Dobrindt U. 2011. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 301:642–647. doi: 10.1016/j.ijmm.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Brisse S, Passet V, Grimont PA. 2014. Description of Klebsiella quasipneumoniae sp. nov., isolated from human infections, with two subspecies, Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and demonstration that Klebsiella singaporensis is a junior heterotypic synonym of Klebsiella variicola. Int J Syst Evol Microbiol 64:3146–3152. doi: 10.1099/ijs.0.062737-0. [DOI] [PubMed] [Google Scholar]
- 31.Moradigaravand D, Martin V, Peacock SJ, Parkhill J. 2017. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. mBio 8:e01976-. doi: 10.1128/mBio.01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brisse S, van Himbergen T, Kusters K, Verhoef J. 2004. Development of a rapid identification method for Klebsiella pneumoniae phylogenetic groups and analysis of 420 clinical isolates. Clin Microbiol Infect 10:942–945. doi: 10.1111/j.1469-0691.2004.00973.x. [DOI] [PubMed] [Google Scholar]
- 33.de Melo ME, Cabral AB, Maciel MA, da Silveira VM, de Souza Lopes AC. 2011. Phylogenetic groups among Klebsiella pneumoniae isolates from Brazil: relationship with antimicrobial resistance and origin. Curr Microbiol 62:1596–1601. doi: 10.1007/s00284-011-9903-7. [DOI] [PubMed] [Google Scholar]
- 34.Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. 2014 Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. mBio 5:e01355-. doi: 10.1128/mBio.01355-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brisse S, Passet V, Haugaard AB, Babosan A, Kassis-Chikhani N, Struve C, Decre D. 2013. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol 51:4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chavda KD, Chen L, Fouts DE, Sutton G, Brinkac L, Jenkins SG, Bonomo RA, Adams MD, Kreiswirth BN. 2016. Comprehensive genome analysis of carbapenemase-producing Enterobacter spp.: new insights into phylogeny, population structure, and resistance mechanisms. mBio 7:e02093-. doi: 10.1128/mBio.02093-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohad S, Block C, Kravitz V, Farber A, Pilo S, Breuer R, Rorman E. 2014. Rapid identification of Enterobacter hormaechei and Enterobacter cloacae genetic cluster III. J Appl Microbiol 116:1315–1321. doi: 10.1111/jam.12439. [DOI] [PubMed] [Google Scholar]
- 38.Matsumura Y, Peirano G, Motyl MR, Adams MD, Chen L, Kreiswirth B, DeVinney R, Pitout JD. 2017. Global molecular epidemiology of IMP-producing Enterobacteriaceae. Antimicrob Agents Chemother 61:e02729-. doi: 10.1128/AAC.02729-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsumura Y, Peirano G, Devinney R, Bradford PA, Motyl MR, Adams MD, Chen L, Kreiswirth B, Pitout JDD. 2017. Genomic epidemiology of global VIM-producing Enterobacteriaceae. J Antimicrob Chemother 72:2249–2258. doi: 10.1093/jac/dkx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zurfluh K, Bagutti C, Brodmann P, Alt M, Schulze J, Fanning S, Stephan R, Nuesch-Inderbinen M. 28 June 2017. Wastewater is a reservoir for clinically relevant carbapenemase- and 16S rRNA methylase-producing Enterobacteriaceae. Int J Antimicrob Agents. doi: 10.1016/j.ijantimicag.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 41.Tafoukt R, Touati A, Leangapichart T, Bakour S, Rolain JM. 2017. Characterization of OXA-48-like-producing Enterobacteriaceae isolated from river water in Algeria. Water Res 120:185–189. doi: 10.1016/j.watres.2017.04.073. [DOI] [PubMed] [Google Scholar]
- 42.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. 2017. Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum-beta-lactamase-producing Escherichia coli in wastewater. Antimicrob Agents Chemother 61:e00564-. doi: 10.1128/AAC.00564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Del Fabbro C, Scalabrin S, Morgante M, Giorgi FM. 2013. An extensive evaluation of read trimming effects on Illumina NGS data analysis. PLoS One 8:e85024. doi: 10.1371/journal.pone.0085024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richter M, Rossello-Mora R, Oliver Glockner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richter M, Rossello-Mora R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gardner SN, Slezak T, Hall BG. 2015. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 31:2877–2878. doi: 10.1093/bioinformatics/btv271. [DOI] [PubMed] [Google Scholar]
- 53.Kaas RS, Friis C, Ussery DW, Aarestrup FM. 2012. Estimating variation within the genes and inferring the phylogeny of 186 sequenced diverse Escherichia coli genomes. BMC Genomics 13:577. doi: 10.1186/1471-2164-13-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forde BM, Ben Zakour NL, Stanton-Cook M, Phan MD, Totsika M, Peters KM, Chan KG, Schembri MA, Upton M, Beatson SA. 2014. The complete genome sequence of Escherichia coli EC958: a high quality reference sequence for the globally disseminated multidrug resistant E. coli O25b:H4-ST131 clone. PLoS One 9:e104400. doi: 10.1371/journal.pone.0104400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hudson CM, Bent ZW, Meagher RJ, Williams KP. 2014. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 9:e99209. doi: 10.1371/journal.pone.0099209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. 2017. Whole-genome analysis of antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in river water. Appl Environ Microbiol 83:e02703-. doi: 10.1128/AEM.02703-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peter-Getzlaff S, Polsfuss S, Poledica M, Hombach M, Giger J, Bottger EC, Zbinden R, Bloemberg GV. 2011. Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J Clin Microbiol 49:2924–2932. doi: 10.1128/JCM.00091-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. 2009. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 60.Jove T, Da Re S, Denis F, Mazel D, Ploy MC. 2010. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet 6:e1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gomi R, Matsuda T, Fujimori Y, Harada H, Matsui Y, Yoneda M. 2015. Characterization of pathogenic Escherichia coli in river water by simultaneous detection and sequencing of 14 virulence genes. Environ Sci Technol 49:6800–6807. doi: 10.1021/acs.est.5b00953. [DOI] [PubMed] [Google Scholar]
- 64.Hoffmann H, Roggenkamp A. 2003. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol 69:5306–5318. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.