Abstract
Purpose
The purpose of this study was to investigate the therapeutic mechanism(s) of Clematis chinensis Osbeck/Notopterygium incisum K.C. Ting ex H.T (CN).
Methods
A network pharmacology approach integrating prediction of ingredients, target exploration, network construction, module partition and pathway analysis was used.
Results
This approach successfully helped to identify 12 active ingredients of CN, interacting with 13 key targets (Akt1, STAT3, TNFsf13, TP53, EPHB2, IL-10, IL-6, TNF, MAPK8, IL-8, RELA, ROS1 and STAT4). Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis indicated that CN-regulated pathways were mainly classified into signal transduction and immune system.
Conclusion
The present work may help to illustrate the mechanism(s) of action of CN, and it may provide a better understanding of antirheumatic effects.
Keywords: targets prediction, pathways analysis, action mechanism, Clematis chinensis Osbeck, Notopterygium incisum K.C. Ting ex H.T. Chang
Introduction
RA is a chronic autoimmune disease influenced by genetic factors, environmental factors and interaction.1 The prevalence of RA is ~1% in the adult population, with a higher incidence in the elderly and women.2 In addition to disability and joint destruction,3 patients with RA have a higher risk of dying prematurely from cardiovascular diseases.4 Consequently, prevention and treatment of RA are critical in clinical therapy. Therapeutic agents for RA include NSAIDs, glucocorticoids, DMARDs, biologic DMARDs and, most recently, small molecular signal inhibitors.5 However, most of current drugs, which play an important role in treating RA, have severe adverse effects, including gastrointestinal irritation, kidney injury, cardiovascular risk and even the so-called Cushing’s syndrome.6 Consequently, TCMs, with clinical application for thousands of years, have recently attracted more and more attention due to prominent effectiveness and less side effects.7 CC and NI, as TCMs, have been frequently used to treat RA for their anti-inflammatory activity.8–10 Our previous study has reported that CN has evident anti-rheumatic effects in adjuvant-induced arthritis in rats.11 However, the molecular mechanism(s) of CN in the treatment of RA remains to be elucidated. Network pharmacology is an efficient tool to clarify targets and mechanisms of TCMs.12 The methodologies of network pharmacology highlight the paradigm shift from “one drug, one target” to “multicomponent therapeutics, biological network”.13 TCMs have the advantages of multiple components and targets, which correspond to the methodologies of network pharmacology. Thus, network pharmacology is desirable for exploring the mechanisms of TCMs. In the present study, we respectively collected the information of targets from active ingredients in CN and targets of RA from several databases for the first time. In order to uncover the rationality of CN, network construction and topological structural analysis were established, which offered underlying synergistic mechanisms of CN for treating RA.
Methods
Building database of ingredients
All the chemical ingredients’ data of CC and NI were derived from Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (http://lsp.nwu.edu.cn/tcmsp.php). TCMSP is a unique system pharmacology platform of TCMs that is capable of providing the relationship between drugs, targets and diseases.
Screening of active ingredients
The active constituents from CC and NI were filtered by integrating OB and DL. DL helps to describe pharmacokinetic and pharmaceutical properties of compounds, such as solubility and chemical stability. Usually, the selection criterion for the “drug-like” compounds in TCMs is 0.18.14 OB represents the relative amount of an oral drug that is absorbed into the blood circulation.15 Since low OB is the primary reason responsible for the development of TCMs into therapeutic drugs, it is vital to conduct OB screening criterion. Based on literatures and suggestions in TCMSP, we selected OB≥30% and DL≥0.18 as a screening threshold.16,17 The ingredients conforming to both standards mentioned earlier will be preserved for further analysis.
TCM-associated target prediction
Three databases are combined to predict relevant targets of active ingredients in CC and NI comprehensively. GeneCards database (http://www.genecards.org/) automatically integrates gene-centric data from ~125 web sources, while BATMAN-TCM (http://bionet.ncpsb.org/batman-tcm) ranks potential drug–target interactions based on their similarity to the known drug–target interactions. STITCH database (http://stitch.embl.de/) integrates many sources of experimental and manually curated evidence with text-mining information and interaction predictions. First, the active constituents were severally entered into GeneCards, BATMAN-TCM and STITCH. Then, duplications and unified names were removed from the targets obtained from the aforementioned three tools. Noteworthy, only the targets of Homo sapiens were kept for further study.
RA-associated target prediction
Different genes associated with RA were collected from DisGeNET (http://www.disgenet.org/web/DisGeNET). DisGeNET is a useful platform providing the search of the molecular underpinnings of diseases, the analysis of disease genes, the validation of predicted genes and so on.
Network construction and node screening
The different targets from CC, NI and RA were submitted to Agilent Literature Search 3.1.1 (LitSearch version 2.69). Based on the human targets, we set “Max Engine Matches” as 10 and searched through the whole text. Then, the protein–protein interaction network was visualized by Cytoscape 3.5.1 software. Finally, we severally selected the top 30 targets of high-node degree as key targets for further analysis.
Module partition and KEGG pathway analysis
MCODE was applied to identify the molecular network for module identification according to the clustering of genes in the network. Then, main modules obtained from CC, NI and RA were submitted to DAVID Bioinformatics Resources 6.8 software (https://david.ncifcrf.gov/) to carry out GO functional enrichment analysis. “Homo Sapiens” was also limited to identify KEGG pathways that were significantly enriched in the identification module. Of note, P-value was implemented to explore the statistical significance of the modules. KEGG pathways with P<0.05 (P-values were corrected using the Benjamini–Hochberg procedure) are significant signaling pathways.
Results
Active ingredients of CC and NI
A total of 484 ingredients of CN were retrieved from TCMSP, including 114 ingredients of CC and 370 ingredients of NI. In this study, 21 active compounds from 484 compounds met both the requirements, OB≥30% and DL≥0.18 (Table 1). It has been validated experimentally that some ingredients possess pharmacological activities. For example, β-sitosterol (OB=36.91, DL=0.75) plays a significant role in anti-inflammation,18 anti-tumor19 and anti-hyperlipidemia.20 Moreover, nodakenin (OB=57.12, DL=0.69) plays an important therapeutic effect in inflammatory disorders and has been regarded as one of the standard ingredients of NI in Chinese Pharmacopoeia.10,21
Table 1.
Active ingredients of CC and NI
| Molecule ID | Molecule name | Structure | OB (%) | DL | Herb |
|---|---|---|---|---|---|
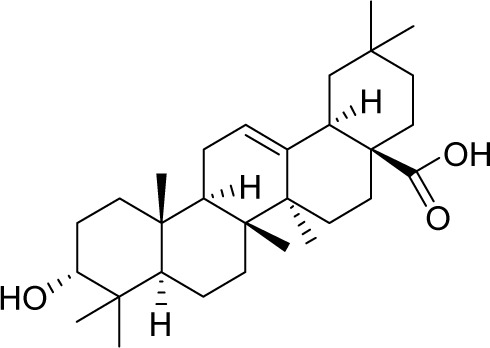
| MOL001663 | (4aS,6aR,6aS,6bR,8aR,10R,12aR,14bS)-10-hydroxy-2,2,6a,6b,9,9,12a-heptamethyl-1,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydropicene-4a-carboxylic acid |
|
32.03 | 0.76 | CC |
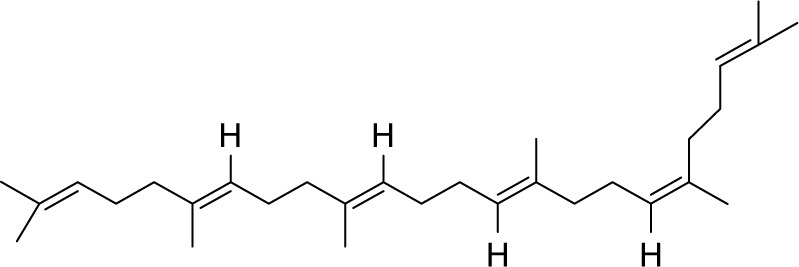
| MOL002372 | (6Z,10E,14E,18E)-2,6,10,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaene |
|
33.55 | 0.42 | CC |
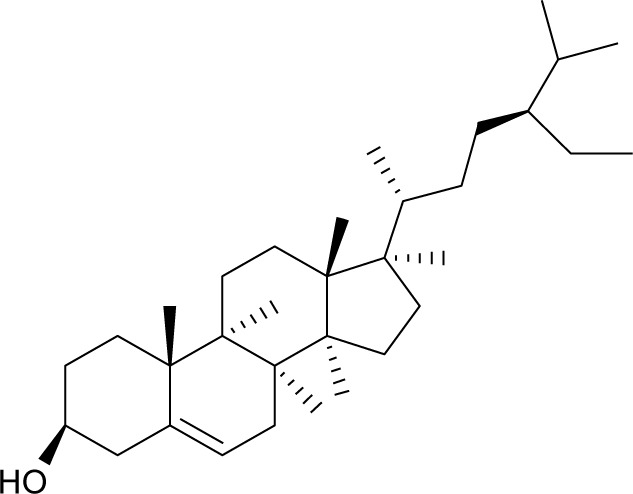
| MOL000358 | β-Sitosterol |
|
36.91 | 0.75 | CC, NI |
| MOL005594 | Clematoside A′_qt |
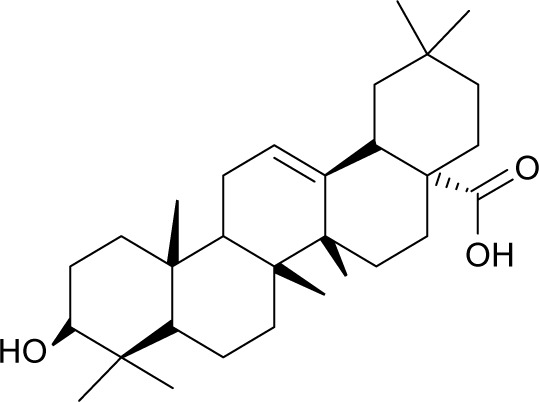
|
37.51 | 0.76 | CC |
| MOL005598 | Embinin |
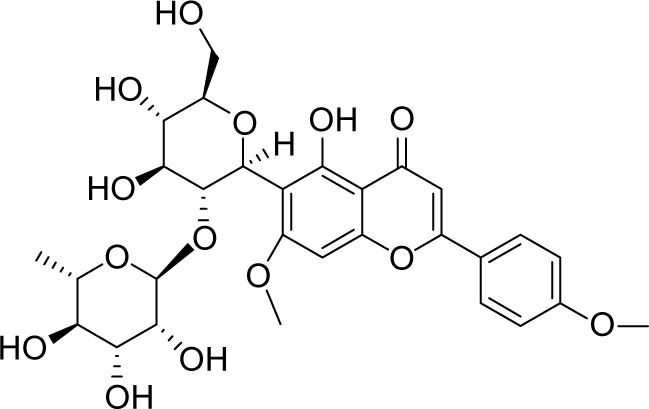
|
33.91 | 0.73 | CC |
| MOL005603 | Heptyl phthalate |
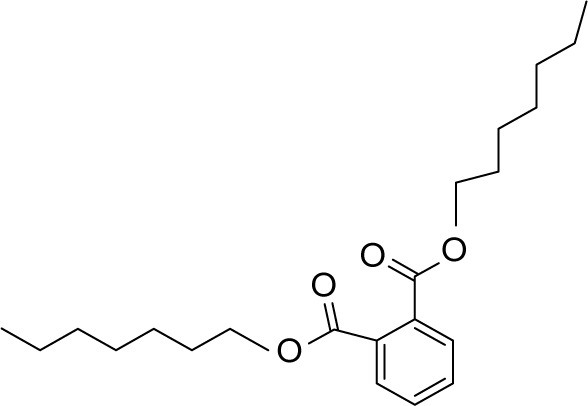
|
42.26 | 0.31 | CC |
| MOL000449 | Stigmasterol |
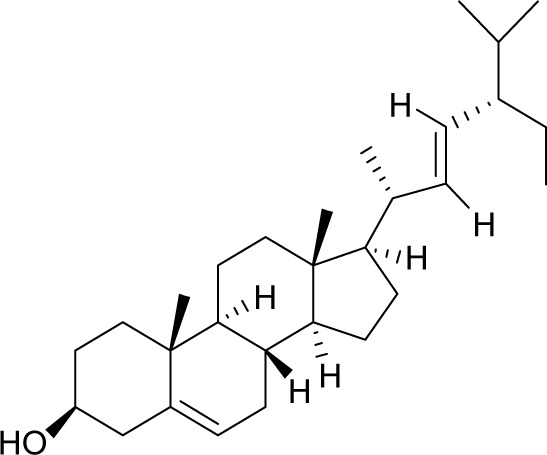
|
43.83 | 0.76 | CC |
| MOL001941 | Ammidin |
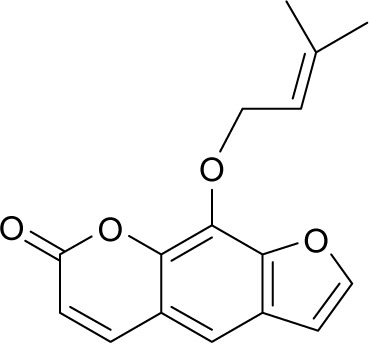
|
34.55 | 0.22 | NI |
| MOL011962 | 6′-Feruloylnodakenin |
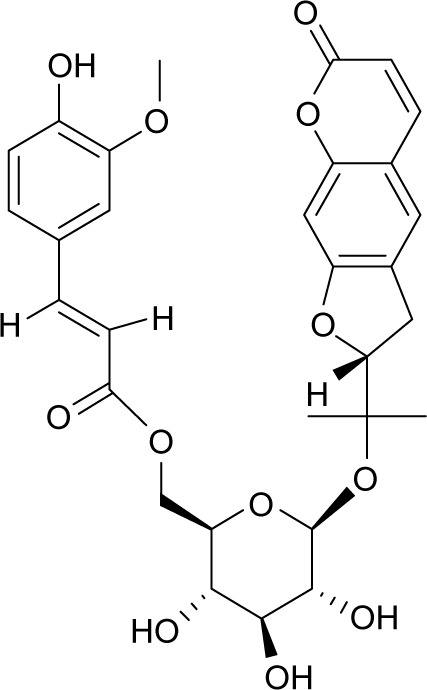
|
32.02 | 0.67 | NI |
| MOL011963 | 8-Geranoxy-5-methoxypsoralen |
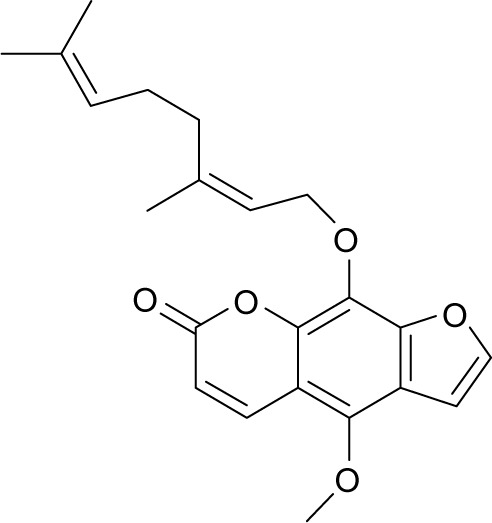
|
40.97 | 0.50 | NI |
| MOL011968 | Coumarin glycoside |
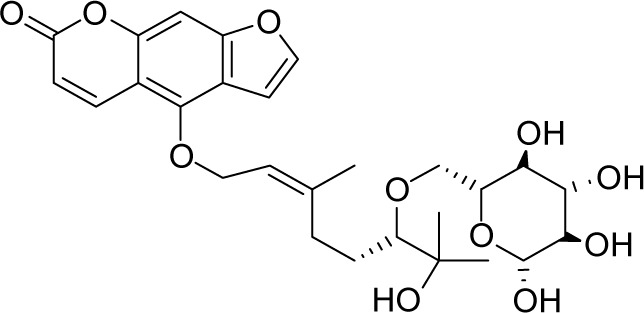
|
33.07 | 0.78 | NI |
| MOL011969 | Demethylfuropinnarin |
|
41.31 | 0.21 | NI |
| MOL011971 | Diversoside_qt |
|
67.57 | 0.31 | NI |
| MOL011975 | Notoptol |
|
62.97 | 0.48 | NI |
| MOL001951 | Bergapten |
|
41.73 | 0.42 | NI |
| MOL001956 | Cnidilin |
|
32.69 | 0.28 | NI |
| MOL000359 | Sitosterol |
|
36.91 | 0.75 | NI |
| MOL004792 | Nodakenin |
|
57.12 | 0.69 | NI |
| MOL001942 | Isoimperatorin |
|
45.46 | 0.23 | NI |
| MOL002644 | Phellopterin |
|
40.19 | 0.28 | NI |
| MOL002881 | Diosmetin |
|
31.14 | 0.27 | NI |
Abbreviations: CC, Clematis chinensis Osbeck; NI, Notopterygium incisum K.C. Ting ex H.T. Chang; OB, oral bioavailability; DL, druglikeness.
Target prediction
TCMs give play to their pharmacological effects through multiple ingredients and targets. Thus, besides predicting ingredients, it is also necessary for the exploration of targets. However, searching for targets through literatures is time consuming and labor intensive. In the present work, predictive models including GeneCards, BATMAN-TCM and STITCH were used to predict 301 targets, which interacted with 12 active ingredients. It is interesting that another nine ingredients were removed for having no relevant targets. In addition, the DisGeNET database was also used to predict 1,869 targets associated with RA.
Network construction and node screening
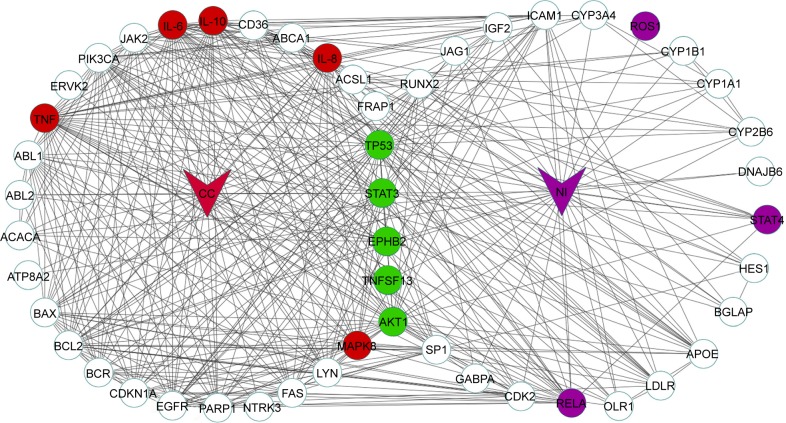
Network construction was automatically performed after searching by Agilent Literature Search. Molecular network of CC, NI and RA separately consisted of 781, 267 and 746 nodes and was connected by 2,873, 633 and 2,303 edges, respectively. The topological parameters of CC, NI and RA show that node-degree distribution obeys the power law distribution. To further investigate the synergistic mechanisms of herb couple, the top 30 targets of high-node degree were severally chosen. As shown in Figure 1, the herb couple shares 13 targets (Akt1, STAT3, TNFsf13/APRIL, TP53, EPHB2, IL-10, IL-6, TNF, MAPK8/JNK, IL-8, RELA, ROS1 and STAT4) with RA, in which five targets (STAT3, Akt1, TP53, TNFSF13 and EPHB2) are the overlapped targets in CC and NI and another five targets (IL-10, IL-6, TNF, MAPK8 and IL-8) and three targets (RELA, ROS1 and STAT4) are separate in CC and NI.
Figure 1.
Top 30 targets of CC and NI.
Note: Green nodes are shared targets from RA and herb couple, red ones are shared targets from RA and CC and purple ones are shared targets from RA and NI.
Abbreviations: CC, Clematis chinensis Osbeck; NI, Notopterygium incisum K.C. Ting ex H.T. Chang; RA, rheumatoid arthritis.
Module partition and KEGG pathway analysis
Module partition
MCODE software was used to analyze the molecular network. A total of 122 modules were identified from original networks with 47 modules from CC, 25 modules from NI and 50 modules from RA. Top 10 modules with more nodes were respectively selected for KEGG pathway analysis.
KEGG pathway analysis
In order to deduce the potential pathways affected by herb couple, DAVID Bioinformatics Resources 6.8 software was used to perform pathway enrichment analysis. Since diseases arise from the dysfunctions of basic biological functions, we removed the KEGG pathway section of human diseases. We found that herb couple could totally affect 35 signal pathways, including 16 pathways from both CC and NI, 17 pathways solely from CC and 2 pathways solely from NI.
As shown in Table 2A, herb couple acts on six pathways in signal transduction, such as PI3K–Akt signaling pathway and JAK–STAT signaling pathway. Immune system and endocrine system, respectively, cover three pathways such as NOD-like receptor signaling pathway and GnRH signaling pathway. Furthermore, the herb couple also regulates other pathways in cell process, development and nervous system. Signal pathways solely from CC are shown in Table 2B and indicated that CC has the possibility of being associated with the immune system, signal transduction, endocrine system and cell process. Compared with the signal pathways in CC, NI solely regulates cytosolic DNA-sensing pathway and cell process, which are associated with signal transduction and cell process, respectively (Table 2C).
Table 2.
(A) Signal pathways of herb couple, (B) individual signal pathways of CC and (C) individual biological pathways of NI
| Pathway class | Pathway name | CC’s targets on pathway | NI’s targets on pathway |
|---|---|---|---|
| A | |||
| Signal transduction | FoxO signaling pathway | IL-6, HRAS, SGK1, TGFB3, IGF1, FOXO1, IL-7R, IL-10, STAT3, PCK1, AKT1, MAPK1, NRAS, G6PC, CDKN1A, KRAS, CDKN2B, MAPK9, PIK3CA, MAPK8, CAT, EGF | AKT1, IL-6, SMAD4, PIK3CA, IL-10, STAT3, CDK2 |
| PI3K–Akt signaling pathway | FGFR1, HRAS, KITLG, NFKB1, COL2A1, BCL2L1, KIT, IL-7R, ATF2, AKT1, KRAS, BCL2, PIK3CA, EGF, CSF1R, IL-4, IL-3, IL-6, SGK1, IL-2RB, HSP90AA1, RELA, TP53, IGF1, KDR, PCK1, NRAS, MAPK1, CDKN1A, G6PC, IFNB1, VEGFA, PDGFRA, JAK1, PDGFRB, JAK2, EPOR | IBSP, AKT1, IL-6, IFNA1, BCL2, RELA, TP53, KITLG, PIK3CA, COL2A1, TLR4, CDK2, SPP1 | |
| NF-κB signaling pathway | MAP3K7, IRAK1, TNF, TNFSF13B, LYN, BCL2, RELA, BCL2A1, IL-1B, NFKB1, BCL2L1, BIRC3, TRAF6, CXCL12, BLNK | ICAM1, TNF, TNFSF11, PTGS2, RELA, BCL2, TLR4 | |
| TNF signaling pathway | CSF2, IL-6, TNF, SOCS3, RELA, NFKB1, CX3CL1, BIRC3, MMP3, ATF2, MAP3K7, AKT1, MAPK1, FOS, IL-1B, PIK3CA, MAPK9, MAPK8, FAS, MAP2K7 | AKT1, ICAM1, IL-6, TNF, CCL2, PTGS2, RELA, PIK3CA | |
| Sphingolipid signaling pathway | HRAS, TNF, RELA, TP53, NFKB1, AKT1, NRAS, MAPK1, KRAS, BAX, BCL2, PIK3CA, MAPK9, MAPK8, DEGS1 | AKT1, TNF, RELA, BAX, BCL2, TP53, PIK3CA | |
| HIF-1 signaling pathway | IL-6, ERBB2, RELA, IGF1, NFKB1, STAT3, AKT1, MAPK1, CDKN1A, BCL2, VEGFA, CAMK2D, PIK3CA, NOS2, EGF | AKT1, IL-6, RELA, BCL2, PIK3CA, TLR4, NOS2, STAT3, RBX1 | |
| Endocrine system | Prolactin signaling pathway | HRAS, SOCS3, RELA, NFKB1, STAT3, AKT1, NRAS, MAPK1, FOS, KRAS, SLC2A2, MAPK9, PIK3CA, MAPK8, JAK2 | AKT1, TNFSF11, RELA, PIK3CA, STAT3 |
| Insulin resistance | PPARA, IL-6, TNF, SOCS3, RELA, FOXO1, NFKB1, PPARGC1A, CPT1A, STAT3, PTPN11, PCK1, AKT1, G6PC, CD36, SLC2A2, MAPK9, PIK3CA, MAPK8 | AKT1, PPARA, IL-6, TNF, CD36, RELA, PIK3CA, STAT3 | |
| Adipocytokine signaling pathway | PPARA, TNF, SOCS3, RELA, NFKB1, PPARGC1A, CPT1A, STAT3, PCK1, PTPN11, AKT1, G6PC, ACSL1, CD36, MAPK9, MAPK8, JAK2 | AKT1, PPARA, TNF, CD36, RELA, ADIPOQ, STAT3 | |
| Cell process | Apoptosis | AKT1, IL-3, TNF, AIFM1, BCL2, RELA, NTRK1, BAX, TP53, PIK3CA, NFKB1, FAS, BCL2L1, BIRC3 | AKT1, TNF, RELA, BAX, BCL2, TP53, PIK3CA |
| Cytokine–cytokine receptor interaction | CSF2, TNF, TGFB3, KITLG, IL-13, TNFSF13, KIT, CX3CL1, IL-7R, CXCL12, IL-10, IL-1B, FAS, EGF, CSF1R, IL-4, IL-3, IL-2RB, IL-6, FLT3, KDR, TNFSF8, ACVR2A, TNFSF13B, IFNB1, VEGFA, PDGFRA, PDGFRB, EPOR | TNFRSF11B, IL-6, IFNA1, TNF, CCL2, TNFSF11, KITLG, TNFSF13, IL-10 | |
| Development | Osteoclast differentiation | TNF, SOCS3, RELA, PPARG, NFKB1, MAP3K7, TYK2, AKT1, FOS, MAPK1, IFNB1, MAPK9, JAK1, IL-1B, PIK3CA, MAPK8, TRAF6, MAP2K7, BLNK, CSF1R | AKT1, TYK2, TNFRSF11B, TNF, TNFSF11, SQSTM1, RELA, PIK3CA |
| Immune system | Toll-like receptor signaling pathway | IRAK1, IL-6, TNF, RELA, NFKB1, MAP3K7, AKT1, MAPK1, FOS, IFNB1, IL-1B, PIK3CA, MAPK9, MAPK8, TRAF6, MAP2K7 | AKT1, IL-6, IFNA1, TNF, IRF5, RELA, IRF7, PIK3CA, TLR4, SPP1, TLR9 |
| RIG-I-like receptor signaling pathway | MAP3K7, TNF, IFNB1, RELA, MAPK9, NFKB1, MAPK8, TRAF6 | IFNA1, TNF, ATG5, RELA, IRF7 | |
| JAK–STAT signaling pathway | IL-4, CSF2, IL-3, IL-6, IL-2RB, SOCS3, IL-13, BCL2L1, IL-24, IL-7R, IL-10, STAT3, PTPN11, TYK2, AKT1, STAT4, IFNB1, JAK1, PIK3CA, EPOR, JAK2 | AKT1, TYK2, IL-6, STAT4, IFNA1, PIK3CA, IL-10, STAT3 | |
| Nervous system | Neurotrophin signaling pathway | IRAK1, HRAS, RELA, TP53, NFKB1, PTPN11, NTRK3, AKT1, NRAS, MAPK1, BDNF, KRAS, BCL2, BAX, NTRK1, CAMK2D, SH2B3, MAPK9, PIK3CA, MAPK8, ABL1, TRAF6, MAP2K7 | AKT1, RELA, BAX, BCL2, TP53, PIK3CA |
| B | |||
| Immune system | NOD-like receptor signaling pathway | MAP3K7, MAPK1, IL-6, TNF, HSP90AA1, RELA, MAPK9, IL-1B, NFKB1, MAPK8, TRAF6, BIRC3 | |
| B-cell receptor signaling pathway | AKT1, MAPK1, NRAS, FOS, HRAS, KRAS, LYN, RELA, PIK3CA, NFKB1, BLNK | ||
| Chemokine signaling pathway | ITK, HRAS, LYN, RELA, NFKB1, CX3CL1, CXCL12, STAT3, AKT1, NRAS, MAPK1, KRAS, PTK2B, PIK3CA, JAK2 | ||
| Intestinal immune network for IgA production | IL-4, IL-6, TNFSF13B, TNFSF13, CXCL12, IL-10 | ||
| Fc epsilon RI signaling pathway | IL-4, CSF2, IL-3, HRAS, TNF, LYN, IL-13, AKT1, NRAS, MAPK1, KRAS, MAPK9, PIK3CA, MAPK8, MAP2K7 | ||
| Hematopoietic cell lineage | IL-4, CSF2, IL-3, IL-6, TNF, FLT3, KITLG, ANPEP, KIT, IL-7R, CD36, MS4A1, IL-1B, EPOR, CSF1R | ||
| T-cell receptor signaling pathway | IL-4, PTPRC, ITK, CSF2, HRAS, TNF, RELA, CBL, NFKB1, IL-10, MAP3K7, AKT1, NRAS, MAPK1, FOS, KRAS, PAK4, PIK3CA, MAP2K7 | ||
| Endocrine system | Progesterone-mediated oocyte maturation | AKT1, MAPK1, HSP90AA1, KRAS, PIK3CA, MAPK9, IGF1, MAPK8 | |
| GnRH signaling pathway | MAPK1, NRAS, HRAS, KRAS, PTK2B, CAMK2D, MAPK9, MAPK8, MAP2K7 | ||
| Estrogen signaling pathway | AKT1, MAPK1, NRAS, FOS, HRAS, HSP90AA1, KRAS, SP1, FKBP5, PIK3CA, ATF2 | ||
| Signal transduction | Rap1 signaling pathway | FGFR1, HRAS, KITLG, IGF1, CDH1, KIT, KDR, CTNNB1, AKT1, NRAS, MAPK1, KRAS, VEGFA, PDGFRA, PIK3CA, PDGFRB, EGF, CSF1R | |
| MAPK signaling pathway | FGFR1, HRAS, TNF, TGFB3, NFKB1, ATF2, MAP3K7, AKT1, FOS, BDNF, KRAS, IL-1B, FAS, EGF, TRAF6, MAP2K7, RELA, NF1, TP53, DUSP5, NRAS, MAPK1, NTRK1, MAPK8IP2, PDGFRA, MAPK9, PDGFRB, MAPK8 | ||
| VEGF signaling pathway | AKT1, MAPK1, NRAS, HRAS, KRAS, VEGFA, PIK3CA, KDR | ||
| ErbB signaling pathway | HRAS, ERBB2, CBL, AKT1, NRAS, MAPK1, CDKN1A, KRAS, PAK4, CAMK2D, MAPK9, PIK3CA, MAPK8, ABL1, EGF, MAP2K7, ABL2 | ||
| Ras signaling pathway | FGFR1, HRAS, KITLG, NFKB1, KIT, BCL2L1, AKT1, KRAS, PAK4, PIK3CA, EGF, CSF1R, RELA, NF1, IGF1, KDR, PTPN11, NRAS, MAPK1, RASSF1, VEGFA, PDGFRA, PDGFRB, MAPK9, MAPK8, ABL1, ABL2 | ||
| Cell process | Focal adhesion | HRAS, ERBB2, IGF1, COL2A1, BIRC3, KDR, CTNNB1, AKT1, MAPK1, BCL2, PAK4, VEGFA, PDGFRA, MAPK9, PIK3CA, PDGFRB, MAPK8, EGF | |
| Signaling pathways regulating pluripotency of stem cells | FGFR1, HRAS, TBX3, IGF1, STAT3, CTNNB1, AKT1, ACVR2A, INHBA, WNT1, NRAS, MAPK1, KRAS, PIK3CA, JAK1, JAK2, APC | ||
| C | |||
| Signal transduction | Cytosolic DNA-sensing pathway | IL-6, IFNA1, RELA, IRF7, IL-33 | |
| Cell process | Cell cycle | HDAC1, TP53, SMAD4, CHEK1, CUL1, CDK2, RBX1 |
Abbreviations: CC, Clematis chinensis Osbeck; NI, Notopterygium incisum K.C. Ting ex H.T. Chang.
Discussion
RA is a chronic autoimmune disease that is implicated in inflammation, angiogenesis, bone destruction and the immune regulation. Therapeutic agents for RA, including NSAIDs, glucocorticoids and DMARDs, are limited due to their side effects.22 TCMs are common drugs for the treatment of RA, with clinical effectiveness and less adverse effects. Our prior study has noted that herb couple of CC and NI was experimentally validated possessing anti-rheumatic effects.11 However, TCM, as a multi-component synergistic system agent, is comprehensive and abstruse. Therefore, their research method is different from chemical drugs. In the present study, to better recognize the drug combination of CN, we proposed a network pharmacology approach integrating prediction of ingredients and pathway analysis strategy of targets for CN. The method was applied to explore the potential regulation of inflammatory response, immune system and angiogenesis of CN and provide a new sight for the treatment of RA. Nonetheless, our method still has some limitations and needs to further improve. The approach just predicts and analyzes the potential synergetic mechanism of CC and NI from the perspective of biological network. Clinical and experimental trials are required to be further validated.
Targets analysis of herb couple
Targets related to RA
Synovial inflammation is a basic pathological change in RA and results in swelling and pain in the joints of RA patients. Thus, anti-inflammation is critical in the treatment of RA. Results of key targets’ analysis found that RA and CN shared a total of 13 targets. Among these targets, IL-6, IL-8 and IL-10 all belonged to the IL family. In RA, IL-6 can be released by monocytes, macrophages and endothelial cells and influences T-cell development, which indirectly promotes the production of Th1, Th2 and Th17 cells with proinflammatory properties. IL-6 can also increase the level of VEGF in synovial fibroblasts, aggravating joint inflammation and damage.23 In addition to inflammation, IL-6 increases osteoclast recruitment by acting on hematopoietic stem cells, leading to joint damage in RA.24,25 Therefore, blockade of IL-6 action is effective to reduce both inflammation and joint destruction in RA.26 The anti-inflammatory response is essential to control the degree and duration of the inflammatory response in RA. In macrophages, the anti-inflammatory response relies on IL-10/JAK/STAT3 signaling pathway. IL-10 signaling cascade starts upon IL-10 binding to IL-10R and activates STAT3 via the JAK1 kinase.27 STAT3 stimulates the transcription of specific genes and in turn represses proinflammatory cytokines such as IL-1, IL-6, IL-12 and TNF-α. Moreover, MAPK8, also known as JNK, is activated in RA synovium and mediates joint destruction in adjuvant arthritis of rats.28 MAPK8 signalosome represents a target to prevent joint destruction.29 TNFSF13 sustains B-cell activation and thus enhances autoimmune diseases. It also regulates synovial inflammation in RA.30 Therefore, TNFSF13 could also be a therapeutic strategy aimed at downregulating synovial inflammation. Furthermore, other targets are also involved in the process of RA, such as STAT4,31 EphB2,32 TP53,33 Akt134 and RELA.35 Upregulation or downregulation of abovementioned targets contributes to treating RA. On the other hand, ROS1, highly expressed in a variety of tumor cell lines, is used as a drug target to suppress tumors clinically.36 Based on our predictions, we speculate that ROS1 may have some relevance with RA, and the result would be validated in our future study.
Pathway analysis of herb couple
Pathways related to immune response
As shown in Table 2, many signal pathways are classified into immune system and regulate the balance of the immune system. Innate immune is the first line of defense against foreign pathogens and is the basis and initiator of adaptive immunity. B and T cells are well known to be related to adaptive immune response.37 Table 2B shows that CC is associated with B-cell receptor signaling pathway and T-cell receptor signaling pathway, indicating that CC may have a play in the adaptive immune response. In addition, we also found that CN could regulate some proinflammatory molecule-involved pathways such as chemokine signaling pathway and Fc epsilon RI signaling pathway. In addition, cytosolic DNA-sensing pathway, MAPK signaling pathway and apoptosis are highly associated with the function of immune response, although not classified into the immune system.38,39 Considering the effects of immune pathways on disease progress and joint destruction, modulation of these pathways may have important implications for treating RA.
Pathways related to inflammation
Another large category of signal pathways is signal transduction, including a number of well-known signal pathways that are related to inflammation, such as JAK–STAT signaling pathway, NF-κB signaling pathway and TNF signaling pathway. For example, canonical NF-κB signaling pathway is critical for the regulation of the inflammation response. Although less extensively studied, non-canonical pathway plays an indirect role in synovial inflammation via the high expression of its activators such as CD40L, CD40, BAFF/BAFF-R and RANKL in RA synovium.40 Since both canonical and non-canonical NF-κB signaling pathways participate in inflammatory response and the pathogenesis of RA, inhibitors of these two pathways can play a role in the treatment of RA. TNF, as the upstream target of the NF-κB pathway, has already been regarded as a therapeutic target in RA.41 Moreover, the JAK–STAT pathway is an important pathway for the transduction of cytokines associated with RA and is regarded as a target in inflammatory and autoimmune diseases.42 Tofacitinib, a JAK inhibitor, proves effective in the treatment of RA through reducing the expression of metalloproteinase and interferon-regulated gene in RA synovium.43
Pathways related to angiogenesis
Angiogenesis is a complex process involving the growth of new blood vessels and plays an important role in the growth, metastasis and prognosis of tumor. It is accompanied by the entire process of RA and can foster the infiltration of inflammatory cells into the joints, leading to synovial hyperplasia and progressive bone destruction.44 Ras signaling pathway and VEGF signaling pathway regulated solely by CC are related to angiogenesis. VEGF signaling pathway plays an important role in promoting the proliferation of vascular endothelial cells and the formation of new blood vessels,45,46 while Ras signaling pathway is related to tumor angiogenesis and vascular permeability.47 Certainly, inhibiting VEGF signaling is a feasible antiangiogenic and anti-inflammatory therapeutic strategy in RA.48
Conclusion
TCMs usually exert a multicomponent and multi-pathway synergetic efficacy in the treatment of various diseases. Therefore, the research approach applied to TCMs should correspond to the mechanisms of synergy. In this study, we applied a network pharmacology approach to identify the RA-related targets and signal pathways of CN, making it possible to connect genomic space to pharmacological space. In summary, we predicted the action mechanism(s) of herb couple for treating RA through the analysis of key targets and KEGG pathways.
Acknowledgments
This work was supported by the Major Research Plan of Shandong Province (No. 2016GSF202041), National Natural Science Foundation of China (No. 81403080), Overseas Expertise Introduction Center for Discipline Innovation (“111 Center”; No. B16046) and Foundation for Innovative Research Groups of the National Natural Science Foundation of China (No. 81421005).
Abbreviations
- RA
rheumatoid arthritis
- CC
Clematis chinensis Osbeck
- NI
Notopterygium incisum K.C. Ting ex H.T. Chang
- CN
Clematis chinensis Osbeck/Notopterygium incisum K.C. Ting ex H.T. Chang
- TCM
traditional Chinese medicine
- NSAIDs
nonsteroidal anti-inflammatory drugs
- DMARDs
disease-modifying antirheumatic drugs
- TCMSP
traditional Chinese medicine systems pharmacology database and analysis platform
- OB
oral bioavailability
- DL
druglikeness
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Akt1/PKB
protein kinase B
- TNFsf13/APRIL
proliferation-induced ligand
- TP53
tumor protein p53
- EPHB2
EPH receptor B2
- IL-10
interleukin-10
- IL-8
interleukin-8
- NF-κB3
nuclear factor-κB3
- ROS1
c-ros oncogene 1 receptor tyrosine kinase
- GnRH
gonadotropin-releasing hormone
- CD40L
CD40 ligand
- CD40
CD40 molecule
- BAFF
B-cell-activating factor belonging to the TNF family
- RANKL
receptor activator for nuclear factor-κB ligand
- JAK
janus kinase
- STAT
signal transducing activator of transcription
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor-kappaB
- MAPK8
mitogen-activated protein kinase 8
- TNF
tumor necrosis factor
- IL-6
interleukin-6
- VEGF
vascular endothelial growth factor
- BATMAN-TCM
bioinformatics analysis tool for molecular mechanism of TCM
- GO
gene ontology
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lerner A, Matthias T. Rheumatoid arthritis–celiac disease relationship: joints get that gut feeling. Autoimmun Rev. 2015;14(11):1038–1047. doi: 10.1016/j.autrev.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Costenbader KH, Chang SC, Laden F, Puett R, Karlson EW. Geographic variation in rheumatoid arthritis incidence among women in the United States. Arch Intern Med. 2008;168:1664. doi: 10.1001/archinte.168.15.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Markusse IM, Dirven L, Gerards AH, et al. Disease flares in rheumatoid arthritis are associated with joint damage progression and disability: 10-year results from the BeSt study. Arthritis Res Ther. 2015;17(1):232. doi: 10.1186/s13075-015-0730-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindhardsen J, Ahlehoff O, Gislason GH, et al. The risk of myocardial infarction in rheumatoid arthritis and diabetes mellitus: a Danish nationwide cohort study. Ann Rheum Dis. 2011;70(6):929–934. doi: 10.1136/ard.2010.143396. [DOI] [PubMed] [Google Scholar]
- 5.Burmester GR, Bijlsma JWJ, Cutolo M, McInnes IB. Managing rheumatic and musculoskeletal diseases – past, present and future. Nat Rev Rheumatol. 2017;13(7):443–448. doi: 10.1038/nrrheum.2017.95. [DOI] [PubMed] [Google Scholar]
- 6.Obiri DD, Osafo N, Ayande PG, Antwi AO. Xylopia aethiopica (Annonaceae) fruit extract suppresses Freund’s adjuvant-induced arthritis in Sprague-Dawley rats. J Ethnopharmacol. 2014;152(3):522–531. doi: 10.1016/j.jep.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Venkatesha SH, Berman BM, Moudgil KD. Herbal medicinal products target defined biochemical and molecular mediators of inflammatory autoimmune arthritis. Bioorgan Med Chem. 2011;19:21–29. doi: 10.1016/j.bmc.2010.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng C, Perera PK, Li Y, Fang W, Liu L, Li F. Anti-inflammatory effects of Clematis chinensis Osbeck extract(AR-6) may be associated with NF-κB, TNF-α, and COX-2 in collagen-induced arthritis in rat. Rheumatol Int. 2012;32(10):3119–3125. doi: 10.1007/s00296-011-2083-8. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Zhang X, Xie W, et al. Identification of UQCRB as an oxymatrine recognizing protein using a T7 phage display screen. J Ethnopharmacol. 2016;193:133–139. doi: 10.1016/j.jep.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Committee CP, editor. Pharmacopoeia of the People’s Republic of China. Beijing: China Medical Science and Technology Press; 2015. [Google Scholar]
- 11.Pan T, Cheng TF, Jia YR, Li P, Li F. Anti-rheumatoid arthritis effects of traditional Chinese herb couple in adjuvant-induced arthritis in rats. J Ethnopharmacol. 2017;205:1–7. doi: 10.1016/j.jep.2017.04.020. [DOI] [PubMed] [Google Scholar]
- 12.Trame MN, Biliouris K, Lesko LJ, Mettetal JT. Systems pharmacology to predict drug safety in drug development. Eur J Pharm Sci. 2016;94:93–95. doi: 10.1016/j.ejps.2016.05.027. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Zhang B, Jiang D, Wei Y, Zhang N. Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinformatics. 2010;11:11. doi: 10.1186/1471-2105-11-S11-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tao W, Xu X, Wang X, et al. Network pharmacology-based prediction of the active ingredients and potential targets of Chinese herbal Radix Curcumae formula for application to cardiovascular disease. J Ethnopharmacol. 2013;145(1):1–10. doi: 10.1016/j.jep.2012.09.051. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci. 2012;13:6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yue S, Liu J, Feng W, et al. System pharmacology-based dissection of the synergistic mechanism of Huangqi and Huanglian for diabetes mellitus. Front Pharmacol. 2017;8:694. doi: 10.3389/fphar.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao Y, Zhang X, Wang Z, et al. Deciphering the combination principles of Traditional Chinese Medicine from a systems pharmacology perspective based on Ma-huang Decoction. J Ethnopharmacol. 2013;150(2):619–638. doi: 10.1016/j.jep.2013.09.018. [DOI] [PubMed] [Google Scholar]
- 18.Kim K, Lee I, Gu W, Hyam SR, Kim D. β-Sitosterol attenuates high-fat diet-induced intestinal inflammation in mice by inhibiting the binding of lipopolysaccharide to toll-like receptor 4 in the NF-κB pathway. Mol Nutr Food Res. 2014;58(5):963–972. doi: 10.1002/mnfr.201300433. [DOI] [PubMed] [Google Scholar]
- 19.Bin SM, Ameen SS. Beta-sitosterol: a promising but orphan nutraceutical to fight against cancer. Nutr Cancer. 2015;67(8):1214–1220. doi: 10.1080/01635581.2015.1087042. [DOI] [PubMed] [Google Scholar]
- 20.Liu Y, Lei L, Wang X, et al. Plasma cholesterol-raising potency of dietary free cholesterol versus cholesteryl ester and effect of β-sitosterol. Food Chem. 2015;169:277–282. doi: 10.1016/j.foodchem.2014.07.123. [DOI] [PubMed] [Google Scholar]
- 21.Lee NY, Chung KS, Jin JS, Lee YC, An HJ. The inhibitory effect of nodakenin on mast-cell-mediated allergic inflammation via downregulation of NF-κB and Caspase-1 activation. J Cell Biochem. 2017;118(11):3993–4001. doi: 10.1002/jcb.26055. [DOI] [PubMed] [Google Scholar]
- 22.Burmester GR, Pope JE. Targeted treatments for rheumatoid arthritis 2 Novel treatment strategies in rheumatoid arthritis. Lancet. 2017;389:2338–2348. doi: 10.1016/S0140-6736(17)31491-5. [DOI] [PubMed] [Google Scholar]
- 23.Nakahara H, Song J, Sugimoto M, et al. Anti-interleukin-6 receptor antibody therapy reduces vascular endothelial growth factor production in rheumatoid arthritis. Arthritis Rheum. 2003;48:1521–1529. doi: 10.1002/art.11143. [DOI] [PubMed] [Google Scholar]
- 24.Otsuka T, Thacker JD, Hogge DE. The effects of interleukin 6 and interleukin 3 on early hematopoietic events in long-term cultures of human marrow. Exp Hematol. 1991;19:1042–1048. [PubMed] [Google Scholar]
- 25.Yoshitake F, Itoh S, Narita H, Ishihara K, Ebisu S. Interleukin-6 directly inhibits osteoclast differentiation by suppressing receptor activator of NF-kappaB signaling pathways. J Biol Chem. 2008;283:11535–11540. doi: 10.1074/jbc.M607999200. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka T, Narazaki M, Ogata A, Kishimoto T. A new era for the treatment of inflammatory autoimmune diseases by interleukin-6 blockade strategy. Semin Immunol. 2014;26:88–96. doi: 10.1016/j.smim.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Hutchins AP, Poulain S, Miranda-Saavedra D. Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Mol Immunol. 2015;64(1):90–98. doi: 10.1182/blood-2011-09-381483. [DOI] [PubMed] [Google Scholar]
- 28.Schett G, Tohidast-Akrad M, Smolen JS, et al. Activation, differential localization, and regulation of the stress-activated protein kinases, extracellular signal-regulated kinase, c-JUN N-terminal kinase, and p38 mitogen-activated protein kinase, in synovial tissue and cells in rheumatoid arthritis. Arthritis Rheum. 2000;43(11):2501–2512. doi: 10.1002/1529-0131(200011)43:11<2501::AID-ANR18>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 29.Sundarrajan M, Boyle DL, Chabaud-Riou M, Hammaker D, Firestein GS. Expression of the MAPK kinases MKK-4 and MKK-7 in rheumatoid arthritis and their role as key regulators of JNK. Semin Immunol. 2014;26(1):88–96. doi: 10.1002/art.11228. [DOI] [PubMed] [Google Scholar]
- 30.Seyler TM, Park YW, Takemura S, et al. BLyS and APRIL in rheumatoid arthritis. J Clin Invest. 2005;115(11):3083–3092. doi: 10.1172/JCI25265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen HN, Noss EH, Mizoguchi F, et al. Autocrine loop involving IL-6 family member LIF, LIF receptor, and STAT4 drives sustained fibroblast production of inflammatory mediators. Immunity. 2017;46(2):220–232. doi: 10.1016/j.immuni.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu H. The Role of EphB/ephrinB in Inflammation. [dissertation] University of Hedelberg; 2013. [Google Scholar]
- 33.Tak PP, Zvaifler NJ, Green DR, Firestein GS. Rheumatoid arthritis and p53: how oxidative stress might alter the course of inflammatory diseases. Immunol Today. 2000;21(2):78–82. doi: 10.1016/s0167-5699(99)01552-2. [DOI] [PubMed] [Google Scholar]
- 34.Pope RM. Apoptosis as a therapeutic tool in rheumatoid arthritis. Nat Rev Immunol. 2002;2(7):527–535. doi: 10.1038/nri846. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Ni S, Yang L. Expression of immune-related cytokine IL-1 β in non-neoplastic epithelial disorders of the vulvar tissue and its clinical significance. Int J Clin Exp Pathol. 2017;10(5):4988–5000. [Google Scholar]
- 36.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19(15):4040–4045. doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bauch A, Superti-Furga G. Charting protein complexes, signaling pathways, and networks in the immune system. Immunol Rev. 2006;210(1):187–207. doi: 10.1111/j.0105-2896.2006.00369.x. [DOI] [PubMed] [Google Scholar]
- 38.Mayadas TN, Cullere X. Neutrophil β2 integrins: moderators of life or death decisions. Trends Immunol. 2005;26(7):388–395. doi: 10.1016/j.it.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Lee MS, Kim Y. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu Rev Biochem. 2007;76:447–480. doi: 10.1146/annurev.biochem.76.060605.122847. [DOI] [PubMed] [Google Scholar]
- 40.Noort AR, Tak PP, Tas SW. Non-canonical NF-κB signaling in rheumatoid arthritis: Dr Jekyll and Mr Hyde? Arthritis Res Ther. 2015;17(1):15. doi: 10.1186/s13075-015-0527-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umiċeviċ Mirkov M, Cui J, Vermeulen SH, et al. Genome-wide association analysis of anti-TNF drug response in rheumatoid arthritis patients. Ann Rheum Dis. 2013;72:1375–1381. doi: 10.1136/annrheumdis-2012-202405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banerjee S, Biehl A, Gadina M, Hasni S, Schwartz DM. JAK–STAT signaling as a target for inflammatory and autoimmune diseases: current and future prospects. Drugs. 2017;77(5):521–546. doi: 10.1007/s40265-017-0701-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyle DL, Soma K, Hodge J, Kavanaugh A, Mandel D. Extended report: the JAK inhibitor tofacitinib suppresses synovial JAK1-STAT signalling in rheumatoid arthritis. Ann Rheum Dis. 2015;74:1311–1316. doi: 10.1136/annrheumdis-2014-206028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elshabrawy HA, Chen Z, Volin MV, Ravella S, Virupannavar S, Shahrara S. The pathogenic role of angiogenesis in rheumatoid arthritis. Angiogenesis. 2015;18(4):433–448. doi: 10.1007/s10456-015-9477-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438(7070):932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 46.Olsson A, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling – in control of vascular function. Nat Rev Mol Cell Bio. 2006;7(5):359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 47.Li W, Liang RR, Zhou C, et al. The association between expressions of Ras and CD68 in the angiogenesis of breast cancers. Cancer Cell Int. 2015;15(1):17. doi: 10.1186/s12935-015-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Biniecka M, Kennedy A, Fearon U, Ng CT, Veale DJ. VEGF as an activity marker in rheumatoid arthritis. Int J Clin Rheumatol. 2010;5(3):287–289. [Google Scholar]